Abstract
Metastasis involves the dissemination of single or small clumps of cancer cells through blood or lymphatic vessels and their extravasation into distant organs. Despite the strong regulation of metastases development by a cell dormancy phenomenon, the dormant state of cancer cells remains poorly characterized due to the difficulty of in vivo studies. We have recently shown in vitro that clonogenicity of prostate cancer cells is regulated by a dormancy phenomenon that is strongly induced when cells are cultured both at low cell density and in a slightly hypertonic medium. Here, we characterized by RT-qPCR a genetic expression signature of this dormant state which combines the presence of both stemness and differentiation markers. We showed that both TFGβ/BMP signaling and redox imbalance are required for the full induction of this dormancy signature and cell quiescence. Moreover, reconstruction experiments showed that TFGβ/BMP signaling and redox imbalance are sufficient to generate a pattern of genetic expression displaying all characteristic features of the dormancy signature. Finally, we observed that low cell density was sufficient to activate TGFβ/BMP signaling and to generate a slight redox imbalance thus priming cells for dormancy that can be attained with a co-stimulus like hypertonicity, most likely through an increased redox imbalance. The identification of a dual regulation of dormancy provides a framework for the interpretation of previous reports showing a restricted ability of BMP signaling to regulate cancer cell dormancy in vivo and draws attention on the role of oxidative stress in the metastatic process.
Keywords: cell proliferation, cell signaling, prostate cancer, redox regulation, SMAD transcription factor
Abbreviations
- BSO
buthionine sulfoximine
- CE
cloning efficiency
- DMEM-FCS
Dulbecco Modified Essential Medium supplemented with 10% Fetal Calf Serum and penicillin-streptomycin
- LD_hyper
low cell density condition in DMEM-FCS medium
- LD_hypo
low cell density condition in hypotonic medium made by addition of 25% water to DMEM-FCS
- MD_hypo
medium cell density condition in hypotonic medium made by addition of 25% water to DMEM-FCS
- MD_hyper
medium cell density condition in DMEM-FCS medium (slightly hypertonic)
- NAM
Normalized Averaging Method
- ARM
Averaging Ratio Method
Introduction
Metastases originate from the dissemination of single or possibly small clumps of cancer cells through blood or lymphatic vessels and are clonal cell populations resulting from the proliferation of an individual cancer cell. Experimental models suggest that, following dissemination, cancer cells that survive can enter into a reversible dormant state that strongly limits the development of metastases. 1-5 In humans, the phenomenon of dormancy also accounts for metastatic recurrences appearing up to several decades after the cure of the primary cancer, as observed in breast and prostate cancers, some leukemia and melanoma.6-11 These recurrences are also in agreement with the observation that dormant cells are more resistant to chemotherapy than the primitive cancer cells, which makes them very difficult to eradicate.12-14 Dormancy thus appears as both a barrier to metastasis and as a mechanism of resistance to chemotherapy, and a better understanding of the underlying mechanisms is warranted in order to control the metastatic process. Several signaling pathways have been implicated in the regulation of disseminated cancer cell dormancy in vivo including BMP/TGFβ signaling. These pathways could allow determining how cell intrinsic features and environmental interactions can lead to metastasis development.15-22 However, a detailed analysis of dormancy has been hampered by the challenge of identifying and studying dormant cells in vivo. Therefore, models of dormancy in cell culture have been developed which rely on the parallel between metastasis in vivo and the clonogenic capacity in vitro, i.e. the ability of isolated cells to give rise to a clonal population. Two major in vitro models have been set up with breast cancer cell lines, which differ by the cell culture conditions.23,24 These studies have focused on the relationship between the dormant state and distinctive features linked to epithelial-mesenchymal transition (EMT) including cytoskeleton rearrangement, E cadherin expression and the influence of extracellular matrix components.23,25-28 These studies linked dormancy escape in cell culture to acquisition of mesenchymal traits by breast cancer cells including loss of E-cadherin expression, but the mechanisms involved in dormancy entry and maintenance remain largely elusive. Additional investigations are required to characterize cancer cell dormancy and to distinguish it from other forms of quiescence in a G0 phase of cell cycle such as those induced by contact inhibition and/or restricted nutrient availability.
We have recently developed a new model of cell dormancy in vitro based on the LNCaP prostate cancer cell line, a prevalent paradigm for the study of androgen-dependent human prostate cancers. In this model, about 99% of the cultured cells can be induced to enter into a reversible dormant state when they are cultured at low cell density and in a slightly hypertonic growth medium such as Dulbecco Modified Essential Medium supplemented with 10% fetal calf serum.29 We showed that dormancy drastically limits prostate cancer cell clonogenicity. Indeed, changing the cell culture medium from RPMI to DMEM decreased the clonogenicity by up to 3 orders of magnitude without significantly altering cell viability. During this initial characterization we observed that 2 of the signaling pathways that are involved in normal stem cell dormancy, TP53 and TGFβ/BMP, also played a role in our cellular model but could not fully account for the control of dormancy. In the present work, we exploited our model to generate a gene expression signature of dormant cells in order to explore in more details the signaling pathways regulating dormancy.
Results
Characterization of a genetic expression signature of the dormant state by RT-qPCR
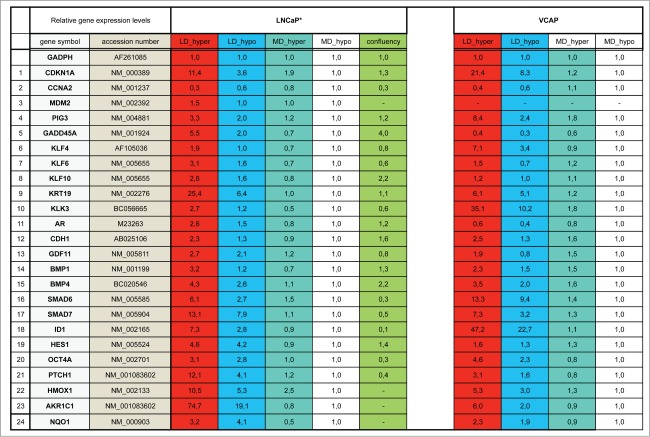
We have previously shown that the association of a low cell density (< 160 cells/cm2) and a slightly hypertonic medium such as Dulbecco Modified Essential Medium supplemented with fetal calf serum (DMEM-FCS) efficiently induces the entry into dormancy of LNCaP* cells.29 To characterize the biological state of these cells we used RT-qPCR to analyze the expression of an initial panel of 84 genes involved in cell cycle regulation, prostate epithelial cell differentiation, Epithelial-Mesenchymal Transition and prostate cancer progression (the complete list of genes investigated in this study is presented in Fig. S1). We first compared the expression profile of dormant cells (low cell density in DMEM-FCS abbreviated LD_hyper) with that of cells in exponential growth conditions (medium cell density in DMEM-FCS+25% water, abbreviated MD_hypo). We also measured variations of mRNA species levels for intermediate conditions (at low cell density in hypotonic medium (LD_hypo), and at medium cell density in hypertonic medium (MD_hyper) to assess the effects of cell density and medium tonicity separately.
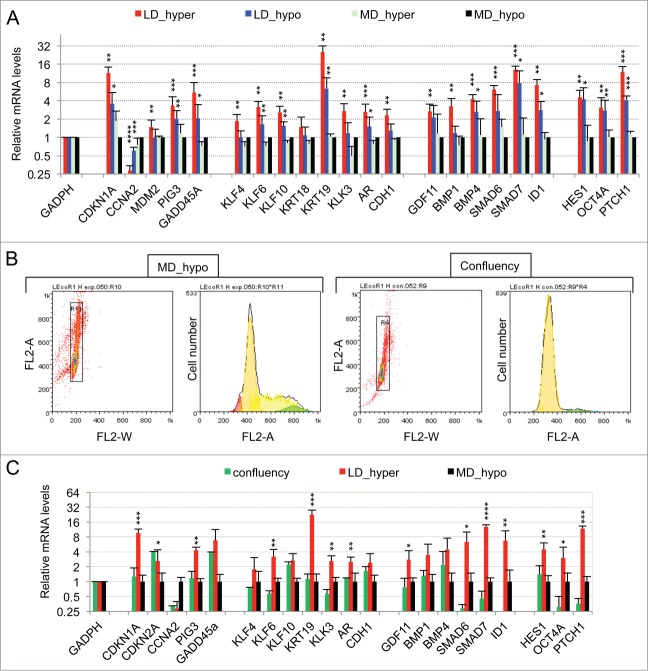
As displayed in Figure 1A (red bars) 21 genes were initially found to display a significant (p value of Student's t test inferior to 0.05) and more than a twofold change in their mRNA levels in dormant cells (LD_hyper) as compared to exponentially growing cells (MD_hypo). Notably, we observed in dormant cells a 3-fold decrease in the mRNA level of CCNA2 encoding cyclin A2 and a 11 fold increase in the mRNA level of CDKN1A mRNA encoding the p21Cip1/Waf1 cyclin-dependent kinase inhibitor, in agreement with their growth-arrested state. Another salient feature was the increased mRNA levels of Krüppel-like factors (KLF4, 6, 10) -that are involved in normal prostate cell growth inhibition and differentiation- and of markers of differentiation including the androgen receptor (AR), the prostate specific antigen (KLK3), the cytokeratin 19 (KRT19) and E-Cadherin (CDH1). By contrast, KRT18 mRNA level was unchanged, further indicating that the dormant LNCaP* cells resemble normal prostate epithelial luminal cells at an intermediate or late differentiation stage. mRNA for cytokeratin 5 (KRT5), CD44 and TP63 levels were below the detection threshold and only extremely low levels could be detected for CD133 (data not shown). Interestingly, several of the up-regulated genes were associated with stemness like KLF4, OCT4A, ID1, HES1 and BMP4. It is noteworthy that up-regulation of stemness markers was not linked to EMT since only an extremely low and stable level of CDH2 mRNA encoding N-cadherin protein (that was undetectable by Flow Cytometry analysis; data not shown), a slight CDH1 mRNA increase (Fig. 1A) or non-significant changes in the mRNA levels of vimentin, SNAI2 and ZEB1 were observed in dormant cells (data not shown). Finally, this signature showed the induction of genes associated with cellular stress (PIG3, MDM2, GADD45A and also CDKN1A) or activation of the TGFβ/BMP signaling pathways (BMP1, BMP4, GDF11, SMAD6, SMAD7, ID1) or repression of the Hedgehog signaling pathway (PTCH1). It is noteworthy that BMP4 can activate SMAD1, 5, 8 whereas GDF11 can activate SMAD2 and 3 and BMP1 is an astacin protease that can activate both BMP4 and GDF11 proteins.30 The subsequent analysis of transfected cells and their control populations confirmed these initial observations and established their robustness (see Fig. 4B).
