Genome-wide association studies have implicated the C-type lectin gene CLEC16A in several autoimmune diseases. Van Luijn et al. reveal upregulation of CLEC16A in multiple sclerosis, and show that the protein acts as a key regulator of the HLA class II antigen presentation pathway by controlling late endosomal maturation.
Keywords: C-type lectin, human leukocyte antigen, antigen presentation, immunogenetics, autoimmune disease
Genome-wide association studies have implicated the C-type lectin gene CLEC16A in several autoimmune diseases. Van Luijn et al. reveal upregulation of CLEC16A in multiple sclerosis, and show that the protein acts as a key regulator of the HLA class II antigen presentation pathway by controlling late endosomal maturation.
Abstract
C-type lectins are key players in immune regulation by driving distinct functions of antigen-presenting cells. The C-type lectin CLEC16A gene is located at 16p13, a susceptibility locus for several autoimmune diseases, including multiple sclerosis. However, the function of this gene and its potential contribution to these diseases in humans are poorly understood. In this study, we found a strong upregulation of CLEC16A expression in the white matter of multiple sclerosis patients (n = 14) compared to non-demented controls (n = 11), mainly in perivascular leukocyte infiltrates. Moreover, CLEC16A levels were significantly enhanced in peripheral blood mononuclear cells of multiple sclerosis patients (n = 69) versus healthy controls (n = 46). In peripheral blood mononuclear cells, CLEC16A was most abundant in monocyte-derived dendritic cells, in which it strongly co-localized with human leukocyte antigen class II. Treatment of these professional antigen-presenting cells with vitamin D, a key protective environmental factor in multiple sclerosis, downmodulated CLEC16A in parallel with human leukocyte antigen class II. Knockdown of CLEC16A in distinct types of model and primary antigen-presenting cells resulted in severely impaired cytoplasmic distribution and formation of human leucocyte antigen class II-positive late endosomes, as determined by immunofluorescence and electron microscopy. Mechanistically, CLEC16A participated in the molecular machinery of human leukocyte antigen class II-positive late endosome formation and trafficking to perinuclear regions, involving the dynein motor complex. By performing co-immunoprecipitations, we found that CLEC16A directly binds to two critical members of this complex, RILP and the HOPS complex. CLEC16A silencing in antigen-presenting cells disturbed RILP-mediated recruitment of human leukocyte antigen class II-positive late endosomes to perinuclear regions. Together, we identify CLEC16A as a pivotal gene in multiple sclerosis that serves as a direct regulator of the human leukocyte antigen class II pathway in antigen-presenting cells. These findings are a first step in coupling multiple sclerosis-associated genes to the regulation of the strongest genetic factor in multiple sclerosis, human leukocyte antigen class II.
Introduction
Recent genome-wide association studies demonstrate that single nucleotide polymorphisms at the 16p13 locus containing C-type lectin domain family 16, member A (CLEC16A) increase the risk of developing multiple sclerosis (Hafler et al., 2007; Hoppenbrouwers et al., 2009; Sawcer et al., 2011; Beecham et al., 2013). This risk locus is also associated with several other autoimmune diseases, such as type I diabetes (Hakonarson et al., 2007), primary biliary cirrhosis (Mells et al., 2011) and coeliac disease (Dubois et al., 2010). Hence, CLEC16A likely plays a key role in the pathogenesis of autoimmune diseases, but its biological function in humans is completely unknown.
CLEC16A is a family member of the C-type lectins, which are broadly expressed by antigen-presenting cells and share innate immune functions in recognition of pathogens or (altered) self molecules via their carbohydrate recognition domains (Geijtenbeek et al., 2004; Geijtenbeek and Gringhuis, 2009). Certain C-type lectins, such as DC-SIGN (now known as CD209), DEC-205 (now known as LY75) and Dectin-1 (now knwon as CLEC7A), contain specific internalization motifs to target antigens into the endosomal pathway required for human leukocyte antigen class II (HLA-II)-mediated antigen presentation to T cells (Geijtenbeek et al., 2004). However, the short carbohydrate recognition domain of CLEC16A is predicted to be inactive (Zelensky and Gready, 2005; Todd et al., 2007), suggesting a non-classical C-type lectin function.
The CLEC16A gene is associated with a set of other immune genes located on the short arm of chromosome 16, which is considered to be an important locus in the pathogenesis of multiple sclerosis (Zuvich et al., 2011; Berge et al., 2013). CLEC16A is localized adjacent to CIITA, the master transcription factor of HLA-II (Choi et al., 2011). HLA-II is long established as the strongest genetic risk factor in multiple sclerosis, but its role in multiple sclerosis pathogenesis is still unclear (Ramagopalan et al., 2009a). There is growing interest in proteins serving as direct regulators of the HLA-II antigen presentation pathway and revealing linkage to autoimmune disease (Paul et al., 2011).
In Drosophila, the human CLEC16A orthologue Ema has been identified as a protein directly involved in endosomal maturation (Kim et al., 2010). The formation of late endosomal compartments is an important step in the HLA-II antigen presentation pathway. HLA-II molecules synthesized in the endoplasmic reticulum are transported to specialized mature late endosomes called MIIC (MHC class II-containing compartment) (Neefjes et al., 1990; Peters et al., 1995). In these MIIC, HLA-II molecules are loaded with peptides for presentation at the plasma membrane. Therefore, we hypothesize that CLEC16A plays a key role in this process.
Here, we demonstrate that CLEC16A expression directly links to multiple sclerosis and controls the HLA-II antigen presentation pathway via late endosomal maturation. CLEC16A levels were strongly elevated in multiple sclerosis patients compared to control subjects. CLEC16A knockdown in various antigen-presenting cells showed that CLEC16A is a key regulator of late endosomal biogenesis to drive HLA-II expression, a novel function for C-type lectins. Based on these findings, we propose that CLEC16A promotes late endosomal maturation to disrupt the HLA-II antigen presentation pathway in multiple sclerosis.
Material and methods
Clinical samples
Multiple sclerosis patients were between 18 and 65 years of age and diagnosed according to the McDonald criteria (Polman et al., 2005). Gender-matched healthy control subjects were aged between 18 and 55 years and were relatives or family members of patients included in this study. Exclusion criteria were prior symptoms suggestive of or diagnosis with multiple sclerosis as well as the use of immunomodulatory agents for other autoimmune diseases. All patients and controls gave written informed consent. Post-mortem brain tissue of patients with multiple sclerosis and non-demented control subjects was obtained from the Netherlands Brain Bank (NBB; Amsterdam, The Netherlands). Samples and clinical information was collected from donors who gave written informed consent for brain autopsy. Cryopreserved brain tissues were stored at −80°C until use. This study was approved by the Medical Ethical Committee of the Erasmus MC, University Medical Center.
Human cell cultures
The melanoma cell line MelJuSo was cultured in Iscove's modified Dulbecco's medium (IMDM) (Lonza) supplemented with 8% foetal calf serum. Rab7-GFP+MelJuSo cells were generated as reported previously (Jordens et al., 2001). The HEK293T cell line was grown in Dulbecco’s modified Eagle’s medium (Lonza) containing 8% foetal calf serum and 1% penicillin-streptomycin. Peripheral blood samples of patients and controls were collected in Vacutainer CPT tubes (BD Biosciences). Peripheral blood mononuclear cells from buffy coats (Sanquin) were isolated using standard Ficoll-Paque (Amersham Pharmacia Biotech) density-gradient centrifugation. A Percoll density gradient or CD14 microbeads (Miltenyi Biotec) was used to isolate monocytes from peripheral blood mononuclear cell fractions, resulting in a purity of CD14+ cells of >90% as determined by flow cytometry. For differentiation into monocyte-derived dendritic cells, the isolated monocyte fraction was cultured in the presence of GM-CSF (600 U/ml; BioSource) and IL4 (400 U/ml; R&D Systems). Cells were treated with or without the active form of vitamin D (10−7 M) After washing and seeding, monocyte-derived dendritic cells were matured by lipopolysaccharide (LPS) treatment (Sigma-Aldrich; E. coli 026:B6, 1 µg/ml) for 16 h. Macrophages were differentiated from primary monocytes by culturing in RPMI (Lonza) with 5% normal human serum for 5 days under non-adherent conditions using Teflon conical flasks.
DNA isolation and genotyping
DNA was isolated from blood pellets obtained in EDTA tubes according to standard laboratory practice as previously reported (Aulchenko et al., 2008). Multiple sclerosis risk single nucleotide polymorphism rs7200786 in CLEC16A was assessed on the Illumina 610 K array or with a custom-made Sequenom array, both according to the manufacturers’ protocols.
Production of DNA constructs
CLEC16A cDNA (GenBank no BC112897.1) was cloned into the Bgl2-EcoR1 sites of 2HA-C1 and mGFP-C1. CLEC16A cDNA ligation was checked using classical PCR and CLEC16A mRNA overexpression in cells transfected with these constructs was validated using quantitative PCR (Supplementary Fig. 5A). GFP-, mRFP- and HA-RILP as well as HA-VPS41 DNA constructs were produced as previously shown (Jordens et al., 2001; van der Kant et al., 2013). Constructs were transformed into Escherichia coli DH5α competent cells, which were plated out on Luria broth agar plates containing the appropriate antibiotics to select for single colonies at 37°C. A single colony was further grown overnight in 250–500 ml Luria broth selection medium and plasmid DNA was isolated using Midiprep or Maxiprep kits (Roche Applied Science).
