Abstract
Zinc finger E-box binding homeobox 1 (ZEB1) is a transcription factor that promotes tumor invasion and metastasis by inducing epithelial-mesenchymal transition (EMT) in carcinoma cells. EMT not only plays an important role in embryonic development and malignant progression, but is also implicated in cancer therapy resistance. It has been hypothesized that carcinoma cells that have undergone EMT acquire cancer stem cell properties including self-renewal, chemoresistance and radioresistance. However, our recent data indicate that ZEB1 regulates radioresistance in breast cancer cells through an EMT-independent mechanism. In this Perspective, we review different mechanisms by which ZEB1 regulates tumor progression and treatment resistance. Based on studies by us and others, we propose that it is specific EMT inducers like ZEB1, but not the epithelial or mesenchymal state itself, that dictate cancer stem cell properties.
Keywords: epithelial-mesenchymal transition, drug resistance, metastasis, radioresistance, ZEB1
Introduction
Epithelial-mesenchymal transition (EMT), originally observed during embryogenesis,1 plays important roles in early developmental processes, including neural crest development, heart valve development, mesoderm formation and secondary palate formation.2-4 It has been recognized that the EMT program can be resurrected in adult tissues under several disease conditions, including wound healing, fibrosis and cancer.2-4 EMT and its reverse process, mesenchymal-epithelial transition (MET), are involved in different stages of metastasis. The induction of EMT in epithelial cancer cells promotes migration, invasion and dissemination, whereas MET facilitates metastatic colonization of distant sites by disseminated tumor cells.5-7 Recently, mesenchymal-like tumor cells generated by EMT have been shown to exhibit characteristics of cancer stem cells, including self-renewal, radioresistance and drug resistance.8-12 Thus, research on EMT and MET will not only advance our understanding of tumor progression, but also shed light on improving cancer treatment.
Over the past decade, extensive studies have been performed to investigate the role and regulation of EMT and MET in tumor progression.5-7,13 A growing list of EMT regulators has been identified, including extracellular factors (such as TGF-β, HGF, FGF, IGF and Notch ligands), transcription factors (such as Twist, Snail, Slug and ZEB1), microRNAs (such as the miR-200 family, miR-205 and miR-9) and microenvironment.5,13-15 Although the association between EMT and cancer stem cell properties has been recognized by many recent studies, new evidence suggests that EMT-independent mechanisms exist and that all EMT inducers are not equal.16,17 For instance, while the transcription factor ZEB1 can promote migration and invasion by inducing EMT, this protein enhances tumor radioresistance independent of EMT.16 In this article, we review EMT-dependent and EMT-independent mechanisms by which ZEB1 regulates development, tumor progression and therapy resistance.
ZEB1 Protein and Its Physiological Functions
Expression of ZEB1 (also named TCF8 or DeltaEF1) is regulated by multiple signaling pathways, such as WNT,18 NF-κB,19 TGF-β,20 COX2,21 HIF signaling22 and microRNAs.23-25 ZEB1 belongs to the ZEB family of transcription factors characterized by the presence of 2 zinc finger clusters, which are responsible for DNA binding, and a centrally located homeodomain. In addition, ZEB1 contains other protein binding domains including the Smad interaction domain (SID), CtBP interaction domain (CID) and p300-P/CAF binding domain (CBD)26-29 (Fig. 1A). Through zinc finger clusters, ZEB1 can bind to specific DNA sequences named E-boxes.28 By recruiting co-suppressors or co-activators through CID, SID or CBD, ZEB1 can either downregulate or upregulate the expression of its target genes.27,30 For instance, ZEB1 directly binds to the E-box located in the promoter of CDH1, the gene encoding E-cadherin,30 and recruits the CtBP transcriptional co-repressors31 and/or the SWI/SNF chromatin-remodeling protein BRG1,32 leading to repression of CDH1 transcription and induction of EMT. On the other hand, by recruiting p300-P/CAF and Smad proteins, ZEB1 can activate the transcription of TGF-β-responsive genes and promote osteoblastic differentiation.26,27 In MDA-MB-231 human breast cancer cells, knockdown of ZEB1 resulted in upregulation of ∼200 genes and downregulation of ∼30 genes, most of which are determinants of epithelial differentiation and cell-cell adhesion.
Figure 1.