Figure 1.
Characterization of a specific genetic expression signature of dormant LNCaP* cells. (A) RT-qPCR analysis of the variations of mRNA species levels according to cell culture conditions. Relative mRNA levels were averaged using the Averaging Ratio Method (ARM), with all mRNA levels reported to those measured under MD_hypo conditions in parallel experiments. Three to 5 independent experiments were performed with cells cultured in parallel under LD_hyper, LD_hypo, MD_hypo and 2 under MD_hyper conditions. (B) Cell cycle analysis of LNCaP* cells cultured under exponential growth (MD_hypo) or highly confluent conditions (confluency; see Experimental procedures). Cells were analyzed by Flow Cytometry after propidium iodide staining. A two-step gating procedure using Side SCattering-Height versus Forward SCattering-Height (data not shown) followed by pulse Area vs. pulse Width FLuorescence (FL2-A-versus FL2-W) was performed to eliminate cell doublets. Gated region is indicated on the FL2-A-versus FL2-W dot plot representation. Next to dot plots are fluorescence histograms. One representative experiment is shown. (C) Comparison of the patterns of mRNA level variations between confluent and dormant cells. Relative mRNA levels were averaged using the Normalized Averaging Method (NAM with levels under MD_hypo conditions taken as unity) from 2, 3, and 5 or 6 cell cultures experiments for confluent, dormant (LD_hyper conditions) and exponentially growing (MD_hypo) cells, respectively. Data are presented as mean ± s.d., with statistical significance indicated for the differences in mRNA levels between cells cultured under exponentially growing (MD_hypo) and under dormancy-inducing (LD_hyper) or permissive (LD_hyper) conditions (A), or between cells cultured under dormancy-inducing and confluent conditions (B); *, P < 0.1; **, P < 0.05; ***, P < 0.01; ****, P < 0.002. See the Materials and Methods section for more details on the averaging methods.
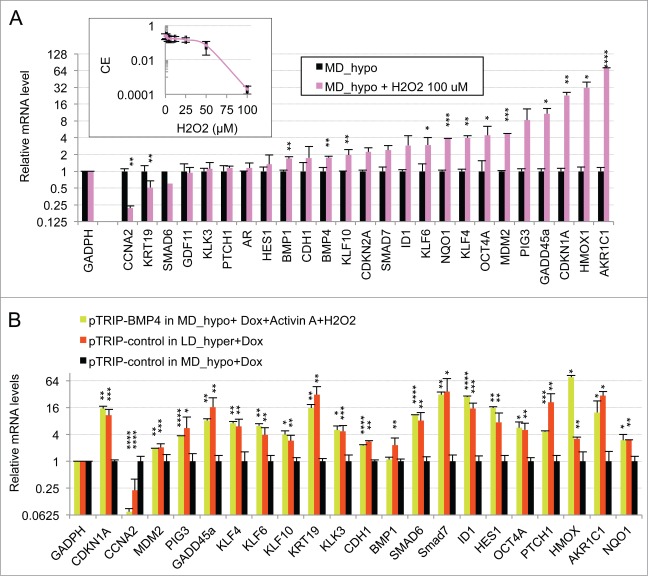
Figure 4.
Oxidative stress mediated by hydrogen peroxide cooperates with BMP4/activinα signaling to induce the dormancy gene expression signature in LNCaP* cells cultured at middle cell density in hypotonic medium (MD_hypo). (A) Effect of H2O2-induced oxidative stress on mRNA species accumulation in LNCaP* cells cultured under MD_hypo conditions. Inset: Dose response of the cloning efficiency of LNCaP* cells to H2O2. Data are average of 2 cloning efficiency assays made with 103 and 2 × 103 cells in 6 cm diameter dish, each in duplicate. Histogram: Effects of hydrogen peroxide treatment on mRNA species accumulation. Relative mRNA species levels were averaged with NAM from 2 parallel cell culture experiments. Statistical significance of the effects of H2O2 addition is indicated. All Data are presented as mean ± s.d., with significance indicated as in Figure 1. (B) Comparison of the mRNA accumulation patterns generated by activation of BMP4/ activin α signaling plus H2O2 treatment and by dormancy. Relative mRNA levels were averaged with NAM from 2 independent cell populations transduced with pTRIP-BMP4 and cultured under MD_hypo conditions in the presence of doxycycline (Dox; 0.5 μg/ml) + activin α (50 ng/ml) + H2O2 100 μM, 4 independent cell populations transduced with pTRIP-control and cultured under LD_hyper conditions in the presence of doxycycline and 4 to 6 independent cell populations transduced with pTRIP-control and cultured under MD_hypo conditions in the presence of doxycycline. Data were calculated using the normalized averaging method and are presented as mean ± s.d., and stars indicate statistical significance of mRNA level variations compared to basal levels measured for pTRIP-control transduced cell populations cultured under MD_hypo conditions in the presence of doxycycline.
Interestingly, many of the mRNA changes observed in dormant cells were already present, albeit on a smaller scale, in cells under growth permissive conditions but at low density LD_hypo; blue bars in Fig. 1A. By contrast, hypertonicity had a limited impact on gene expression in cells at medium density (aquamarine bars) but strongly increased the changes associated with low cell density. These results are in agreement with our previous observation that LNCaP* cells have similar growth rates in DMEM-FCS and DMEM-FCS + 25% water when cells were cultured at medium cell density.29
Specificity of the dormancy signature
To assess whether the pattern of expression of the 21 genes characterized above could constitute a signature of the dormant state we first compared it with the corresponding pattern of growth-arrested LNCaP*cells obtained by culturing cells in DMEM-FCS up to confluence without culture medium change for the last 3 d. Growth arrest was confirmed by cell cycle analysis with propidium iodide staining and under these conditions the G0/G1 population increased from 60,6 ± 3.2 % (average ± s.d.; n = 4) in exponentially growing cells to 94.0 ± 2.9 % (p value of Student's t test <10−5; see Fig. 1B). As shown in Figure 1C (green bars), confluent growth-inhibited cells displayed a strong decrease of CCNA2 mRNA level in agreement with the growth arrest. We also observed a significant induction of GADD45A and CDKN2A (encoding p16INK4A) genes but a complete lack of induction for all others genes of the dormancy signature. This indicated that the proposed genetic signature reflected cellular physiology beyond the growth-arrest.
Extension of the dormancy signature to VCaP cancer cells
As a second example of prostate cancer cell line, we chose the VCaP cell line that was established from a vertebral metastatic lesion. Like LNCaP cells, VCaP cells harbor a functional androgen receptor and markers of the luminal epithelial phenotype as in majority of prostate cancers in human. However, at variance with LNCaP cells, VCaP express a mutated TP53 gene and are more metastatic. As shown in Figs. S2A and B, clonogenicity of VCaP cells was strongly dependent on the tonicity of the culture medium with about one thousand fold difference in cloning efficiency between cells cultures under LD_hyper and LD_hypo conditions. RT-qPCR analysis revealed a strong homology in the pattern of gene expression between LNCaP* and VCaP cultured under dormancy inducing conditions (LD_hyper) (Figs. S2C and S3). In particular we observed a down regulation of CCNA2 and an up regulation of CDKN1A. Markers of cell differentiation like KLK3, KRT19, CDH1 were upregulated along with KLF4. OCT4A as well as genes involved in TGFβ/BMP signaling were also induced under culture in LD_hyper conditions. Most salient differences were a lack of GADD45A, AR and KLF10 induction along with reduced inductions of HES1 and PTCH1, but a stronger induction of KLF4 and ID1. Finally, the same relationship was observed between gene expression, cell density and medium tonicity as in LNCaP* cells: nearly all of the mRNA changes observed in dormant cells were present at a smaller scale in cells under growth permissive conditions but at low density (LD_hypo). Hypertonicity had a limited impact on gene expression in cells at medium density but strongly increased the changes associated with low cell density. Overall, VCaP cells displayed the same essential features as observed in LNCaP* cells.