Silencing RNA and DNA transfections
One day before transfection, fresh culture medium without penicillin/streptomycin was added. For each transfection, siGENOME silencing small interfering RNA (siRNA) duplexes (500 nM; Thermo Fisher Scientific) were mixed with DharmaFECT® reagent in IMDM in 96 - or 24-well plates (flat bottomed). After 20 min incubation at room temperature, cells were added to the mixture and cultured for 3 days. For immunofluorescence analysis, cells were grown on cover slides (Thermo Fisher Scientific). We used three different CLEC16A siRNA duplexes: #1, UCACAGGUCUUCUUAAUUA; #2, UGUCUGAGAUGUACGCUAA; and #3, CGUAAAUUCUAUCAUCGUU. Scrambled and HLA-DMβ siRNA duplexes were used as internal controls in each experiment. To exclude off-target effects, we tested the effects of combinations of CLEC16A siRNA duplexes on both CLEC16A and HLA-DRA mRNA expression.
To overexpress CLEC16A, rab7-interacting lysosomal protein (RILP) or homotypic fusion and vacuole protein sorting (HOPS) complex proteins, we transfected the MelJuSo cell line with DNA constructs using either FuGENE 6 or X-tremeGENE 9 reagent (Roche Applied Science). In short, DNA was mixed with transfection reagent supplemented in serum-free IMDM and incubated for 20 min at room temperature. The mixture was then added drop-wise to the cells 1 or 2 days after CLEC16A siRNA transfection. HEK293T cells were seeded 60–70% confluent. For these cells, a mixture of serum-free Dulbecco’s modified Eagle’s medium, polyethylenimine [1 mg/ml diluted in phosphate-buffered saline (PBS)] and DNA was added after 30 min incubation at room temperature. Cells were harvested and analysed 24–48 h after incubation.
Lentivirus production and transduction into monocytes
We obtained lentiviral pLKO.1 constructs containing specific CLEC16A short hairpin RNA (shRNA) from Open Biosystems (Thermo Fisher Scientific). Two different CLEC16A shRNA were used: #1, CAGCTCTGTATTTGACTTCTT; and #2, GCTAAGACTGAACAGGATATT. For lentivirus production, we transfected HEK293T cells with packaging constructs (pRSV-Rev, pCMV-VSV-G and pMDLg/pRRE) and shRNA using polyethylenimine (PEI, MW 25 kDa, Polysciences). In each experiment, cotransfection with a pEGFP-C1 construct revealed 70–80% GFP+ cells. After 2–3 days of cell culture, the supernatants were filtered and lentivirus was concentrated using ultracentrifugation. Viral particles were taken up in medium, snap-frozen and stored at −80°C.
Thawed monocytes were transduced with lentivirus in CellGro® serum-free medium (CellGenix) supplemented with polybrene (4 µg/ml) and cultured in the presence of IL4 (800 U/ml) and GM-CSF (1000 U/ml) for differentiation into dendritic cells. Dendritic cells were split 6 days after transduction and one part was matured by addition of LPS (1 µg/ml) and IFNG (1000 U/ml) for 24 h. Dendritic cell differentiation and maturation was determined by flow cytometric analysis of CD11c, HLA-DR, CD40, CD80, CD83 and CD86 expression.
Co-immunoprecipitation and immunoblotting
HEK293T cells transfected with HA- and GFP-tagged CLEC16A, RILP and/or HOPS complex proteins were lysed for 30 min in 0.8% NP-40 lysis buffer containing 50 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2 and a protease inhibitor cocktail (EDTA-free; Roche Applied Science). After spinning (15 min at 14 000g), the supernatants were incubated with anti-HA or -GFP antibodies coupled to Protein G-Sepharose 4 Fast Flow beads (GE Healthcare) for 1 h. Beads were washed in 8% NP-40 lysis buffer and three times in 0.08% NP-40 buffer containing 50 mM Tris-HCl, 500 mM NaCl and 5 mM MgCl2 before adding sample buffer and incubation for 5 min at 95°C. Immune-precipitated proteins were detected via sodium dodecyl sulphate polyacrylamide gel electrophoresis followed by immunoblotting on polyvinylidene difluoride membranes for 2 h at 300 mA at 4°C. Membranes were probed with an HRP-conjugated rat anti-HA (Roche Applied Science) or rabbit anti-GFP (Rocha et al., 2009) antibody. Protein bands were visualized using ECL (Supersignal West Dura Extended Duration Substrate; Thermo Fisher Scientific).
Electron microscopy
Either peripheral blood mononuclear cells containing around 40% of monocytes or stable MelJuSo CLEC16A or scrambled shRNA transductants were fixed for 2 h in a mixture of 2% paraformaldehyde and 0.2% glutaraldehyde in 60 mM PIPES, 25 mM HEPES, 2 mM MgCl2, 10 mM EGTA, pH 6.9 and further processed for ultrathin cryosectioning as described (Calafat et al., 1997). For immunolabelling, the sections were incubated for 10 min with 0.15 M glycine in PBS and for 10 min with 1% bovine serum albumin (BSA) in PBS to block free aldehyde groups and prevent aspecific antibody binding, respectively. Sections were incubated with either rabbit anti-CLEC16A antibody (Sigma-Aldrich) followed by swine anti-rabbit IgG as a signal strengthening step or with rabbit anti-HLA-II antibody (made by J. Neefjes) and 10 nm protein-A conjugated colloidal gold (EM Lab, University of Utrecht, The Netherlands), all in 1% BSA in PBS. Finally, the cryosections were embedded in uranylacetate and methylcellulose and assessed by a Philips CM 10 electron microscope (FEI).
For electron microscopy quantification, internal vesicle-poor multivesicular bodies were defined as late endosomal compartments containing <50% of internal vesicles. Only HLA-II+ multivesicular bodies were analysed. Each electron microscopy image represented a 70 nm section of a single cell, in which the number of HLA-II molecules was related to that of multivesicular bodies to calculate the relative number of HLA-II per multivesicular body. Multivesicular body and HLA-II numbers were counted in triplicate in 12–15 different cells and were confirmed by a second independent observer.
Fluorescence microscopy and imaging analysis
We used a fluorescence (Zeiss Axioplan 2) and confocal laser-scanning (Leica TCS SP2) microscope together with Leica LAS AF and ImageJ software to analyse cells. To quantify co-localization, we calculated the Pearson’s correlation coefficient for each cell using the co-localization tool in LAS AF. Co-localization analysis was done on representative images. For the quantification of late endosome localization, first the outermost cytoplasmic region of each cell was determined using HLA-DR as plasma membrane marker. Then, a line scan was performed by taking the shortest distance from the nuclear rim (DAPI+) to this area. Finally, fluorescence intensities for each line region of interest were assessed using the line profile tool in LAS AF.
Brain lesion staging
Staging of multiple sclerosis lesions in brain white matter tissue was performed as previously reported (van der Valk and De Groot, 2000). In short, lesions were characterized using Oil Red O to detect myelin degradation products and monoclonal antibodies against HLA-II and PLP or MOG. Lesion stages included normal-appearing white matter as well as pre-active, active demyelinating, chronic active and chronic inactive lesions.
Statistical analysis
Statistical analyses were performed in SPSS 20.0 and GraphPad Prism, version 5.04 software. The two-tailed Mann-Whitney U-test was used to compare two groups. For comparison of more than two groups, we performed the Kruskal-Wallis followed by the Dunn’s multiple comparison test. Correlations were evaluated using Spearman’s rank test or linear regression analysis. P-values < 0.05 were considered as statistically different. Mean values and standard errors of the mean are indicated, unless stated otherwise.
Results
CLEC16A is abundant in monocytic cells and monocyte-derived dendritic cells and localizes with HLA class II
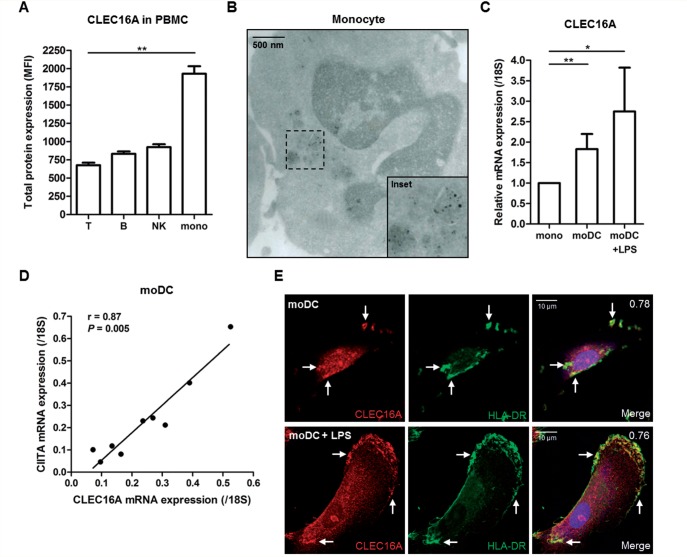
We first assessed CLEC16A expression levels in selected peripheral blood leukocyte subsets of healthy donors (n = 3–4). CLEC16A protein expression was low in T cells, moderate in natural killer and B cells and highest in monocytes (Fig. 1A and Supplementary Fig. 1A), consistent with the reported mRNA expression levels of CLEC16A for these subsets (Gene Expression Atlas; www.ebi.ac.uk/gxa). In primary monocytes (n = 2), CLEC16A was mainly seen on vesicular membranes near the nuclear indentation (Fig. 1B and Supplementary Fig. 1B). CLEC16A levels were not altered in monocyte-derived macrophages (Supplementary Fig. 1C), but were ∼2-fold higher in monocyte-derived dendritic cells than in the monocytes from the same donor (Fig. 1C and Supplementary Fig. 1D, n = 7). We obtained comparable results after lipopolysaccharide-induced maturation of macrophages (Supplementary Fig. 1C) and monocyte-derived dendritic cells (Fig. 1C and Supplementary Fig. 1D and E, n = 7).
