Abstract
The nucleolus is a well-organized site of ribosomal gene transcription. Moreover, many DNA repair pathway proteins, including ATM, ATR kinases, MRE11, PARP1 and Ku70/80, localize to the nucleolus (Moore et al., 2011). We analyzed the consequences of DNA damage in nucleoli following ultraviolet A (UVA), C (UVC), or γ-irradiation in order to test whether and how radiation-mediated genome injury affects local motion and morphology of nucleoli. Because exposure to radiation sources can induce changes in the pattern of UBF1-positive nucleolar regions, we visualized nucleoli in living cells by GFP-UBF1 expression for subsequent morphological analyses and local motion studies. UVA radiation, but not 5 Gy of γ-rays, induced apoptosis as analyzed by an advanced computational method. In non-apoptotic cells, we observed that γ-radiation caused nucleolar re-positioning over time and changed several morphological parameters, including the size of the nucleolus and the area of individual UBF1-positive foci. Radiation-induced nucleoli re-arrangement was observed particularly in G2 phase of the cell cycle, indicating repair of ribosomal genes in G2 phase and implying that nucleoli are less stable, thus sensitive to radiation, in G2 phase.
Keywords: DNA damage, live cells, nucleolus, nuncleoli tracking, UBF1
Abbreviations
- 53BP1
p53-binding protein
- ACT-D
actinomycin D
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3-related protein
- AR
area ratio (area of nucleolus/area of nucleus)
- BrdU
5-bromo-2′-deoxy-uridine
- BER
base excision repair
- CBs
Cajal bodies
- DDR
DNA damage response
- CPDs
cyclobutane pyrimidine dimers
- DMEM
Dulbecco's modified Eagle's medium
- DNA-PK
DNA-dependent protein kinase
- DSBs
double-strand breaks
- FA
foci area
- NF
number of foci
- FI
fluorescence intensity
- FRAP
fluorescence recovery after photobleaching
- GGR
global genome repair
- GFP
green fluorescence protein
- HP1
heterochromatin protein 1
- HR
homologous recombination
- HRR
homologous recombination repair
- iMEFs
immortalized mouse embryonic fibroblasts
- LR
local radius
- NBs
nuclear bodies
- NER
nucleotide excision repair
- NHEJ
non-homologous end-joining
- nA
nucleolus area
- NA
nucleus area
- PBS
phosphate-buffered saline
- P2A
compactness
- PCR
polymerase chain reaction
- PML
promyelocytic leukemia bodies
- rDNA
ribosomal DNA
- RNA Pol II
RNA polymerase II
- ROI
region of interest
- RPA
replication-related protein A
- SEM
standard error of the mean
- SDS
sodium dodecyl sulfate
- TCR
transcription-coupled NER
- UBF1
upstream binding factor 1
- UV
ultraviolet.
Introduction
DNA lesions are considered deleterious to the genome because their incorrect repair can lead to chromosome instability. Single-stranded DNA lesions are corrected by a mechanism called nucleotide excision repair (NER), which can proceed as transcription-coupled NER (TCR) or global genome repair (GGR). Depending on the genome injury, base excision repair (BER) or mismatch repair mechanisms also may be initiated. Together, these processes are responsible for the repair of single-stranded DNA lesions induced by genotoxic factors.1
Homologous recombination repair (HRR) and non-homologous end-joining (NHEJ), two fundamental cell cycle-dependent mechanisms, recognize and repair double-strand breaks (DSBs) (summarized by2,3). Standard NHEJ, which can appear at all cell cycle phases, involves the mobilization of the catalytic subunit of DNA-dependent protein kinase (DNA-PK) and the activation of the XRCC4/DNA ligase IV complex.4,5 In contrast, HRR is initiated during late S phase and proceeds into G2 phase; this repair mechanism is considered less prone to errors. As an initial step, HRR involves the activation of MRE11, RAD50, and NBS1 proteins, which together are called the MRN complex. The MRN complex localizes to eukaryotic DNA lesions together with the BRCA2-DSS1 proteins.2 HRR requires DNA degradation by nucleases to form 3′ single-stranded DNA ends. This event is accompanied by replication-related protein A (RPA) activation to eliminate secondary structures. Moreover, RAD-related proteins (RAD51, RAD52, RAD54, or RAD59) are recruited to DNA lesions and this process is accompanied by structural changes (Holliday junctions in prokaryotes and D-loops in eukaryotes). This well-organized nuclear event finally permits the repair of damaged DNA.2 Interestingly, protein accumulation at DNA lesions has a certain kinetic hierarchy. For example, RAD52-positive protein foci are formed spontaneously as a consequence of replication collision or telomere dysfunction and are rapidly dispersed at DNA lesions within 10 min6. Similarly, the accumulation of heterochromatin-related proteins, such as HP1β or BMI1, is maintained at locally induced DNA lesions for several minutes.7 Therefore, understanding of DNA repair mechanisms requires studies on protein kinetics at DNA lesions and analysis of localized repair foci movement at radiation-damaged chromatin.
From the view of DNA repair processes, the nucleolus compartment is an underexplored nuclear region. Only Shav-Tal et al.8 previously demonstrated that the primary nucleolar regions, such as the fibrillar center (FC), dense fibrillar component (DFC), and granular component (GC), become separated after genome injury. Moreover, nucleolar protein foci, including Upstream Binding Factor 1 (UBF1)-positive foci, morphologically reorganize after cell exposure to UVA irradiation.9,10 Recently, we showed that the UBF1 protein is recruited to cyclobutane pyrimidine dimer (CPD)-positive DNA lesions in parallel with heterochromatin protein 1 β (HP1β).11 Moreover, similar to UBF1, HP1β and HP1γ are involved in ribosomal gene transcription.11-13 Thus, highly transcriptionally active ribosomal genes represent an excellent example to study DNA repair mechanisms in euchromatin.
According to Foltánková et al.,14 the nucleolar DNA damage response (DDR) is strikingly different in UV-irradiated and γ-irradiated genomes compared to non-irradiated cells. Thus, information regarding the radiation-specific reorganization of DNA damage-related nucleolar proteins is valuable. To this end, DNA repair protein mobility can be analyzed in individual nuclear compartments. The following methods can be applied for such analyses: 1] FRAP analysis of protein diffusion at DNA lesions, 2] advanced image processing algorithms to follow DNA repair foci trajectories, and 3] image analysis to describe morphology of protein-abundant DNA lesions.15-18 Critical parameters also include the particle dynamics and fluorescence signal intensity of radiation-damaged chromatin, which can be monitored in 3 dimensions over time. Thus, the use of above mentioned experimental approaches can allow for the proper characterization of DNA lesion behavior in space and time.
Here, we focused on local motion and morphology of nucleoli especially after cell irradiation by γ-rays. We analyzed if the damage and subsequent repair in ribosomal genes is accompanied by nucleoli re-arrangement and if expected morphological changes are cell cycle specific. We studied several morphological parameters in nucleoli exposed to γ-radiation, UV radiation, or actinomycin D (ACT-D) treatment. In addition, using the HeLa-Fucci cellular model system, we examined the localized movement and morphology of UBF1-positive regions of the nucleoli in G1 and G2 cell cycle phases. DNA lesions occur spontaneously (as a consequence of replication collision or telomere dysfunction) or in response to diverse genotoxic stresses, including radiation. This activates checkpoint pathways that regulate specific DNA-repair mechanisms in different phases of the cell cycle. As revised by Branzei and Foiani,19 DSBs that appear in the S or G2 cell cycle phases are repaired by HRR. However, DSBs in G1-cells are corrected by non-homologous end joining. The NER repair pathway in budding yeast is initiated after UV-irradiation in G1 cells. Similarly, the BER pathway recognizing chemical alterations of nucleotide bases can appear in the G1 phase.19,20 Based on this knowledge, the cell cycle phases are decisive factors in the control of DNA repair mechanisms and we addressed this aspect from the view of ribosomal genes. Indeed, we observed that radiation-induced morphological changes on the level of nucleoli are cell cycle specific, appearing in the G2 phase of the cell cycle. Thus, our observations explain repair processes in ribosomal genes.
Results
We tested if genome injury by genotoxic factors can affect local motion and morphology of nucleoli, which likely reflect a response of ribosomal genes to radiation. Moreover, we wanted to know if radiation-induced changes in nucleoli are cell cycle specific and what happens to the main transcription factor of ribosomal genes, UBF1, when the cells are irradiated.