SMAD signaling pathway is activated in the dormant state
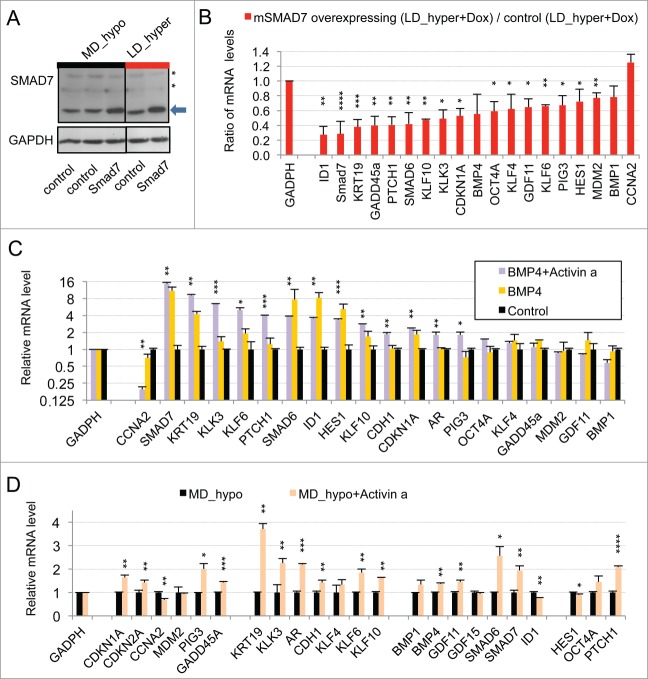
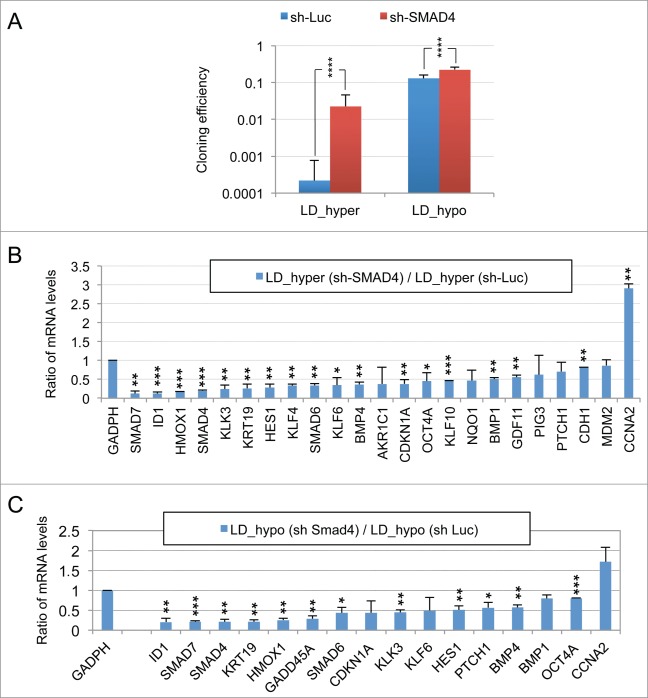
An important role of the TGFβ/BMP signaling pathways in dormancy was previously suggested by our observation that constitutive overexpression of murine Smad7, an inhibitory Smad, rendered LNCaP* cells partially refractory to dormancy induction by low cell density and hypertonicity.29 Similar results were observed by expressing a small hairpin RNA targeting SMAD4 (sh-SMAD4), encoding a binding partner of regulatory SMAD or by treating LNCaP* cells with pharmacologic inhibitors of BMP and, to a lesser extent, TGFβ receptors (Figs. S4 and S5). To further investigate the role of TGFβ/BMP signaling pathways in gene expression, we transduced LNCaP* cells with pTRIP vectors allowing the doxycycline-inducible expression of murine inhibitory Smad7 or human BMP4 cDNA, the empty vector being used as a control. In the presence of doxycycline, the intensity of the band detected by Western blot using a pan-species antibody and corresponding to SMAD7 and Smad7 was 2.4 ± 0.5 times higher (p = 0.02) in Smad7 transduced cells, (Fig. 2A left panel). As pTRIP is a potent expression vector yielding a high level of mRNA accumulation (data not shown), this suggested that Smad7 expression is tightly regulated at a post transcriptional level, in line with the observation that the ten-fold increase of endogenous SMAD7 mRNA in dormant cells (Fig. 1A) did not result in a significant increase in SMAD7 protein level (Fig. 2A). Note that the RT-qPCR assay is specific of the human SMAD7 and does not detect the transduced murine Smad7 mRNA. Nonetheless, murine Smad7 expression under dormancy-inducing conditions increased CCNA2 mRNA accumulation and decreased mRNA accumulation of most others genes of the dormancy signature (Fig. 2B). Notably, mRNA levels for well-characterized direct targets of the TGFβ/BMP signaling pathways such as SMAD6, 7 and ID1 were decreased by more than 50%, as were those for KRT19, GADD45A, PTCH1, KLF10 and KLK3. Similar results were obtained upon depletion of endogenous SMAD4 using sh-SMAD4 under dormancy-inducing (LD_hyper) conditions (Fig. S4B).
Figure 2.
Implication of SMAD signaling in LNCaP* dormancy. (A) Western Blot analysis of total smad7 expression (endogenous human SMAD7 and transduced murine Smad7) in cell populations transduced with pTRIP-control and pTRIP-mSmad7 cultured under exponentially growing (MD_hypo) or dormancy inducing (LD_hyper) conditions. Cells were grown in the presence of doxycycline to induce pTRIP expression and a polyspecific monoclonal antibody was used. The arrow indicates the common expected size for SMAD7 and Smad7 proteins (46 kDa) while a star indicates bands resulting from non-specific antibody hybridizations. Two identically processed parts of the same gel have been regrouped for the figure. (B) Regulation of mRNA species accumulation under dormancy-inducing conditions by ectopic Smad7 overexpression. Ratios of mRNA species levels measured by RT-qPCR between Smad7 and control-transduced cell populations cultured under LD_hyper conditions were derived from 2 couples of cell populations. Statistical significance of the effects of Smad7 overexpression is indicated. (C) Regulation of mRNA species accumulation by BMP4 and Activin α signaling in cells cultured under MD_hypo conditions. Relative mRNA levels determined by RT-qPCR were averaged with NAM (with levels under MD_hypo conditions taken as unity) from 2 and 4 independent pTRIP-BMP4 and pTRIP-control transduced cell populations respectively, cultured under MD_hypo conditions in the presence of doxycycline and activin α at 50 ng/ml as indicated. Statistical significance of the difference in mRNA levels between cells stimulated by BMP4 plus activin α and control cells is indicated. D) Regulation of mRNA species accumulation by activin α signaling in LNCaP* cells cultured under MD_hypo conditions. Relative mRNA levels were measured by RT-qPCR from 2 control and 2 treated cell populations respectively. Note that mRNA level of ID1, a target gene of BMP signaling, was decreased by activin α addition. Data are presented as mean ± s.d., with significance of mRNA levels variations indicated as in Figure 1.
To further address the role of TGFβ/BMP signaling in the generation of the dormancy signature, we analyzed the effect of ectopic expression of BMP4 in exponentially growing pTRIP-BMP4 transduced cells (MD_hypo). As BMP4 signaling activates only SMAD1, 5 and 8 but not SMAD2 and 3 like GDF11 or TGFβ, we also analyzed its synergy with activin α, taken as a surrogate for SMAD2 and 3 activation, that was previously shown to induce cellular quiescence of LNCaP* cells treated at low cell density.29 Most of the genes that were robustly repressed upon Smad7 or sh-SMAD4 overexpression under dormancy-inducing conditions were strongly induced upon BMP4 or BMP4+activin α cell stimulation under exponential growth conditions (Fig. 2C). These included the classical targets of BMP signaling (SMAD6, 7 and ID1) and others genes like KRT19 and KLK3. CCNA2 was strongly repressed only by co-stimulation with BMP4+activin α, activin α at 50 ng/ml having only a small effect on mRNA accumulations under MD_hypo conditions (Fig. 2D). However, induction of CDKN1A, GADD45A and PTCH1 expression by BMP4+ activin α at medium cell density was significantly lower than in dormant cells suggesting the implication of other signaling pathways, besides SMAD signaling in the regulation of these 3 genes (see below). Overall, these data showed that SMAD signaling was a major contributor to the dormancy signature.
Redox imbalance is a component of the dormant state
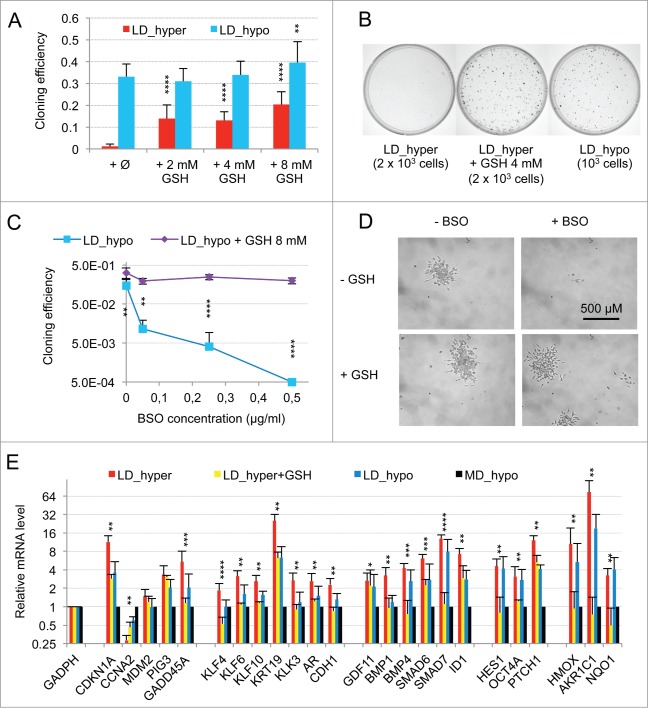
The limited induction of PTCH1, CDKN1A and GADD45A genes suggested that hypertonicity activated other signaling pathways beside SMAD. The previous observation of an induction of PIG3, GADD45A, CDKN1A, KLF4 and 6 in response to cellular oxidative stress suggested the involvement of a redox imbalance in the establishment of cellular dormancy. Although we initially reported that glutathione alone only partially suppressed dormancy in LNCaP* cells,29 subsequent studies with glutathione (2 to 8 mM) and with others thiol-based antioxidants like N-acetylcysteine (NAC, 2 to 8 mM) or dithiothreitol (200 μM) showed that they induced a 40–100-fold increase in cloning efficiency under LD_hyper conditions when added at the beginning of the assay (data not shown and Fig. 3A and B). Thus, thiol-based antioxidant supplementation effectively blunted the effects of hypertonicity, enabling similar cloning efficiencies in LD_hyper and LD_hypo. Inversely, we found that treatment with buthionine sulfoximine (0.05 to 0.5 μg/ml), an inhibitor of glutathione synthesis strongly decreased cloning efficiency under LD_hypo conditions (Fig. 3C). For doses inferior or equal to 0.25 μg/ml, isolated cells remained alive for at least 2 weeks suggesting an induction of cell quiescence in a large fraction of the seeded cells (Fig. 3D). Importantly, this inhibitory effect of BSO was reversed if glutathione (8 mM) was simultaneously added (Fig. 3C and D). Altogether, these experiments showed that glutathione levels strongly determined cell fate. We therefore investigated the effects of glutathione supplementation on the establishment of the dormancy signature for cells cultured under LD_hyper growth conditions. As shown in Figure 3E, early glutathione treatment increased mRNA accumulation for CCNA2 and decreased mRNA accumulation for all others genes of the dormancy signature, their mRNA levels becoming similar or even lower to those obtained at low cell density in hypotonic conditions. Thus the effects of hypertonicity on the dormancy signature could be suppressed by an early antioxidant treatment.
