Abstract
Non-small cell lung cancer (NSCLC) is a common malignancy with a poor prognosis. Despite progress targeting oncogenic drivers, there are no therapies targeting tumor suppressor loss. Smad4 is an established tumor suppressor in pancreatic and colon cancer, however, the consequences of Smad4 loss in lung cancer are largely unknown. We evaluated Smad4 expression in human NSCLC samples and examined Smad4 alterations in large NSCLC datasets and found that reduced Smad4 expression is common in human NSCLC and occurs through a variety of mechanisms including mutation, homozygous deletion, and heterozygous loss. We modeled Smad4 loss in lung cancer by deleting Smad4 in airway epithelial cells and found that Smad4 deletion both initiates and promotes lung tumor development. Interestingly, both Smad4−/− mouse tumors and human NSCLC samples with reduced Smad4 expression demonstrated increased DNA damage while Smad4 knockdown in lung cancer cells reduced DNA repair and increased apoptosis after DNA damage. In addition, Smad4 deficient NSCLC cells demonstrated increased sensitivity to both chemotherapeutics that inhibit DNA topoisomerase and drugs that block double strand DNA break repair by non-homologous end joining. In sum, these studies establish Smad4 as a lung tumor suppressor and suggest that the defective DNA repair phenotype of Smad4 deficient tumors can be exploited by specific therapeutic strategies.
Keywords: Smad4, lung cancer, TGFβ, DNA damage, genotoxic stress, chemosensitivity
INTRODUCTION
Although over 175,000 cases of non-small cell lung cancer (NSCLC) are diagnosed annually in the United States the five-year survival rate remains less than 20% (1). Despite advances in targeting oncogenic drivers, most NSCLC are not amenable to the currently available small molecule inhibitors and there is a paucity of approaches targeting tumor suppressor loss (2). Smad4 mediates signaling of transforming growth factor beta (TGFβ) and bone morphogenic protein (BMP) ligands (3) and is a well-defined tumor suppressor in pancreatic and colon cancer (4). Smad4 loss and mutation have also been described in NSCLC (5, 6) where they are associated with lymph node metastases, increased angiogenesis, and more aggressive cellular behavior in vitro (7, 8), though other consequences of Smad4 alterations in lung cancer are unknown.
In animal models, Smad4 deletion can initiate tumor formation in keratinized epithelial layers and can promote the development of tumors initiated by other oncogenic events (9-16). Smad4 loss increases proliferation secondary to loss of TGFβ-mediated growth suppression and also increases the proportion of well-differentiated tumors, presumably because Smad4 loss blocks TGFβ-driven epithelial to mesechymal transition (10, 12, 14, 17). However, essentially nothing is known about whether Smad4 loss initiates or promotes lung tumor development in vivo or the characteristics of Smad4 deficient lung tumors.
TGFβ1 deletion increases genomic instability and sensitivity to ionizing radiation (18, 19), and we previously reported that Smad4 deletion increases genomic instability in a mouse model of head and neck cancer (11). These findings suggest that TGFβ signaling may regulate genomic stability through DNA repair. DNA repair is a complex process that requires detecting DNA damage and activating appropriate repair pathways (20). Single strand DNA damage is repaired through a combination of mismatch repair, base excision repair, and nucleotide excision repair, while double strand breaks (DSB) are repaired by homologous recombination repair (HRR) or non-homologous end joining (NHEJ) (20). DSB are sensed by ataxia telangiectasia mutated (ATM) which then activates HRR (21, 22) and by Ku family proteins that then activate DNA dependent protein kinase (DNA-PK) and repair through NHEJ (23). HRR is more accurate as it relies on homologous sequences while NHEJ has a lower fidelity and is responsible for the majority of tumorigenic chromosomal rearrangements (23). In this study we sought to evaluate the role of Smad4 loss in human lung cancer, to determine whether Smad4 is a lung tumor suppressor, and to assess DNA repair in Smad4 deficient lung cancer cells, with the ultimate goal of improving treatment strategies for Smad4 deficient tumors.
RESULTS
Reduced Smad4 expression is common in human NSCLC
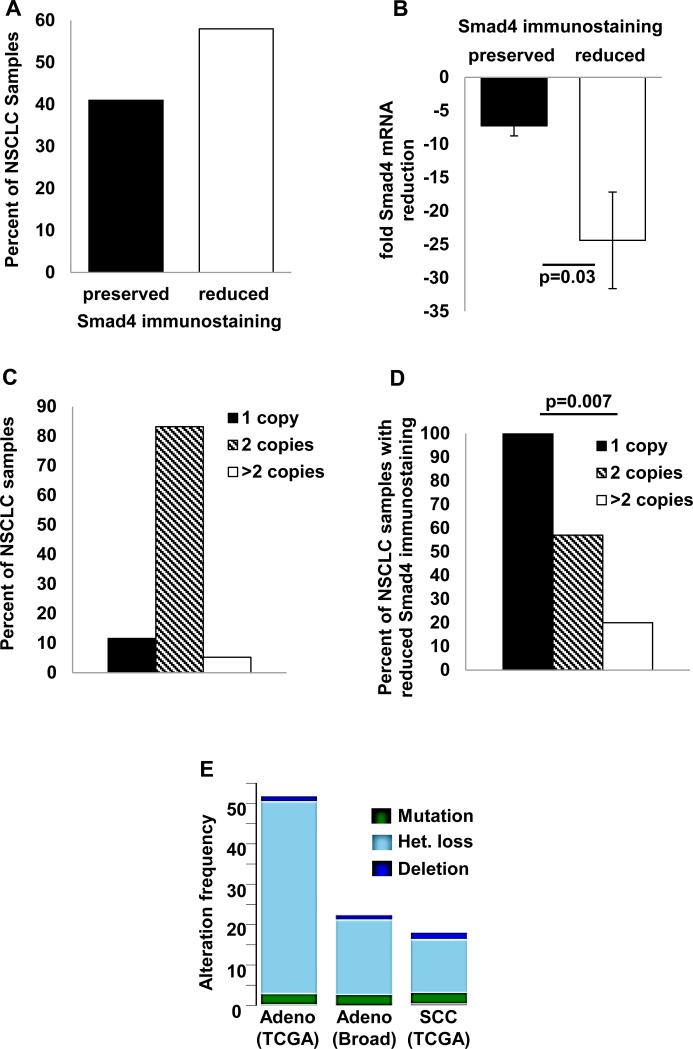
Smad4 loss through a combination of mutation and homozygous deletion has been reported in 4-6% of NSCLC (24, 25). However, other molecular events can also reduce Smad4 expression as the frequency of reduced Smad4 immunostaining far exceeds that of Smad4 mutation in most aerodigestive cancers (4). We found reduced Smad4 immunostaining in 58% (98/168) of human NSCLC samples (Fig. 1A) and observed that reduced Smad4 immunostaining was associated with reduced Smad4 mRNA expression (Fig. 1B), suggesting that Smad4 down-regulation occurs pre-translationally. Consistent with data from The Cancer Genome Atlas (TCGA) (25), we did not detect Smad4 promoter methylation by pyrosequencing (data not shown). When we assessed Smad4 copy number in NSCLC samples and paired non-malignant lung by qPCR of genomic DNA, we observed reduced Smad4 copy number in 9% (9/95) of samples (Fig. 1C) and all samples with Smad4 copy loss demonstrated reduced Smad4 immunostaining (Fig. 1D). In our dataset, we did not detect associations between reduced Smad4 immunostaining, mRNA expression, or copy loss and clinical or pathologic variables (data not shown). We also examined the frequency of Smad4 alterations in publically available NSCLC datasets using the cBioPortal web site (http://www.cbioportal.org/public-portal/) and found that while Smad4 mutation or homozygous deletion were relatively rare, each occurring in <5% of NSCLC, heterozygous Smad4 loss was seen in 13% of lung squamous cell carcinomas and up to 47% of lung adenocarcinomas (Fig. 1E).
Figure 1. Reduced Smad4 expression in human NSCLC.