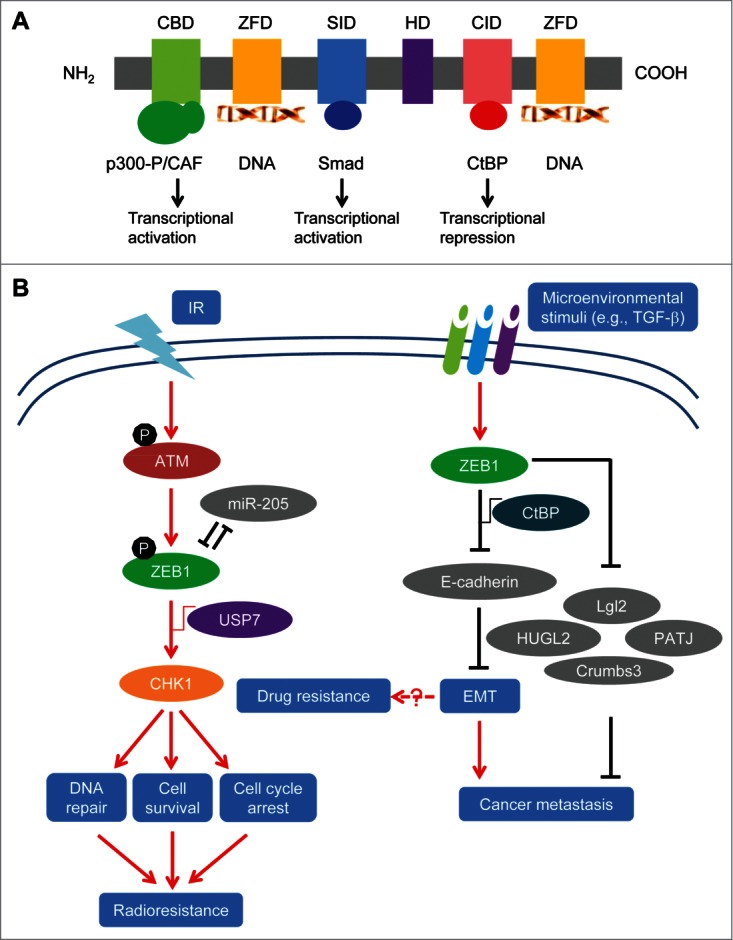
ZEB1 protein domains and mechanisms of action. (A) Schematic representation of ZEB1 protein. CBD: coactivator p300 and P/CAF binding domain; ZFD: zinc finger domain; SID: Smad interaction domain; HD: homeodomain; CID: CtBP interaction domain. (B) The working model of regulation of EMT, metastasis and therapy resistance by ZEB1. (i) Upon ionizing radiation (IR), ZEB1 is phosphorylated and stabilized by activated ATM kinase, and ZEB1 in turn recruits USP7 which deubiquitinates and stabilizes CHK1, leading to induction of DNA repair, cell survival and radioresistance. In parallel, ZEB1 represses its own negative regulator, miR-205. (ii) Microenvironmental stimuli such as TGF-β upregulate ZEB1 expression, and ZEB1 can promote cancer metastasis by repressing E-cadherin expression and inducing EMT, or by repressing other target genes (such as Lgl2, PATJ, HUGL2 and Crumbs3). In addition, ZEB1 can promote drug resistance through EMT-dependent and EMT-independent mechanisms. Gray color indicates ZEB1's transcriptional targets.
When Zeb1 was first identified as a repressor of δ1-crystallin enhancer in the chicken embryo in early 1990s, it was thought to be involved in embryogenesis because Zeb1 is specifically expressed in mesodermal tissues, the nerve system and the lens in the chicken embryo.34 Later, mouse models with Zeb1 deletion revealed the role of Zeb1 in mammalian development. While heterozygous Zeb1 mouse mutants are viable and fertile, Zeb1 null mice die perinatally exhibiting respiratory failure, severe T cell deficiency of the thymus, and various skeletal defects including craniofacial abnormalities, limb and sternum defects, and malformed ribs.35 Interestingly, these developmental defects are associated with a mesenchymal-epithelial transition, as evidenced by re-expression of E-cadherin and loss of vimentin in several embryonic tissues (including the palate and nasal mesenchyme, the perichondrial region of forming cartilage, the embryonic eye and the ventricular zone of the brain) and in embryonic fibroblasts.36 This finding underscores the critical role of Zeb1 in developmental EMT. In addition, either heterozygous or homozygous inactivation of Zeb1 in mouse embryos results in corneal defects, which mimics human posterior polymorphous corneal dystrophy.37 In humans, ZEB1 mRNA expression levels vary significantly among different adult tissues, from barely detectable expression in the pancreas and liver, to moderate expression in the mammary gland and ovary, and to high expression in the bladder and uterus.38 However, the function of ZEB1 in adult tissues is still largely unknown. Therefore, mouse models with conditional Zeb1 deletion or overexpression will not only shed light on the physiological functions of Zeb1 in adult tissues, but also provide tools to investigate the role of Zeb1 in cancer and other diseases.