Figure 3.
Modulation of dormancy by glutathione (GSH). (A) Glutathione supplementation reverse the effect of hypertonicity on the cloning efficiency of LNCaP* cells. Statistical significance of the effects of glutathione addition is indicated. (B) Pictures of representative cloning efficiency assays for the indicated cell culture conditions. (C) LNCaP* cell cloning inhibition by Buthionine Sulfoximine (BSO) under hypotonic conditions and its reversal by glutathione supplementation. Cloning efficiency of LNCaP* cells cultured in hypotonic conditions (LD_hypo) in the presence of BSO and/or Glutathione as indicated were determined. Data are averaged from 2 independent cell experiments and are presented as mean ± s.d., with statistical significance indicated for the effects of glutathione supplementation. (D) Pictures of cells after 13 d of culture under LD_hypo conditions in the presence of none, GSH 8mM, BSO 0.25 mM and BSO plus GSH. (E) Glutathione supplementation reverses the gene expression signature of dormancy. Relative mRNA levels were calculated as in Fig. 1A as the ratio of mRNA levels species measured in LNCaP* cell populations cultured under LD_hyper conditions in the presence of 4 mM glutathione and in MD_hypo conditions. Data are averaged from 3 cell culture experiments. Relative mRNA levels under LD_hyper and LD_hypo conditions are those of Figure 1. Statistical significance of the differences between dormant and glutathione-treated cells is indicated as in Figure 1.
As ROS are also involved in physiological cell signaling without implying any true redox imbalance, we searched for an activation of the NF-E2 p45-related factor 2 (NRF2) transcription factor, since it is inactive under homeostatic conditions but is activated in response to redox imbalance through thiol modification of its inhibitor KEAP1.31 Accordingly, we measured the mRNA levels of 3 well-characterized targets of NRF2, the Aldo-keto reductase family 1 member C1 (AKR1C1), the Heme Oxygenase 1 (HMOX1), and the NAD(P)H dehydrogenase quinone 1 (NQO1) genes. A 74, 10 and 3 fold change in the mRNA levels of these genes was observed in cells plated under LD_hyper conditions, respectively (Fig. 3E, right). An induction of these genes was also observed in VCaP cells plated under LD_hyper conditions (Figs. S2C and S3). As expected, early glutathione treatment strongly decreased the mRNA accumulations of these genes confirming the dependence of their induction on redox imbalance (Fig. 3E, right). This confirmed that a redox imbalance played an essential role in the setting-up of the dormant state.
To further assess the role of redox imbalance in regulation of gene expression in dormant cells, we investigated the effects of an oxidative stress on gene expression in cells cultured under exponential growth conditions in hypotonic medium (MD_hypo). Here we chose to treat cells with hydrogen peroxide since it is a natural oxidant produced by cellular O2 metabolism, and it readily oxidizes thiols to their disulfide or sulfonic acid derivatives. Under LD_hypo conditions a strong inhibition of LNCaP* cell clonogenicity was observed when 100 μM H2O2 was added at the beginning of the assay (Fig. 4A, inset). We therefore performed RT-qPCR analyses on LNCaP* cells cultured under MD_hypo conditions in the presence of 100 μM H2O2. After two days the mRNA levels of all cellular stress markers (CDKN1A, PIG3, GADD45A, KLF4, KLF6, HMOX1, AKR1C1, NQO1) were strongly induced while the CCNA2 mRNA was repressed (Fig. 4A). In addition, expression of BMP1, BMP4, KLF10, OCT4A, MDM2 was increased while that of KRT19 was down-regulated. Thus in exponentially growing cells a redox imbalance induced the expression of a subset of the dormancy signature genes.
Reconstitution of the dormancy signature by reconstruction experiments
Comparison of mRNA accumulation patterns in hydrogen peroxide-treated cells (Fig. 4A) and pTRIP-BMP4 transduced cell populations plus exogenous activin α (Fig. 2C) suggested that the association of these stimuli could generate a complete dormancy signature. This was tested by analyzing the mRNA accumulation in pTRIP-BMP4 transduced cells populations cultured under MD_hypo conditions in the presence of activin α and hydrogen peroxide. Remarkably, cell stimulation by activin α, BMP4 and hydrogen peroxide induced a pattern of mRNA accumulation almost indistinguishable from that of dormant cells (Fig. 4B) in cells under growth-permissive conditions (MD_hypo). This response included enhanced mRNA accumulation of both differentiation and stemness markers and a strong down regulation of CCNA2 mRNA. The main discrepancies were BMP1, PTCH1 and HMOX1. The lack of BMP1 mRNA up regulation by BMP4 + Activin α + H2O2 might be related to the artificial stimulation by BMP4 + activin α as this mRNA species seemed to be decreased under these conditions (Fig. 2C). The high induction of PTCH1 mRNA in dormant cells was cell-specific as it was lower in VCaP cells (see Fig. S2C). Finally the lower accumulation of HMOX1 mRNA in dormant cells is clearly linked to some quantitative differences in the effects of hydrogen peroxide and hypertonicity (see Fig. 4A). Thus, the similarity in gene expression signatures suggested that signaling elicited by members of the TGFβ/BMP family and redox imbalance could account for the key features of the dormant state.
SMAD signaling and redox imbalance induction by low cell density
As noted in Figure 1A, and Figures S2A and S3, many features of the dormancy signature were already present, albeit at a lower level, in cells cultured at low density without hypertonic conditions. As several of these genes, including SMAD6, 7, ID1 and KRT19 were also regulated by the BMP/TGFβ signaling pathways during dormancy this suggested that this pathway was at least partially activated by culture at low density. To investigate this point, we tested the effects of over-expressing the inhibitory murine Smad7 on gene expression of cell plated at low density under hypotonic conditions (LD_hypo). As shown in Figure 5A, Smad7 overexpression inhibited the expression of endogenous SMAD6, 7, ID1, PTCH1 and KRT19 genes normally observed in cells at low cell density. Depletion of SMAD4 by expressing sh-SMAD4 confirmed the requirement of SMAD signaling in the induction of these genes by the mere culture at low density and further suggested that induction of HMOX1, GADD45A, KLK3, HES1, BMP4 and to a lesser extent OCT4A was also dependent upon this signaling pathway (Fig. S4C). Overall, this supported that SMAD signaling is activated by cell culturing at low cell density.
Figure 5.