(A) Smad4 immunostaining in human NSCLC samples was graded as either preserved (staining equal to normal airway) or reduced (less than normal airway) by two independent observers; reduced Smad4 immunostaining was observed in 58% (98/168) of NSCLC. Multiple examples of preserved and reduced Smad4 immunostaining are shown in Fig. 3C-D. (B) Reduced Smad4 immunostaining correlates with reduced Smad4 mRNA expression. Smad4 expression (normalized to paired non-malignant lung) is expressed as fold reduction in NSCLC samples with preserved or reduced Smad4 immunostaining. (C) Smad4 copy number in NSCLC samples and paired nonmalignant lung was assessed by qPCR of Smad4 exons 5 and 10 and scored using CopyCaller v1.0 software (Life Technologies); Smad4 copy loss was observed in 9/95 (9%) of NSCLC samples. (D) Smad4 copy loss was associated with reduced Smad4 immunostaining; all samples with copy loss exhibited reduced immunostaining. (E) Frequency of Smad4 alterations in lung cancer datasets. Data were obtained from the cBioPortal web site (http://www.cbioportal.org/public-portal/) by searching “Smad4: mutation homdel hetloss” in published NSCLC datasets.
Smad4 deletion initiates lung tumor formation and increases the size and malignant conversion of Kras-initiated lung tumors
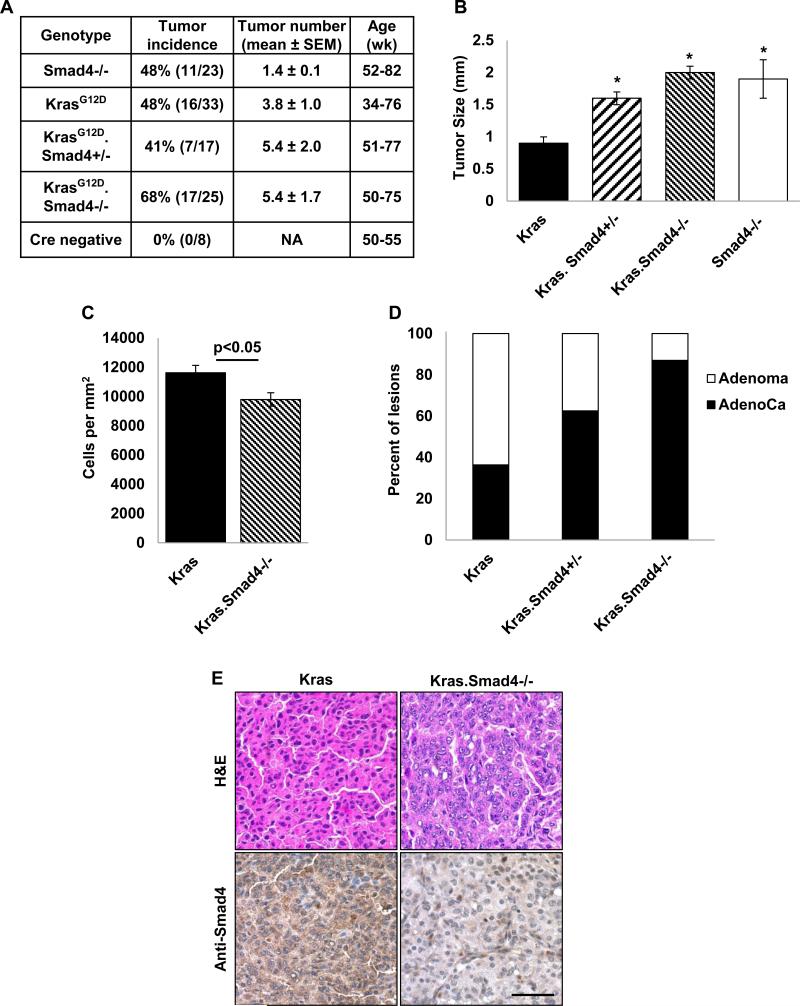
Because it was unknown whether Smad4 functions as a lung tumor suppressor, we created several novel NSCLC mouse models to directly evaluate the role of Smad4 loss in lung tumorigenesis. First, we induced Smad4 deletion in airway basal cells using either the K5CrePR* or K14CrePR transgene (26, 27). Both constructs use a keratin promoter to direct expression of an RU486-inducible Cre recombinase-progesterone receptor (CrePR) fusion protein; recombination is limited to the lung by tracheal RU486 instillation (28). This strategy induced tumor formation in 48% (11/23) animals (Fig. 2A, Smad4−/−). These animals typically had a single large adenoma or adenocarcinoma (not shown) that occurred between 12-18 months of age.
Figure 2. Smad4 deletion initiates lung tumor formation and increases size and malignant conversion of KrasG12D-initiated lung tumors.
(A) Tumor incidence, number, and age in mice with the indicated genotypes. Animals were treated and monitored as described in Methods. Tumors were observed in 48% of animals with K5CrePR* (n=13) or K14CrePR (n=10) directed Smad4 deletion (hereafter referred to as Smad4−/− animals). Tumors were seen in 48% of K5CrePR*.LSL-KrasG12D mice, 41% of K5CrePR*.LSL-KrasG12D.Smad4fl/+ mice, and 68% of K5CrePR*.LSL-KrasG12D.Smad4fl/fl mice (hereafter referred to as Kras, Kras.Smad4+/−, and Kras.Smad4−/−, respectively). Smad4−/− animals exhibited fewer tumors compared to Kras animals (p=0.05), however the increased tumor number seen in Kras.Smad4+/− and Kras.Smad4−/− animals (compared to Kras animals) did not reach statistical significance. (B) Deletion of one or both Smad4 alleles increased the size of Kras initiated lung tumors (*p<0.05 vs Kras). Smad4−/− tumors were also larger than Kras tumors (*p<0.05 vs Kras). (C) Smad4 deletion increased the average cell size of Kras initiated tumors as demonstrated by fewer cells per mm2. (D) Smad4 deletion promotes malignant conversion of Kras-initiated lung tumors as evidenced by the larger proportion of adenocarcinomas with deletion of one or two copies of Smad4 (p=0.0022 for trend). (E) An example of a Kras adenoma and a Kras.Smad4−/− adenocarcinoma (top) and an example of reduced Smad4 immunostaining in Kras.Smad4−/− tumor (bottom); scale bar is 50μm. Positive (brown) cells in the Kras.Smad4−/− tumor are infiltrating stromal cells.
Since Kras mutations are common in human lung cancer and can occur in conjunction with both Smad4 mutation and copy loss (24, 25), we also assessed the effect of Smad4 deletion on KrasG12D-initiated lung tumor formation. Similar to what we previously observed {Malkoski, 2012 #1553}, KrasG12D activation directed by K5CrePR* induced lung tumor formation in 48% (16/33) animals. To confirm Smad4 deletion we extracted DNA from tumor and grossly normal lung and detected the recombinant Smad4 allele only in tumors (Supplemental Fig. 1), we also confirmed that tumors with Smad4 deletion also had reduced Smad4 immunostaining (Fig. 2E, bottom panel). Although Smad4 deletion did not significantly increase tumor incidence or multiplicity (Fig. 2A), it almost doubled KrasG12D initiated tumor size (Fig. 2B). An increase in tumor size was also seen when a single Smad4 allele was deleted (Fig. 2B) consistent with Smad4 haploinsufficiency promoting tumor growth. We also observed that Kras.Smad4−/− tumors had ~20% fewer cells per mm2 than Kras tumors, suggesting that these tumors have larger individual cell size (Fig. 2C). Moreover, deletion of either one or both Smad4 alleles increased malignant conversion as evidenced by a larger fraction of adenocarcinomas relative to adenomas (Fig. 2D). These data are consistent with Smad4 functioning as a lung tumor suppressor.