ZEB1 in Tumor Progression and Metastasis
E-cadherin, a major cell-cell adhesion molecule, has long been recognized as a tumor suppressor protein.39 Loss of E-cadherin leads to tumor cell dissociation and enhanced ability to migrate, invade and metastasize, which is associated with poor prognosis in human cancer patients.5,40-42 Due to the pivotal role of ZEB1 in the downregulation of E-cadherin, ZEB1 acts as a driver of EMT and cancer progression.16,28,33,34,44 Aberrant expression of ZEB1 has been observed in many human cancers, such as uterine cancer,45 pancreatic cancer,29,46 osteosarcoma,47 lung cancer,48,49 liver cancer,50 gastric cancer,51,52 colon cancer53 and breast cancer.30 In these tumors, ZEB1 expression correlates with loss of E-cadherin and is associated with advanced diseases or metastasis, which indicates the relevance of ZEB1 induction of EMT and tumor progression in clinical cancers. This notion is further supported by functional studies using human cancer cell lines, in which overexpression of ZEB1 can induce EMT and promote metastasis in multiple cell lines, such as breast cancer,30 colon cancer43 and lung cancer54 cell lines.
In addition to repressing E-cadherin expression, ZEB1 also regulates other target genes involved in tumor progression (Fig. 1B). For example, ZEB1 binds to the promoters of epithelial polarity genes including Crumbs3, HUGL2 and PATJ (Pals1-associated tight junction) and represses their transcription, leading to reduced adhesion and increased invasiveness of breast cancer cells.33 In colorectal cancer cells, ZEB1 promotes metastasis and loss of cell polarity by repressing the expression of Lgl2 (lethal giant larvae homolog 2),43 a cell polarity factor. Moreover, ZEB1 enhances the tumorigenicity of pancreatic cancer cells by inhibiting the expression of stemness-repressing microRNAs, including miR-203 and the miR-200 family.55 It is not clear, however, what relationship these various ZEB1 target genes have: synergistic, additive or independent?
ZEB1 in Cancer Therapy Resistance
Cancer therapeutic resistance, including radioresistance and drug resistance, is a major challenge in cancer research and treatment.9,56 Drug resistance has become a key obstacle in developing targeted therapies and limited the efficacy of these therapies in treating cancer patients.56-58 Cancer stem cells, a small subpopulation of cancer cells with tumor-seeding and self-renewing ability, have been found to contribute to therapy resistance in different cancer types through preferential activation of the DNA damage checkpoint response and repair, thereby protecting cells from DNA damage, cleaning reactive oxygen species (ROS) and activating the cell survival signaling pathways.9,56,59 Since tumor cells that have gone EMT can acquire cancer stem cell properties,8 the EMT program has been implicated in therapy resistance in cancer, including radioresistance, chemoresistance, HER2, EGFR, endocrine and other targeted therapy resistance.11,12,44,60-64 A provocative study by Weinberg and colleagues demonstrated that basal, but not luminal breast cancer cells can readily convert from non-cancer stem cell state to cancer stem cell state in response to microenvironmental stimuli such as TGF-β; such conversion is driven by activated expression of ZEB1.65 Interestingly, ZEB1 overexpression is frequently observed in mesenchymal-like carcinoma cells. These findings underscore the importance of elucidating the functional role that ZEB1 plays in the acquisition of cancer stem cell properties. Moreover, what is the mechanism by which these mesenchymal-like carcinoma cells acquire treatment resistance: EMT-dependent or EMT-independent?