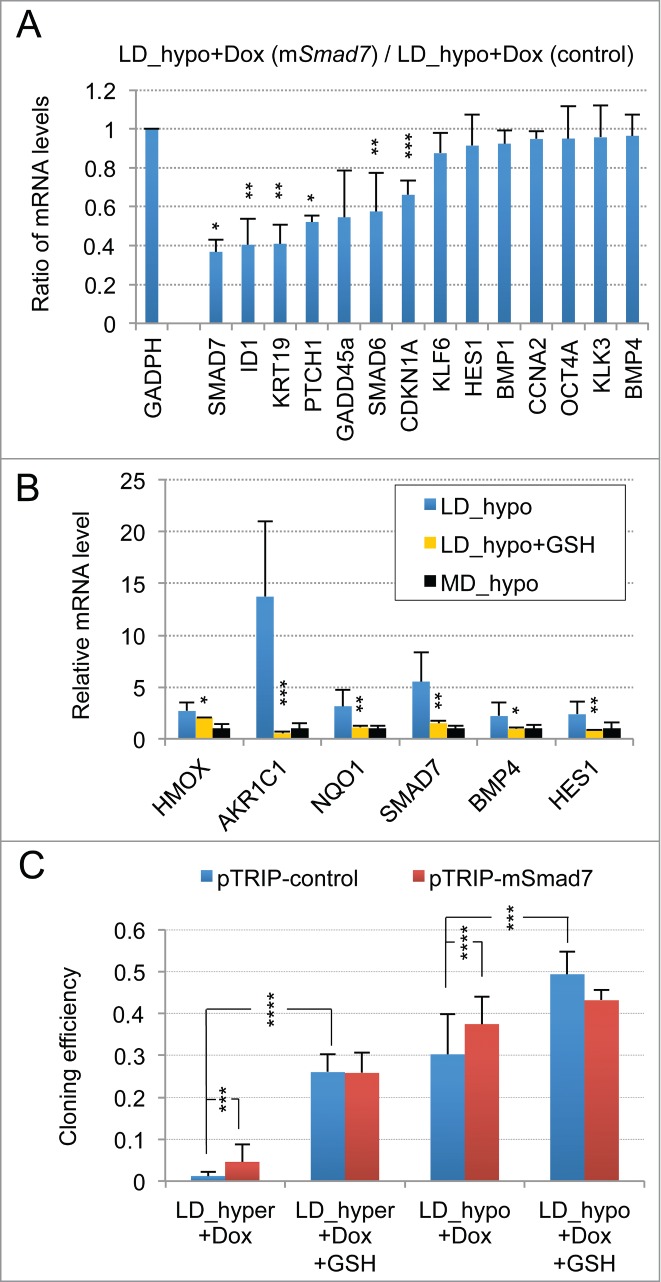
Activation of SMAD signaling at low cell density and contribution of ROS signaling. (A) Smad7 overexpression changes the gene expression profile of LNCaP* cells under LD_hypo conditions. Ratios of mRNA species levels measured by RT-qPCR in pTRIP-mSMAD7 and pTRIP-control transduced cell populations cultured under LD_hypo conditions were averaged from 2 experiments using independent cell populations. Stars indicate significance of the decreased mRNA species accumulation upon murine Smad7 overexpression. (B) Glutathione supplementation reduces the activation of SMAD signaling at low density. Relative mRNA levels measured by RT-qPCR under LD_hypo, LD_hypo+glutathione and MD_hypo conditions were averaged from 4–5, 2 and 7–9 cell culture experiments, respectively. Stars indicate statistical significance of the difference in mRNA levels between cells cultured in LD_hypo and LD_hypo+glutathione. (C) Lack of additive effects between Smad7 overexpression and glutathione treatment on cloning efficiency. Cloning efficiency of pTRIP-control and pTRIP-mSmad7 transduced cell populations were measured under the indicated cell culture conditions. Data were averaged from 2 to 5 experiments made with 2 independent control and 2 independent Smad7 transduced cell populations processed in parallel. Stars indicate significance of the differences in cloning efficiency. Data are presented as mean ± s.d.
We also observed that glutathione treatment under LD_hyper cell culture conditions decreased mRNA accumulation of a few genes including HES1, SMAD7, BMP4 and also NRF2-targets to their minimal levels (corresponding to MD_hypo conditions) suggesting that their mRNA induction required cell signaling triggered by redox imbalance. Since these genes were also induced by culture at low cell density (LD_hypo), this suggested that low cell density was sufficient to promote some redox imbalance. To assess this possibility, we analyzed the effects of glutathione treatment on mRNA accumulation of these genes in cells cultured under LD_hypo conditions. Data on Figure 5B indicates that glutathione treatment significantly inhibited mRNA accumulation of these genes under LD_hypo conditions. This strongly suggested that the mere culture at low cell density could induce some redox imbalance leading to the expression of these genes. Moreover, the strong inhibition of BMP4 and SMAD7 expression by glutathione suggested that the redox imbalance associated with a low cell density could be in part responsible for the activation of TGFβ/BMP signaling. Accordingly, either glutathione addition and murine Smad7 overexpression significantly increased cloning efficiency under both LD_hyper and LD_hypo conditions but their association had no additive effects (Fig. 5C).
To further ascertain the above conclusions, we reasoned that if TGFβ/BMP and redox imbalance signaling were the main signaling pathways, both activated under LD_hyper and LD_hypo conditions although at different intensities, then the patterns of mRNA accumulation for cells cultured under these 2 conditions should be correlated. Therefore, we calculated the Pearson linear correlation coefficients between the patterns of mRNA level variations measured in cells grown under LD_hyper, LD_hypo, MD_hyper (exponential) and confluent growth conditions as described in the Methods section using expression data in Figure S3. A high degree of correlation (r = 0.94; n = 24 genes; p < 0.0001) was indeed observed for LNCaP* cultured under LD_hyper and LD_hypo. By contrast, correlation coefficient values did not reach significance when comparing mRNA level variation patterns for cells cultured under LD_hyper and MD_hyper (r = 0.05; n = 24; p = 0.81) or LD_hyper and confluent (r = 0.07; n = 20; p = 0.76) conditions. This also held true for VCaP cells for which the correlation coefficient reached 0.95 (n = 23 genes; p< 0.001) between LD_hyper and LD_hypo conditions but did not reach significance when comparing LD_hyper and MD_hyper conditions. Of note, because this test measured linear correlations, this indicated that cellular states reached under LD_hyper and LD_hypo conditions were highly related. And indeed, we calculated that there is on average a 1.90 ± 0.82 (n = 24) and 1.93 ± 0.67 (n = 23) fold change in the mRNA levels between LD_hypo and LD_hyper conditions for genes of the dormancy signature in LNCaP* and VCaP cells, respectively (geometric means ± s. d. calculated from data in Fig. S3).
Discussion
In this study we exploited our ability to induce a stable but reversible dormant state in cultured prostate cancer cells29 to generate a genetic expression signature of the dormant state in order to identify the signaling pathways regulating dormancy.
Respective contribution of low cell density and hypertonicity to the induction of the dormant state
There are 2 distinct requirements to induce dormancy in our cellular model: low cell density and exposure to a slightly hypertonic medium (Fig. 6). A requirement for low cell density has been previously observed in a model of breast cancer cell dormancy in culture,26 and at least several dormant cells in vivo, including normal haematopoietic stem cells,32 cancer cells,2-5,33 or circulating memory B or T lymphocytes display very few if any homotypic cell interactions. Here we observed that low cell density was sufficient to activate the BMP/TGFβ signaling pathway and to modulate the mRNA level of most genes of the dormancy signature including ID1, SMAD7, KRT19, HES1 and BMP4 (Fig. 5A and Fig. S4C). The BMP/TGFβ pathway has been previously implicated in restricting cancer metastasis development.15-20 Thus, it has been reported that BMP7 could inhibit formation of bone but not loco-regional lymph node metastases by human prostate cancer cells inoculated in nude mice. 17 In a subsequent study, expression of the BMPR2 receptor (that can bind BMP4 or BMP7) in prostate cancer cells was inversely correlated with bone metastasis and recurrence in men and required for the quiescence of cancer stem cells.15 Finally, BMP signaling was shown to modulate dormancy of human breast cancer cells.16,19 These observations and those, numerous, implicating the BMP/TGFβ signaling pathway in the control of normal stem cells in different tissues indicate the relevance of our in vitro experimental approach to decipher the control of dormancy.
Figure 7.
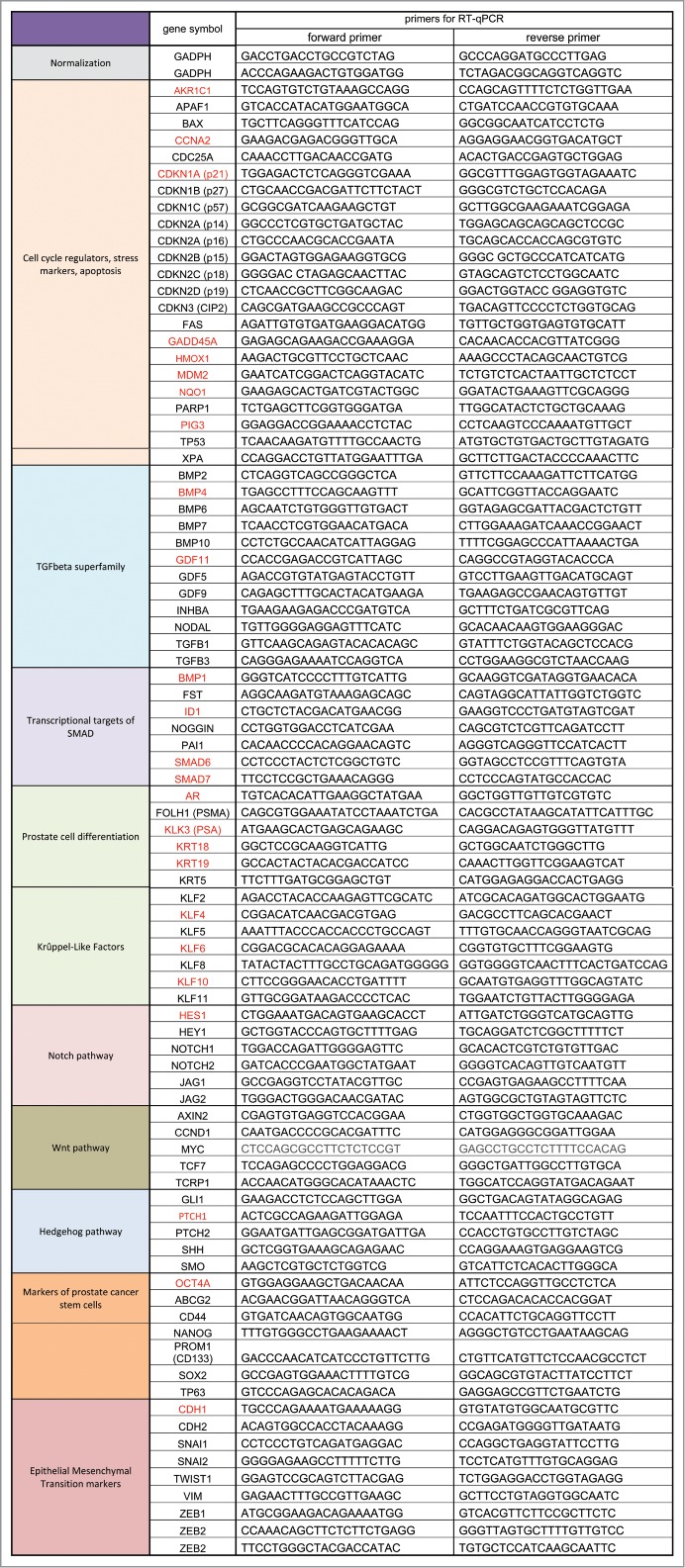
Figure 8.