Reduced Smad4 expression is associated with increased DNA damage
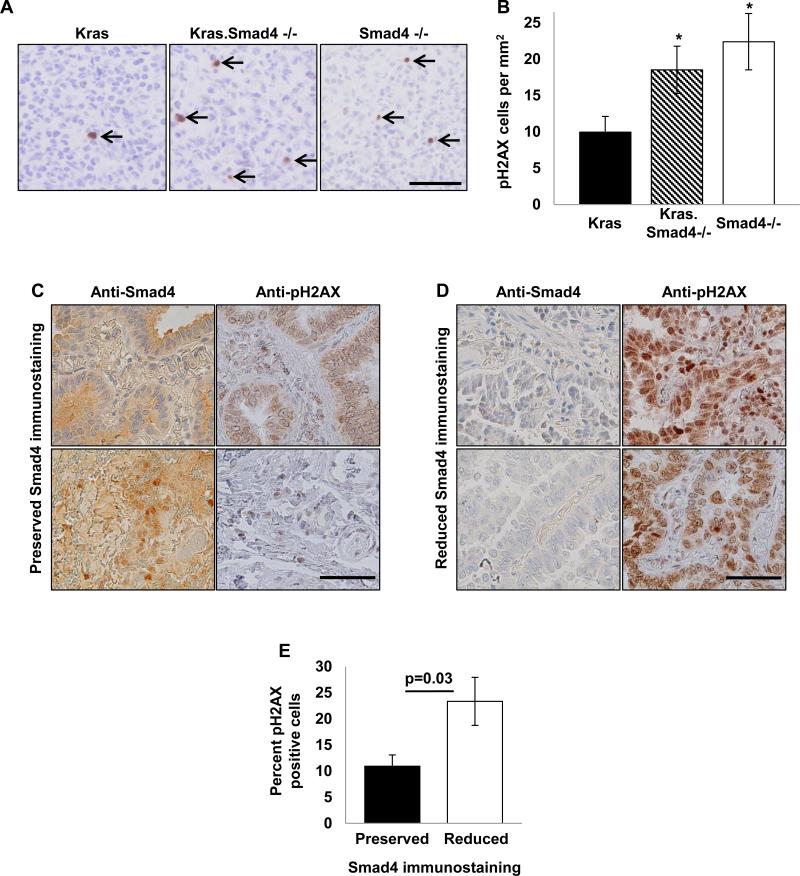
Because TGFβ/Smad signaling regulates genomic stability (11, 18), we assessed DNA damage in our mouse Smad4−/− lung tumors using pH2AX immunostaining to mark DSB. Compared to Kras tumors, both Kras.Smad4−/− and Smad4−/− tumors demonstrated increased pH2AX staining (Fig. 3A-B). When we assessed whether human NSCLC, we found that reduced Smad4 immunostaining was associated with increased DSB (Fig. 3C-E). These data demonstrate that reduced Smad4 expression is associated with increased DNA damage in vivo and suggest genomic instability as a potential mechanism for tumor initiation by Smad4 deletion.
Figure 3. Reduced Smad4 expression is associated with increased DNA damage.
(A) Tumors from at least five individual animals of each genotype (Kras, Kras.Smad4−/−, Smad4−/−) were immunostained for pH2AX to mark DSB. pH2AX positive cells are designated by arrows; scale bar is 50μm. (B) Quantification of pH2AX staining in murine tumors; * p<0.05 vs Kras. (C-D) Human NSCLC samples that were previously immunostained for Smad4 were stained for pH2AX to mark DSBs. Two examples of preserved (panel C) and reduced (panel D) Smad4 staining and the corresponding pH2AX staining of the same samples are shown; scale bar is 50μm in both panels. (E) Quantification of pH2AX staining from 27 human NSCLC sample as a function of Smad4 immunostaining. pH2AX staining ranged from 1-17% of cells in tumors with preserved Smad4 staining and 5-56% of cells in tumors with reduced Smad4 staining.
Reduced Smad4 expression reduces DNA repair and increases apoptosis after DNA damage
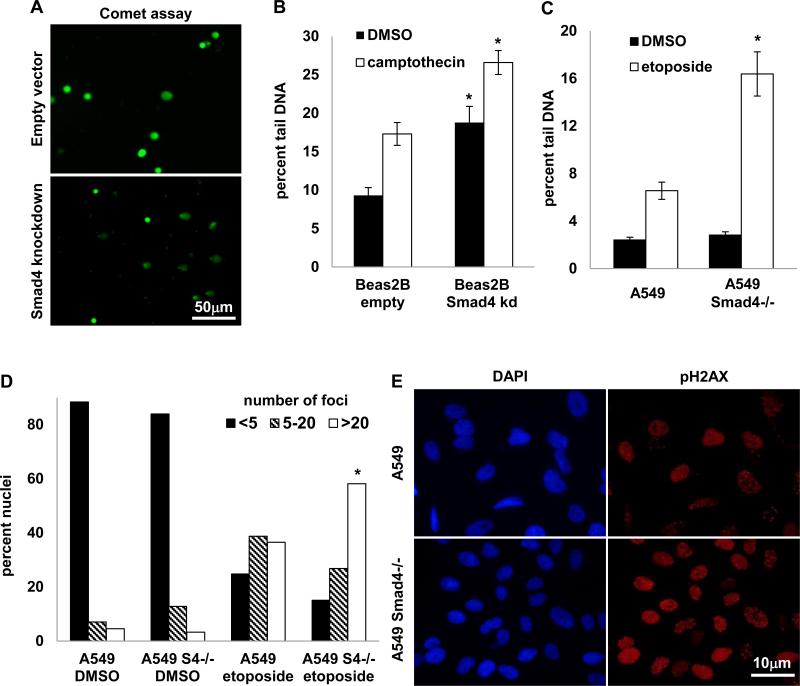
Because Smad4 deficient tumors had increased DNA damage, we hypothesized that Smad4 deficient cells might exhibit reduced DNA repair. We stably knocked down Smad4 in Beas2B (SV40 T antigen immortalized) airway epithelial cells and purchased A549 lung adenocarcinoma cells with a zinc finger engineered Smad4 deletion (Sigma). Reduced Smad4 expression was confirmed at the mRNA and protein level (Supplemental Fig. 2). Both at baseline and after treatment with the DNA topoisomerase I inhibitor, camptothecin, Beas2B cells with Smad4 knockdown exhibited increased DNA damage as measured by the comet assay (Fig. 4A-B). Although A549 Smad4−/− cells did not exhibit increased DNA damage at baseline (Fig. 4C), when challenged with etoposide (a DNA topoisomerase II inhibitor) for 4h and then allowed to recover for 24h, A549 Smad4−/− cells also exhibited increased DNA damage by comet assay (Fig. 4C). Similarly, A549 Smad4−/− cells also had increased DNA damage as measured by an increased number of nuclei with large number of pH2AX foci 12h after an etoposide challenge (Fig. 4D-E), suggesting that Smad4 deficient cells have a reduced ability to resolve DNA damage.
Figure 4. Reduced Smad4 expression reduces DNA repair.
(A-B) Beas2B cells were treated with 5μM camptothecin for 24h were assayed for DNA damage using the comet assay where damaged DNA appears in the comet “tail”; * p<0.05 vs empty vector control cells. (C) A549 Smad4−/− cells were treated with 100μM etoposide for 4h then assayed for DNA damage 24h later using the comet assay; * p<0.05 vs A549. (D) A549 Smad4−/− cells were treated with 25μM etoposide for 4h then assayed 12h later for DSB by counting pH2AX nuclear foci detected by indirect immunofluorescence; * p<0.001 vs A549. (E) Example of pH2AX (red) and DAPI (blue) immunostaining in A549 and A549 Smad4−/− cells 12h after etoposide treatment.