ZEB1 regulates radioresistance through an ATM−ZEB1−CHK1 signaling axis
Recently, we and others reported that ZEB1 plays a critical role in regulating tumor radiosensitivity.16,66 Knockdown of ZEB1 in SUM159-P2 (a radioresistant subline of the SUM159 breast cancer cells derived from ionizing radiation), U2OS (osteosarcoma), H460 (non-small cell lung cancer) and H1299 (non-small cell lung cancer) cell lines increased radiosensitivity, while ectopic expression of ZEB1 in radiosensitive mammary epithelial cell lines, HMLE and MCF7, led to elevated radioresistance.16,66 Since ZEB1 is a well-established EMT-inducing transcription factor, we were led to question whether ZEB1 regulates radiosensitivity through EMT. First, we overexpressed Snail, Twist or ZEB1 in HMLE cells. Each of these transcription factors induced EMT and increased radioresistance, and in each case, expression of Snail, Twist and ZEB1 was upregulated, regardless of which factor was used to induce EMT.16 These findings demonstrate an association between EMT and radioresistance, but do not reveal whether it is EMT itself or a specific EMT inducer that causes radioresistance. When we silenced each of the 3 transcription factors in HMLE cells overexpressing Snail, Twist or ZEB1, it did not cause reversal of EMT. Notably, only knockdown of ZEB1 increased radiosensitivity.16 We also overexpressed these 3 transcription factors in the MCF7 human breast cancer cell line, and none of them induced EMT due to the expression of intact p53 protein, a barrier to EMT induction. Interestingly, only ZEB1, but not Snail or Twist, conferred radioresistance on MCF7 cells even without inducing EMT.16 These results suggest that it is a specific EMT regulator, ZEB1, but not EMT itself, that induces radioresistance.
Mechanistically, ZEB1 regulates radiosensitivity in cancer cells by promoting homologous recombination (HR)-mediated DNA damage repair and the clearance of DNA breaks (Fig. 1B).16 CHK1 (but not CHK2) mediates, at least in part, the effect of ZEB1 on DNA damage repair and radioresistance. Interestingly, ZEB1 regulates CHK1 levels by interacting with the deubiquitinase USP7. This interaction promotes USP7's ability to deubiquitinate and stabilize CHK1 protein without affecting CHK1 gene transcription.16
Upon ionizing radiation, ZEB1, but not Twist and Snail, is upregulated in SUM159 cells.16 Radiation causes DNA damage which leads to activation of ATM and ATR kinases. Activated ATM, but not ATR, phosphorylates ZEB1 at serine 585 and stabilizes ZEB1, resulting in elevated ZEB1 protein levels.16 How ATM-mediated phosphorylation of ZEB1 leads to ZEB1 stabilization remains to be determined. Similarly, others also reported that radiation causes upregulation of ZEB1 in the A549 lung cancer cell line67 and in the CNE1 and CNE2 nasopharyngeal cancer cell lines.68 Taken together, these findings by us and others suggest that radiation treatment can give rise to increased ZEB1 levels and therapy-induced radioresistance. Cancer cells with treatment resistance including radioresistance are most likely to be the source of local and distant recurrence. Indeed, in a cohort of 286 human breast cancer patients (87% of them received radiotherapy), patients with high ZEB1 expression in their mammary tumors had much worse distant relapse-free survival compared with those with low ZEB1 expression.16
Since silencing ZEB1 expression in cancer cells leads to increased tumor radiosensitivity in vitro and in vivo,16,66 microRNAs targeting ZEB1, such as miR-205 and the miR-200 family, have the potential to be used as radiosensitizers in cancer treatment. Recently, we found that miR-205 can radiosensitize SUM159-P2 (breast cancer), MDA-MB-231 (breast cancer), A549 (lung cancer) and U2OS (osteosarcoma) cells by targeting ZEB1 and Ubc13, leading to inhibition of DNA damage repair; we also demonstrated the therapeutic utility of nanoparticle-encapsulated miR-205 mimics as a tumor radiosensitizer in a preclinical model.69 Similarly, another group reported that therapeutic delivery of a miR-200 family member, miR-200c, can sensitize lung cancer cells to radiation in xenograft models.66 Therefore, ZEB1-targeting agents like miR-205 and miR-200c microRNAs represent a new class of radiosensitizers.
ZEB1 regulates drug resistance: EMT-dependent or EMT-independent?
Over the past few years, emerging evidence has implicated ZEB1 in drug resistance in multiple cancers, but it remains unclear whether ZEB1-induced drug resistance is EMT-dependent or EMT-independent. In glioblastoma (GBM), ZEB1 is preferentially expressed at the invasion front of xenografts generated by cell lines derived from primary GBM specimens. Silencing ZEB1 expression in these cell lines reduced both invasion and the resistance to temozolomide (TMZ), a standard of care chemotherapeutic drug for glioblastoma.63 Mechanistically, ZEB1 regulates the expression of MGMT, a gene promoting TMZ resistance, through a ZEB1−miR-200c−c-MYB loop.63 In pancreatic cells, the EMT status (gauged by E-cadherin and vimentin expression levels) and the expression level of ZEB1 correlate with the resistance to chemotherapeutic agents including gemcitabine, 5-fluorouracil (5-FU) and cisplatin.61 Interestingly, knockdown of ZEB1 in mesenchymal-like pancreatic cancer cell lines not only reversed the EMT phenotype, but also sensitized these cells to gemcitabine, 5-FU and cisplatin treatment.61 Similarly, ZEB1, but not other EMT-inducing transcription factors including Snail, Slug and Twist, is upregulated in docetaxel-resistant human lung adenocarcinoma cells.44 Ectopic expression of ZEB1 conferred docetaxel resistance on the SPC-A1 human lung adenocarcinoma cell line, while silencing ZEB1 in SPC-A1/DTX cells, which are mesenchymal-like and docetaxel-resistant cells, reversed EMT and chemoresistance.44 These studies led the authors to conclude that ZEB1-induced EMT contributes to drug resistance. However, an alternative interpretation is that ZEB1 induces EMT and treatment resistance separately. Despite the association, it may not be the EMT status itself that dictates drug sensitivity.
