Abstract
Nuclear pore complexes (NPCs) form the gateway to the nucleus, mediating virtually all nucleocytoplasmic trafficking. Assembly of a nuclear pore complex requires the organization of many soluble sub-complexes into a final massive structure embedded in the nuclear envelope. By use of a LacI/LacO reporter system, we were able to assess nucleoporin (Nup) interactions, show that they occur with a high level of specificity, and identify nucleoporins sufficient for initiation of the complex process of NPC assembly in vivo. Eleven nucleoporins from different sub-complexes were fused to LacI-CFP and transfected separately into a human cell line containing a stably integrated LacO DNA array. The LacI-Nup fusion proteins, which bound to the array, were examined for their ability to recruit endogenous nucleoporins to the intranuclear LacO site. Many could recruit nucleoporins of the same sub-complex and a number could also recruit other sub-complexes. Strikingly, Nup133 and Nup107 of the Nup107/160 subcomplex and Nup153 and Nup50 of the nuclear pore basket recruited a near full complement of nucleoporins to the LacO array. Furthermore, Nup133 and Nup153 efficiently targeted the LacO array to the nuclear periphery. Our data support a hierarchical, seeded assembly pathway and identify Nup133 and Nup153 as effective “seeds” for NPC assembly. In addition, we show that this system can be applied to functional studies of individual nucleoporin domains as well as to specific nucleoporin disease mutations. We find that the R391H cardiac arrhythmia/sudden death mutation of Nup155 prevents both its subcomplex assembly and nuclear rim targeting of the LacO array.
Keywords: cardiac arrhythmia, ELYS, LacO array, LacI, nuclear pore assembly, nuclear rim localization, Nup133, Nup153, Nup160/107 complex, Nup155 R391H
Abbreviations
- FG
phenylalanine-glycine
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- LacO
Lac operon
- LacI
Lac repressor gene
Introduction
The nuclear pore complex (NPC) is a large multi-protein complex that mediates virtually all macromolecular traffic across the nuclear envelope. Nuclear pores are composed of multiple copies of ∼30 different nucleoporin proteins that contribute to its structure and function.1–4 Nucleoporins can be categorized into groups based on their location within the NPC. These groups include transmembrane nucleoporins (such as Pom121), scaffold nucleoporins such those in the large Nup107/160 or “Y” subcomplex, central scaffold spoke-ring nucleoporins (such as Nup53, Nup93, and Nup155), transport channel nucleoporins (such as Nup62 and Nup98), nuclear basket nucleoporins (such as Nup153 and Nup50), and cytoplasmic ring and filament nucleoporins (such as Nup214 and Nup358, respectively) (Fig. 1A).2,3, 5-12 Notably, approximately a third of all nucleoporins contain phenylalanine-glycine (FG) repeats, which are critical for the interaction with the soluble import and export receptors that mediate nuclear transport.13-15
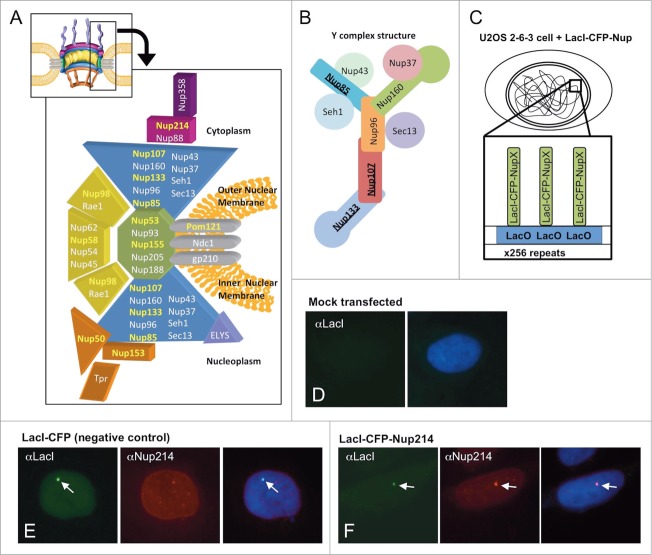
Figure 1.