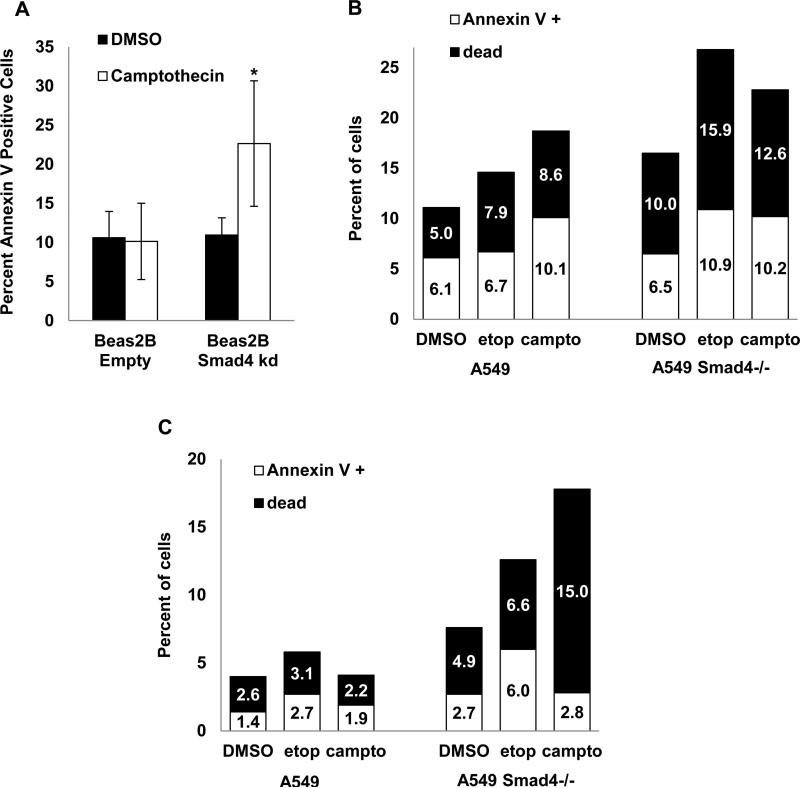
We then tested whether increased DNA damage resulted in increased apoptosis or cell death in Smad4 deficient cells and found that Beas2B cells with Smad4 knockdown treated with camptothecin for 4h demonstrated increased apoptosis as measured by Annexin V staining (Fig. 5A). A549 Smad4−/− cells also exhibited increased apoptosis and cell death after a 4h treatment with camptothecin or etoposide when assayed either immediately after treatment or 24h later (Fig. 5B-C, respectively), suggesting that Smad4 deficient cells have increased apoptosis when faced with a genotoxic challenge. To assess whether apoptotic cells exhibited DNA damage, we simultaneously stained for both Annexin V to mark apoptotic cells and pH2AX to mark DSB. In both A549 and A549 Smad4−/− cells, treatment with camptothecin or etoposide caused a large increase in the percentage of Annexin V positive cells that also stained positive for pH2AX (Supplemental Fig. 3).
Figure 5. Reduced Smad4 expression is associated with increased apoptosis after DNA damage.
(A) Increased apoptosis (Annexin V staining quantified by flow cytometry) in Beas2B cells with Smad4 knockdown treated with 10μM camptothecin for 4h then assayed immediately; * p<0.05 vs Beas2B empty vector control. (B-C) Increased apoptosis (Annexin V staining) and cell death in A594 Smad4−/− cells treated with 5μM camptothecin or 100μM etoposide for 4h then assayed immediately (panel B) or 24h after drug treatment (panel C).
Reduced Smad4 expression increases sensitivity to DNA topoisomerase inhibitors and inhibitors of NEHJ
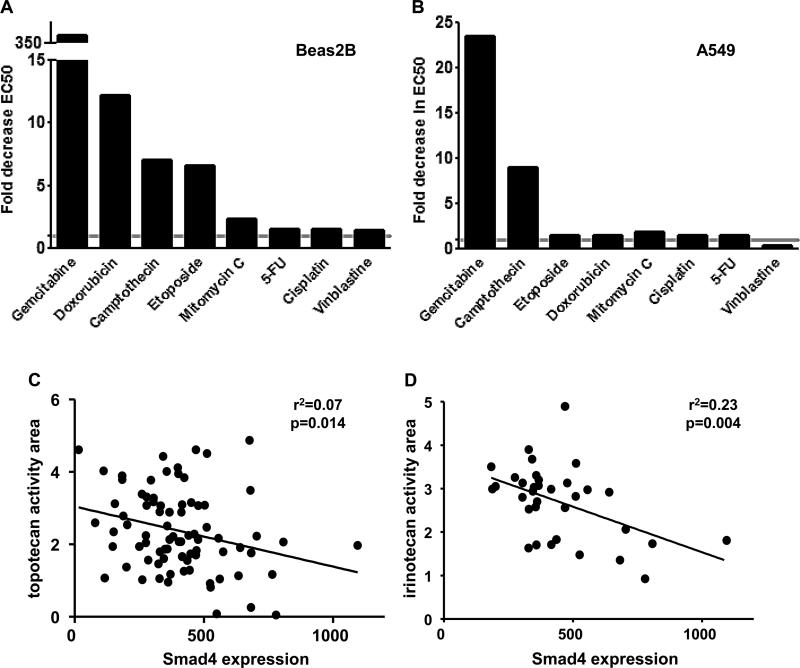
Because reduced Smad4 expression increased apoptosis in response to DNA damage, we hypothesized that cells with low Smad4 expression would be more sensitive to DNA damaging agents. We performed a series of cell viability assays in Beas2B cells with Smad4 knockdown and A549 Smad4−/− cells and found that reduced Smad4 expression sensitizes to numerous DSB causing drugs (gemcitabine, camptothecin, etoposide, doxorubicin, Fig. 6A-B). In contrast, reduced Smad4 expression had a more modest effect on sensitivity to alkylating-like agents (cisplatin) and DNA crosslinkers (mitomycin C), perhaps because the DNA adducts formed by these agents are predominantly intrastrand crosslinks not DSB (29, 30). H1299 lung cancer cells with Smad4 knockdown also demonstrated increased sensitivity to both gemcitabine and etoposide in chemosensitivity assays (not shown). Corroborating these observations, in the Cancer Cell Line Encyclopedia (CCLE) (31), low Smad4 expression correlates with increased sensitivity of NSCLC lines to the camptothecin derivatives, irinotecan and topotecan (Fig. 6C-D).
Figure 6. Reduced Smad4 expression increases sensitivity to DNA topoisomerase inhibitors.
(A-B) Drug sensitivity was assessed in Beas2B immortalized airway epithelial cells with stable Smad4 knock down and A549 Smad4−/− cells using a cell viability assay. Data are expressed as the average fold decrease in EC50 (Smad4 deficient to control). All drugs were tested in at least two independent experiments. H1299 lung cancer cells with Smad4 knockdown also demonstrated increased sensitivity to gemcitabine and etoposide (data not shown). (C-D) Reduced Smad4 expression correlates with increased sensitivity (activity area) to camptothecin derivatives NSCLC cell lines in the CCLE. Although the association of reduced Smad4 expression and activity area was modest, it was confirmed by analysis of residuals.