First, we studied the appearance of irradiation-induced foci in cells after exposure to γ-radiation or UV light. We specifically analyzed DNA repair foci inside and around nucleoli because nucleoli represent an important stress sensor of the nucleus.9,21 We also inhibited ribosomal transcription by treating the cells with ACT-D and verified that ACT-D is a DNA damaging agent. We confirmed that ACT-D treatment and selected radiation treatments induce not only DNA damage but also subsequent apoptotic events.22,23 Thus, we characterized the morphology of apoptotic cells by image analysis methods designed for actual experimental data. Then, we used irradiation conditions that did not induce apoptosis to continue our studies on the localized movement and morphology of GFP-UBF1-positive nucleolar regions in G1 and G2 cell cycle phases.
Basic nuclear and nucleolar patterns in cells exposed to γ-radiation or ribosomal gene transcription inhibition
To examine nucleolar morphology and to monitor nucleolar responses to radiation, we analyzed the appearance of 53BP1-positive nuclear bodies (NBs) in whole cell nuclei, particularly around or inside nucleoli (compare 1–3 robust, spontaneously occurring 53BP1-positive NBs in Fig. 1Aa, Ba, Ca with 53BP1-positive irradiation-induced foci in Fig. 1Bb, Cb). Spontaneous 53BP1-positive NBs usually appear in in vitro cultivated tumor cells and γ-radiation induces so-called irradiation-induced foci (IRIF) throughout the entire genome. Here, we found that IRIF also appeared at the periphery or inside the nucleoli as visualized by an antibody directed against fibrillarin or by visualization of GFP-UBF1 (Fig. 1Bb, Cb). In total, ∼50% of 53PB1-positive NBs were associated (co-localized) with nucleoli in untreated control cells, ∼26% in γ-irradiated cells, and ∼8% in ACT-D-treated cells (example of association is shown in Fig. 1Bb, Bc and 1Cb; arrows). We confirmed the results described previously by others that ACT-D treatment also has the ability to induce DNA damage-related foci (see Fig. 1Ab, Bc, Cc and Ref.24). We found that ACT-D treatment increased the number of 53BP1-positive NBs 8–14x in comparison to non-treated control cells. For example, 1–3 NBs were observed in control cell nuclei and 14–24 NBs in ACT-D treated cells (Fig. 1Ca, Cc). Intriguingly, after γ-irradiation and ACT-D-treatment, a lower percentage of 53BP1 positive NBs associated with nucleoli (∼26% and ∼8%, respectively; see explanation above). This could mean that nucleoli, and thus ribosomal genes, are less sensitive to DNA damage. Alternatively, the different number and morphology of 53BP1-positive NBs may reflect different DNA lesions that must be repaired by different mechanisms.
Figure 1.
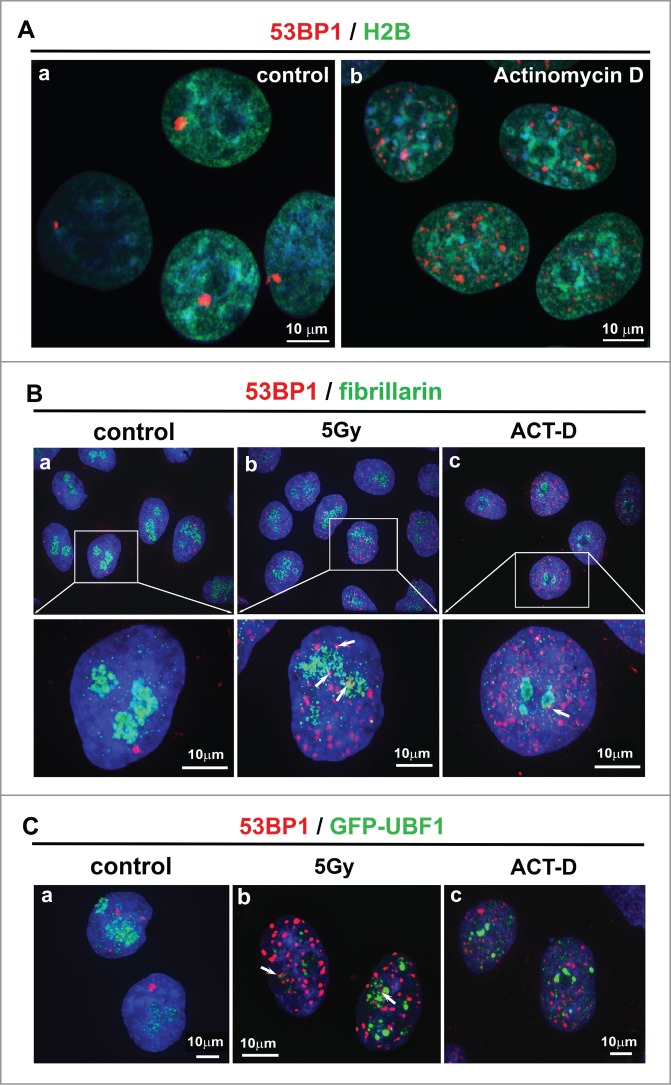
(A) Pronounced DNA damage by ACT-D treatment was confirmed by the appearance of 53BP1-positive NBs (red), which were visualized in (a) control untreated cells and (b) in ACT-D-treated cells which were characterized by an increased number of 53BP1 NBs. HeLa cell nuclei were analyzed according to GFP-H2B expression (green) following DAPI staining (blue). (B) Nuclear patterns of 53BP1 (red) and fibrillarin (green) in (a) control, (b) γ-irradiated, and (c) ACT-D treated HeLa cells. (C) Nuclear patterns of 53BP1 (red) and GFP-UBF1 (green) in (a) control, (b) γ-irradiated, and (c) ACT-D treated HeLa cells. 53BP1-positive nuclear bodies (NBs) were evaluated the following way: 1) foci outside fibrillarin-positive region of nucleolus; 2) foci which co-localized (exact overlapping) with fibrillarin-positive region or 3) foci associated with fibrillarin region (foci were in close proximity to the nucleolus). The following numbers of NBs were inspected: 550 NBs in control cells, 737 irradiation induced foci (IRIF) in γ-irradiated cells, and 458 NBs in ACT-D treated cells. Scale bars are 10 µm.
Changes in nucleolar morphology and local motion after radiation exposure or ribosomal gene transcription inhibition
We analyzed the localized movement of the UBF1-positive nucleolar regions in untreated control immortalized mouse embryonic fibroblasts (iMEFs; Fig. 2Aa–Ac) and in iMEFs that were exposed to 5 Gy γ-irradiation (Fig. 2Ba–Bc), UVC irradiation (Fig. 2Ca–c), or ACT-D treatment (Fig. 2Da–Dc). We monitored nucleolar movement (Fig. 2A–D; all experimental events) at 15-s intervals over 2 h by time-lapse confocal microscopy. We performed analyses of nucleolar movement and studies on nucleolar morphology after compensating for global nuclear motion. The development of contours around UBF1-positive regions showed the movement of nucleoli hubs from the beginning to the end of image acquisition (the c panels in Fig. 2A–D). Images representing the minimal enclosing ellipses around the tracks of the UBF1-positive region centroids are shown in the b panels of Fig. 2A–D. By this advanced image analysis approach, we revealed that γ-irradiation altered localized nucleolar movement, which is visible as a pronounced shift in the nuclear contour overlays (Fig. 2Bc, white arrows). Compared to untreated control cells, UVC irradiation and ACT-D treatment did not change the localized movement of UBF1-positive nucleolar regions (compare Fig. 2Ac, Cc, Dc).
Figure 2.
Single-particle tracking analysis shows localized movement of the GFP-UBF1-positive nucleolar compartment in iMEFs. Tracking of individual nucleoli (a panels) was visualized as the trajectories of the centroids of UBF1-positive regions of nucleoli and their minimal enclosing ellipses (b panels). We constructed the evolution of contours (panel c) of UBF1-positive regions of nucleoli with time overlaid over the first frame. Blue contours correspond to the start of scanning, and red contours correspond to the end of scanning (panels labeled as for c). Analysis was performed for (A) control non-irradiated and untreated cells, (B) γ-irradiated cells treated with 5 Gy of γ-irradiation (white arrows show changes in the contour overlays over time), (C) whole nuclei irradiated by UVC lamp, and (D) cells after ACT-D treatment. GFP-UBF1-positive regions were monitored every 15 s for 2 h. Scale bars are 5 µm. Five nuclei for each event (control and each treatment) were analyzed.
Tracking of the nucleus and UBF1-positive nucleolar regions in apoptotic irradiated cells
We exposed the cells to 3 different irradiation sources, UVA, UVC, and γ-rays, to analyze which type of irradiation induced apoptosis. By western blot analysis, we observed that both UVA and UVC irradiation induced lamin B fragmentation, which is an important apoptotic marker (Fig. 3A). In addition, local micro-irradiation of defined regions of interest (ROIs) by the UVA laser induced γH2AX- and CPD-positivity, which was accompanied by apoptosis (Fig. 3B, C). However, cell irradiation with 5 Gy of γ-rays did not induce lamin B fragmentation; thus, we used this treatment for additional studies (Figs. 4–6).