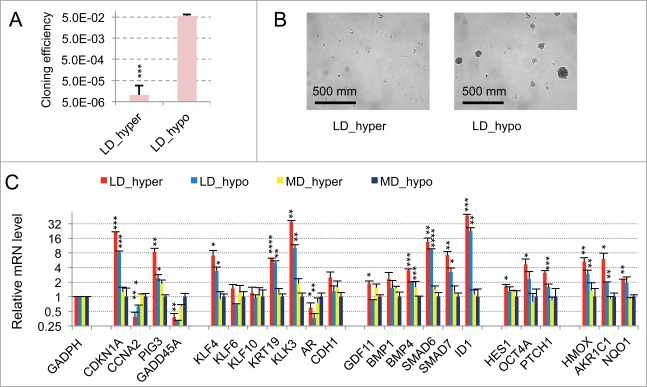
Figure 9.
Figure 10.
Figure 11.
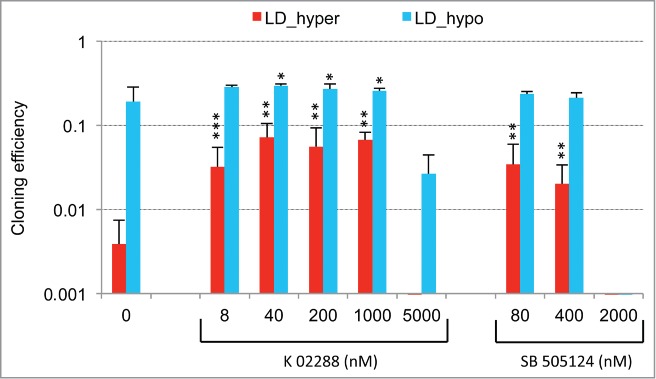
Figure 6.
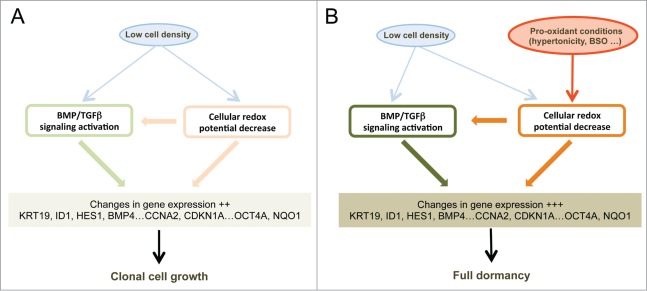
Schematic representation of the signaling pathways involved in cell dormancy. (A) Cell signaling pathways activated by low cell density under clonogenic conditions (i. e. in isotonic or slightly hypotonic culture medium). Under these conditions, a mild activation of BMP/TGFβ (Fig. 1A, 5A and Fig. S4C) and redox imbalance (Fig. 1A and 5B) signaling pathways is observed, redox imbalance also contributing to the activation of BMP/TGFβ signaling (Fig. 5B). (B) Reinforcements of these signaling pathways under dormancy-inducing conditions in slightly hypertonic medium. The main effect of hypertonicity is to amplify redox imbalance and BMP/TGFβ signaling (Fig. 3, Fig. 2B and Fig. S4B and C) thus increasing the expression level of the dormancy gene expression signature that is already activated in cells plated at low density but in hypotonic or isotonic medium.
Importantly, we report that another role of low cell density is to generate a slight redox imbalance that is important for gene induction at low cell density. Notably, activation of the BMP/TGFβ pathway by culture at low density is markedly reduced in the presence of glutathione as indicated by the expression of BMP4, SMAD7 and HES1 (Fig. 5B) establishing that these 2 pathways are coupled (Fig. 6A). As to the role of the second stimulus, hypertonicity (Fig. 6B), one striking result of our analysis is that it mainly enhances the redox imbalance of cells cultured at low density while having no significant effect on cells at medium density. This enhanced redox imbalance reinforces a program already expressed by cells plated at low density. This translates into highly similar patterns of gene expression between LD_hypo and LD_hyper cells, which is apparent through a Pearson correlation coefficient of 0.94. Consequently, genes of the dormancy signature are expressed on average twice as much in LD_hyper than in LD_hypo cells. These observations could mean that entry into dormancy requires a threshold level of activation of the dormancy inducing pathways that is reached in the overwhelming majority of the cells only when low cell density is associated with pro-oxidant condition (Fig. 6B). This also suggests the possible presence of dormant cells in cultures at low density under permissive conditions (LD-hypo; see Fig. 6A). This is compatible with our observations that conditions that can induce exit from dormancy in hypertonic medium (glutathione treatment, Smad7 or sh-SMAD4 ectopic expression or CDKN1A down regulation) increase modestly but significantly cloning efficiency under permissive conditions (Figs. 3A, 5C, Fig. S4A, and data not shown). In any case, the similarity between the gene expression profiles in LD_hypo and LD_hyper conditions indicates a crucial role of low cell density in priming cells for dormancy. Moreover, the increased redox imbalance provides a molecular basis for the role of hypertonicity which will warrant further investigation. As several thiol-based antioxidants but not sodium ascorbate, tocopherol acetate or phenol derivatives are effective to counterbalance the effects of hypertonicity (Fig. 3A, B and data not shown) oxidation of cellular thiols should be involved. TP53 is one of the multiple cellular targets regulated by thiol modification 34,35 and we have previously observed a significant role of TP53 of dormancy.29 However, TP53 alone cannot account for the importance of the redox imbalance since its impact on the number of dormant cells is only modest. Moreover, VCaP cells which express a dominant-negative TP53 protein (R248W mutation, ref.36) enter into dormancy like LNCaP cells and express a very similar gene expression signature (Figs. S2 and S3) indicating that the p53 pathway is not essential for dormancy. Others signaling pathways modulated by hypertonicity in a thiol sensitive manner are currently investigated.
BMP/TGFβ and redox imbalance signaling are necessary and sufficient to establish a dormant state
We found that both signaling pathways were required for establishment of the dormant state since their disruption upon Smad7 or sh-SMAD4 overexpression (Fig. 5C and Fig. S4A), treatment with pharmacologic inhibitors of BMP/TGFβ receptors (Fig. S5) or antioxidant treatment (Fig. 3A, B and data not shown) increases cloning efficiency under dormancy-inducing conditions. Inversely, we showed that activin α or redox imbalance induced by BSO or H2O2 treatment could decrease cloning efficiency of cells cultured at low cell density under permissive conditions (Fig. 3C, inset in Fig. 4A and ref.29). Association of a redox imbalance with BMP4 is likely to further strengthen this growth-inhibitory activity as suggested by CCNA2 mRNA down regulation and CDKN1A induction at medium cell density (Figs. 2C, 4A and 4B), ectopic overexpression of CDKN1A having also been shown to be sufficient to induce a complete growth arrest at low and medium cell density (data not shown).
We further demonstrated that BMP/TGFβ signaling pathways and redox imbalance are sufficient to account for most features of the dormancy signature. Activation of these 2 signaling pathways seems specific to dormant cells since the dormancy signature can be easily distinguished from that of cells rendered quiescent by culture at high confluence. Interestingly, the genetic signature of the dormant state includes mRNA of genes involved in epithelial differentiation as well as markers of stemness. It is interesting to note that expression of markers like HES1 and PTCH1 in dormant cells were not affected by the addition of Notch or Hedgehog signaling inhibitors (cyclopamine and DAPT; data not shown) suggesting that these signaling pathways are not involved in their induction. Thus dormant cells do not readily conform to a standard phenotype and, at least in the 2 cell lines derived from prostate cancer that we have studied, dormancy is associated with epithelial traits. We suggest that in addition to replicative senescence, cell cycle arrest induced by cell contact-inhibition/nutrient starvation and terminally differentiated cellular states this dormant state constitutes a fourth state of cellular quiescence that is actively maintained through both a decreased cellular redox potential and BMP signaling.
Predictions of the model
Our observation that, in association with BMP signaling, an oxidative stress can induce dormancy could provide an overlooked rationale for some reports on the control of the metastatic process. Expression of COCO, an inhibitor of BMP signaling, was shown to be sufficient to revert dormancy of breast cancer cells disseminated in the lung but not in others organs like bones or brain.16 The authors related this restricted activity of COCO to a lower paracrine activity of BMP in these latter organs. However, if the major findings of our model hold true for breast cancer cells, the pro-oxidant conditions encountered by cancer cells in the lung could account the ability of BMP to induce dormancy specifically in this organ and not in those with a lower exposure to oxygen. Finally, our observations lead us to predict that the prevailing conditions in the blood circulation should entice Circulating Tumor Cells (CTCs) to enter into a dormant state before their extravasation into the stroma of distant organs. Indeed, CTCs in the blood are exposed to a higher oxygen concentration than in the initial tumor. This could contribute to the poor efficiency of the metastatic process when compared with the number of circulating cells.4,37,38 Additionally, the observation that the number of metastases correlates with the number of CTC clusters of 4 cells or more rather than with individual CTC might reflect a role of homotypic intercellular contacts in preventing dormancy.39 Thus, by highlighting the role of 2 different biological phenomena, cell density and redox imbalance, our in vitro analysis of dormancy suggests further avenues of investigation of the metastatic process.