Additional studies provided further insight into ZEB1-induced drug resistance. In a Kras/p53-mutant lung adenocarcinoma mouse model, high expression of ZEB1 drives metastasis which can be suppressed by a PI3K inhibitor.70 Mechanistically, ZEB1 activates PI3K by derepressing miR-200 and miR-183 targets, including amphiregulin (AREG), betacellulin (BTC), the transcription factor GATA6 and friend of GATA 2 (FOG2).70 FOG2 mediates ZEB1-induced expression of the p110α catalytic subunit of PI3K, but surprisingly, FOG2 is not required for ZEB1-induced EMT,70 suggesting that ZEB1-mediated activation of PI3K and sensitization to the PI3K inhibitor is independent of EMT. On the other hand, knockdown of ZEB1 in erlotinib (an EGFR inhibitor)-resistant head and neck squamous cell carcinoma cell lines (1386LN and UMSCC1) increased erlotinib sensitivity in an E-cadherin-dependent manner,64 indicating that ZEB1's effect on erlotinib resistance may be EMT-dependent.
Conclusions
As a driver of EMT, ZEB1 plays an important role in tumor progression and metastasis and correlates with poor clinical outcomes in cancer patients. In addition, ZEB1 is involved in therapy resistance in multiple cancers through both EMT-dependent and EMT-independent mechanisms. We propose that ZEB1 employs multiple different mechanisms to regulate therapy resistance, depending on the specific cancer type and treatment type. This can be mediated by distinct transcriptional targets and different interacting proteins of ZEB1. Therefore, the underlying mechanisms of therapy resistance should be carefully examined in individual situations. Despite the apparent association between EMT and therapy resistance, we propose that it is not the epithelial or mesenchymal state itself that dictates cancer stem properties such as radioresistance and drug resistance; instead, it depends on the functions and mechanisms of action of specific EMT regulators. All EMT inducers are not equal.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The authors’ research is supported by the US National Institutes of Health grants R01CA166051 and R01CA181029 (to L.M.) and a Cancer Prevention and Research Institute of Texas Scholar Award R1004 (to L.M.). L.M. is an R. Lee Clark Fellow (supported by the Jeanne F. Shelby Scholarship Fund) of The University of Texas MD Anderson Cancer Center.
References
- 1. Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995; 154:8-20; PMID:8714286; http://dx.doi.org/ 10.1159/000147748 [DOI] [PubMed] [Google Scholar]
- 2. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7:131-42; PMID:16493418; http://dx.doi.org/ 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- 3. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139:871-90; PMID:19945376; http://dx.doi.org/ 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 4. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119:1420-8; PMID:19487818; http://dx.doi.org/ 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13:97-110; PMID:23344542; http://dx.doi.org/ 10.1038/nrc3447 [DOI] [PubMed] [Google Scholar]
- 6. Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 2013; 27:2192-206; PMID:24142872; http://dx.doi.org/ 10.1101/gad.225334.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer cell 2012; 22:725-36; PMID:23201165; http://dx.doi.org/ 10.1016/j.ccr.2012.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. . The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133:704-15; PMID:18485877; http://dx.doi.org/ 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer 2008; 8:545-54; PMID:18511937; http://dx.doi.org/ 10.1038/nrc2419 [DOI] [PubMed] [Google Scholar]
- 10. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444:756-60; PMID:17051156; http://dx.doi.org/ 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 11. Sui H, Zhu L, Deng W, Li Q. Epithelial-mesenchymal transition and drug resistance: role, molecular mechanisms, and therapeutic strategies. Oncol Res Treat 2014; 37:584-9; PMID:25342509; http://dx.doi.org/ 10.1159/000367802 [DOI] [PubMed] [Google Scholar]