Figure 1.
CLEC16A is abundant in monocytes and monocyte-derived cells from peripheral blood of healthy donors. (A) Flow cytometric analysis of total CLEC16A expression levels in peripheral blood mononuclear cells (PBMC) of four healthy individuals (age/gender: 54/F, 48/M, 28/M, 56/F). Cells were stained with monoclonal antibodies against CD3, CD19, CD56 or CD14 and CLEC16A to evaluate CLEC16A mean fluorescence intensity in T, B and NK cells and monocytes (P = 0.0049, Kruskal-Wallis test). (B) Vesicular localization of CLEC16A around the nuclear indentation of monocytes from buffy coats, as determined by immunogold-labelling of rabbit anti-CLEC16A antibody and electron microscopy. This was observed for monocytes from two different donors. Scale bar = 500 nm. (C) Relative CLEC16A mRNA expression levels in monocytes and both unstimulated and LPS-stimulated monocyte-derived dendritic cells (moDC) of the same healthy donors (n = 7). (D) Correlation between CLEC16A and CIITA mRNA expression levels in immature monocyte-derived dendritic cells from similar donors (n = 9). (E) Representative immunofluorescence co-staining for CLEC16A and HLA-DR in immature and mature monocyte-derived dendritic cells, as analysed by confocal microscopy. Rabbit anti-CLEC16A and mouse anti-HLA-DR (L243 clone) antibodies were used. Scale bars = 10 μm. *P < 0.05; **P < 0.01.
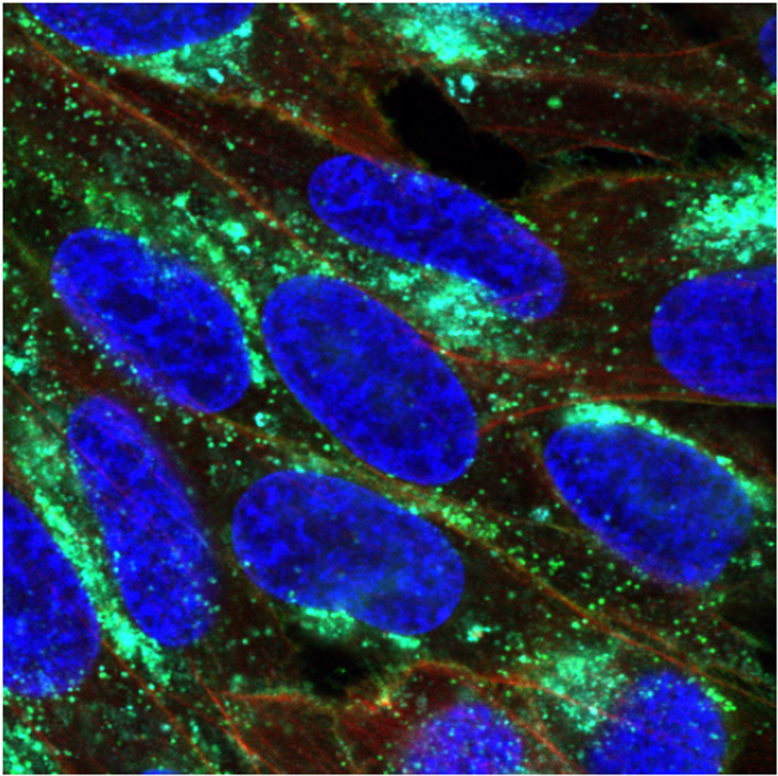
Interestingly, in monocyte-derived dendritic cells, mRNA expression levels of CLEC16A strongly correlated with those of its neighbouring gene, CIITA (r = 0.87, P = 0.005; Fig. 1D, n = 9). This was also found for CLEC16A and HLA-DRA mRNA levels (Supplementary Fig. 1F), suggesting coordinate expression of CLEC16A and HLA-II. Immunofluorescence assessment of immature monocyte-derived dendritic cells (n = 3) showed partial co-localization of CLEC16A with HLA-DR, particularly in perinuclear regions (Pearson’s correlation coefficient = 0.78; Fig. 1E). LPS-induced maturation of monocyte-derived dendritic cells (n = 3) resulted in a shift of CLEC16A and HLA-DR co-localization towards peripheral regions near or at the plasma membrane (Pearson’s correlation coefficient = 0.76; Fig. 1E). This co-localization was seen for different monocyte-derived dendritic cells (Supplementary Fig. 2). The strong expression of CLEC16A in primary monocytes and its association with HLA-II in monocyte-derived dendritic cells suggest a role of CLEC16A in late endosomal processing of HLA-II by antigen-presenting cells.
CLEC16A knockdown impairs the biogenesis of HLA class II-positive late endosomal compartments
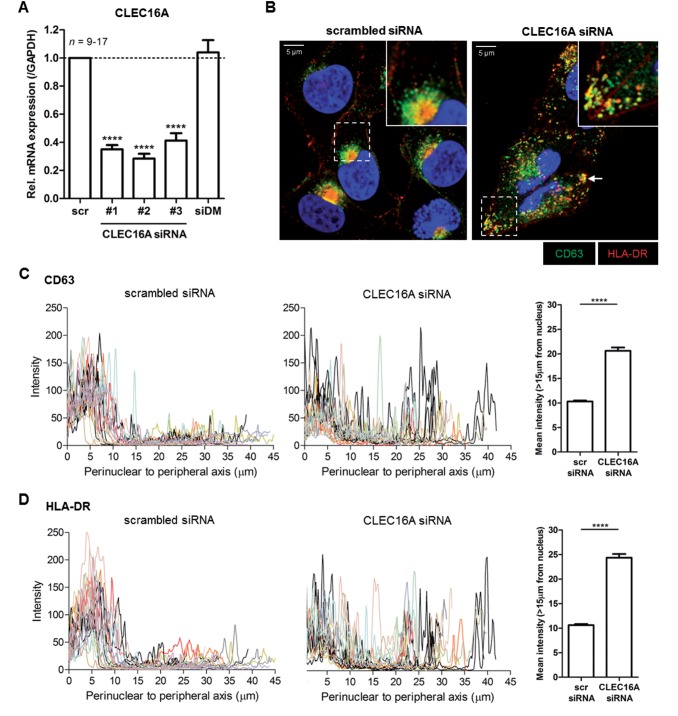
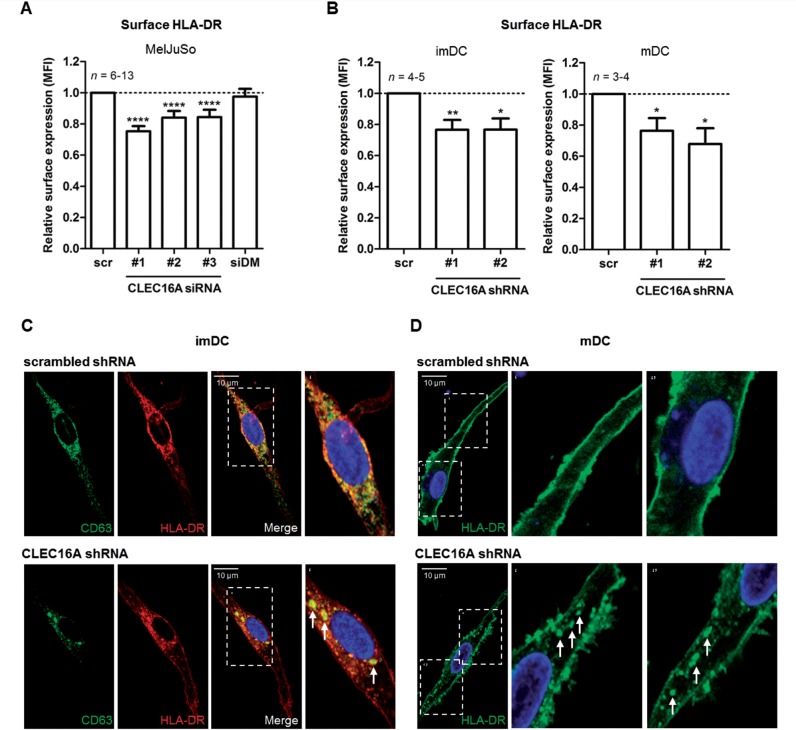
Drosophila mutants of CLEC16A orthologue Ema fail to form mature late endosomes and lysosomes (Kim et al., 2010). Additionally, several signalling motifs targeting antigens to HLA-II+ late endosomal compartments or MIIC (Mahnke et al., 2000) are present in CLEC16A (e.g. EDE triads; see Supplementary Fig. 3). To explore a role for CLEC16A in the HLA-II antigen presentation pathway in human antigen-presenting cells, we first used the melanoma MelJuSo cell line, which is a frequently used antigen-presenting cell model to study late endosomal and HLA-II biology (Wubbolts et al., 1996). CLEC16A siRNA transfections caused a 59–72% reduction in CLEC16A mRNA levels compared to scrambled siRNA (Fig. 2A). When analysing late endosomal distribution, both CD63+ and HLA-DR+ late endosomes were strongly dispersed in CLEC16A siRNA-treated cells, in contrast to the perinuclear localization in controls (Fig. 2B–D). These observations were made using several duplexes of CLEC16A siRNA (Supplementary Fig. 4A) and confirmed by assessing another late endosomal marker, Rab7, in co-localization with HLA-DR in Rab7-GFP+ MelJuSo cells (Supplementary Fig. 4B). The late endosomal scattering was reversed after re-expression of CLEC16A (Supplementary Fig. 5A and B). These observations provide strong evidence that CLEC16A silencing disrupts late endosomal biogenesis.