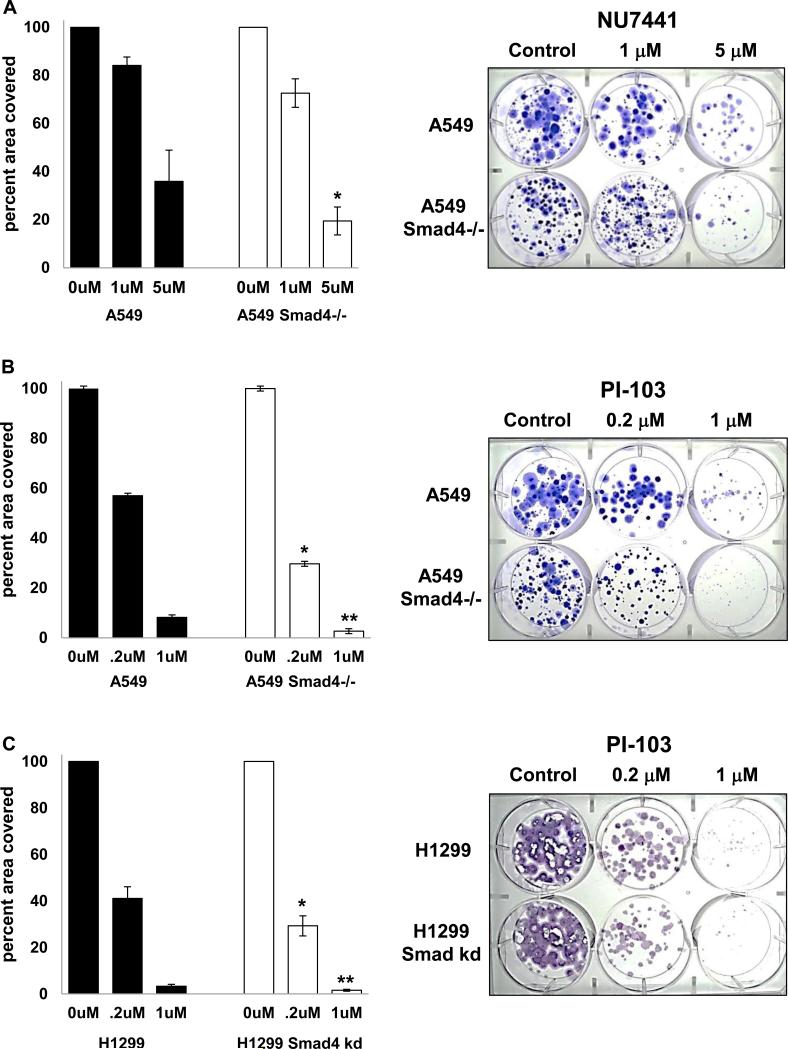
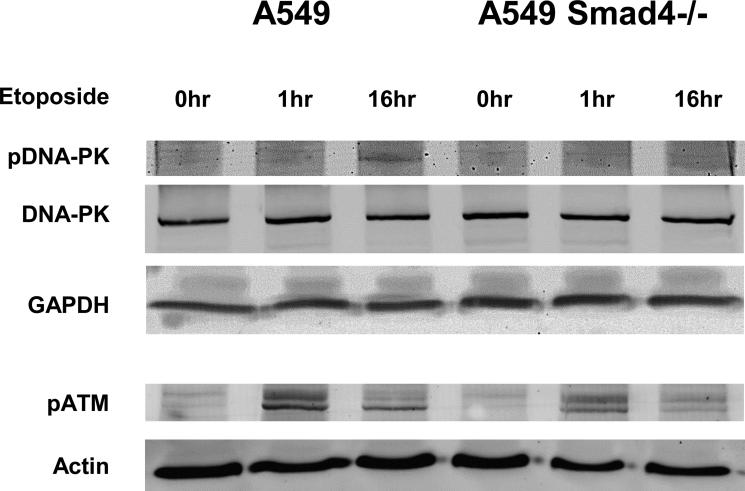
Since Smad4 deficient cells had defective DNA repair and increased apoptosis after DNA damage, we tested whether these cells were more sensitive to DNA repair inhibitors using a long term proliferation assay and found that A549 Smad4−/− cells were more sensitive to NU7441, a DNA-PK inhibitor (Fig. 7A) and to PI-103 a dual DNA-PK/mTOR inhibitor (Fig. 7B) while H1299 cells with Smad4 knockdown were more sensitive to PI-103 (Fig. 7C). We also assessed activation of DNA repair pathways after DNA damage and found that A549 Smad4−/− cells demonstrated reduced activation of both pDNA-PK and pATM after etoposide treatment (Fig. 8).
Figure 7. Reduced Smad4 expression increases sensitivity to DNA-PK inhibitors.
(A) A549 Smad4−/− cells were grown in NU7441 at the indicated concentrations for 17-23 days then stained with crystal violet. * p=0.05 vs A549. (B) A549 Smad4−/− cells (100/well) were grown in PI-103 at the indicated concentrations for 12-23 days and then stained with crystal violet. * p=0.02 vs A549; ** p=0.08 vs A549. (C) H1299 cells with Smad4 knockdown (100/well) were grown in PI-103 at the indicated concentrations for 12-17 days and then stained with crystal violet; * p=0.07 vs H1299; ** p=0.02 vs H1299. Examples of long term proliferation assays are shown at right.
Figure 8. Smad4 loss decreases DNA repair activity after DNA damage.
A549 Smad4−/− cells were treated with etoposide for the indicated times then protein harvested for analysis. A549 Smad4−/− cells demonstrate reducted pDNA-PK and pATM after exposure to etoposide. Experiments were repeated at least twice.
DISCUSSION
Reduced Smad4 expression is common in human NSCLC
Reduced Smad4 expression was seen in 58% of our NSCLC samples (Fig. 1A) and is likely a result of a combination of mutation, homozygous deletion, and heterozygous loss. Our data are relatively consistent with genome-wide datasets demonstrating Smad4 alterations in 18-51% of NSCLC (Fig. 1E). Although TCGA data demonstrate heterozygous Smad4 loss in 13% of lung squamous cell carcinomas and 47% of lung adenocarcinomas (24, 25), we detected Smad4 copy loss in only 9% of NSCLC. This difference is likely related to our conservative criteria for determining Smad4 copy loss with the CopyCaller v1.0 software: with less stringent criteria, we likely would have identified many more samples as having heterozygous Smad4 loss. Although Smad4 mutations have been previously associated with lymph node metastases, increased angiogenesis, and more aggressive cellular behavior in vitro (7, 8), we did not observe associations between reduced Smad4 expression and clinical or pathologic parameters. Reasons for the discrepancy between our data and a previous report (8) are not entirely clear. It could be that Smad4 loss is an early or tumor initiating event and is not associated with more aggressive tumor behavior, however, even in pancreatic and colon cancer where Smad4 loss has been better characterized, relationships between Smad4 loss and more aggressive tumor behavior have not been consistently observed (4).
Targeting Smad4 deletion to airway basal cells initiates and promotes lung tumor formation
One of our most significant findings is that Smad4 deletion both initiates spontaneous lung tumor formation and promotes the growth of Kras-initiated lung tumors. In other models, Smad4 deletion promotes tumor development initiated by Kras activation or PTEN deletion (10-16), however Smad4-initiated tumor formation has been previously described only in keratinized epithelial layers (9-11, 17). In our model, targeting Smad4 deletion (with or without Kras activation) to keratin positive airway basal cells produced predominantly adenomas and adenocarcinomas and ~5% squamous cell carcinomas (not shown). This is consistent with our previous experience targeting oncogenic alterations to basal cells via a keratin promoter (32, 33) and demonstrates that airway basal cells can give rise to multiple tumor types in the same way that they can differentiate into multiple airway epithelial cell types (34-36). We also observed that deletion of a single Smad4 allele increased both tumor size and malignant conversion (Fig. 2), consistent with the Smad4 haploinsufficiency seen in other models (10, 11). Importantly, this suggests that the heterozygous Smad4 loss seen commonly in human NSCLC likely promotes tumor growth.
Smad4 loss increases DNA damage and reduces DNA repair
Smad4−/− murine lung tumors and human NSCLC with reduced Smad4 expression demonstrate increased DNA damage (Fig. 3) and Smad4 deficient cells have reduced DNA repair and reduced activation of DNA repair pathways after DNA damage (Figs. 4 and 8). Genomic instability provides a potential mechanism for tumor initiation by Smad4 deletion and was observed in our previous model where Smad4 deletion initiated tumor formation but was not seen in another model where Smad4 deletion did not initiate tumor formation (11, 12). It is possible that reduced DNA repair in Smad4 deficient cells facilitates the accumulation of oncogenic mutations in vivo leading to tumor initiation after Smad4 loss.
The phenotype of Smad4 deficient cells reveals potential therapeutic strategies
Our study demonstrates that Smad4 cells are more sensitive to certain DNA damaging agents, specifically the DNA topoisomerase inhibitors, suggesting that Smad4 deficient tumors may be preferentially susceptible to similar treatments. Our observation that Smad4 knockdown sensitizes to DSB causing drugs contrasts with previous reports demonstrating that Smad4 knockdown in colon or breast cancer cells decreases sensitivity to 5-fluorouracil (5-FU) (37, 38). This is likely related to the repair of 5-FU related lesions by single strand DNA repair pathways {Wyatt, 2009 #1457} and the repair of gemcitabine, etoposide, and camptothecin related DNA damage by DSB repair pathways (39-43). Smad4 deficient cells have reduced activation of both pDNA-PK and pATM after DNA damage by etoposide (Fig. 8). Because there is substantial cross talk between DSB repair by NHEJ and HRR (44, 45), additional studies will be required to mechanistically define the precise double strand DNA repair defects present in Smad4 deficeint cells.
In summary, our study suggests that reduced Smad4 expression produces a potentially targetable tumor phenotype: one that will be more sensitive to treatment with DNA topoisomerase inhibitors and specific DNA repair inhibitors. It remains to be determined whether reduced Smad4 expression or Smad4 mutation correlates with increased responsiveness to the DNA topoisomerase inhibitors in human cancers. This would be highly relevant since most chemotherapy regimens for malignancies that frequently have Smad4 loss (NSCLC, head and neck cancer, colon cancer, pancreatic cancer) often do not include the agents to which Smad4 deficient cells are most sensitive. In addition, our findings pave the way for future studies using reduced Smad4 expression as a biomarker for selecting treatment approaches that exploit the defective DNA repair phenotype of Smad4 deficient cells.