Figure 3.
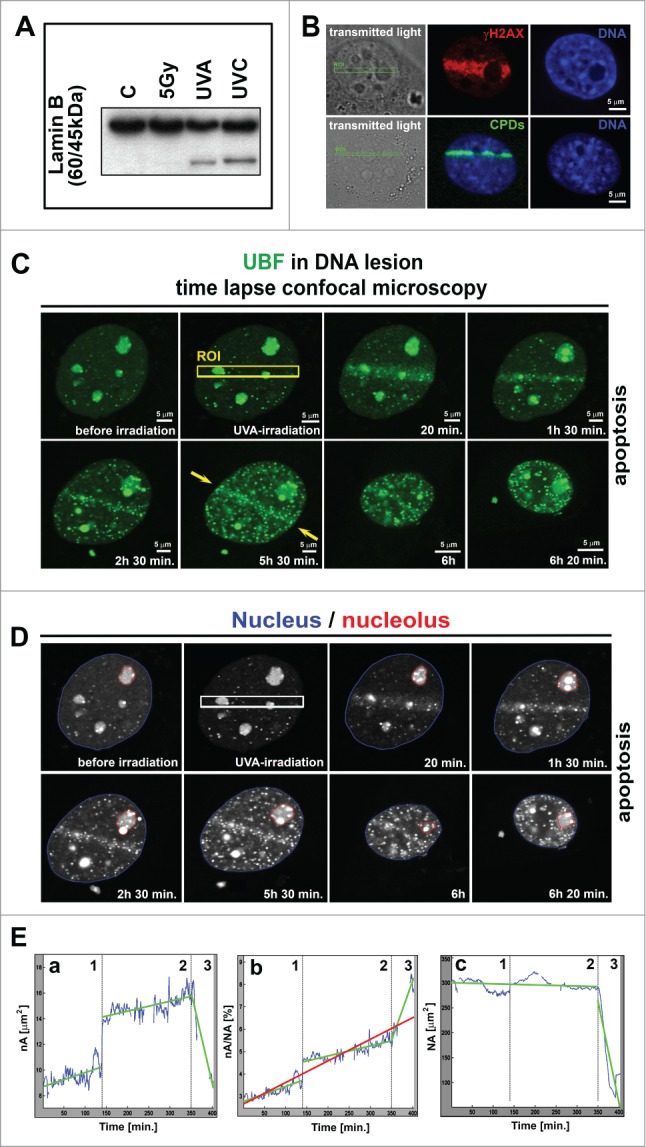
Time-lapse microscopy of apoptotic UVA-irradiated cells. (A) Western blot studies of the apoptotic marker lamin B (60 kDa), fragmented during apoptosis into a 45 kDa fragment. Cells were irradiated by γ-rays and by UVA and UVC lamps. (B) Verification of DNA injury induction by UVA laser (355 nm of excitation). Induced lesions were positive for the phosphorylation of γH2AX (red) and CPDs (green). The same results were found in iMEFs (panel B) and HeLa cells (not shown). (C) Monitoring of MEFs exposed to local micro-irradiation by UVA laser (355 nm wavelength and BrdU pre-sensitization) (˜6 h of observation). The UVA-irradiated region is indicated by the yellow frame (ROI), and yellow arrows indicate the maintenance of accumulated GFP-tagged UBF1 protein at UVA-irradiated ROI during the observation time. Scale bars are 5 µm. (D) Contours of cell nucleus (blue) and selected nucleolus (red) are shown for images from panel C. (E) Cell morphology parameters: (a) changes in nA (blue curve), (b) nA/NA, and (c) NA over time. The interruptions in blue curves correspond to unreliable portions of the image sequence where the cell was out of focus or out of the imaging plane. The data were estimated by linear regression analysis for 3 separate cellular stages [(1) adherent state, (2) cell during apoptosis-related detachment, and (3) terminal apoptotic shrinkage]. Green lines represent the regression line for individual stages 1–3, and the red line in panel b is the regression line for the entire cellular event when calculating nA/NA.
Figure 4.
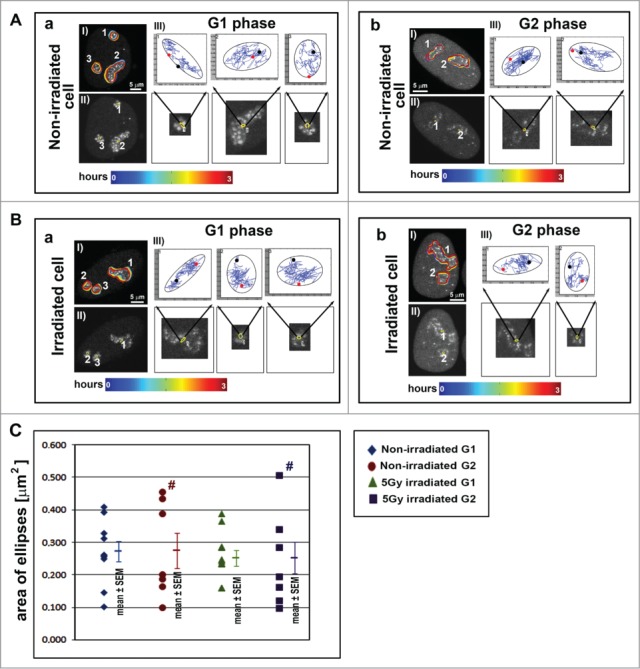
Tracking of UBF1-positive regions during cell cycle phases. Tracking of GFP-UBF1-positive nucleolar regions in representative images: (A) non-irradiated HeLa-Fucci cells and (B) γ-irradiated HeLa-Fucci cells, in (a) G1 and (b) G2 phases. Panels labeled I show the evolution of contours surrounding the UBF1-positive regions of nucleoli over time. Panels labeled II show the ellipses plotted over the first frame (see also enlarged images indicated by black arrows). Panels labeled III show ellipses (black) enclosing particular tracks of the nucleolus centroid. The start of the track (red) and the end of the track (black) are shown. (C) The average areas of the enclosing ellipses observed in G1 and G2 phases in non-irradiated and γ-irradiated HeLa-Fucci cells are plotted. The data are shown as the mean ± SEM. Number signs indicate a wider data distribution in G2 phase compared to G1 phase. In total, 3 to 4 nuclei were analyzed for each case containing different number of nucleoli, resulting in 7 to 11 total nucleoli. Scale bars are 5 µm.
Figure 5.
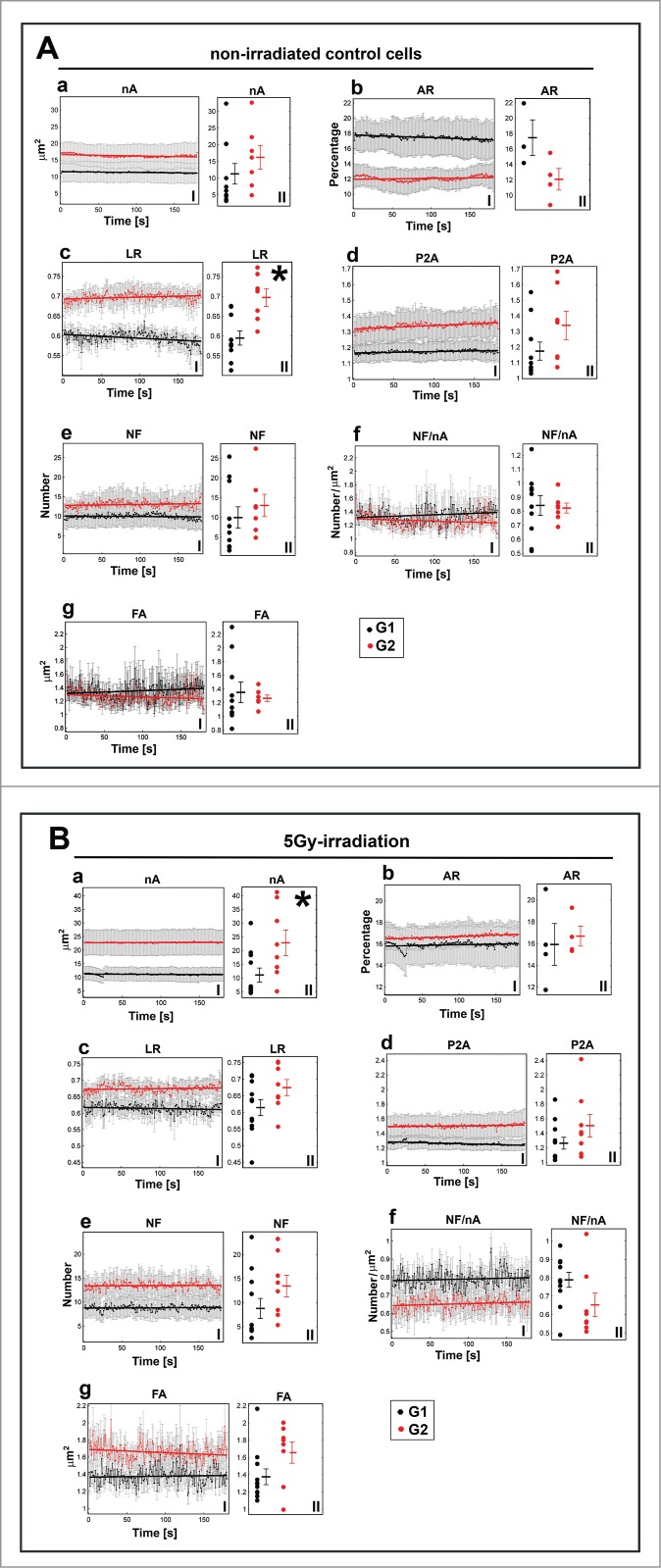
Comparison of the morphological parameters of nucleoli in (A) non-irradiated and (B) γ-irradiated G1 and G2 HeLa-Fucci cells. I panels show average values ± SEM over nucleoli in time, and II panels show the mean values over time for individual nucleoli. The following data are shown: (a) the nA, (b) AR, (c) LR, (d) P2A, (e) NF, (f) NF/nA, and (g) FA. In total, 3 to 4 nuclei were analyzed for each case containing different number of nucleoli, resulting in 7 to 11 total nucleoli. The error bars correspond to the SEM. The bold lines correspond to the increasing or decreasing trends obtained by linear regression. Asterisks show statistically significant differences (see also Table S1).