Figure 2.
CLEC16A knockdown in antigen-presenting cells causes dispersed localization of HLA-II-positive late endosomes. The human melanoma cell line MelJuSo was transfected with three distinct CLEC16A siRNA duplexes (#1, #2 and #3) and analysed after 3 days. (A) Relative CLEC16A mRNA levels in CLEC16A siRNA- compared to scrambled siRNA-treated cells (n = 9–17). Cells were also transfected with a pool of irrelevant HLA-DMβ siRNA as additional control. Immunofluorescence analysis (B) and quantification of the cytoplasmic localization of CD63+HLA-DR+ late endosomes (C and D) in CLEC16A siRNA- and scrambled siRNA-treated MelJuSo cells (n = 17–21). The images are representative of five experiments. Late endosomal scattering was quantified by analysing fluorescence intensities from the perinuclear area to the outermost peripheral regions (>15 μm from the nucleus; 15 μm cut-off based on scrambled siRNA histograms) for each cell using a line scan (LAS AF software). Scale bars = 5 μm. ****P < 0.0001.
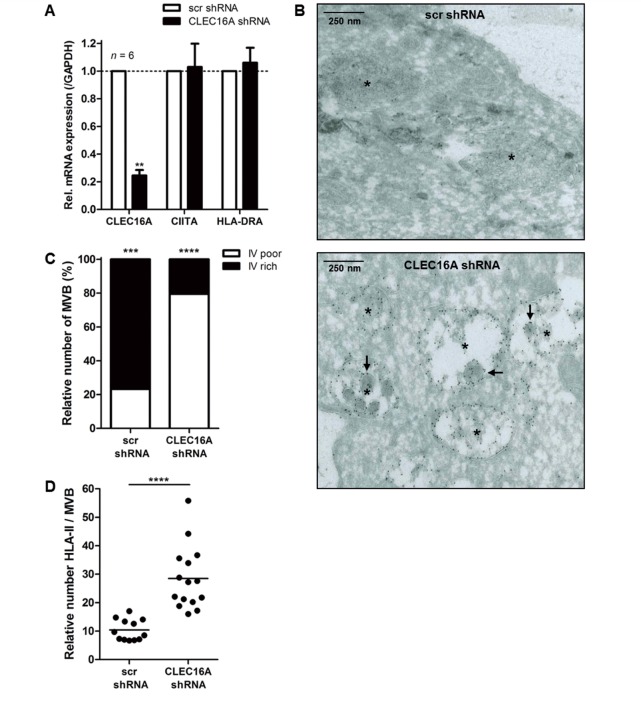
To verify these findings, we generated stable MelJuSo transductants containing scrambled or CLEC16A shRNA. CLEC16A mRNA expression levels were markedly decreased in CLEC16A shRNA transductants (mean reduction of 74%), whereas both CIITA and HLA-DRA mRNA levels remained unaffected (Fig. 3A). Electron microscopy revealed multivesicular bodies that completely lacked internal vesicles or consisted of a few remaining internal vesicles with an enlarged phenotype in CLEC16A shRNA transductants (Fig. 3B), indicating abnormal maturation of late endosomes. Three-fold more internal vesicle-poor than internal vesicle-rich multivesicular bodies were found (Fig. 3C). In addition, the number of HLA-II molecules in multivesicular bodies of CLEC16A shRNA transductants were increased 3-fold (Fig. 3B and D), without affecting multivesicular body size (Supplementary Fig. 5C). Together, this suggests that CLEC16A has an important role in the cytoplasmic localization and formation of MIIC, thereby regulating HLA-II expression in antigen-presenting cells.
Figure 3.
CLEC16A-silenced antigen-presenting cells contain abnormally matured late endosomes with elevated numbers of HLA-II. (A) Stable MelJuSo transductants with scrambled shRNA or CLEC16A shRNA were analysed for CLEC16A, CIITA and HLA-DRA mRNA levels by quantitative PCR (n = 6). In these cells, characteristic features of HLA-II+ late endosomes were assessed using electron microscopy (B). Both internal vesicle (IV)-poor and -rich multivesicular bodies (MVB) (C) and gold-labelled HLA-II molecules per MVB (D) were quantified. An asterisk is depicted in the centre of each multivesicular body and arrows indicate abnormally enlarged multivesicular bodies. Scale bars = 250 nm. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Silencing of CLEC16A leads to intracellular accumulation and reduced plasma membrane expression of HLA class II
In CLEC16A siRNA-treated MelJuSo cells, HLA-DR surface expression was reduced by 16–25% compared to controls (Fig. 4A). HLA-DRA mRNA expression levels were not reduced after CLEC16A siRNA transfection (Supplementary Fig. 5D), demonstrating that the reduction of HLA-DR surface levels by CLEC16A siRNA is independent of HLA-DR transcription. Simultaneous transfections with siRNA duplexes against HLA-DMβ caused a 2-fold upregulation of class II-associated invariant chain peptide (CLIP) surface expression (Supplementary Fig. 5E), which confirmed the efficacy of siRNA transfection. In addition, several foci of HLA-DR+ late endosomes were seen after CLEC16A siRNA treatment (Fig. 2B and Supplementary Fig. 4). This is consistent with the reduced HLA-DR cell surface expression (Fig. 4A), and the accumulation of HLA-II in multivesicular bodies as observed by electron microscopy (Fig. 3B and D) in MelJuSo cells.
Figure 4.
CLEC16A silencing disrupts both intracellular and surface expression of HLA-II in antigen-presenting cells. Relative cell surface expression levels of HLA-DR for (A) CLEC16A siRNA-treated MelJuSo cells (n = 6–13) as well as (B) CLEC16A shRNA-treated immature (imDC; n = 4–5) and mature (mDC; n = 3–4) monocyte-derived dendritic cells compared to scrambled controls, as determined by flow cytometry. (C and D) Intracellular distribution of late endosomes in scrambled and CLEC16A shRNA-treated (C) immature monocyte-derived dendritic cells (mouse anti-CD63 and rabbit anti-HLA-DR antibody) and (D) mature monocyte-derived dendritic cells (mouse anti-HLA-DR L243 antibody), as analysed by confocal microscopy. Images are representative for three different donors. Scale bars = 10 μm. *P < 0.05; **P < 0.01; ****P < 0.0001.
To substantiate this in primary antigen-presenting cells, CLEC16A expression was silenced in monocyte-derived dendritic cells using CLEC16A shRNA lentivirus (Supplementary Fig. 6A). In line with MelJuSo cells, immature and mature monocyte-derived dendritic cells showed a reduced HLA-DR surface expression (mean reduction of 20–38%; Fig. 4B, n = 3–5). In parallel, intracellular analysis revealed several foci of HLA-DR+ late endosomes in immature monocyte-derived dendritic cells (Fig. 4C and Supplementary Fig. 6B and C) and a more immature HLA-DR phenotype for maturation-induced monocyte-derived dendritic cells (Fig. 4D and Supplementary Fig. 6D and E) after transduction with CLEC16A shRNA. In conclusion, CLEC16A regulates intracellular and surface expression of HLA-DR also in primary antigen-presenting cells, likely by controlling the biogenesis of HLA-II+ late endosomes (Figs 2 and 3).