(A) A cut-away schematic representation of the massive nuclear pore complex, which has 8 radial spokes, is shown (inset). The subcomplexes that comprise one radial spoke and their predicted localization within that spoke are shown in the enlarged portion of (A). The nucleoporins used as LacI-CFP fusions in this study are highlighted in yellow. Each separate shape indicates a different subcomplex that must assemble with others to form a nuclear pore. The different colors are used to demarcate distinct regions of the formed nuclear pore, such as the large Y complex subunits (blue), ELYS (lavender), the central scaffold (green), the central transporter regions (gold), the cytoplasmic ring (purple), the cytoplasmic filaments (deep purple), and the nuclear basket (light brown). The three transmembrane Nups are shown in gray. (A few nucleoporins such as Aladin and hCG1 have been omitted for clarity). (B) Details of the Nup107–160 or “Y” complex, with the nucleoporins tested as LacI-CFP fusions underlined. (C) Diagram of the LacI-LacO system used in this study. A diagram of a U2OS 2–6–3 cell is shown, with the LacO array integrated into one location in the genome. The enlargement shows the expected binding of a fusion LacI-CFP-NupX (green) to each of the multiple copies of the LacO array (blue). (D–F) U2OS 2–6–3 cells were mock transfected (D), transfected with the negative control fusion LacI-CFP (E), or transfected with LacI-CFP-Nup fusion such as LacI-CFP Nup214 (F). To test for LacI-CFP-NupX binding to the LacO array, cells were visualized with anti-LacI to detect the fusion protein (left panel in each set), or a specific anti-nucleoporin antibody (middle panel in E and F). DNA was detected with DAPI stain. The right-hand panel in each set shows a merge of all 3 stains. The arrows point to the position of the LacO array.
In metazoans, assembly of nuclear pores occurs at 2 distinct phases of the cell cycle: during interphase into an intact, double-membrane nuclear envelope, and at the end of mitosis when the nuclear envelope reforms around chromatin, termed post-mitotic assembly.16-20
Post-mitotic NPC assembly starts with the binding of several specific nucleoporins to chromatin. First, the DNA-binding nucleoporin ELYS binds via its DNA-binding AT-hook domain to chromatin,21,22 where it is required for the proper recruitment of the large Nup107/160 pore subcomplex to chromatin.19, 23–28 Indeed, ELYS depletion by RNAi or from in vitro nuclear reconstitution reactions results in nuclei devoid of NPCs, demonstrating that ELYS is essential for nuclear pore assembly on chromatin.22,23,26,29 Live imaging experiments of GFP-tagged nucleoporins in cultured cells have also revealed early recruitment of the Nup107/160 complex to mitotic chromosomes.30 In a separate study, using Xenopus nuclear reconstitution extracts with either lowered temperature or lipid inhibitors to manipulate the membrane-membrane fusion event of nuclear pore assembly, we found that the recruitment of ELYS, the Nup107/160 complex, and the transmembrane nucleoporin Pom121 occur as early steps in post-mitotic pore assembly and precede inner/outer nuclear membrane fusion and recruitment of the FG nucleoporins.31
The critical Nup107/160 subcomplex of the nuclear pore is formed in vertebrates from 9 nucleoporins (Fig. 1B; Nup160, Nup133, Nup107, Nup96, Nup85, Sec13, Seh1, Nup37 and Nup43).32-36 This subcomplex is a close relative of the S. cerevisiae subcomplex, also called the “Y” complex due to its overall shape.37-39 The Y complex is by far the largest subcomplex of both yeast and vertebrate nuclear pores. Depletion of Xenopus extracts with antibody to Y complex members (Nup133, Nup85 or Nup107), like ELYS depletion, leads to nuclei devoid of NPCs.40-42 The Y complex was recently shown to form 4 head-to-tail rings of 8 Y's each, with 2 rings situated on either face of the nuclear pore.6,43
Nup153, which comprises much of the nuclear basket of the NPC, has also been shown to potentially have an early role in NPC assembly.30, 44–47 The association of Nup153 with the forming nuclear pore in mitosis is biphasic: in live imaging studies, 10% of GFP-tagged Nup153 associates with the telophase chromosomes before many of the other Nups, suggesting a possible role in the initiation of NPCs (∼90% of Nup153 then associates with the pore later in the assembly process).30 Interestingly, the association of Nup153 with a fully-formed, interphase NPC is highly dynamic, with a residence time between 1 and 13 min.34,48 This is unlike the Y complex, which can be stably associated with the NPCs of non-dividing somatic cells literally for years.49–51 Thus, the proteins of the Y complex and Nup153, while quite different from one another in a large number of structural and functional aspects, both showed evidence for a role in early steps in nuclear pore assembly.30 Disparate findings such as these showed a clear need for further and more comprehensive analysis of the role of nucleoporins in NPC assembly.
Here we developed a direct in vivo approach to study the role of individual nucleoporins in NPC assembly. We used a previously characterized cell line, U2OS 2–6–3, which contains tandem copies of an E. coli LacO-containing DNA array stably integrated at a single locus in the human genome.52,53 We transfected these cells with constructs expressing individual nucleoporins tagged with both E. coli Lac repressor (Lac I) and cyan fluorescent protein (CFP) sequences. The LacI tag, binding with high affinity to the stably integrated LacO array, has the ability to target the tagged nucleoporin specifically to the LacO site. To study steps in pore assembly, we then examined the immobilized nucleoporin: (1) for its ability to recruit endogenous nucleoporins to the intranuclear LacO array and, separately, (2) for the ability to target the LacO/nucleoporin assemblage to the nuclear rim. This type of approach was used previously to investigate the de novo formation of intranuclear bodies such as Cajal bodies54 and paraspeckles.55,56 The LacI-Nup/LacO approach developed here has allowed us to detect intra- and inter-NPC subcomplex interactions at an ectopic nuclear site, and to compare the ability of different nucleoporins to initiate NPC assembly. An advantage of this system is that we can analyze Nup-Nup interactions in vivo without having to disrupt the endogenous nuclear pores by RNAi, a disruption that can lead to unwanted cell cycle defects, metabolic stress, and cell death.