Figure 6.
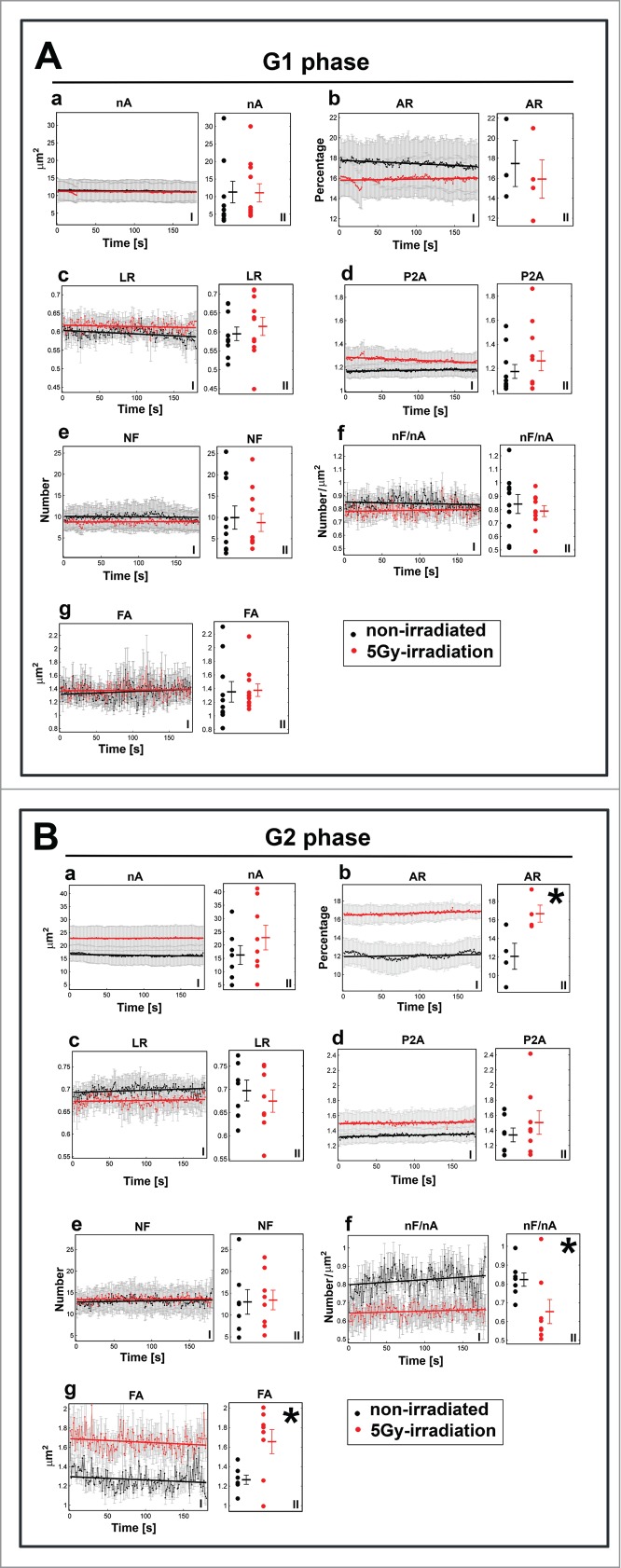
Comparison of morphological parameters of nucleoli in (A) G1 and (B) G2 of γ-irradiated and non-irradiated HeLa-Fucci cells. I panels show average values over nucleoli in time, and II panels show the mean values over time for individual nucleoli. In panels: (a) the nA, (b) AR, (c) LR, (d) P2A, (e) NF, (f) NF/nA, and (g) FA. In total, 3 to 4 nuclei were used for each case containing different number of nucleoli, resulting in 7 to 11 total nucleoli. The error bars correspond to the SEM. The bold lines correspond to increasing or decreasing trends obtained by linear regression. Asterisks show statistically significant differences (see also Table S1.).
To study the apoptotic event, we examined UVA-irradiated cells for 6–7 h. During irradiation induced-apoptosis, we observed nuclear shrinkage and UBF1-positive (body-like) structure formation (Fig. 3C, D). Interestingly, increased GFP-UBF1 levels at UVA-irradiated DNA lesions (ROIs) were maintained for approximately 5.5 h after irradiation (Fig. 3C, yellow arrows). Then, continuous nuclear shrinkage was observed (Fig. 3C, D). Next, we segmented the nucleus and the nucleolus in the image sequences (Fig. 3D). Image segmentation allowed us to analyze the time-dependent changes in the nucleolus area (nA), the ratio of the nucleolus area to the nucleus area (nA/NA), and the change in nuclear area (NA) (Fig. 3Ea–Ec). Interestingly, the profiles of the graphs in Fig. 3Ea–Ec can be divided into 3 cellular stages that describe the nuclear kinetics of apoptotic events (see dashed gray vertical lines in Fig. 3Ea–Ec). From a biological point of view, the first stage corresponds to the period when the adherent cells exhibited no morphological signs of apoptosis and remained attached to the cultivation dish. During the second stage, the cells detach from the cultivation dish, and the third stage likely corresponds to terminal apoptotic shrinkage of cell nucleus. Interruptions in the blue curves (in Fig. 3Ea–Ec) represent the unreliable portion of the image sequences when the cell was out of focus or out of the imaging plane and the nucleolus or nucleus area could not be estimated.
We analyzed the nA using linear regression for each of the 3 stages (see green lines in Fig. 3Ea). The growth rates for stages 1 and 2 were 0.0109 µm2 and 0.0077 µm2 per minute, with p-values of 4.10 × 10−13 and 1.36 × 10−25, respectively, whereas the decrease rate in the third stage was 0.143 µm2 per minute, with a p-value of 7.64 × 10−15.
In Fig. 3Eb, the red line corresponds to the linear regression results for the whole image sequence (stages 1–3) and demonstrates that the ratio increased by an average of 0.0096% per minute (p-values = 1.11 × 10−145). The green lines in Fig. 3Eb correspond to linear regression results for each stage separately and show the growth rates of 0.0066% for stage 1, 0.0045% for stage 2, and 0.0512% for stage 3 per minute, with p-values of 1.25 × 10−28, 3.22 × 10−49, and 5.73 × 10−14, respectively.
We analyzed the data in Fig. 3Ec using linear regression for stages 1 and 2 together and stage 3 separately. The green lines correspond to the linear regression results for stages 1 and 2 and show a decrease rate of 0.0209 µm2 per minute (p-value = 0.00083). For stage 3, the linear regression indicated a decrease rate of 4.059 µm2 per minute (p-value = 4.29 × 10−14). Together, our data demonstrate differences in nuclear and nucleolar kinetics during UVA radiation-induced apoptosis. Advanced image analysis methods demonstrated how the morphological phases of apoptosis can be characterized in detail (Fig. 3Ea–Ec).