Materials and Methods
Plasmids and chemicals
Doxycycline-inducible pTRIP vectors for murine Smad7 (mSmad7) and human BMP4 cDNA expression were constructed by replacing the RFP-shRNA cassette downstream the TET/ON promoter in pTRIPZ-shp21 (Open Biosystems, Thermofisher Scientific Biosciences) with the corresponding cDNA. Excision of the RFP-shRNA cassette was achieved by Age1-Mlu1 digestion of pTRIPZ-shp21 followed by Klenow treatment. mSmad7 cDNA was recovered from pBabepuro-mSmad7 (previously constructed by T.T. from murine F9 cell cDNA) by BamH1-SphI digestion and Klenow blunting, BMP4 cDNA was derived from pCMV-Sport6-hBMP4 (sequence verified cDNA clone IMAGE n° 4399276 from RZPD; German Science Center for Genome Research) by Xho1-Age1 digestion and Klenow blunting. pTRIP-control vector refers to pTRIPZ-shp21 following excision of the RFP-shRNA cassette. pSuperRetro-sh-SMAD4 was constructed as previously described.40 Recombinant human Activin A was from R&D systems (Catalog No 338-AC, lot BNV3211101). K-0228841 was from Tocris (Catalog No 4986 Batch No 1A/158861), SB-50512442 was from Abcam (Catalog No ab144402; Lot No GR204729–1), reduced L-glutathione (Catalog No G6013–5G; Lot No 088K07621V) and L-Buthionine sulfoximine (Catalog No B2515) were from Sigma-Aldrich.
Cells and retroviruses
DMEM-FCS refer to DMEM (containing Glutamax-I and 4.5 g/l glucose without pyruvate, ref 61965 Life Technologies) supplemented with 10% fetal calf serum (PAA Laboratories, Pasching, Austria) and penicillin plus streptomycin solution (Life Technologies). Cloning efficiency assays were mostly performed in 6 cm-diameter dishes seeded with 103 to 4 × 103 cells. LNCaP* designates a cell population derived from LNCaP cells (a gift from Dr Florence Cabon, Institut André Lwoff, Villejuif, France) through transfection of plasmid pBabeBleo-EcoR (encoding the murine ecotropic leukemia retrovirus receptor)43 and phleomycin selection (10 μg/ml). These cells were routinely cultured in DMEM-FCS as described previously.29 VCaP cells (from Dr A. Chauchereau, Institut Gustave Roussy, France) were routinely passaged in DMEM-FCS supplemented with 15% water to make medium isotonic at a 1:4 dilution after detachment of highly confluent cells with trypsin-EDTA (Life Technologies). Recombinant lentiviruses were produced by DNA transfection of 0.7 × 106 293T cells, plated in 6-wells plates the day before, with 2 μg of retroviral vectors and 2 μg of trans-complementing plasmids (1.3 μg of pCMV-ΔR8.74 plus 0.7 μg of pMD.G) using Lipofectamine 2000 (Gibco Invitrogen Corporation). Viral transductions were performed as previously.29
Flow cytometry and western blot analysis
Western Blot analysis were carried out as previously described,29 except for a modified Bradford procedure to determine protein concentration.44 Smad7 antibody was from R&D systems (Catalog No MAB2029, lot No YHN02). Bands intensity on pictures of scanned films in TIFF format were determined using GelQuant.NET software (V 1.7.8 from http://biochemlabsolutions.com). Flow Cytometry and cytometric analysis were performed as previously except for a 2 step gating procedure to eliminate G1 cell doublets using SSC-H versus FSC-H and then FL2-A vs. FL2-W fluorescence dot plot representations.45
RT-qPCR assays
For low cell density culture conditions, 10 or 20 dishes (10 cm diameter) were seeded with 104 cells in the indicated medium and incubated for 7 d at 37°C and 5% CO2. At that time, supernatants were discarded and cells were harvested by vigorous pipeting in PBS1X supplemented with 0.5% FCS and 1 mM EDTA at 4°C, pelleted by centrifugation, rinsed once with cold PBS1X and total RNA extracted using a RNeasy extraction kit (Qiagen). Amounts of RNA extracted per cell were similar for different cell culture conditions. For medium cell density conditions, cells were seeded at 4 × 105 cells in 6 cm diameter dish and lysed 3 d later directly in the cell dishes after one cold PBS1X wash. For high-density growth-arrested cell culture, cells were seeded at 6 × 105 cells in 6 cm diameter dish and grown for 7 days, with no medium change in the last 3 d before cell lysis. For single strand cDNA synthesis, 2 μg of RNA were used with the Maxima first strand cDNA synthesis kit (Thermofisher Scientific Biosciences) or with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems Life Technologies). Real-time PCR analysis were performed on about 20 ng of cDNA using the Maxima SYBR Green qPCR Master Mix (2X) (Thermofisher Scientific Biosciences) and the primers listed in Fig. S1 in a Lightcycler-01 or a LightCycler-1.5 machine (ROCHE) with LCS3.5 or LCS4 software respectively. Background signal was determined by analyzing negative controls generated by omitting reverse transcriptase at the mRNA reverse transcription step. Genes which generated a signal not exceeding 8 times the background were excluded from further analysis. For each experimental condition, amplification signals were normalized to that of GAPDH and relative expression level for gene X was calculated as 2[Ct(GADPH)- Ct(gene X)], Ct being the cutting threshold calculated by the LCS3.5 or LCS4 qPCR software. Importantly, GADPH expression levels differed by less than a factor 2 between different experiments. For averaging the mRNA levels, one of 2 calculation methods was used depending on the experimental design and the relative importance of stochastic and non-stochastic (such as use of different RT-qPCR kit or machines) errors in mRNA levels measurements. In the normalized averaging (NA) method, the relative expression levels obtained from independent experiments were averaged and the corresponding standard deviation calculated. Values were then divided by a normalization factor chosen for each gene to set expression level at medium cell density under hypotonic conditions (MD_hypo) to unity. In the averaging ratio (AR) method used when experiments involved comparisons between paired cell populations processed in parallel, the ratios between the GADPH-normalized mRNA levels measured for each gene in the paired cell populations were calculated, averaged and standard deviation of the ratios derived. When both applicable, these 2 methods led to very similar results (Pearson linear correlation coefficient greater than 0.99 for the dormancy signature of LNCaP* cells; compare Fig. 1A and C) as they correspond to calculation of the ratio of the averaged mRNA levels and the average of the mRNA level ratios respectively.
Statistical analysis
Significance of mRNA levels variations was calculated using a paired or a heteroscedatic Student's t test with unilateral distribution. Significance (p value) was indicated by stars as **** for P < 0.002; *** for P < 0.01; ** for P < 0.05 and * for P < 0.1. For correlation analysis between patterns of mRNA accumulation measured under different cell culture conditions, the linear correlation coefficient of Pearson was calculated for genes of the dormancy signature. For this analysis, we used for each gene the relative mRNA levels normalized to unity under MD_hypo cell culture condition because large variations between the levels of different mRNA species countermanded mRNA level variations originating in different cell culture conditions for a given gene (data not shown). Significance of the correlation coefficients was calculated on the VassarStats web site (http://vassarstats.net/rsig.html).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors gratefully thank Christian Auclair for his generous support of this project and C. Ohanna for performing flow cytometry analysis of LNCaP* cells for CDH1 and CDH2 expression. The authors also thank Paul Tchénio and Claude Boucheix for their gift of K-02288 and SB-505124 inhibitors.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Aguirre-Ghiso JA, Ossowski L, Rosenbaum SK. Green fluorescent protein tagging of extracellular signal-regulated kinase and p38 pathways reveals novel dynamics of pathway activation during primary and metastatic growth. Cancer Res 2004; 64:7336-45; PMID:15492254; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-0113 [DOI] [PubMed] [Google Scholar]
- 2. Cameron MD, Schmidt EE, Kerkvliet N, Nadkarni KV, Morris VL, Groom AC, Chambers AF, MacDonald IC. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res 2000; 60:2541-6; PMID:10811137 [PubMed] [Google Scholar]
- 3. Goodison S, Kawai K, Hihara J, Jiang P, Yang M, Urquidi V, Hoffman RM, Tarin D. Prolonged dormancy and site-specific growth potential of cancer cells spontaneously disseminated from nonmetastatic breast tumors as revealed by labeling with green fluorescent protein. Clin Cancer Res 2003; 9:3808-14; PMID:14506175 [PubMed] [Google Scholar]
- 4. Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, Groom AC. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol 1998; 153:865-73; PMID:9736035; http://dx.doi.org/ 10.1016/S0002-9440(10)65628-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naumov GN, MacDonald IC, Weinmeister PM, Kerkvliet N, Nadkarni KV, Wilson SM, Morris VL, Groom AC, Chambers AF. Persistence of solitary mammary carcinoma cells in a secondary site: a possible contributor to dormancy. Cancer Res 2002; 62:2162-8; PMID:11929839 [PubMed] [Google Scholar]
- 6. Wiedswang G, Borgen E, Karesen R, Qvist H, Janbu J, Kvalheim G, Nesland JM, Naume B. Isolated tumor cells in bone marrow three years after diagnosis in disease-free breast cancer patients predict unfavorable clinical outcome. Clin Cancer Res 2004; 10:5342-8; PMID:15328170; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-0245 [DOI] [PubMed] [Google Scholar]