- 12. Dave B, Mittal V, Tan NM, Chang JC. Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res 2012; 14:202; PMID:22264257; http://dx.doi.org/ 10.1186/bcr2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15:178-96; PMID:24556840; http://dx.doi.org/ 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jung HY, Fattet L, Yang J. Molecular pathways: linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin Cancer Res 2014; PMID:25107915; http://dx.doi.org/10.1158/1078-0432.CCR-13-3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev 2012; 31:653-62; PMID:22684369; http://dx.doi.org/ 10.1007/s10555-012-9368-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, Yuan J, Wang M, Chen D, Sun Y, et al. . ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol 2014; 16:864-75; PMID:25086746; http://dx.doi.org/ 10.1038/ncb3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen D, Sun Y, Yuan Y, Han Z, Zhang P, Zhang J, You MJ, Teruya-Feldstein J, Wang M, Gupta S, et al. . miR-100 induces epithelial-mesenchymal transition but suppresses tumorigenesis, migration and invasion. PLoS genetics 2014; 10:e1004177; PMID:24586203; http://dx.doi.org/ 10.1371/journal.pgen.1004177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kahlert UD, Maciaczyk D, Doostkam S, Orr BA, Simons B, Bogiel T, Reithmeier T, Prinz M, Schubert J, Niedermann G, et al. . Activation of canonical WNT/beta-catenin signaling enhances in vitro motility of glioblastoma cells by activation of ZEB1 and other activators of epithelial-to-mesenchymal transition. Cancer Lett 2012; 325:42-53; PMID:22652173; http://dx.doi.org/ 10.1016/j.canlet.2012.05.024 [DOI] [PubMed] [Google Scholar]
- 19. Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene 2007; 26:711-24; PMID:16862183; http://dx.doi.org/ 10.1038/sj.onc.1209808 [DOI] [PubMed] [Google Scholar]
- 20. Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, et al. . TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene 2012; 31:3190-201; PMID:22037216; http://dx.doi.org/ 10.1038/onc.2011.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dohadwala M, Yang SC, Luo J, Sharma S, Batra RK, Huang M, Lin Y, Goodglick L, Krysan K, Fishbein MC, et al. . Cyclooxygenase-2-dependent regulation of E-cadherin: prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res 2006; 66:5338-45; PMID:16707460; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-3635 [DOI] [PubMed] [Google Scholar]
- 22. Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH, Semenza GL. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res 2006; 66:2725-31; PMID:16510593; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-3719 [DOI] [PubMed] [Google Scholar]
- 23. Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283:14910-4; PMID:18411277; http://dx.doi.org/ 10.1074/jbc.C800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10:593-601; PMID:18376396; http://dx.doi.org/ 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- 25. Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22:894-907; PMID:18381893; http://dx.doi.org/ 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J 2003; 22:2443-52; PMID:12743038; http://dx.doi.org/ 10.1093/emboj/cdg225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J 2003; 22:2453-62; PMID:12743039; http://dx.doi.org/ 10.1093/emboj/cdg226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7:415-28; PMID:17508028; http://dx.doi.org/ 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- 29. Wellner U, Brabletz T, Keck T. ZEB1 in Pancreatic Cancer. Cancers (Basel) 2010; 2:1617-28; PMID:24281177; http://dx.doi.org/ 10.3390/cancers2031617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005; 24:2375-85; PMID:15674322; http://dx.doi.org/ 10.1038/sj.onc.1208429 [DOI] [PubMed] [Google Scholar]
- 31. Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003; 422:735-8; PMID:12700765; http://dx.doi.org/ 10.1038/nature01550 [DOI] [PubMed] [Google Scholar]
- 32. Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010; 29:3490-500; PMID:20418909; http://dx.doi.org/ 10.1038/onc.2010.102 [DOI] [PubMed] [Google Scholar]
- 33. Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, et al. . The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 2007; 26:6979-88; PMID:17486063; http://dx.doi.org/ 10.1038/sj.onc.1210508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development 1993; 119:433-46; PMID:7904558 [DOI] [PubMed] [Google Scholar]
- 35. Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development 1998; 125:21-31; PMID:9389660 [DOI] [PubMed] [Google Scholar]