CLEC16A is directly involved in RILP-mediated recruitment of late endosomes to regulate HLA class II
In Drosophila, Ema directly interacts with the HOPS complex (Kim et al., 2010), a Rab7-activating complex consisting of different vacuolar protein sorting subunits that play a critical role in late endosomal biogenesis in yeast (Rink et al., 2005; Plemel et al., 2011). RILP also binds to the HOPS and dynein motor complex to transport Rab7+ late endosomes along microtubuli to the perinuclear microtubule-organizing centre (Jordens et al., 2001; van der Kant et al., 2013; Lin et al., 2014). As MIICs are localized directly around the microtubule-organizing centre (Wubbolts et al., 1999), we explored whether CLEC16A is involved in this late endosomal machinery to control HLA-II processing.
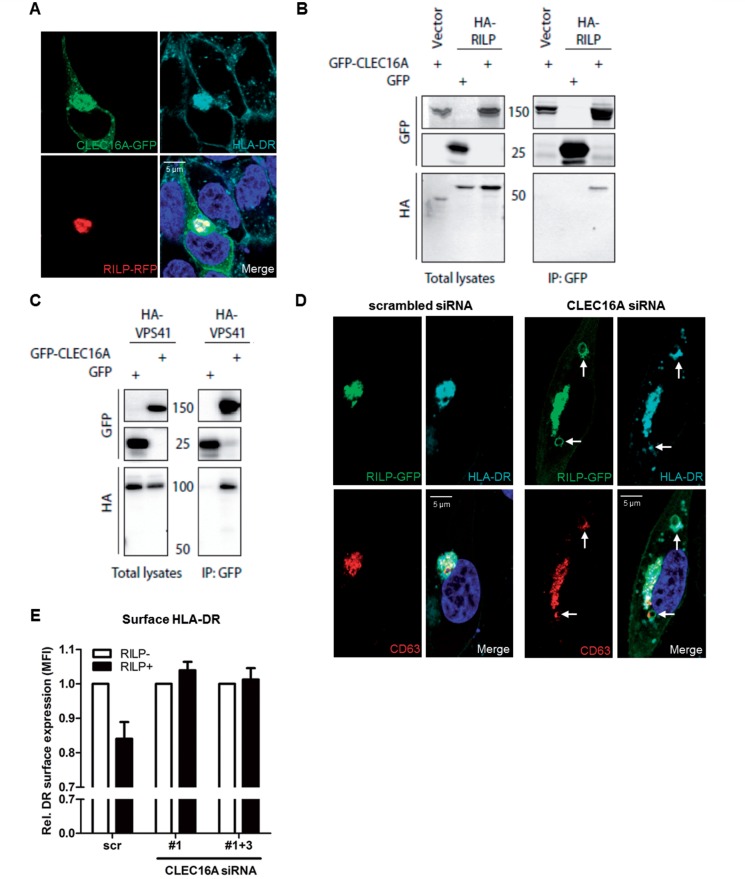
In CLEC16A-GFP treated MelJuSo cells, a large part of GFP+ vesicles co-localized with HLA-DR in perinuclear areas (Supplementary Fig. 7A). HLA-DR was further recruited to these regions after RILP-GFP transfection (Supplementary Fig. 7B). Cotransfections of MelJuSo cells with CLEC16A-GFP and RILP-RFP revealed recruitment of both CLEC16A and HLA-DR to the same perinuclear region as RILP (Fig. 5A and Supplementary Fig. 7C). This probably reflects the recruitment of HLA-DR+CLEC16A+ late endosomes to the microtubule-organizing centre by RILP.
Figure 5.
CLEC16A is required for RILP-mediated perinuclear recruitment of HLA-II-positive late endosomes. (A) MelJuSo cells transfected with CLEC16A-GFP and RILP-RFP constructs were assessed for intracellular localization with HLA-DR by confocal microscopy. (B and C) GFP coimmunoprecipitations of HEK293T cells cotransfected with CLEC16A-GFP and (B) RILP-HA or (C) VPS41-HA constructs. GFP pull-downs were analysed for HA expression on immunoblots. Immunoprecipitated empty GFP (‘GFP’) constructs served as negative controls. These data are representative for 2–3 experiments. (D and E) MelJuSo cells treated with scrambled or CLEC16A siRNA were transfected with a RILP-GFP construct. (D) Intracellular localization (n = 3) and (E) surface expression (n = 3) of HLA-DR was evaluated in RILP-GFP+and GFP− cells 3 days after siRNA transfection [RILP-GFP+ versus GFP− scrambled siRNA-treated cells (scr): P = 0.06]. Scale bars = 5 μm.
Next, we performed co-immunoprecipitation assays using cotransfected HEK293T cells to explore the association of CLEC16A with molecules of the late endosomal processing machinery. In pull-down reactions, CLEC16A-GFP associated with RILP-and VPS41-HA (Fig. 5B and C). Both RILP and VPS41 did not interact with GFP alone, confirming the specificity of all co-immunoprecipitations. The analyses of total lysates derived from cotransfectants showed a similar or higher expression of control GFP-only relative to CLEC16A-GFP (Fig. 5B and C).
To assess a functional role of CLEC16A in this machinery, we silenced CLEC16A expression and assessed the perinuclear accumulation of HLA-DR+ late endosomes mediated by RILP. After transfection with RILP-GFP, perinuclear localization of HLA-DR was markedly disturbed in CLEC16A siRNA-treated MelJuSo cells. Perinuclear foci of HLA-DR were elongated instead of round-shaped, in co-localization with RILP and CD63 (Fig. 5D and Supplementary Fig. 7D). Additional HLA-DR+CD63+ loci were found in other areas, of which the enlarged ones were positive for RILP (Fig. 5D and Supplementary Fig. 7E). HLA-DR surface expression decreased up to 16% in RILP-GFP+ as compared to GFP− cells treated with scrambled siRNA (Fig. 5E). This effect was abolished by CLEC16A silencing, since HLA-DR surface expression was not more reduced in RILP-GFP+ than in GFP− cells treated with CLEC16A siRNA (Fig. 5E). CLEC16A, CIITA and HLA-DRA mRNA levels were not altered by RILP upregulation (Supplementary Fig. 7F).
This reveals that CLEC16A is essential for the molecular machinery regulating the trafficking of HLA-II+ late endosomes. We argue that CLEC16A knockdown impairs late endosomal transport to perinuclear regions, thereby affecting MIIC formation and HLA-II processing and subsequent expression at the plasma membrane.
Vitamin D down-modulates CLEC16A and HLA class II expression in monocyte-derived dendritic cells
Interestingly, expression of the strongest genetic risk factor in multiple sclerosis, HLA-DRB1*1501, is modulated by vitamin D (Ramagopalan et al., 2009b), of which high serum levels associate with reduced multiple sclerosis risk (Munger et al., 2006). As vitamin D is able to limit the antigen presentation capacity of antigen-presenting cells (Penna et al., 2007; Mora et al., 2008), we explored whether vitamin D modulates CLEC16A expression in parallel with HLA-II in primary antigen-presenting cells.
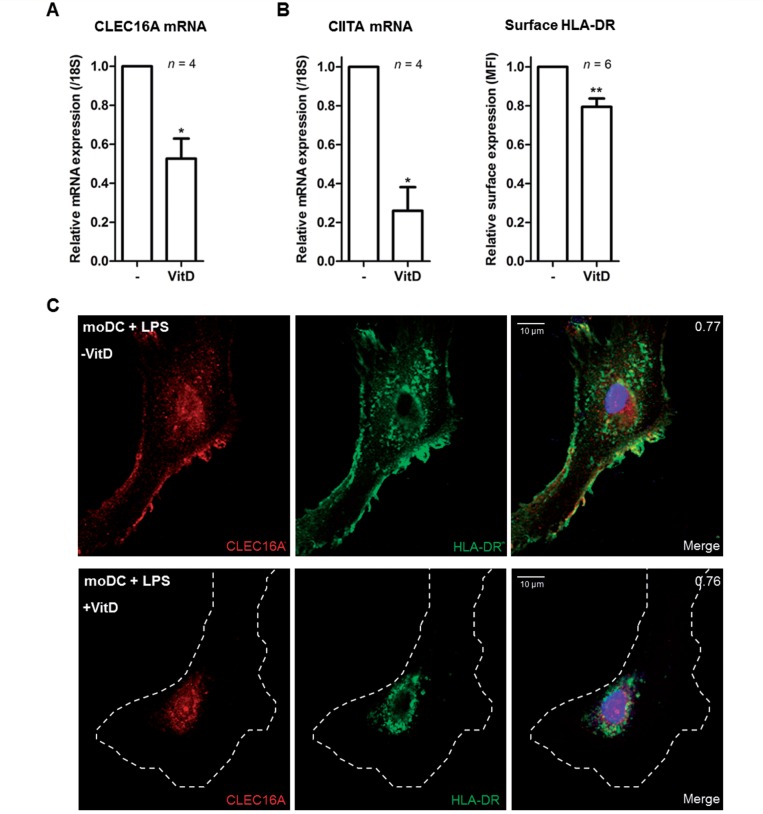
We cultured monocytes from healthy individuals with the active form of vitamin D (1,25[OH]2D) during differentiation into monocyte-derived dendritic cells. CLEC16A mRNA expression levels were significantly reduced in vitamin D-treated as compared to untreated monocyte-derived dendritic cells (Fig. 6A, n = 4). These differences positively correlated with CIITA mRNA and HLA-DR surface expression levels in monocyte-derived dendritic cells (Fig. 6B, n = 4–6). Vitamin D stimulation of immature monocyte-derived dendritic cells did not affect the perinuclear localization of CLEC16A and HLA-DR (data not shown). Vitamin D treatment of monocyte-derived dendritic cells did impair maturation-induced relocalization of both CLEC16A and HLA-DR towards peripheral areas and the plasma membrane (Fig. 6C; representative for three different donors). These findings show that one of the prime environmental factors in multiple sclerosis, vitamin D, strongly attenuates co-expression of CLEC16A and HLA-II in monocyte-derived dendritic cells, which is consistent with the presence of a vitamin D receptor binding site in their genes (Ramagopalan et al., 2010).