MATERIALS AND METHODS
Analysis of human NSCLC samples
All studies were IRB approved. Human samples were obtained from three sources: the Oregon Health & Sciences University Department of Pathology Tumor Bank (n=37), the Colorado Lung SPORE Tissue Bank (n=87), and a commercially available tissue microarray (n=64, T8235724, lot A606098, Biochain, Hayward, CA). Clinical and pathologic data were extracted from the medical record or provided with the tissue array. Survival data was obtained by searching the social security death index database. Complete pathologic and demographic data is shown in Supplemental Table 1. Immunostaining for Smad4 (1:100 #7154 Santa Cruz Biotechnology, Santa Cruz, CA) was performed as previously described (32). Staining intensity was graded as preserved or reduced by two independent observers (SPM and SMH) and differences in immunostaining frequency between groups analyzed by chi-squared or Fisher's exact test. RT-qPCR with a Smad4 probe (Hs00232068, Life Technologies, Grand Island, NY) was performed and analyzed by non-parametric methods (as previously described (32). Smad4 copy number at exons 5 and 10 was assessed in tumor and matched normal lung DNA samples with Smad4 Copy Number Assays Hs01715545 and Hs02847956, an internal RNase P control and CopyCaller v1.0 software (Life Technologies) per the manufacture's recommendations (ΔCt<0.15, z<1.75). Double strand DNA breaks were analyzed by immunostaining for pH2AX (1:100 #9718, Cell Signaling Technology, Danvers, MA), quantified as the percent pH2AX positive nuclei, and differences compared with an unpaired t test.
Generation and analysis of NSCLC mouse model
All animal studies were IACUC approved. The Smad4 conditional knockout allele, lox-stop-lox-KrasG12D conditional knock-in allele, and K5Cre*PR transgene, and K14CrePR transgene have been previously described (26, 27, 46-48). Lung tumors were initiated with 500μg tracheal RU486 at 4-6 weeks of age. Mice were euthanized between 12-18 months of age or if they lost >15% of their body weight; tumors were measured, enumerated, collected, then classified according to consensus criteria (49). PCR for recombinant Kras and Smad4 alleles was performed as previously described (50, 51). Smad4 and pH2AX immunostaining was performed as described above. Tumor cell size was analyzed by counting the number of cells in a 40X field from H&E stained sections from at least 5 animals of each genotype. All images were acquired on a Nikon Eclipse 80i microscope with Nikon Elements Software and analyzed with ImageJ (rsbweb.nih.gov). Differences between groups were compared with by one-way ANOVA, unpaired t test, or Fisher's exact test as appropriate.
Generation and validation of cell lines
Beas2B (ATCC, Manassas, VA) were grown in BEGM (Lonza, Basel, Switzerland). A549 cells and A549 cells with Smad4 deletion (Sigma-Aldrich, St. Louis, MO) were grown in F12 supplemented with 10% FBS and 2mM L-glutamine. H1299 cells (ATCC) were grown in RPMI supplemented with 10% FBS. Cell lines with stable Smad4 knockdown were created by transfecting cells with pcPUR+U6 containing an anti-Smad4 shRNA (52) (kindly provided by Hideaki Ijichi, University of Tokyo, Japan) followed by puromycin selection; the pcPUR+U6 cassette (iGene Therapeutics, Tokyo, Japan) was used as a control. Reduced Smad4 expression was confirmed by RT-qPCR and western blot against Smad4 (1:200 #7966 Santa Cruz) with GAPDH control (1:40000, ab8245, Abcam, Cambridge, MA). Cell line identity was validated by DNA profiling at the University of Colorado DNA Sequencing & Analysis Core.
Comet assay
Beas2B cells (125,000) and A549 cells (100,000) were plated into 12 well plates and allowed to attach overnight. Beas2B cells were treated with camptothecin (5μM for 24h); A549 cells were treated with etoposide (100μM for 4h, assayed 24h later). Cells were washed with PBS and removed from the plates by trypsinization, then 15,000 cells in 15μL PBS were combined with 85μl of agarose; 75μL of this mixture was placed in each well of the OxiSelect Comet Assay Kit (Cell Biolabs, San Diego, CA). Cells were then subject to alkaline electrophoresis per the manufacturer's instructions. Images were acquired on Nikon Eclipse 80i microscope and analyzed using CaspLab Comet Assay Software (http://casplab.com). Differences between groups were compared by unpaired t test.
pH2AX immunofluorescence
A549 Smad4−/− cells (7500/well) were plated into glass chamber slides (BD Biosciences) and allowed to attach overnight. Cells were treated with etoposide (25μM for 4h) then harvested at various time points post treatment. Cells were washed with PBS, fixed with 4% paraformaldehyde (15min at room temperature), washed with PBS, permeabilized with 100% ice-cold methanol, then washed again with PBS. After blocking (5% normal goat serum/1% BSA/0.1% Triton X-100 for 1h at room temperature) cells were stained overnight at 4°C with pH2AX antibody (1:400, Cell Signaling, in 5% NGS/1% BSA), rinsed with PBS-T then incubated with anti-rabbit Alexa Fluor 594 (1:500, Life Technologies) for 1h at room temperature. Cells were then rinsed with PBS-T and mounted using DAPI Fluoromount-G (Southern Biotech, Birmingham, AL). pH2AX immunostaining was quantified by counting at least 100 nuclei per treatment condition; differences were compared by Chi-squared test.
Annexin V flow cytometry
Beas2B cells with Smad4 knockdown were treated with 10μM camptothecin for 4h then harvested with TrypLE (Life Technologies) and assayed with the Dead Cell Apoptosis Kit (Life Technologies) per the manufacturer's instructions using a FC500 flow cytometer (BD Biosciences, San Jose, CA). A549 Smad4−/− cells were treated with camptothecin or etoposide, were harvested with TrypLE (Life Technologies) then washed with cold PBS. Cells were assayed with the Fixable Viability Dye eFluor®506, Annexin V PE Apoptosis Detection Kit, and Anti-Human/mouse pH2AX (Ser139) PerCP-eFluor® 710 (all from eBioscience) per the manufacturer's protocol (http://media.ebioscience.com/data/pdf/best-protocols/annexin-v-staining.pdf) using a Gallios flow cytometer.
Chemosensitivity Assay
Cells were seeded at 1500/well (A549) or 3000/well (Beas-2B and H1299) into 96-well plates and allowed to attach overnight. Cells were treated with chemotherapeutic agents (0.001-10μM, depending on solubility) for 48-72h then assayed using the CellTiter-Glo kit (Promega) per the manufacturer's instructions. Gemcitabine was from LC Laboratories (Wobern, MA); doxorubicin, etoposide, mitomycin C, 5-fluorouracil, cisplatin, and vinblastine were from Sigma; camptothecin was from Fisher Scientific (Pittsburg, PA). Drug stocks were prepared in DMSO (doxorubicin, etoposide, 5-fluorouracil, cisplatin, vinblastine, camptothecin) or PBS (gemcitabine, mitomycin C) and stored at −80°C. Each drug was tested at least 3 times and IC50 determined using Prism software (GraphPad, La Jolla, CA).
Clonogenic growth assay
A549 and H1299 cells (100/well) were allowed to attach overnight then treated with NU7441 or PI-103 (Selleckchem.com, Houston, TX) at the indicated concentrations for 17-22 days. Cell were then rinsed with PBS then stained with 0.5% crystal violet in 25% methanol, photographed and quantified using ImageJ as percent area covered. Differences between groups were compared by unpaired t test.