Analysis of UBF1-positive foci mobility in G1 and G2 phases of non-irradiated and γ-irradiated cells
For this analysis, we used HeLa-Fucci cells expressing RFP-tagged-cdt1 in G1 phase and GFP-tagged geminin in G2 phase (Fig. S1A). The cell cycle profile was altered after 5 Gy of γ-radiation exposure (Fig. S1Ba, Bb) (γ-radiation did not induce apoptosis; Fig. 3A). Six hours after γ-irradiation, the cells were blocked in G2-M phase of the cell cycle when compared to control non-irradiated cells (Fig. S1Ba; see also Fig. S1Bb for the quantification of individual cell cycle phases). After the cell cycle analyses by flow cytometry, we determined how γ-radiation-induced cell cycle arrest altered nucleolar trajectory and morphology (Figs. 4–6, Tables S1–2). We performed analyses on nucleolar morphology and movement after compensating for global nuclear motion (see the Materials and Methods section).
To quantify the mobility of individual nucleoli, we plotted the enclosing ellipses of the trajectories of UBF1-positive region centroids. The average area of the enclosing ellipses did not significantly change in G1 cells compared to G2 cells (Fig. 4Aa, Ab and 4Ba, Bb). However, the ellipses area was more diverse in G2 cells, as shown by wider data distribution in non-irradiated G2 cells (Fig. 4C, brown number sign). We observed a similar trend for G2 cells after γ-irradiation (Fig. 4C, dark blue number sign).
Comparison of morphological parameters in G1 and G2 phase of the cell cycle in non-irradiated and γ-irradiated cells
We studied how some morphological parameters differ between G1 and G2 in non-irradiated and γ-irradiated cells (Fig. 5Aa–Ag for non-irradiated cells; Fig. 5Ba–Bg for γ-irradiated cells). We studied the following morphological parameters: nA, the ratio of UBF1-positive nucleoli area and nuclear area (AR, see Appendix 1 for formal definition), the average local radius of individual UBF1 foci within each nucleolus (LR, see Appendix 1 for formal definition), nucleolar compactness (P2A, see Appendix 1 for formal definition), the number of internal UBF1 foci inside each nucleolus (NF), the number of internal UBF1 foci inside each nucleolus normalized to nucleolar area (NF/nA), and the foci area averaged over each nucleolus (FA; Fig. 5Aa–Ag and 5Ba–Bg). We averaged these parameters for all nucleoli in all nuclei of each group to examine their changes over time (Fig. 5Aa-g and 5Ba–Bg; I panels). We analyzed the time dependency using linear regression, as described later.
We also averaged the parameters independent of time for each nucleolus to visualize the differences in their values for irradiated cells in G1 and G2 and non-irradiated cells in G1 and G2 phases (Fig. 5Aa–Ag and 5Ba-Bg; II panels). We only observed a statistically significant difference (p-value ≤ 0 .05) between the LR in G1 and the LR in G2 of non-irradiated cells (Fig. 5Ac, black asterisk; Table S1). These data suggest that, in non-irradiated cells, the UBF1 internal foci tend to be closer to the periphery of the nucleoli in G2 phase, whereas they are closer to the center of nucleoli in G1 phase. In γ-irradiated cells, a significant difference (p-value ≤ 0 .05) was observed between the nucleoli areas in G1 and G2; the nucleoli area in G2 cells was larger than that in G1 cells (Fig. 5Ba, black asterisk; Table S1).
Comparison of morphological parameters in non-irradiated and γ-irradiated cells according to G1 and G2 cell cycle phases
We used the same parameters described above to study the difference between non-irradiated and γ-irradiated cells in G1 and G2 phases of the cell cycle (Fig. 6Aa–Ag and 6Ba–Bg). We observed different relations between the parameters when we compared G1 (Fig. 6Aa–Ag) and G2 (Fig. 6Ba–Bg). Notably, the differences in AR were opposite in G1 and G2 non-irradiated and irradiated cells. For example, in G1 phase (Fig. 6Ab) the AR values were higher in non-irradiated cells than in γ-irradiated cells. However, in the G2 phase (Fig. 6Bb), the relation between AR values was reversed; the AR was lower in non-irradiated cells than in γ-irradiated cells.
We performed a similar analysis for time-averaged parameters to determine whether a difference existed between non-irradiated and irradiated cells in G1 (Fig. 6A, panel II) and G2 (Fig. 6B, panel II). In G1 cells we did not observe any statistically significant differences between non-irradiated and γ-irradiated cells. However, in G2, we calculated statistically significant differences for AR, nF/nA, and FA (Fig. 6Bb, Bf, and Bg, black asterisks; Table S1). This observation is consistent with radiation-induced changes in the cell cycle because γ-radiation primarily causes cell cycle arrest in G2 phase (e.g.,25). We also observed G2 arrest in the HeLa-Fucci model used here (Fig. S1Ba, Bb).
The changes in AR (Fig. 6Bb) showed that the nucleoli occupy a larger space within the nucleus after γ-irradiation. In contrast, the nF/nA parameter, which indicates the relative density of foci within a nucleolus, was higher in non-irradiated cells compared to γ-irradiated cells (Fig. 6Bf).
Together, our data show that repair of ribosomal genes is linked to the G2 phase, which is not a consequence of increased DNA content in the G2 phase compared to G1. We studied G2 non-irradiated and G2 irradiated cells, characterized by an identical DNA content. In this case, statistically significant differences were found for the morphological parameters shown in Fig. 6Bb, Bf, and Bg. We also compared G1 and G2 non-irradiated cells with different DNA content (Fig. 5A). This analysis showed only one significantly different parameter, LR (Fig. 5A). Thus, it is unlikely that the observed morphological changes in the nucleus were caused by higher amounts of DNA in the G2 phase. We unambiguously show that morphology of nucleoli is affected by γ-radiation, especially in the G2 phase of the cell cycle (Fig. 5Ba, 6Bb, Bf, and Bg).
Time dependency of UBF1-positive nucleolar morphology
Regression analysis showed an additional criterion that must also be considered (see Table S2 and red or black lines in Figs. 5, 6; I panels). We only considered the parameters that demonstrated a stable increasing or decreasing tendency for both irradiated and non-irradiated cells in both G1 and G2 phases (growth coefficient p-value ≤ 0 .05 for all 4 cases in Table S2). Namely, we studied the differences in the time-dependency trends of nA, AR, and P2A (see increasing/decreasing trends in Figs. 5 and 6; I panels). For example, Fig. 5Ad (panel I) shows the regression lines for P2A in non-irradiated G1 and G2 cells. These regression lines indicate an identical increasing tendency. However, in γ-irradiated G1 and G2 cells, the regression lines for P2A parameters diverge (Fig. 5Bd; panel I). The decreasing tendency of the regression line indicates that the cell nucleoli became more round after γ-irradiation in G1 phase (see Fig. 5Bd, black line), whereas in all the other cases, the regression line demonstrates an increasing trend, meaning that the compactness of nucleoli compartments became more relaxed. Next, we examined AR for non-irradiated and irradiated G1 (Fig. 6Ab) and non-irradiated and irradiated G2 cells (Fig. 6Bb). The regression lines for AR in Fig. 6Ab converged, whereas an upward tendency was found for both AR-related regression lines in Fig. 6Bb. These data suggest that, in G2 phase, the area of nucleus occupied by nucleoli increases regardless of irradiation. However, in G1 phase, the area of the nucleus occupied by nucleoli increased only after γ-irradiation but decreased in non-irradiated cells.
Discussion
The nucleolus holds the largest transcription machinery in the cell and is highly sensitive to genotoxic factors, including radiation.26-28 Moore et al.9 showed radiation-induced morphological changes in the UBF1-positive regions of nucleoli. After γ-irradiation, we observed a shift in nucleoli contours, which we monitored in live cells over time (Fig. 2Bc, white arrows). We studied nucleolar morphology and localized movement by advanced image analysis approaches. However, notably, single-particle tracking analyses vary between laboratories because of the specific nature of the data being analyzed and because of the various algorithms used.16 We used several image processing algorithms for our data analysis. We applied methods similar to other studies that revealed the movement of nuclear bodies or protein foci.17,27, 29,30 We used contour-based image registration techniques31 and specifically tailored tracking and segmentation algorithms (Figs. 2A–D, 3D and 4A, B). These image analysis approaches allowed us to describe nucleolar behavior after radiation exposure in detail. Using the aforementioned experimental approaches, we analyzed protein recruitment and function at DNA lesions and the mobility of accumulated proteins after cell irradiation (Figs. 2A–D, 4A, 4B, or Ref.14,32). In this study, we analyzed the GFP-UBF1-positive regions of nucleoli after exposure to UVA or γ-radiation. In our first report, we showed that UBF1 is recruited to DNA lesions simultaneously with heterochromatin protein 1β (HP1β). Moreover, the UBF1-HP1β interaction appeared in CPD-positive DNA lesions. Thus, UBF1 likely functions not only during transcription as a transcription factor for ribosomal genes, but also during nucleotide excision repair.12
This study is the first demonstration that the localized movement of UBF1-positive nucleolar regions (enclosing ellipses around tracks of nucleoli hubs) changed after γ-irradiation (Fig. 2Bc, white arrows). Previous studies made similar conclusions related to the movement of the nuclear domain, for example, different classes of PML bodies or Cajal bodies (CBs) which are highly mobile in non-irradiated cells.27,29, 33 Moreover, we recently demonstrated that local CB motion decreases after γ-irradiation, which is in agreement with data that radiation induces changes in nuclear domain trajectories (compare32 with Fig. 2Bc, white arrows). This phenomenon also should be tested in other nuclear compartments, including replication foci, transcription factories, or nuclear speckles. Accumulated protein mobility in the nucleus also can be addressed by additional biophysics-related computational methods, such as FRAP analysis of protein diffusion or computation of mean-square displacement (MSD).18 However, diffusive motion analysis via MSD curve construction presumes a small size for the analyzed objects, and this presumption is not the case for large UBF1-positive nucleolar regions. Therefore, we could not apply this analysis for our data.