Figure 6.
CLEC16A and HLA-II are co-regulated by vitamin D in monocyte-derived dendritic cells. (A) Quantitative PCR to determine CLEC16A mRNA expression levels in unstimulated monocyte-derived dendritic cells and monocyte-derived dendritic cells stimulated with the active form of vitamin D (1,25[OH]2D). Treated and untreated monocyte-derived dendritic cells were analysed 7 days after monocyte differentiation. (B) CIITA mRNA and HLA-DR cell surface expression levels in untreated compared to vitamin D-treated monocyte-derived dendritic cells (moDC), as determined by quantitative PCR and flow cytometry, respectively. (C) Representative confocal image showing the localization of CLEC16A and HLA-DR in untreated and vitamin D-treated monocyte-derived dendritic cells after LPS treatment. The white dashed line represents the plasma membrane. The images are representative for three different donors and at least 20 monocyte-derived dendritic cells per donor were analysed. Scale bars = 10 μm. *P < 0.05; **P < 0.01.
CLEC16A expression is elevated in multiple sclerosis patients and localizes with HLA class II
To determine whether this novel mechanism of CLEC16A plays a role in autoimmune disease, we compared CLEC16A expression in primary cells and tissue between multiple sclerosis patients and control subjects.
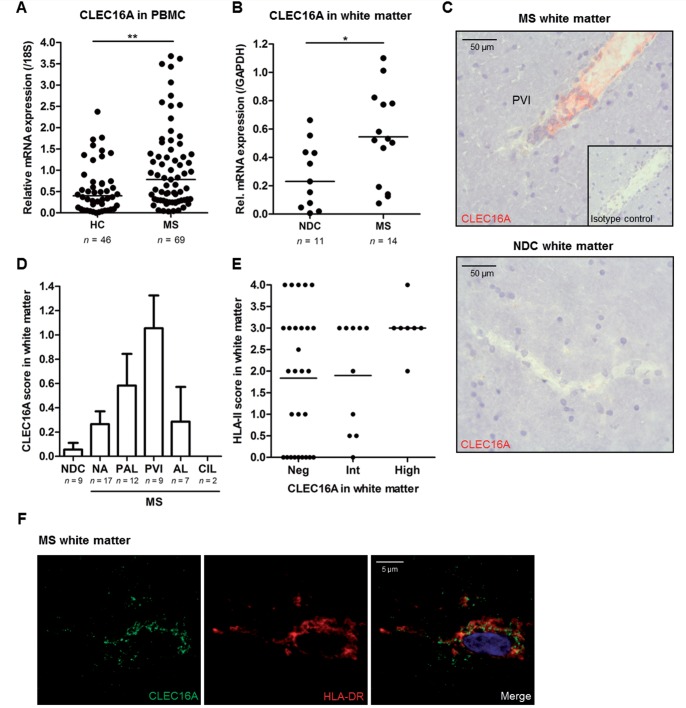
We first assessed CLEC16A expression levels in peripheral blood mononuclear cells from multiple sclerosis patients (n = 69) and healthy controls (n = 46; Supplementary Table 1). A 2-fold increase in CLEC16A mRNA expression was found for multiple sclerosis patients as compared to healthy controls (P = 0.003; Fig. 7A). The expression levels in multiple sclerosis patients did not significantly change after correcting for age and gender (P = 0.26 and P = 0.45, respectively; Supplementary Fig. 8A and B). In healthy controls, we found no association with gender (P = 0.09) and a biologically irrelevant correlation with age (R2 = 0.11, P = 0.03; Supplementary Fig. 8A and B), which was abrogated when corrected for gender (adjusted R2 = 0.09, P = 0.05; data not shown). After correction for treatment, CLEC16A mRNA levels remained significantly increased in multiple sclerosis patients compared to controls (P = 0.03; Supplementary Fig. 8C). Stratification according to multiple sclerosis risk SNP rs7200786[A] (Sawcer et al., 2011) did not affect CLEC16A mRNA levels in multiple sclerosis patients and healthy controls (P > 0.09; Supplementary Fig. 8D). When comparing these levels to monocyte, B cell and both CD4+ and CD8+ T cell frequencies in blood, only a weak correlation was found with CD4+ T cells in multiple sclerosis (R2 = 0.09, P = 0.02; Supplementary Fig. 9).
Figure 7.
CLEC16A expression is elevated in peripheral immune cells and brain tissue of multiple sclerosis patients. (A) Expression levels of CLEC16A mRNA in peripheral blood mononuclear cells (PBMC) of multiple sclerosis patients (MS; n = 69) and healthy controls (HC; n = 46). (B) CLEC16A mRNA expression levels in brain white matter tissues of multiple sclerosis (n = 14) and non-demented controls (NDC; n = 11). (C) Representative image of CLEC16A within a perivascular infiltrate (PVI) of a pre-active multiple sclerosis white matter lesion. Scale bars = 50 μm. (D) Quantification of CLEC16A expression in white matter of non-demented controls and in different regions of multiple sclerosis white matter. We used established arbitrary immunohistochemical scores defined by the number of positive cells in a particular area. AL = active lesion; CIL = chronic inactive lesion; PAL = pre-active lesion; PVI = perivascular infiltrate; NA = normal appearing. (E) Immunohistochemistry scores for HLA-II expression in multiple sclerosis white matter regions with negative, intermediate or high CLEC16A expression. Each dot represents a separate white matter region and a total of 18 multiple sclerosis patients were assessed. (F) Immunofluorescence analysis of CLEC16A and HLA-DR in multiple sclerosis white matter tissue using confocal microscopy. Co-expression of CLEC16A and HLA-DR was observed in three different patients. *P < 0.05, **P < 0.01.
In white matter tissue, CLEC16A mRNA levels were 4-fold elevated in multiple sclerosis cases (n = 14) compared to non-demented controls (n = 11, Supplementary Table 1; P = 0.03; Fig. 7B). Additionally, immunohistochemistry for CLEC16A and HLA-II in white matter tissue from 18 patients with multiple sclerosis revealed increased frequencies of CLEC16A-positive cells in pre-active lesions (n = 12), primarily in perivascular lymphocytic infiltrates (n = 9), in contrast to multiple sclerosis normal-appearing and non-demented control white matter (n = 17 and n = 9, respectively; Fig. 7C and D). Multiple sclerosis white matter regions that were strongly positive for CLEC16A also had high expression of HLA-II (Fig. 7E). This expression did not always correlate with CLEC16A, since HLA-II was also present in some CLEC16A-negative regions (Fig. 7E). Subsequent immunofluorescence analysis showed co-expression of CLEC16A with HLA-DR in multiple sclerosis white matter (Fig. 7F, representative for three patients). The increased expression of CLEC16A in multiple sclerosis patients and its direct role in the HLA-II pathway strongly suggest that CLEC16A-mediated regulation of HLA-II antigen presentation is an underlying mechanism in multiple sclerosis.
Discussion
The majority of novel multiple sclerosis risk genes identified in genome-wide association studies are immune-related and are shared with many other autoimmune diseases (Zhernakova et al., 2009; Sawcer et al., 2011; Cotsapas and Hafler, 2012). To unravel the contribution of these novel risk genes in the cascade that leads to autoimmune demyelination in multiple sclerosis, it is imperative to determine their biological functions. In silico analysis of findings from genome-wide association studies in multiple sclerosis suggested CD4+T helper cell activation and differentiation as the principal pathogenic mechanism (Sawcer et al., 2011). Recent studies by our group and others have shown functional alterations of multiple sclerosis risk genes that are primarily expressed by T cells (Gregory et al., 2012; Kreft et al., 2012a, b). Here, we reveal that the expression of CLEC16A, a gene located at the 16p13 risk locus, is associated with multiple sclerosis and has a previously unknown role in the HLA-II antigen presentation pathway.
Knockdown experiments in different antigen-presenting cells demonstrate a clear role of CLEC16A in the machinery regulating both the cytoplasmic localization and maturation of HLA-II+ late endosomal compartments. The localization of endosomes is critical for the efficiency of HLA-II antigen presentation and is directly associated with their maturation (Kleijmeer et al., 2001; Huotari and Helenius, 2011). During maturation, HLA-II+ late endosomes are transported to perinuclear areas to undergo acidification, activating proteolytic enzymes that process antigens for peptide loading onto HLA-II molecules (Rocha and Neefjes, 2008). This endosomal maturation and transport to perinuclear regions is controlled by the dynein motor complex, which forms a bridge between the microtubuli and Rab7+ late endosomes (Huotari and Helenius, 2011). The association of CLEC16A with two main regulators of this machinery, RILP and HOPS (Fig.5), suggests that CLEC16A uses this mechanism to control HLA-II antigen presentation via the MIIC. This is supported by our recent identification of RILP as a molecule that controls CLIP surface expression (Paul et al., 2011).