Western Blotting
Cells were washed with PBS then lysed in modified RIPA buffer (50mM Tris pH 8.0, 150mM NaCl, 1mM EDTA pH 8.0, 0.5% NP-40, 5% glycerol) containing protease and phosphatase inhibitors (cOmplete, Roche, Indianapolis, IN). Lysates were rocked (4°C for 30min) then clarified (16,000g for 10min). Protein concentration was determined with the DC Bio-Rad Protein assay kit (Bio-Rad Laboratories, Hercules, CA). Samples prepared for loading in 1X Laemmli buffer with 5% beta-mercaptoethanol and stored at −20°C. Following gel electrophoresis, samples were transferred to Immun-Blot PVDF membranes (Bio-Rad) then probed with the following antibodies phospho-DNA-PK (pSer2056 1:200, Abcam), DNA-PK (1:50 Santa Cruz), GAPDH (1:20,000 Abcam), actin (1:200 Santa Cruz), pATM (pSer1981, 1:500 Millipore), and detected by probing with Irdye-700 or Irdye-800 (1:10,000 Rockland, Limerick, PA) and detected on an Odyssey system (LiCor, Lincoln, NE).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the NIH/NCI (K08 CA131483 to S.P.M.) and NIH/NIDCR (R01 DE015953 to X.J.W.). S.P.M. was also supported by the National Lung Cancer Partnership and a Career Development Award from the Colorado Lung Cancer SPORE (P50 CA058187). The OHSU BioLibrary is supported by P30 CA069533. The Colorado Lung Cancer SPORE Tissue Bank is supported by P30 CA046934 and P50 CA058187. We would like to thank OHSU BioLibrary staff for their assistance with patient samples. We would like to thank Jerry Haney, Christina Nall, Marina Lewis, and Wilbur Franklin at the Colorado Lung SPORE for assistance acquiring patient samples and data.
This work was supported by the NIH/NCI (K08 CA131483 to S.P.M.) and NIH/NIDCR (R01 DE015953 to X.J.W.). S.P.M. was also supported by the National Lung Cancer Partnership, a Career Development Award from the Colorado Lung Cancer SPORE (NIH/NCI P50 CA058187) and an American Cancer Society Institutional Research Grant (ACS IRG 57-001-53).
Footnotes
CONFLICT OF INTEREST
This work has been funded by the NIH (Drs. Malkoski and Wang), the National Lung Cancer Partnership (Dr. Malkoski), and the American Cancer Society (Dr. Malkoski). The other authors declare no conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com.onc).
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 009 Jul-Aug;59(4):225–49. doi: 10.3322/caac.20006. PubMed PMID: 19474385. Epub 2009/05/29. eng. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, et al. Non-small-cell lung cancer. Lancet. 2011 Nov 12;378(9804):1727–40. doi: 10.1016/S0140-6736(10)62101-0. PubMed PMID: 21565398. [DOI] [PubMed] [Google Scholar]
- 3.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012 Oct;13(10):616–30. doi: 10.1038/nrm3434. PubMed PMID: 22992590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malkoski SP, Wang XJ. Two sides of the story? Smad4 loss in pancreatic cancer versus head-and-neck cancer. FEBS Lett. 2012 Jul 4;586(14):1984–92. doi: 10.1016/j.febslet.2012.01.054. PubMed PMID: 22321641. Epub 2012/02/11. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagatake M, Takagi Y, Osada H, Uchida K, Mitsudomi T, Saji S, et al. Somatic in vivo alterations of the DPC4 gene at 18q21 in human lung cancers. Cancer Res. 1996 Jun 15;56(12):2718–20. PubMed PMID: 8665501. [PubMed] [Google Scholar]
- 6.Yanagisawa K, Uchida K, Nagatake M, Masuda A, Sugiyama M, Saito T, et al. Heterogeneities in the biological and biochemical functions of Smad2 and Smad4 mutants naturally occurring in human lung cancers. Oncogene. 2000 May 4;19(19):2305–11. doi: 10.1038/sj.onc.1203591. PubMed PMID: 10822381. [DOI] [PubMed] [Google Scholar]
- 7.Gemma A, Takenaka K, Hosoya Y, Matuda K, Seike M, Kurimoto F, et al. Altered expression of several genes in highly metastatic subpopulations of a human pulmonary adenocarcinoma cell line. Eur J Cancer. 2001 Aug;37(12):1554–61. doi: 10.1016/s0959-8049(01)00154-x. PubMed PMID: 11506965. [DOI] [PubMed] [Google Scholar]
- 8.Ke Z, Zhang X, Ma L, Wang L. Expression of DPC4/Smad4 in non-small-cell lung carcinoma and its relationship with angiogenesis. Neoplasma. 2008;55(4):323–9. PubMed PMID: 18505344. Epub 2008/05/29. eng. [PubMed] [Google Scholar]
- 9.Qiao W, Li AG, Owens P, Xu X, Wang XJ, Deng CX. Hair follicle defects and squamous cell carcinoma formation in Smad4 conditional knockout mouse skin. Oncogene. 2006 Jan 12;25(2):207–17. doi: 10.1038/sj.onc.1209029. PubMed PMID: 16170355. [DOI] [PubMed] [Google Scholar]
- 10.Teng Y, Sun AN, Pan XC, Yang G, Yang LL, Wang MR, et al. Synergistic function of Smad4 and PTEN in suppressing forestomach squamous cell carcinoma in the mouse. Cancer Res. 2006 Jul 15;66(14):6972–81. doi: 10.1158/0008-5472.CAN-06-0507. PubMed PMID: 16849541. [DOI] [PubMed] [Google Scholar]
- 11.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009 Nov;119(11):3408–19. doi: 10.1172/JCI38854. PubMed PMID: 19841536. Epub 2009/10/21. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007 Mar;11(3):229–43. doi: 10.1016/j.ccr.2007.01.017. PubMed PMID: 17349581. [DOI] [PubMed] [Google Scholar]
- 13.Kojima K, Vickers SM, Adsay NV, Jhala NC, Kim HG, Schoeb TR, et al. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res. 2007 Sep 1;67(17):8121–30. doi: 10.1158/0008-5472.CAN-06-4167. PubMed PMID: 17804724. Epub 2007/09/07. eng. [DOI] [PubMed] [Google Scholar]
- 14.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006 Nov 15;20(22):3130–46. doi: 10.1101/gad.1478706. PubMed PMID: 17114584. Epub 2006/11/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Kobayashi S, Qiao W, Li C, Xiao C, Radaeva S, et al. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J Clin Invest. 2006 Jul;116(7):1843–52. doi: 10.1172/JCI27282. PubMed PMID: 16767220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Ehdaie B, Ohara N, Yoshino T, Deng CX. Synergistic action of Smad4 and Pten in suppressing pancreatic ductal adenocarcinoma formation in mice. Oncogene. 2010 Feb 4;29(5):674–86. doi: 10.1038/onc.2009.375. PubMed PMID: 19901970. Epub 2009/11/11. eng. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Mao C, Teng Y, Li W, Zhang J, Cheng X, et al. Targeted disruption of Smad4 in mouse epidermis results in failure of hair follicle cycling and formation of skin tumors. Cancer Res. 2005 Oct 1;65(19):8671–8. doi: 10.1158/0008-5472.CAN-05-0800. PubMed PMID: 16204035. [DOI] [PubMed] [Google Scholar]
- 18.Glick AB, Weinberg WC, Wu IH, Quan W, Yuspa SH. Transforming growth factor beta 1 suppresses genomic instability independent of a G1 arrest, p53, and Rb [published erratum appears in Cancer Res 1997 May 15;57(10):2079]. Cancer Research. 1996;56(16):3645–50. [PubMed] [Google Scholar]
- 19.Kirshner J, Jobling MF, Pajares MJ, Ravani SA, Glick AB, Lavin MJ, et al. Inhibition of transforming growth factor-beta1 signaling attenuates ataxia telangiectasia mutated activity in response to genotoxic stress. Cancer Res. 2006 Nov 15;66(22):10861–9. doi: 10.1158/0008-5472.CAN-06-2565. PubMed PMID: 17090522. [DOI] [PubMed] [Google Scholar]
- 20.Jalal S, Earley JN, Turchi JJ. DNA repair: from genome maintenance to biomarker and therapeutic target. Clin Cancer Res. Nov 15;17(22):6973–84. doi: 10.1158/1078-0432.CCR-11-0761. PubMed PMID: 21908578. Epub 2011/09/13. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woods D, Turchi JJ. Chemotherapy induced DNA damage response: Convergence of drugs and pathways. Cancer biology & therapy. 2013 Feb 4;14(5) doi: 10.4161/cbt.23761. PubMed PMID: 23380594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012 Dec;12(12):801–17. doi: 10.1038/nrc3399. PubMed PMID: 23175119. [DOI] [PubMed] [Google Scholar]