As summarized by Olson and Dundr,34 the nucleolar component moves via various mechanisms. For example, vectorial movement between the fibrillar center and dense fibrillar components of the nucleolus occurs during ribosomal particle assembly. Nucleolar components also can be rapidly exchanged between the nucleus and the nucleoplasm, particularly after genome irradiation (summarized by21,34). Moreover, dynamic exchange of nucleolar components can occur after RNA polymerase I inhibition by ACT-D.17 Therefore, the movement of nucleolar proteins, ribosomal subunits, or the entire nucleolus is a naturally occurring event that can be affected by genotoxic factors, including UV and ionizing radiation. As Rubbi and Milner35 suggested previously, the nucleolus serves as a major stress sensor that responds in a manner related to p53 function. p53 protein stability/instability is decisive for nucleolar disruption or compactness, which is affected by many stress factors.34,36 This fact also corresponds to our observation of pronounced local motion of the p53-binding protein 53BP1 after cells are exposed to γ-irradiation or UVA laser.14 Interestingly, 53BP1-positive DNA damage-related NBs are visible in G1 phase, whereas some cell types do not exhibit 53BP1 clustering in G2 phase.37 This observation is consistent with our data confirming the cell cycle-specific morphology of accumulated proteins, as shown here for UBF1, an extremely important nucleolar protein (Fig. 5Ac, Ba). Moreover, as shown here for nucleoli, many morphological parameters are altered by γ-irradiation, particularly during G2 phase (Fig. 6Bb, Bf, and Bg, Table S1, and Fig. S1B). Therefore, individual cell cycle phases are clearly characterized by specific distributions and patterns of nuclear and nucleolar components, and these distributions and patterns can be affected by radiation.
Conclusions
We showed that genome injury induced by radiation or by a DNA damaging agent changed many morphological parameters of nucleoli (Figs. 2Bc, 2Da, 5A, 5B, 6B). Local nucleoli motions in G1 and G2 non-irradiated and γ-irradiated cells were nearly identical, but with pronounced variability in G2 phase (Fig. 4A–C). Irradiation by γ-rays induced many morphological changes, including re-location of the nucleoli (Fig. 2Bc, white arrows). Morphological changes, likely reflecting rearrangement of ribosomal genes, preferentially appeared in the G2 phase of the cell cycle (Fig. 6Bb, Bf, and Bg). These conclusions imply that DNA lesions in ribosomal genes activate specific cell-cycle dependent DNA repair mechanisms.19,38 Disorder in these mechanisms can contribute to uncontrolled cell proliferation and pathophysiological processes.2,19 Our studies also show that damage and repair of ribosomal genes can be followed by changes in the morphology of nucleoli in a cell cycle-specific manner. Our data suggest G2 phase as a key cell cycle stage for the repair of ribosomal genes and imply that the nucleolus is less stable in G2 phase, thus most sensitive to ionizing radiation.
Materials and Methods
Cell cultivation and transfection
Immortalize mouse embryonic fibroblasts (iMEFs, a generous gift from Dr. Monika Lachner, Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum and appropriate antibiotics at 37°C in a humidified atmosphere of 5% CO2. Immortalization was induced by prolonged cell culture such that the cells adapted to the cultivation conditions. HeLa cells stably expressing GFP-H2A were a generous gift from Marion Cremer (Ludwig-Maximilian-University of Munich, Germany). For studies of GFP-UBF1-stained nucleoli trajectories in individual cell cycle phases, HeLa-Fucci cells that express RFP-tagged-cdt1 in G1 phase and GFP-tagged geminin in G2 phase were used (39; see also http://www.amalgaam.co.jp/products/advanced/fucci.html). In these cells, S phase can be recognized as an overlay of red and green fluorescence (nuclei appear orange). These cells were not analyzed because of fluorescence transition; only unambiguous cell nuclei that were visualized by red or green fluorescence were studied.
For cell transfection and subsequent time-lapse microscopy, iMEFs were cultivated on glass-bottom tissue culture dishes to 70% confluence, and then the cells were transfected with 1–2 μg of plasmid DNA encoding GFP-UBF1 (#17656; Addgene, Cambridge, MA USA) using Metafectane™Pro reagent (#T040–2.0, Biontex Laboratories GmbH, Germany). GFP-UBF1 fluorescence could be distinguished from GFP-geminin fluorescence because of pronounced GFP-UBF1 accumulation at nucleoli (see Fig. S1A).
For selected experiments, cells were treated with 0.5 μg/ml ACT-D, an RNA polymerase I inhibitor (#A9415, Sigma-Aldrich, Prague, Czech Republic). ACT-D was present in cell cultures throughout the 2 h time lapse microscopy experiments.
Induction of DNA lesions and confocal microscopy
An iMEF population was irradiated with 5 Gy of γ-rays using Chisostat (Chirana, Czech Republic), and the source of radiation was Cobalt-60 (60Co). The cells were fixed in 4% formaldehyde at 2 h after γ-irradiation (Fig. 1Bb, 1Cb, 3A). Time-lapse confocal microscopy was performed from 0 to 2 h after γ-irradiation. For live cell observations, a heating hood (EMBL, Heidelberg) was used to maintain a constant temperature of 37°C and a 5% CO2 humidified environment. Cell irradiation by UVA and UVC lamps was performed for 15 min, and the cells were fixed after 30 min (for Fig. 3A). In Fig. 2C, nuclei of living cells were monitored for 0–2 h after UVC exposure. No pre-sensitization was used for UVC or γ-irradiation. The UVC lamp (Philips, Holland) specifications were as follows: model TUV 30 W T8 (UVC 254 nm wavelength); geometry of irradiation: vertically downwards; lamp distance from the sample: 80 cm; irradiated area: 9.2 cm2; dose: 0.828 J/cm2; cell concentration: 2 × 105 cells/ml; total number of cells per dish: ∼6×105; layer of cultivation medium: was 0.5 cm vertically. A UVA lamp was used in some cases, with the following specifications (results in Fig. 3A): model GESP-15, 15 W (UVA 330–400 nm wavelength); maximal efficiency: 365 nm; geometry of irradiation: vertically downward; distance from the sample: 10 cm; irradiated area: 9.2 cm2 (cultivation plate area); irradiation time: 15 min; dose: 0.828 J/cm2. The cells were seeded at a density of 6.5 × 104 cells/cm2 and irradiated at 24 h after seeding. No pre-sensitization or additional treatment was used.
For local micro-irradiation by UVA laser (wavelength: 355 nm; data related to apoptosis induction in Fig. 3C–D), cells were seeded in a μ-Dish 35 mm Grid-500 (#81166, Ibidi, Germany). Pre-sensitization was induced by 10 µM 5-bromo-2′-deoxy-uridine (BrdU) for 16–18 h before local irradiation as described previously by Šustáčková et al.7
Microscopic analyses were performed using a Leica SP5 X confocal microscope (Leica Microsystems, Mannheim, Germany). For cell observations, an oil objective HCX PL APO, 63×, numerical aperture (N.A.) = 1.4 was used. The following image acquisition settings were used: 1024 × 1024 pixels, 400 Hz, bidirectional scanning mode, zoom 8–12. For fluorescence visualization of selected proteins, the following fluorochromes were used: Alexa Fluor® 594 (max. emission: 590 nm; max. excitation: 617 nm), EGFP (max. emission: 488 nm; max. excitation: 509 nm), and DAPI (max. emission: 358 nm; max. excitation: 461 nm).