This novel function of CLEC16A is different from that of other types of C-type lectins. For instance, CLEC9A supports the cross-presentation to CD8+ T cells, but does not influence classical MHC-II antigen presentation in mice (Zelenay et al., 2012). By contrast, CLECL1, which is localized at another multiple sclerosis risk locus (Sawcer et al., 2011), functions as a cell surface receptor to increase HLA-II expression in dendritic cells (Ryan et al., 2009). DC-SIGN and DEC-205 also regulate HLA-II antigen presentation by targeting antigens to the MIIC (Mahnke et al., 2000; Engering et al., 2002). We now demonstrate that CLEC16A is the first member of the C-type lectin family that controls HLA-II antigen presentation by regulating the biogenesis of MIIC. As Ema is also associated with autophagosome formation in Drosophila (Kim et al., 2012), CLEC16A may also participate in autophagy to mediate HLA-II antigen presentation (Schmid et al., 2007; Soleimanpour et al., 2014).
In addition to its function, we demonstrate an evident link of CLEC16A expression with multiple sclerosis pathogenesis. CLEC16A expression in peripheral blood mononuclear cells and white matter, especially in perivascular lymphocytic infiltrates, of multiple sclerosis cases was strongly elevated (Fig. 7). Conflicting results have been reported on the genetic link of the CLEC16A region to multiple sclerosis. In our study, we did not find an association between CLEC16A mRNA levels in blood and the rs7200786[A] risk SNP, confirming the ENCODE study (Bernstein et al., 2012; Boyle et al., 2012, 2014; Xie et al., 2013). Some studies demonstrate risk genotype effects on the expression of CLEC16A itself (Mero et al., 2011; Soleimanpour et al., 2014), whereas others have reported effects on its neighbouring genes, such as DEXI (Leikfoss et al., 2013; Raj et al., 2014). These findings indicate that expression related to genotypes is probably cell type-specific. For example, the correlation with DEXI is mainly found in CD4+ T cells (Raj et al., 2014), a cell type in which CLEC16A expression is low (Fig. 1A and Supplementary Fig. 1A). In addition, genotype effects on specific transcripts of CLEC16A have been found in the thymus, but not in blood (Mero et al., 2011), indicating that alternative splicing and thereby potentially gene function are affected. Another possible underlying mechanism is the selective regulation of CLEC16A gene expression by vitamin D (Ramagopalan et al., 2010), which also controls the expression of the major risk locus in multiple sclerosis, HLA-DRB1 (Ramagopalan et al., 2009b).
The coupling of CLEC16A to the function of HLA-II indicates that in addition to genetic studies, functional evaluation of non-HLA risk genes in similar biological pathways is another attractive strategy to uncover pathogenic mechanisms in multiple sclerosis (International Multiple Sclerosis Genetics Consortium, 2013). Substantial changes in antigen presentation pathways, which is a generally under-appreciated mechanism in the multiple sclerosis field, are assumed to mediate presentation of cryptic peptides (Vanderlugt and Miller, 2002). The increased expression of CLEC16A in multiple sclerosis points to an autoimmune model in which it promotes the biogenesis of MIIC to affect HLA-II antigen presentation by antigen-presenting cells (Fig. 8). Such a role of CLEC16A in HLA-II regulation and multiple sclerosis is likely restricted to selective antigen-presenting cell subsets, in particular to dendritic cells rather than B cells (Zouk et al., 2014). This may then alter T cell responses in the periphery, e.g. by contributing to impaired T cell quiescence in patients with multiple sclerosis (Corvol et al., 2008). The exact aetiological position of CLEC16A, the way it affects T cell responses, either in the periphery or during thymic T cell development, and its role in establishing self-tolerance should be addressed in further studies.
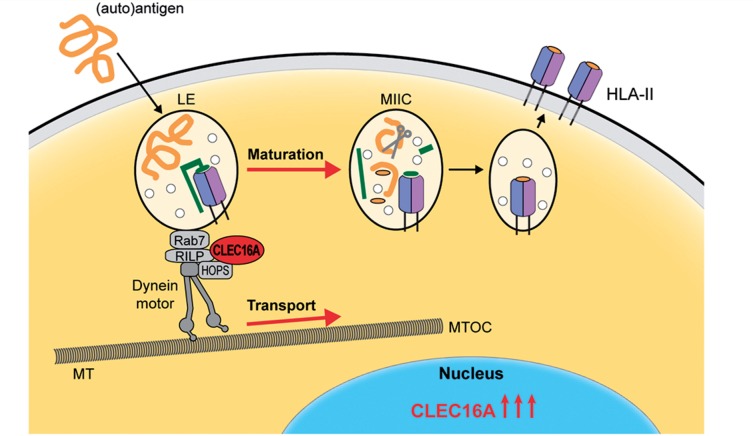
Figure 8.
Key role of CLEC16A in antigen-presenting cells and multiple sclerosis. Based on the results of our study, we postulate that CLEC16A expression is elevated in multiple sclerosis to promote HLA-II antigen presentation by antigen-presenting cells. CLEC16A controls the HLA-II pathway by participating in the molecular machinery of late endosomal biogenesis, which involves the dynein motor complex. Binding of CLEC16A to this complex via RILP and HOPS probably triggers the transport of late endosomes (LE) along microtubuli and stimulate their maturation into specialized antigen-loading compartments, termed MIIC. In these compartments, HLA-II molecules are loaded with peptides for presentation at the plasma membrane. The small circles depicted in late endosomes and the MIIC represent internal vesicles of multivesicular bodies. In green, the invariant chain and its cleavage product, CLIP, are indicated, which are crucial for efficient HLA-II transport to late endosomes and loading with peptides, respectively. MS = multiple sclerosis; MT = microtubuli; MTOC = microtubule-organizing centre; RILP = rab7-interacting lysosomal protein.
In conclusion, we demonstrate that an autoimmunity-related gene, CLEC16A, has a novel C-type lection function by regulating the strongest genetic risk factor in multiple sclerosis, HLA-II. This study takes a first step in coupling the function of non-HLA susceptibility genes to elucidate underlying mechanisms of multiple sclerosis pathogenesis. The identification of multiple sclerosis-associated genes regulating HLA-II function, such as CLEC16A, supports the development of strategies that selectively manipulate HLA-II antigen presentation in antigen-presenting cells (van Kasteren et al., 2014), which will be applicable for multiple sclerosis and a broad spectrum of other autoimmune diseases.
Acknowledgements
We would like to acknowledge R. Huizinga, PhD (Erasmus MC, Rotterdam) and P. Paul, PhD (Netherlands Cancer Institute, Amsterdam) for sharing their experience in working with moDC. We also thank L. Brocks as well as L. Oomen of the Digital Microscopy Facility (Netherlands Cancer Institute, Amsterdam), as well as G. Kremers, PhD, G. van Cappellen, PhD, and A. Nigg, PhD, of the Optical Imaging Centre (Erasmus MC, Rotterdam) for their support in confocal microscopy. We acknowledge B. Wang, PhD (Child and Family Research Institute, Vancouver) for technical assistance.
Glossary
Abbreviations
- CLIP
class II-associated invariant chain peptide
- HLA-II
human leukocyte antigen class II
- HOPS
homotypic fusion and vacuole protein sorting
- MIIC
MHC class II-containing compartment
- siRNA
small interfering RNA
- shRNA
short hairpin RNA
Funding
This work was financially supported by the Dutch MS Research Foundation (grant program no. 10-490 c MS to R.H.) and the Canadian Institute of Health Research (grant MOP-10266 to R.T. and J.P.). We thank the Zabawas Foundation for additional financial support.
Supplementary material
Supplementary material is available at Brain online.