- 23.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008 Jan;18(1):134–47. doi: 10.1038/cr.2007.111. PubMed PMID: 18157161. [DOI] [PubMed] [Google Scholar]
- 24.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012 Sep 14;150(6):1107–20. doi: 10.1016/j.cell.2012.08.029. PubMed PMID: 22980975. Pubmed Central PMCID: 3557932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 Sep 27;489(7417):519–25. doi: 10.1038/nature11404. PubMed PMID: 22960745 Pubmed Central PMCID: 3466113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caulin C, Nguyen T, Longley MA, Zhou Z, Wang XJ, Roop DR. Inducible activation of oncogenic K-ras results in tumor formation in the oral cavity. Cancer Res. 2004 Aug 1;64(15):5054–8. doi: 10.1158/0008-5472.CAN-04-1488. PubMed PMID: 15289303. [DOI] [PubMed] [Google Scholar]
- 27.Berton TR, Wang XJ, Zhou Z, Kellendonk C, Schutz G, Tsai S, et al. Characterization of an inducible, epidermal-specific knockout system: differential expression of lacZ in different Cre reporter mouse strains. Genesis. 2000 Feb;26(2):160–1. PubMed PMID: 10686618. Epub 2000/03/21. eng. [PubMed] [Google Scholar]
- 28.Malkoski SP, Cleaver TG, Lu SL, Lighthall JG, Wang XJ. Keratin promoter based gene manipulation in the murine conducting airway. Int J Biol Sci. 2010;6(1):68–79. doi: 10.7150/ijbs.6.68. PubMed PMID: 20140084. Epub 2010/02/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crul M, van Waardenburg RC, Bocxe S, van Eijndhoven MA, Pluim D, Beijnen JH, et al. DNA repair mechanisms involved in gemcitabine cytotoxicity and in the interaction between gemcitabine and cisplatin. Biochem Pharmacol. 2003 Jan 15;65(2):275–82. doi: 10.1016/s0006-2952(02)01508-3. PubMed PMID: 12504803. Epub 2002/12/31. eng. [DOI] [PubMed] [Google Scholar]
- 30.Bargonetti J, Champeil E, Tomasz M. Differential toxicity of DNA adducts of mitomycin C. J Nucleic Acids. 2010 doi: 10.4061/2010/698960. PubMed PMID: 20798760. Epub 2010/08/28. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012 Mar 29;483(7391):603–7. doi: 10.1038/nature11003. PubMed PMID: 22460905. Pubmed Central PMCID: 3320027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malkoski SP, Haeger SM, Cleaver TG, Rodriguez KJ, Li H, Lu SL, et al. Loss of transforming growth factor beta type II receptor increases aggressive tumor behavior and reduces survival in lung adenocarcinoma and squamous cell carcinoma. Clin Cancer Res. 2012 Apr 15;18(8):2173–83. doi: 10.1158/1078-0432.CCR-11-2557. PubMed PMID: 22399565. Epub 2012/03/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malkoski SP, Cleaver TG, Thompson JJ, Sutton WP, Haeger SM, Rodriguez KJ, et al. Role of PTEN in Basal Cell Derived Lung Carcinogenesis. Mol Carcinog. 2013 Apr 26; doi: 10.1002/mc.22030. PubMed PMID: 23625632. Pubmed Central PMCID: 3906215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011 Oct 28;147(3):525–38. doi: 10.1016/j.cell.2011.10.001. PubMed PMID: 22036562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12771–5. doi: 10.1073/pnas.0906850106. PubMed PMID: 19625615. Epub 2009/07/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004 Feb;164(2):577–88. doi: 10.1016/S0002-9440(10)63147-1. PubMed PMID: 14742263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, et al. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011 Feb 1;71(3):998–1008. doi: 10.1158/0008-5472.CAN-09-3269. PubMed PMID: 21245094. Epub 2011/01/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu SL, Lee DC, Son JW, Park CG, Lee HY, Kang J. Histone deacetylase 4 mediates SMAD family member 4 deacetylation and induces 5-fluorouracil resistance in breast cancer cells. Oncology reports. 2013 Sep;30(3):1293–300. doi: 10.3892/or.2013.2578. PubMed PMID: 23817620. [DOI] [PubMed] [Google Scholar]
- 39.Wachters FM, van Putten JW, Maring JG, Zdzienicka MZ, Groen HJ, Kampinga HH. Selective targeting of homologous DNA recombination repair by gemcitabine. International journal of radiation oncology, biology, physics. 2003 Oct 1;57(2):553–62. doi: 10.1016/s0360-3016(03)00503-0. PubMed PMID: 12957269. [DOI] [PubMed] [Google Scholar]
- 40.Achanta G, Pelicano H, Feng L, Plunkett W, Huang P. Interaction of p53 and DNA-PK in response to nucleoside analogues: potential role as a sensor complex for DNA damage. Cancer Res. 2001 Dec 15;61(24):8723–9. PubMed PMID: 11751391. [PubMed] [Google Scholar]
- 41.Adachi N, So S, Koyama H. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J Biol Chem. 2004 Sep 3;279(36):37343–8. doi: 10.1074/jbc.M313910200. PubMed PMID: 15218034. Epub 2004/06/26. eng. [DOI] [PubMed] [Google Scholar]
- 42.Jin S, Inoue S, Weaver DT. Differential etoposide sensitivity of cells deficient in the Ku and DNA-PKcs components of the DNA-dependent protein kinase. Carcinogenesis. 1998 Jun;19(6):965–71. doi: 10.1093/carcin/19.6.965. PubMed PMID: 9667732. Epub 1998/07/17. eng. [DOI] [PubMed] [Google Scholar]
- 43.Treszezamsky AD, Kachnic LA, Feng Z, Zhang J, Tokadjian C, Powell SN. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 2007 Aug 1;67(15):7078–81. doi: 10.1158/0008-5472.CAN-07-0601. PubMed PMID: 17671173. Epub 2007/08/03. eng. [DOI] [PubMed] [Google Scholar]
- 44.Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res. 2011 Jun 3;711(1-2):73–86. doi: 10.1016/j.mrfmmm.2011.02.010. PubMed PMID: 21376743. Pubmed Central PMCID: 3109507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010 Sep 10;584(17):3703–8. doi: 10.1016/j.febslet.2010.07.057. PubMed PMID: 20691183. Pubmed Central PMCID: 3954739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Qiao W, Chen L, Xu X, Yang X, Li D, et al. Squamous cell carcinoma and mammary abscess formation through squamous metaplasia in Smad4/Dpc4 conditional knockout mice. Development. 2003 Dec;130(24):6143–53. doi: 10.1242/dev.00820. PubMed PMID: 14597578. [DOI] [PubMed] [Google Scholar]
- 47.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001 Apr 26;410(6832):1111–6. doi: 10.1038/35074129. PubMed PMID: 11323676. [DOI] [PubMed] [Google Scholar]
- 48.Wunderlich FT, Wildner H, Rajewsky K, Edenhofer F. New variants of inducible Cre recombinase: a novel mutant of Cre-PR fusion protein exhibits enhanced sensitivity and an expanded range of inducibility. Nucleic Acids Res. 2001 May 15;29(10):E47. doi: 10.1093/nar/29.10.e47. PubMed PMID: 11353092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004 Apr 1;64(7):2307–16. doi: 10.1158/0008-5472.can-03-3376. PubMed PMID: 15059877. [DOI] [PubMed] [Google Scholar]
- 50.Pan D, Schomber T, Kalberer CP, Terracciano LM, Hafen K, Krenger W, et al. Normal erythropoiesis but severe polyposis and bleeding anemia in Smad4 deficient mice. Blood. 2007 Jul 16; doi: 10.1182/blood-2007-02-074393. PubMed PMID: 17638848. [DOI] [PubMed] [Google Scholar]
- 51.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005 Nov 15;65(22):10280–8. doi: 10.1158/0008-5472.CAN-05-2193. PubMed PMID: 16288016. [DOI] [PubMed] [Google Scholar]
- 52.Jazag A, Kanai F, Ijichi H, Tateishi K, Ikenoue T, Tanaka Y, et al. Single small-interfering RNA expression vector for silencing multiple transforming growth factor-beta pathway components. Nucleic Acids Res. 2005;33(15):e131. doi: 10.1093/nar/gni130. PubMed PMID: 16113239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.