Analysis of nucleolar morphology and localized movement of the UBF1-positive nucleolar compartment using time-lapse microscopy data
The nucleolar morphology and motion for untreated, UVC, and γ-irradiated iMEFs and for ACT-D-treated iMEFs were analyzed using the following pipeline. First, global cell motion was compensated for using the rigid point-based algorithm.40 This algorithm uses the nucleoli centroids and the shape of their disposition inside the nucleus to assess the global nucleus motion parameters (rotation and translation). Using the estimated parameters, global motion was compensated, and the nucleolar position in every frame of the image sequence was normalized to their position in the first frame.
The second step included nucleolar segmentation. The nucleoli were segmented using a 2-stage thresholding approach. In the first stage, rough nucleoli masks were generated separately in each frame using a robust multithresholding approach (with 3 threshold levels), followed by morphological post-processing (opening and closing with a circular structure element of a suitable radius). Then, the nucleoli tracks were constructed using the correspondence-finding approach described previously in.40 The nuclei tracks were used to refine each nucleolus segmentation mask by working with the part of the image sequence containing only the specified nucleolus. Thus, the second segmentation stage was performed separately for each nucleolus. The refinement was performed using p-tile thresholding where the number of pixels to retain in the segmentation mask was set such that the area and the compactness of the nucleus did not pronouncedly change compared to the previous frame. The second segmentation stage was followed by morphological post-processing (opening and closing with a circular structure element of a suitable radius).
For the analysis of UBF1-positive structures in untreated HeLa-Fucci cells and γ (5 Gy)-irradiated HeLa-Fucci cells in G1 and G2 phases (Figs. 4–6), a similar image processing pipeline was utilized with a few differences. The first difference was in the global motion compensation approach. The compensation for cell motion was performed using a rigid contour-based algorithm.31 Thus, not only nucleoli segmentation but also proper nucleus segmentation was required to obtain the nucleus contours required for image sequence registration.
The nucleus and nucleoli were segmented in the following way. First, the intensity of the data was normalized, and the images were denoised with Gaussian smoothing. Because the contrast between the nucleoli structures and the nucleus was low, focal structures were suppressed using h-maxima morphological transformation with suitable parameters. Primary nucleus and nucleoli segmentation masks were obtained using multithresholding segmentation (with 3 threshold levels). Then, the nucleus and nucleoli masks were refined by morphological post-processing (opening, closing) with a suitable element size.
Next, the UBF1-positive foci were detected. Foci detection was based on the previously described Hessian-based multiscale approach.14
Nucleolar evolution contours were constructed for both iMEFs and HeLa-Fucci cells (Figs. 2 and 4A, B), in which the colored lines represent the nucleoli contours at different time points (see legend). Blue contours represent the start, and red contours represent the end of nucleolar movement studied by time-lapse microscopy. For this purpose, the nucleoli were segmented as described above in each time frame to obtain their proper contours. Nucleoli movement was interpreted as the movement of their centroids, and the minimal enclosing ellipses were plotted, which are the ellipses of minimal area that enclosed all positions of the nucleoli centroids in a period.17
The following changes in nucleoli morphology were analyzed in HeLa-Fucci cells: the nA, AR, and P2A. Several foci parameters also were analyzed, including the FA, NF, NF/nA, and LR. These values were averaged for all nucleoli in all nuclei of control and treated cells. The formal definitions of these parameters are shown in Appendix 1.
Statistical analysis
Statistical analysis was performed to compare the nA, LR, FA, NF, AR, NF/nA, and P2A parameters in irradiated and non-irradiated HeLa-Fucci cells in G1 and G2 phase. The studied parameters were averaged over all nucleoli. The results are presented in Table S2. Specifically, the growth rates of selected parameters were analyzed by linear regression analysis. Linear regression analysis determined whether a stable increasing or decreasing tendency occurred or whether the parameters demonstrated constant behavior. The F-test was used for this analysis, where the null hypothesis was that the growth coefficient equaled zero. The results of this analysis showed that the nA, AR, and P2A parameters have estimated growth rates that statistically significantly differ from zero in all cases, i.e., irradiated/non-irradiated cells in G1/G2 cell cycle phases (see Table S2).
Although some nucleolar morphological parameters demonstrated statistically significant tendencies (trends), the absolute value of the changes throughout the observation time was extremely low (2.5%). Thus, for estimating the differences in the selected morphological parameters of the nucleoli at different cell cycle stages and irradiation conditions, the dimensionality of the data were reduced and averaged over time (Figs. 5–6, II panels). This manipulation also rendered the distribution of these parameters closer to a normal distribution that allowed for parametric statistical tests, such as Student's t-test. This statistical approach enabled us to determine whether the selected parameters exhibited statistically significant differences in different cell cycle stages or irradiation conditions. Due to the low dimensionality of the data, the resulting p-values were not small (see Table S1); however, some of these parameters had statistically significant differences (see Figs. 5–6 and Table S1).
Western blots
Western blot analyses (Fig. 3A) were performed according to Stixová et al. (2012).18 The anti-lamin B primary antibody (sc-6217, Santa Cruz) was used for these analyses. Total protein levels were measured using a µQuant spectrophotometer (BioTek).
Cell cycle analysis by flow cytometry
Cells were washed twice in PBS, fixed for at least 30 min in ice-cold 70% ethanol, and stored at 4°C until analysis. Fixed cells were washed in PBS and stained with FxCycle™ PI/RNase Staining Solution (#F10797, Molecular Probes®, Eugene, OR USA) for 30 min at 37°C. The DNA content was determined using a BD FACS Canto II (Becton-Dickinson, 488-nm argon laser for excitation). The distribution of the cells in individual cell cycle phases was measured and analyzed using BD FACSDiva software (Becton-Dickinson) and FlowJo software (Tree Star, Ashland, OR USA) (http://www.flowjo.com/). Three independent replicates consisting of 3 samples were analyzed for each event.
Appendix 1
Assume that we have a nucleus N with k nucleoli with centroids defined as ni (i = 1,…, k), each containing m foci with centroids Fj (j = 1,…, m). Let us denote the nucleus area as NA, the nucleoli area as nAi (i = 1,…, k), and the foci area as FAj (j = 1,…, m). Let us denote the nucleus boundary as Nb and the nucleus centroid as Nc.
The local radius LR for each nucleolus was defined by the following formula:
and if all the foci are in the center of the nucleolus, and if all the foci are at the nucleolus boundary.
Compactness (P2A) for each nucleolus was defined as follows:
and when the object is circular, and when the nucleolus has a more complex shape. The higher the value of the compactness parameter, the more complex the nucleolar shape.
The area ratio for each nucleus was defined as follows:
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Funding
This work was supported by the Grant Agency of the Czech Republic (projects P302/12/G157, and 13–07822S). The Czech-Norwegian Research Program CZ09, which is supported by Norway funds and by the Ministry of Education Youth and Sport of the Czech Republic, guaranteed the work of SL (grant number: 7F14369). The postdoctoral fellowships of DVS and LS were supported by the Education for Competitiveness Operational Program (ECOP), CZ.1.07/2.3.00/30.0030.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Authors' Contributions
The authors indicate the following contributions. DVS performed the image analysis and image processing; analyzed nA, NA, LR, FA, NF, AR, NF/nA, and P2A; and provided additional data analysis. LS performed the cell cultivation, cell treatment, plasmid DNA isolation, cell transfection, and time-lapse microscopy. LS also performed the cell cycle analysis by flow cytometry. PŠ performed the cell cycle analysis using FlowJo software. SL and JS performed cell cultivation and immunofluorescence. PS performed the observations using time-lapse microscopy of apoptotic cells. PM performed the statistical analyses. SK and IR coordinated the progress of ongoing projects to support these experiments. MS performed the immunostaining. EB designed the experiments, coordinated the experimental efforts, and wrote this paper. All authors read and approved the final version of this manuscript.