- 36. Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 2008; 135:579-88; PMID:18192284; http://dx.doi.org/ 10.1242/dev.007047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y, Peng X, Tan J, Darling DS, Kaplan HJ, Dean DC. Zeb1 mutant mice as a model of posterior corneal dystrophy. Invest Ophthalmol Vis Sci 2008; 49:1843-9; PMID:18436818; http://dx.doi.org/ 10.1167/iovs.07-0789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hurt EM, Saykally JN, Anose BM, Kalli KR, Sanders MM. Expression of the ZEB1 (deltaEF1) transcription factor in human: additional insights. Mol Cell Biochem 2008; 318:89-99; PMID:18622689; http://dx.doi.org/ 10.1007/s11010-008-9860-z [DOI] [PubMed] [Google Scholar]
- 39. Okegawa T, Li Y, Pong RC, Hsieh JT. Cell adhesion proteins as tumor suppressors. J Urol 2002; 167:1836-43; PMID:11912444; http://dx.doi.org/ 10.1016/S0022-5347(05)65245-7 [DOI] [PubMed] [Google Scholar]
- 40. Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer 2004; 4:118-32; PMID:14964308; http://dx.doi.org/ 10.1038/nrc1276 [DOI] [PubMed] [Google Scholar]
- 41. Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, et al. . Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 2006; 10:437-49; PMID:17097565; http://dx.doi.org/ 10.1016/j.ccr.2006.09.013 [DOI] [PubMed] [Google Scholar]
- 42. Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 2008; 68:3645-54; PMID:18483246; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2938 [DOI] [PubMed] [Google Scholar]
- 43. Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, et al. . The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 2008; 68:537-44; PMID:18199550; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-5682 [DOI] [PubMed] [Google Scholar]
- 44. Ren J, Chen Y, Song H, Chen L, Wang R. Inhibition of ZEB1 reverses EMT and chemoresistance in docetaxel-resistant human lung adenocarcinoma cell line. J Cell Biochem 2013; 114:1395-403; PMID:23255418; http://dx.doi.org/ 10.1002/jcb.24481 [DOI] [PubMed] [Google Scholar]
- 45. Spoelstra NS, Manning NG, Higashi Y, Darling D, Singh M, Shroyer KR, Broaddus RR, Horwitz KB, Richer JK. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res 2006; 66:3893-902; PMID:16585218; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-2881 [DOI] [PubMed] [Google Scholar]
- 46. Bronsert P, Kohler I, Timme S, Kiefer S, Werner M, Schilling O, Vashist Y, Makowiec F, Brabletz T, Hopt UT, et al. . Prognostic significance of Zinc finger E-box binding homeobox 1 (ZEB1) expression in cancer cells and cancer-associated fibroblasts in pancreatic head cancer. Surgery 2014; 156:97-108; PMID:24929761; http://dx.doi.org/ 10.1016/j.surg.2014.02.018 [DOI] [PubMed] [Google Scholar]
- 47. Shen A, Zhang Y, Yang H, Xu R, Huang G. Overexpression of ZEB1 relates to metastasis and invasion in osteosarcoma. J Surg Oncol 2012; 105:830-4; PMID:22213004; http://dx.doi.org/ 10.1002/jso.23012 [DOI] [PubMed] [Google Scholar]
- 48. Zhang J, Lu C, Kang J, Cao C, Li M. Involvement of ZEB1 and E-cadherin in the invasion of lung squamous cell carcinoma. Mol Biol Rep 2013; 40:949-56; PMID:23065281; http://dx.doi.org/ 10.1007/s11033-012-2136-4 [DOI] [PubMed] [Google Scholar]
- 49. Matsubara D, Kishaba Y, Yoshimoto T, Sakuma Y, Sakatani T, Tamura T, Endo S, Sugiyama Y, Murakami Y, Niki T. Immunohistochemical analysis of the expression of E-cadherin and ZEB1 in non-small cell lung cancer. Pathol Int 2014; 64:560-8; PMID:25347933; http://dx.doi.org/ 10.1111/pin.12214 [DOI] [PubMed] [Google Scholar]
- 50. Zhou YM, Cao L, Li B, Zhang RX, Sui CJ, Yin ZF, Yang JM. Clinicopathological significance of ZEB1 protein in patients with hepatocellular carcinoma. Ann Surg Oncol 2012; 19:1700-6; PMID:21584833; http://dx.doi.org/ 10.1245/s10434-011-1772-6 [DOI] [PubMed] [Google Scholar]
- 51. Okugawa Y, Toiyama Y, Tanaka K, Matsusita K, Fujikawa H, Saigusa S, Ohi M, Inoue Y, Mohri Y, Uchida K, et al. . Clinical significance of Zinc finger E-box Binding homeobox 1 (ZEB1) in human gastric cancer. J Surg Oncol 2012; 106:280-5; PMID:22095522; http://dx.doi.org/ 10.1002/jso.22142 [DOI] [PubMed] [Google Scholar]
- 52. Jia B, Liu H, Kong Q, Li B. Overexpression of ZEB1 associated with metastasis and invasion in patients with gastric carcinoma. Mol Cell Biochem 2012; 366:223-9; PMID:22466758; http://dx.doi.org/ 10.1007/s11010-012-1299-6 [DOI] [PubMed] [Google Scholar]