- 7. Uhr JW, Pantel K. Controversies in clinical cancer dormancy. Proc Natl Acad Sci USA 2011; 108:12396-400; PMID:21746894; http://dx.doi.org/ 10.1073/pnas.1106613108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruppender NS, Morrissey C, Lange PH, Vessella RL. Dormancy in solid tumors: implications for prostate cancer. Cancer Metast Rev 2013; 32:501-9; PMID:23612741; http://dx.doi.org/ 10.1007/s10555-013-9422-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ossowski L, Aguirre-Ghiso JA. Dormancy of metastatic melanoma. Pigment Cell Melanoma Res 2010; 23:41-56; PMID:19843243; http://dx.doi.org/ 10.1111/j.1755-148X.2009.00647.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sainaghi PP, Castello L, Bergamasco L, Galletti M, Bellosta P, Avanzi GC. Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J Cell Physiol 2005; 204:36-44; PMID:15605394; http://dx.doi.org/ 10.1002/jcp.20265 [DOI] [PubMed] [Google Scholar]
- 11. Braun S, Pantel K, Muller P, Janni W, Hepp F, Kentenich CR, Gastroph S, Wischnik A, Dimpfl T, Kindermann G, et al. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N Engl J Med 2000; 342:525-33; PMID:10684910; http://dx.doi.org/ 10.1056/NEJM200002243420801 [DOI] [PubMed] [Google Scholar]
- 12. Naumov GN, Townson JL, MacDonald IC, Wilson SM, Bramwell VH, Groom AC, Chambers AF. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast Cancer Res Treat 2003; 82:199-206; PMID:14703067; http://dx.doi.org/ 10.1023/B:BREA.0000004377.12288.3c [DOI] [PubMed] [Google Scholar]
- 13. Najmi S, Korah R, Chandra R, Abdellatif M, Wieder R. Flavopiridol blocks integrin-mediated survival in dormant breast cancer cells. Clin Cancer Res 2005; 11:2038-46; PMID:15756030; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-1083 [DOI] [PubMed] [Google Scholar]
- 14. Braun S, Kentenich C, Janni W, Hepp F, de WJ, Willgeroth F, Sommer H, Pantel K. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J Clin Oncol 2000; 18:80-6; PMID:10623696 [DOI] [PubMed] [Google Scholar]
- 15. Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, Pai SK, Liu W, Fukuda K, Chambers C, et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med 2011; 208:2641-55; PMID:22124112; http://dx.doi.org/ 10.1084/jem.20110840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao H, Chakraborty G, Lee-Lim AP, Mo Q, Decker M, Vonica A, Shen R, Brogi E, Brivanlou AH, Giancotti FG. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell 2012; 150:764-79; PMID:22901808; http://dx.doi.org/ 10.1016/j.cell.2012.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buijs JT, Rentsch CA, van der Horst G, van Overveld PG, Wetterwald A, Schwaninger R, Henriquez NV, Ten DP, Borovecki F, Markwalder R, et al. BMP7, a putative regulator of epithelial homeostasis in the human prostate, is a potent inhibitor of prostate cancer bone metastasis in vivo. Am J Pathol 2007; 171:1047-57; PMID:17724140; http://dx.doi.org/ 10.2353/ajpath.2007.070168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buijs JT, Henriquez NV, van Overveld PG, van der Horst G, ten Dijke P, van der Pluijm G. TGF-beta and BMP7 interactions in tumour progression and bone metastasis. Clin Exp Metast 2007; 24:609-17; PMID:18008174; http://dx.doi.org/ 10.1007/s10585-007-9118-2 [DOI] [PubMed] [Google Scholar]
- 19. Buijs JT, Henriquez NV, van Overveld PG, van der Horst G, Que I, Schwaninger R, Rentsch C, Ten DP, Cleton-Jansen AM, Driouch K, et al. Bone morphogenetic protein 7 in the development and treatment of bone metastases from breast cancer. Cancer Res 2007; 67:8742-51; PMID:17875715; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-2490 [DOI] [PubMed] [Google Scholar]
- 20. Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, Schewe DM, Aguirre-Ghiso JA. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat Cell Biol 2013; 15:1351-61; PMID:24161934; http://dx.doi.org/ 10.1038/ncb2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 2014; 14:611-22; PMID:25118602; http://dx.doi.org/ 10.1038/nrc3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taichman RS, Patel LR, Bedenis R, Wang J, Weidner S, Schumann T, Yumoto K, Berry JE, Shiozawa Y, Pienta KJ. GAS6 receptor status is associated with dormancy and bone metastatic tumor formation. Plos One 2013; 8:e61873; PMID:23637920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korah R, Boots M, Wieder R. Integrin alpha5beta1 promotes survival of growth-arrested breast cancer cells: an in vitro paradigm for breast cancer dormancy in bone marrow. Cancer Res 2004; 64:4514-22; PMID:15231661; http://dx.doi.org/ 10.1158/0008-5472.CAN-03-3853 [DOI] [PubMed] [Google Scholar]
- 24. Barkan D, Green JE. An in vitro system to study tumor dormancy and the switch to metastatic growth. J Visualized Exp: JoVE 2011; 54:e2914; PMID:21860375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wendt MK, Taylor MA, Schiemann BJ, Schiemann WP. Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol Biol Cell 2011; 22:2423-35; PMID:21613543; http://dx.doi.org/ 10.1091/mbc.E11-04-0306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shibue T, Weinberg RA. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci USA 2009; 106:10290-5; PMID:19502425; http://dx.doi.org/ 10.1073/pnas.0904227106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barrios J, Wieder R. Dual FGF-2 and intergrin alpha5beta1 signaling mediate GRAF-induced RhoA inactivation in a model of breast cancer dormancy. Cancer Microenviron 2009; 2:33-47; PMID:19308677; http://dx.doi.org/ 10.1007/s12307-009-0019-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barkan D, El Touny LH, Michalowski AM, Smith JA, Chu I, Davis AS, Webster JD, Hoover S, Simpson RM, Gauldie J, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res 2010; 70:5706-16; PMID:20570886; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Havard M, Dautry F, Tchenio T. A dormant state modulated by osmotic pressure controls clonogenicity of prostate cancer cells. J Biol Chem 2011; 286:44177-86; PMID:22039055; http://dx.doi.org/ 10.1074/jbc.M111.262709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol 2007; 26:508-23; PMID:17560775; http://dx.doi.org/ 10.1016/j.matbio.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baird L, Swift S, Lleres D, Dinkova-Kostova AT. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol Adv 2014; 32:1133-44; PMID:24681086; http://dx.doi.org/ 10.1016/j.biotechadv.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 2010; 7:391-402; PMID:20804974; http://dx.doi.org/ 10.1016/j.stem.2010.06.020 [DOI] [PubMed] [Google Scholar]
- 33. Naumov GN, MacDonald IC, Chambers AF, Groom AC. Solitary cancer cells as a possible source of tumour dormancy? Semin Cancer Biol 2001; 11:271-6; PMID:11513562; http://dx.doi.org/ 10.1006/scbi.2001.0382 [DOI] [PubMed] [Google Scholar]
- 34. Wu HH, Thomas JA, Momand J. p53 protein oxidation in cultured cells in response to pyrrolidine dithiocarbamate: a novel method for relating the amount of p53 oxidation in vivo to the regulation of p53-responsive genes. Biochem J 2000; 351:87-93; PMID:10998350; http://dx.doi.org/ 10.1042/0264-6021:3510087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Augustyn KE, Merino EJ, Barton JK. A role for DNA-mediated charge transport in regulating p53: Oxidation of the DNA-bound protein from a distance. Proc Natl Acad Sci USA 2007; 104:18907-12; PMID:18025460; http://dx.doi.org/ 10.1073/pnas.0709326104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, Nordeen SK, Miller GJ, Lucia MS. Molecular characterization of human prostate carcinoma cell lines. Prostate 2003; 57:205-25; PMID:14518029; http://dx.doi.org/ 10.1002/pros.10290 [DOI] [PubMed] [Google Scholar]
- 37. Tarin D, Vass AC, Kettlewell MG, Price JE. Absence of metastatic sequelae during long-term treatment of malignant ascites by peritoneo-venous shunting. A clinico-pathological report. Invasion Metast 1984; 4:1-12; PMID:6735637 [PubMed] [Google Scholar]
- 38. Butler TP, Gullino PM. Quantitation of cell shedding into efferent blood of mammary adenocarcinoma. Cancer Res 1975; 35:512-6; PMID:1090362 [PubMed] [Google Scholar]
- 39. Liotta LA, Kleinerman J, Saidel GM. Quantitative relationships of intravascular tumor cells, tumor vessels, and pulmonary metastases following tumor implantation. Cancer Res 1974; 34:997-1004; PMID:4841969 [PubMed] [Google Scholar]
- 40. He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 2006; 125:929-41; PMID:16751102; http://dx.doi.org/ 10.1016/j.cell.2006.03.045 [DOI] [PubMed] [Google Scholar]
- 41. Sanvitale CE, Kerr G, Chaikuad A, Ramel MC, Mohedas AH, Reichert S, Wang Y, Triffitt JT, Cuny GD, Yu PB, et al. A new class of small molecule inhibitor of BMP signaling. Plos One 2013; 8:e62721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vogt J, Traynor R, Sapkota GP. The specificities of small molecule inhibitors of the TGFbs and BMP pathways. Cell Signalling 2011; 23:1831-42; PMID:21740966; http://dx.doi.org/ 10.1016/j.cellsig.2011.06.019 [DOI] [PubMed] [Google Scholar]
- 43. Albritton LM, Tseng L, Scadden D, Cunningham JM. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 1989; 57:659-66; PMID:2541919; http://dx.doi.org/ 10.1016/0092-8674(89)90134-7 [DOI] [PubMed] [Google Scholar]
- 44. Zor T, Selinger Z. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem 1996; 236:302-8; PMID:8660509; http://dx.doi.org/ 10.1006/abio.1996.0171 [DOI] [PubMed] [Google Scholar]
- 45. Nunez R. DNA measurement and cell cycle analysis by flow cytometry. Curr Issues Mol Biol 2001; 3:67-70; PMID:11488413 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.