References
- 1.Eppink B, Essers J, Kannar R. Interplay and quality of DNA damage repair mechanism In: Rippe K, ed. Genome organization and function in the cell nucleus. Weinheim, Germany: Wiley-VCH, 2012:395-415 [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071-8; PMID:19847258; http://dx.doi.org/ 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagy Z, Soutoglou E. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol 2009; 19:617-29; PMID:19819145; http://dx.doi.org/ 10.1016/j.tcb.2009.08.010 [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Zeng ZC, Perrault AR, Cheng X, Qin W, Iliakis G. Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic Acids Res 2001; 29:1653-60; PMID:11292837; http://dx.doi.org/ 10.1093/nar/29.8.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem 2000; 275:26196-205; PMID:10854421; http://dx.doi.org/ 10.1074/jbc.M000491200 [DOI] [PubMed] [Google Scholar]
- 6.Bubulya PA, Spector DL. “On the move”ments of nuclear components in living cells. Exp Cell Res 2004; 296:4-11; PMID:15120987; http://dx.doi.org/ 10.1016/j.yexcr.2004.03.018 [DOI] [PubMed] [Google Scholar]
- 7.Sustackova G, Kozubek S, Stixova L, Legartova S, Matula P, Orlova D, Bártová E. Acetylation-dependent nuclear arrangement and recruitment of BMI1 protein to UV-damaged chromatin. J Cell Physiol 2012; 227:1838-50; PMID:21732356; http://dx.doi.org/ 10.1002/jcp.22912 [DOI] [PubMed] [Google Scholar]
- 8.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 2005; 16:2395-413; PMID:15758027; http://dx.doi.org/ 10.1091/mbc.E04-11-0992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, Laiho M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics 2011; 10:M111 009241; PMID:21778410; http://dx.doi.org/ 10.1074/mcp.M111.009241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stixova L, Hruskova T, Sehnalova P, Legartova S, Svidenska S, Kozubek S, Bártová E. Advanced microscopy techniques used for comparison of UVA- and gamma-irradiation-induced DNA damage in the cell nucleus and nucleolus. Folia Biol (Praha) 2014; 60 Suppl 1:76-84; PMID:25369346 [DOI] [PubMed] [Google Scholar]
- 11.Horakova AH, Bartova E, Galiova G, Uhlirova R, Matula P, Kozubek S. SUV39h-independent association of HP1 β with fibrillarin-positive nucleolar regions. Chromosoma 2010; 119:227-41; PMID:20033197; http://dx.doi.org/ 10.1007/s00412-009-0252-2 [DOI] [PubMed] [Google Scholar]
- 12.Stixova L, Sehnalova P, Legartova S, Suchankova J, Hruskova T, Kozubek S, Sorokin DV, Matula P, Raška I, Kovařík A, et al.. HP1beta-dependent recruitment of UBF1 to irradiated chromatin occurs simultaneously with CPDs. Epigenetics Chromatin 2014; 7:39; PMID:25587355; http://dx.doi.org/ 10.1186/1756-8935-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell 2007; 27:585-95; PMID:17707230; http://dx.doi.org/ 10.1016/j.molcel.2007.06.021 [DOI] [PubMed] [Google Scholar]
- 14.Foltankova V, Matula P, Sorokin D, Kozubek S, Bartova E. Hybrid detectors improved time-lapse confocal microscopy of PML and 53BP1 nuclear body colocalization in DNA lesions. Microsc Microanal 2013; 19:360-9; PMID:23410959; http://dx.doi.org/ 10.1017/S1431927612014353 [DOI] [PubMed] [Google Scholar]
- 15.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol 2012; 24:415-22; PMID:22541757; http://dx.doi.org/ 10.1016/j.ceb.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chenouard N, Smal I, de Chaumont F, Maska M, Sbalzarini IF, Gong Y, Cardinale J, Carthel C, Coraluppi S, Winter M, et al.. Objective comparison of particle tracking methods. Nat Methods 2014; 11:281-9; PMID:24441936; http://dx.doi.org/ 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stixova L, Bartova E, Matula P, Danek O, Legartova S, Kozubek S. Heterogeneity in the kinetics of nuclear proteins and trajectories of substructures associated with heterochromatin. Epigenetics Chromatin 2011; 4:5; PMID:21418567; http://dx.doi.org/ 10.1186/1756-8935-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stixova L, Matula P, Kozubek S, Gombitova A, Cmarko D, Raska I, Bártová E. Trajectories and nuclear arrangement of PML bodies are influenced by A-type lamin deficiency. Biol Cell 2012; 104:418-32; PMID:22443097; http://dx.doi.org/ 10.1111/boc.201100053 [DOI] [PubMed] [Google Scholar]
- 19.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 2008; 9:297-308; PMID:18285803; http://dx.doi.org/ 10.1038/nrm2351 [DOI] [PubMed] [Google Scholar]
- 20.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73:39-85; PMID:15189136; http://dx.doi.org/ 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- 21.Antoniali G, Lirussi L, Poletto M, Tell G. Emerging roles of the nucleolus in regulating the DNA damage response: the noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example. Antioxid Redox Signal 2014; 20:621-39; PMID:23879289; http://dx.doi.org/ 10.1089/ars.2013.5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleeff J, Kornmann M, Sawhney H, Korc M. Actinomycin D induces apoptosis and inhibits growth of pancreatic cancer cells. Int J Cancer 2000; 86:399-407; PMID:10760829; http://dx.doi.org/ 10.1002/(SICI)1097-0215(20000501)86:3%3c399::AID-IJC15%3e3.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- 23.Verheij M, Bartelink H. Radiation-induced apoptosis. Cell Tissue Res 2000; 301:133-42; PMID:10928286; http://dx.doi.org/ 10.1007/s004410000188 [DOI] [PubMed] [Google Scholar]
- 24.Mischo HE, Hemmerich P, Grosse F, Zhang S. Actinomycin D induces histone gamma-H2AX foci and complex formation of gamma-H2AX with Ku70 and nuclear DNA helicase II. J Biol Chem 2005; 280:9586-94; PMID:15613478; http://dx.doi.org/ 10.1074/jbc.M411444200 [DOI] [PubMed] [Google Scholar]
- 25.Bartova E, Kozubek S, Kozubek M, Jirsova P, Lukasova E, Skalnikova M, Buchnícková K. The influence of the cell cycle, differentiation and irradiation on the nuclear location of the abl, bcr and c-myc genes in human leukemic cells. Leuk Res 2000; 24:233-41; PMID:10739005; http://dx.doi.org/ 10.1016/S0145-2126(99)00174-5 [DOI] [PubMed] [Google Scholar]
- 26.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell 2010; 40:216-27; PMID:20965417; http://dx.doi.org/ 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muratani M, Gerlich D, Janicki SM, Gebhard M, Eils R, Spector DL. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat Cell Biol 2002; 4:106-10; PMID:11753375; http://dx.doi.org/ 10.1038/ncb740 [DOI] [PubMed] [Google Scholar]
- 28.Scott M, Boisvert FM, Vieyra D, Johnston RN, Bazett-Jones DP, Riabowol K. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucleic Acids Res 2001; 29:2052-8; PMID:11353074; http://dx.doi.org/ 10.1093/nar/29.10.2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 2000; 151:1561-74; PMID:11134083; http://dx.doi.org/ 10.1083/jcb.151.7.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tvarusko W, Bentele M, Misteli T, Rudolf R, Kaether C, Spector DL, Gerdes HH, Eils R. Time-resolved analysis and visualization of dynamic processes in living cells. Proc Natl Acad Sci U S A 1999; 96:7950-5; PMID:10393928; http://dx.doi.org/ 10.1073/pnas.96.14.7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorokin DV, Tektonidis M, Rohr K, Matula P. Non-rigid Contour-based Temporal Registration of 2D Cell Nuclei Images Using the Navier Equation. In IEEE International Symposium on Biomedical Imaging: From Nano to Macro. Beijing: IEEE, 2014:746-749; ISBN 978-1-4673-1959-1. [Google Scholar]
- 32.Bartova E, Foltankova V, Legartova S, Sehnalova P, Sorokin DV, Suchankova J, Kozubek S. Coilin is rapidly recruited to UVA-induced DNA lesions and gamma-radiation affects localized movement of Cajal bodies. Nucleus 2014; 5; PMID:24859326; http://dx.doi.org/ 10.4161/nucl.29229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol 2004; 164:831-42; PMID:15024031; http://dx.doi.org/ 10.1083/jcb.200311121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson MO, Dundr M. The moving parts of the nucleolus. Histochem Cell Biol 2005; 123:203-16; PMID:15742198; http://dx.doi.org/ 10.1007/s00418-005-0754-9 [DOI] [PubMed] [Google Scholar]
- 35.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 2003; 22:6068-77; PMID:14609953; http://dx.doi.org/ 10.1093/emboj/cdg579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci STKE 2004; 2004:pe10; PMID:15026578 [DOI] [PubMed] [Google Scholar]
- 37.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grøfte M, Chan KL, Hickson ID, Bartek J, et al.. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 2011; 13:243-53; PMID:21317883; http://dx.doi.org/ 10.1038/ncb2201 [DOI] [PubMed] [Google Scholar]
- 38.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 2003; 5:255-60; PMID:12598907; http://dx.doi.org/ 10.1038/ncb945 [DOI] [PubMed] [Google Scholar]
- 39.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al.. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008; 132:487-98; PMID:18267078; http://dx.doi.org/ 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- 40.Matula P, Matula P, Kozubek M, Dvorak V. Fast point-based 3-D alignment of live cells. IEEE Trans Image Process 2006; 15:2388-96; PMID:16900692; http://dx.doi.org/ 10.1109/TIP.2006.875209 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