- 53. Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett 2013; 5:564-8; PMID:23420790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y, Zhang N, Wang Y, Xu M, Liu N, Pang X, Cao J, Ma N, Pang H, Liu L, et al. . Zinc finger E-box binding homeobox 1 promotes invasion and bone metastasis of small cell lung cancer in vitro and in vivo. Cancer Sci 2012; 103:1420-8; PMID:22632166; http://dx.doi.org/ 10.1111/j.1349-7006.2012.02347.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. . The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11:1487-95; PMID:19935649; http://dx.doi.org/ 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- 56. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013; 13:714-26; PMID:24060863; http://dx.doi.org/ 10.1038/nrc3599 [DOI] [PubMed] [Google Scholar]
- 57. Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer 2014; 14:455-67; PMID:24957944; http://dx.doi.org/ 10.1038/nrc3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014; 11:473-81; PMID:24981256; http://dx.doi.org/ 10.1038/nrclinonc.2014.104 [DOI] [PubMed] [Google Scholar]
- 59. Peitzsch C, Kurth I, Kunz-Schughart L, Baumann M, Dubrovska A. Discovery of the cancer stem cell related determinants of radioresistance. Radiother Oncol 2013; 108:378-87; PMID:23830195; http://dx.doi.org/ 10.1016/j.radonc.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 60. Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, Sarkar FH. Pancreatic cancer: understanding and overcoming chemoresistance. Nat Rev Gastroenterol Hepatol 2011; 8:27-33; PMID:21102532; http://dx.doi.org/ 10.1038/nrgastro.2010.188 [DOI] [PubMed] [Google Scholar]
- 61. Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res 2009; 69:5820-8; PMID:19584296; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. . miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res 2009; 15:5060-72; PMID:19671845; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Siebzehnrubl FA, Silver DJ, Tugertimur B, Deleyrolle LP, Siebzehnrubl D, Sarkisian MR, Devers KG, Yachnis AT, Kupper MD, Neal D, et al. . The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med 2013; 5:1196-212; PMID:23818228; http://dx.doi.org/ 10.1002/emmm.201302827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Haddad Y, Choi W, McConkey DJ. Delta-crystallin enhancer binding factor 1 controls the epithelial to mesenchymal transition phenotype and resistance to the epidermal growth factor receptor inhibitor erlotinib in human head and neck squamous cell carcinoma lines. Clin Cancer Res 2009; 15:532-42; PMID:19147758; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D'Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013; 154:61-74; PMID:23827675; http://dx.doi.org/ 10.1016/j.cell.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cortez MA, Valdecanas D, Zhang X, Zhan Y, Bhardwaj V, Calin GA, Komaki R, Giri DK, Quini CC, Wolfe T, et al. . Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol Ther 2014; 22:1494-503; PMID:24791940; http://dx.doi.org/ 10.1038/mt.2014.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu W, Huang YJ, Liu C, Yang YY, Liu H, Cui JG, Cheng Y, Gao F, Cai JM, Li BL. Inhibition of TBK1 attenuates radiation-induced epithelial-mesenchymal transition of A549 human lung cancer cells via activation of GSK-3beta and repression of ZEB1. Lab Invest 2014; 94:362-70; PMID:24468793; http://dx.doi.org/ 10.1038/labinvest.2013.153 [DOI] [PubMed] [Google Scholar]
- 68. Chen W, Wu S, Zhang G, Wang W, Shi Y. Effect of AKT inhibition on epithelial-mesenchymal transition and ZEB1-potentiated radiotherapy in nasopharyngeal carcinoma. Oncol Lett 2013; 6:1234-40; PMID:24179501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang P, Wang L, Rodriguez-Aguayo C, Yuan Y, Debeb BG, Chen D, Sun Y, You MJ, Liu Y, Dean DC, et al. . miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nature Commun 2014; 5:5671; PMID:25476932; http://dx.doi.org/ 10.1038/ncomms6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang Y, Ahn YH, Chen Y, Tan X, Guo L, Gibbons DL, Ungewiss C, Peng DH, Liu X, Lin SH, et al. . ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J Clin Invest 2014; 124:2696-708; PMID:24762440; http://dx.doi.org/ 10.1172/JCI72171 [DOI] [PMC free article] [PubMed] [Google Scholar]