References
- Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–3. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge T, Leikfoss IS, Harbo HF. From identification to characterization of the multiple sclerosis susceptibility gene CLEC16A. Int J Mol Sci. 2013;14:4476–97. doi: 10.3390/ijms14034476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Araya CL, Brdlik C, Cayting P, Cheng C, Cheng Y, et al. Comparative analysis of regulatory information and circuits across distant species. Nature. 2014;512:453–6. doi: 10.1038/nature13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat J, Janssen H, Stahle-Backdahl M, Zuurbier AE, Knol EF, Egesten A. Human monocytes and neutrophils store transforming growth factor-alpha in a subpopulation of cytoplasmic granules. Blood. 1997;90:1255–66. [PubMed] [Google Scholar]
- Choi NM, Majumder P, Boss JM. Regulation of major histocompatibility complex class II genes. Curr Opin Immunol. 2011;23:81–7. doi: 10.1016/j.coi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvol JC, Pelletier D, Henry RG, Caillier SJ, Wang J, Pappas D, et al. Abrogation of T cell quiescence characterizes patients at high risk for multiple sclerosis after the initial neurological event. Proc Natl Acad Sci USA. 2008;105:11839–44. doi: 10.1073/pnas.0805065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsapas C, Hafler DA. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends Immunol. 2012;34:22–6. doi: 10.1016/j.it.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engering A, Geijtenbeek TB, van Vliet SJ, Wijers M, van Liempt E, Demaurex N, et al. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J Immunol. 2002;168:2118–26. doi: 10.4049/jimmunol.168.5.2118. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9:465–79. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek TB, van Vliet SJ, Engering A, ‘t Hart BA, van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- Gregory AP, Dendrou CA, Attfield KE, Haghikia A, Xifara DK, Butter F, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–11. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–4. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- Hoppenbrouwers IA, Aulchenko YS, Janssens AC, Ramagopalan SV, Broer L, Kayser M, et al. Replication of CD58 and CLEC16A as genome-wide significant risk genes for multiple sclerosis. J Hum Genet. 2009;54:676–80. doi: 10.1038/jhg.2009.96. [DOI] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium. Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. Am J Hum Genet. 2013;92:854–65. doi: 10.1016/j.ajhg.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–5. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- Kim S, Naylor SA, DiAntonio A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc Natl Acad Sci USA. 2012;109:E1072–81. doi: 10.1073/pnas.1120320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Wairkar YP, Daniels RW, DiAntonio A. The novel endosomal membrane protein Ema interacts with the class C Vps-HOPS complex to promote endosomal maturation. J Cell Biol. 2010;188:717–34. doi: 10.1083/jcb.200911126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijmeer M, Ramm G, Schuurhuis D, Griffith J, Rescigno M, Ricciardi-Castagnoli P, et al. Reorganization of multivesicular bodies regulates MHC class II antigen presentation by dendritic cells. J Cell Biol. 2001;155:53–63. doi: 10.1083/jcb.200103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft KL, Verbraak E, Wierenga-Wolf AF, van Meurs M, Oostra BA, Laman JD, et al. Decreased systemic IL-7 and soluble IL-7Ralpha in multiple sclerosis patients. Genes Immun. 2012a;13:587–92. doi: 10.1038/gene.2012.34. [DOI] [PubMed] [Google Scholar]
- Kreft KL, Verbraak E, Wierenga-Wolf AF, van Meurs M, Oostra BA, Laman JD, et al. The IL-7Ralpha pathway is quantitatively and functionally altered in CD8 T cells in multiple sclerosis. J Immunol. 2012b;188:1874–83. doi: 10.4049/jimmunol.1102559. [DOI] [PubMed] [Google Scholar]
- Leikfoss IS, Mero IL, Dahle MK, Lie BA, Harbo HF, Spurkland A, et al. Multiple sclerosis-associated single-nucleotide polymorphisms in CLEC16A correlate with reduced SOCS1 and DEXI expression in the thymus. Genes Immun. 2013;14:62–6. doi: 10.1038/gene.2012.52. [DOI] [PubMed] [Google Scholar]
- Lin X, Yang T, Wang S, Wang Z, Yun Y, Sun L, et al. RILP interacts with HOPS complex via VPS41 subunit to regulate endocytic trafficking. Sci Rep. 2014;4:7282. doi: 10.1038/srep07282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke K, Guo M, Lee S, Sepulveda H, Swain SL, Nussenzweig M, et al. The dendritic cell receptor for endocytosis, DEC-205, can recycle and enhance antigen presentation via major histocompatibility complex class II-positive lysosomal compartments. J Cell Biol. 2000;151:673–84. doi: 10.1083/jcb.151.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–32. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mero IL, Ban M, Lorentzen AR, Smestad C, Celius EG, Saether H, et al. Exploring the CLEC16A gene reveals a MS-associated variant with correlation to the relative expression of CLEC16A isoforms in thymus. Genes Immun. 2011;12:191–8. doi: 10.1038/gene.2010.59. [DOI] [PubMed] [Google Scholar]
- Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol. 2008;8:685–98. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger KL, Levin LI, Hollis BW, Howard NS, Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA. 2006;296:2832–8. doi: 10.1001/jama.296.23.2832. [DOI] [PubMed] [Google Scholar]
- Neefjes JJ, Stollorz V, Peters PJ, Geuze HJ, Ploegh HL. The biosynthetic pathway of MHC class II but not class I molecules intersects the endocytic route. Cell. 1990;61:171–83. doi: 10.1016/0092-8674(90)90224-3. [DOI] [PubMed] [Google Scholar]
- Paul P, van den Hoorn T, Jongsma ML, Bakker MJ, Hengeveld R, Janssen L, et al. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell. 2011;145:268–83. doi: 10.1016/j.cell.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Penna G, Amuchastegui S, Giarratana N, Daniel KC, Vulcano M, Sozzani S, et al. 1,25-Dihydroxyvitamin D3 selectively modulates tolerogenic properties in myeloid but not plasmacytoid dendritic cells. J Immunol. 2007;178:145–53. doi: 10.4049/jimmunol.178.1.145. [DOI] [PubMed] [Google Scholar]
- Peters PJ, Raposo G, Neefjes JJ, Oorschot V, Leijendekker RL, Geuze HJ, et al. Major histocompatibility complex class II compartments in human B lymphoblastoid cells are distinct from early endosomes. J Exp Med. 1995;182:325–34. doi: 10.1084/jem.182.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemel RL, Lobingier BT, Brett CL, Angers CG, Nickerson DP, Paulsel A, et al. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–63. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58:840–6. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–23. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–60. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Knight JC, Ebers GC. Multiple sclerosis and the major histocompatibility complex. Curr Opin Neurol. 2009a;22:219–25. doi: 10.1097/WCO.0b013e32832b5417. [DOI] [PubMed] [Google Scholar]
- Ramagopalan SV, Maugeri NJ, Handunnetthi L, Lincoln MR, Orton SM, Dyment DA, et al. Expression of the multiple sclerosis-associated MHC class II Allele HLA-DRB1*1501 is regulated by vitamin D. PLoS Genet. 2009b;5:e1000369. doi: 10.1371/journal.pgen.1000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–49. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185:1209–25. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N, Neefjes J. MHC class II molecules on the move for successful antigen presentation. EMBO J. 2008;27:1–5. doi: 10.1038/sj.emboj.7601945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan EJ, Magaletti D, Draves KE, Clark EA. Ligation of dendritic cell-associated lectin-1 induces partial maturation of human monocyte derived dendritic cells. Hum Immunol. 2009;70:1–5. doi: 10.1016/j.humimm.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–19. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, et al. The diabetes susceptibility gene clec16a regulates mitophagy. Cell. 2014;157:1577–90. doi: 10.1016/j.cell.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant R, Fish A, Janssen L, Janssen H, Krom S, Ho N, et al. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci. 2013;126:3462–74. doi: 10.1242/jcs.129270. [DOI] [PubMed] [Google Scholar]
- van der Valk P, De Groot CJ. Staging of multiple sclerosis (MS) lesions: pathology of the time frame of MS. Neuropathol Appl Neurobiol. 2000;26:2–10. doi: 10.1046/j.1365-2990.2000.00217.x. [DOI] [PubMed] [Google Scholar]
- van Kasteren SI, Overkleeft H, Ovaa H, Neefjes J. Chemical biology of antigen presentation by MHC molecules. Curr Opin Immunol. 2014;26:21–31. doi: 10.1016/j.coi.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Vanderlugt CL, Miller SD. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat Rev Immunol. 2002;2:85–95. doi: 10.1038/nri724. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Fernandez-Borja M, Jordens I, Reits E, Dusseljee S, Echeverri C, et al. Opposing motor activities of dynein and kinesin determine retention and transport of MHC class II-containing compartments. J Cell Sci. 1999;112(Pt 6):785–95. doi: 10.1242/jcs.112.6.785. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Fernandez-Borja M, Oomen L, Verwoerd D, Janssen H, Calafat J, et al. Direct vesicular transport of MHC class II molecules from lysosomal structures to the cell surface. J Cell Biol. 1996;135:611–22. doi: 10.1083/jcb.135.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Boyle AP, Wu L, Zhai J, Kawli T, Snyder M. Dynamic trans-acting factor colocalization in human cells. Cell. 2013;155:713–24. doi: 10.1016/j.cell.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S, Keller AM, Whitney PG, Schraml BU, Deddouche S, Rogers NC, et al. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J Clin Invest. 2012;122:1615–27. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- Zouk H, D'Hennezel E, Du X, Ounissi-Benkalha H, Piccirillo CA, Polychronakos C. Functional evaluation of the role of C-type lectin domain family 16A at the chromosome 16p13 locus. Clin Exp Immunol. 2014;175:485–97. doi: 10.1111/cei.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuvich RL, Bush WS, McCauley JL, Beecham AH, De Jager PL, Ivinson AJ, et al. Interrogating the complex role of chromosome 16p13.13 in multiple sclerosis susceptibility: independent genetic signals in the CIITA-CLEC16A-SOCS1 gene complex. Hum Mol Genet. 2011;20:3517–24. doi: 10.1093/hmg/ddr250. [DOI] [PMC free article] [PubMed] [Google Scholar]