Our data indicate that this system can detect specific Nup-Nup interactions, and that nucleoporins vary widely in their ability to promote extensive assembly. In addition, we find that specific Nup subcomplexes promote nuclear rim targeting of the LacO chromatin array, whereas others do not. Furthermore, the system allows study of the effect of specific nucleoporin disease mutations or truncations within an in vivo context of complex assembly and nuclear rim targeting.
Results
Development of a LacI/LacO system for analyzing nucleoporin interaction and assembly
To investigate the role of different nucleoporins in NPC assembly, we anchored individual LacI-CFP-tagged Nups to a specific ectopic nuclear site, with the thought that this nucleoporin/LacO-containing site would promote a potential “seed” site for the recruitment of other nucleoporins. We used the previously described human U2OS 2–6–3 cell line, which contains ∼200 copies of a cassette of 256 LacO repeats (see Methods) integrated into the euchromatic chromosome region 1p36.52
In order to specifically target different Nups to the LacO array, we created 11 fusion proteins of individual nucleoporins that were N-terminally tagged with both the E. coli Lac repressor (LacI) and cyan fluorescent protein (CFP) (Fig. 1C). The LacI protein is known to tightly and specifically bind to LacO sequences. Therefore, a LacI-CFP-nucleoporin fusion protein should localize intranuclearly to the LacO site and could be readily identified by fluorescence microscopy. The nucleoporins chosen for testing were Nups from different subcomplexes and regions of the nuclear pore (Fig. 1A). They included proteins of the cytoplasmic ring (Nup214), the Y complex (Nup133, Nup107, and Nup85), the central scaffold complex (Nup155 and Nup53), the central transporter region (Nup58 and Nup98), and the nuclear basket (Nup153 and Nup50).3,8,9,35,44, 57–59 We also created a soluble form of the transmembrane nucleoporin Pom121, which we term LacI-CFP-sPom121.60 All the LacI-CFP-Nup clones were sequenced to verify correctness, then transfected individually into U2OS 2–6–3 cells. We initially analyzed the different fusion proteins expressed post-transfection by immunoblot analysis and were able to verify the production of intact fusion proteins of the correct size for each construct (Supp. Fig. 1).
Next, immunofluorescence was performed on the transfected U2OS 2–6–3 cells. The LacI-CFP-Nup proteins were detected using either CFP fluorescence or indirect immunofluorescence with anti-LacI antibodies. We found that all the LacI-CFP-Nups tested were efficiently targeted to the LacO array, as visualized by appearance of a new discrete bright intranuclear focus after each transfection (Figs. 1E–F, green; Supp. Fig. 2). When cells were instead stained with antibodies against the transfected nucleoporin, if available, the nucleoporin signal always colocalized with the bright LacI-CFP spot marking the LacO array (Fig. 1F, right panel; Supp. Fig. 3). These results indicate that the LacI-Nup/LacO system can be used to immobilize individual nucleoporins onto the LacO chromatin site. When the negative control LacI-CFP fusion clone which has no added nucleoporin sequence was transfected into U2OS 2–6–3 cells, it targeted to the LacO array as expected (Fig. 1E, green), but did not recruit any of the nucleoporins we subsequently examined (Fig. 1E, red; Supp. Fig. 4). This result indicates that endogenous Nups do not bind to the LacO DNA array, to the LacI protein domain, or to the CFP protein domain. Thus, the LacI-Nup/LacO system is a powerful and highly specific tool for studying the ability of individual nucleoporins to bind and recruit other nucleoporins.
Detection of intra- and inter-NPC subcomplex interactions using the LacI/LacO system
We asked whether the individual LacI-Nup nucleoporins were able to recruit endogenous nucleoporins of their same subcomplex to the LacO chromatin location. This was done via immunofluorescence using antibodies to the different endogenous nucleoporins.
We found that the majority of nucleoporins tested were indeed able to recruit endogenous nucleoporins of their same subcomplex. For example, LacI-tagged Nup53, a member of the central scaffold subcomplex8,61,62 (Fig. 1A, green complex), recruited endogenous Nup93, a member of the same subcomplex, to the LacO array, as seen in 63% of the transfected cells (Fig. 2A; Table 1). LacI-tagged Nup155 from the same subcomplex,63,64 when transfected into U2OS-2–6–3 cells, could be seen to recruit endogenous Nup53 (46% of cells; Fig. 2C; Table 1). Thus, both LacI-Nups showed the ability to recruit known members of their own subcomplex.
Figure 2.
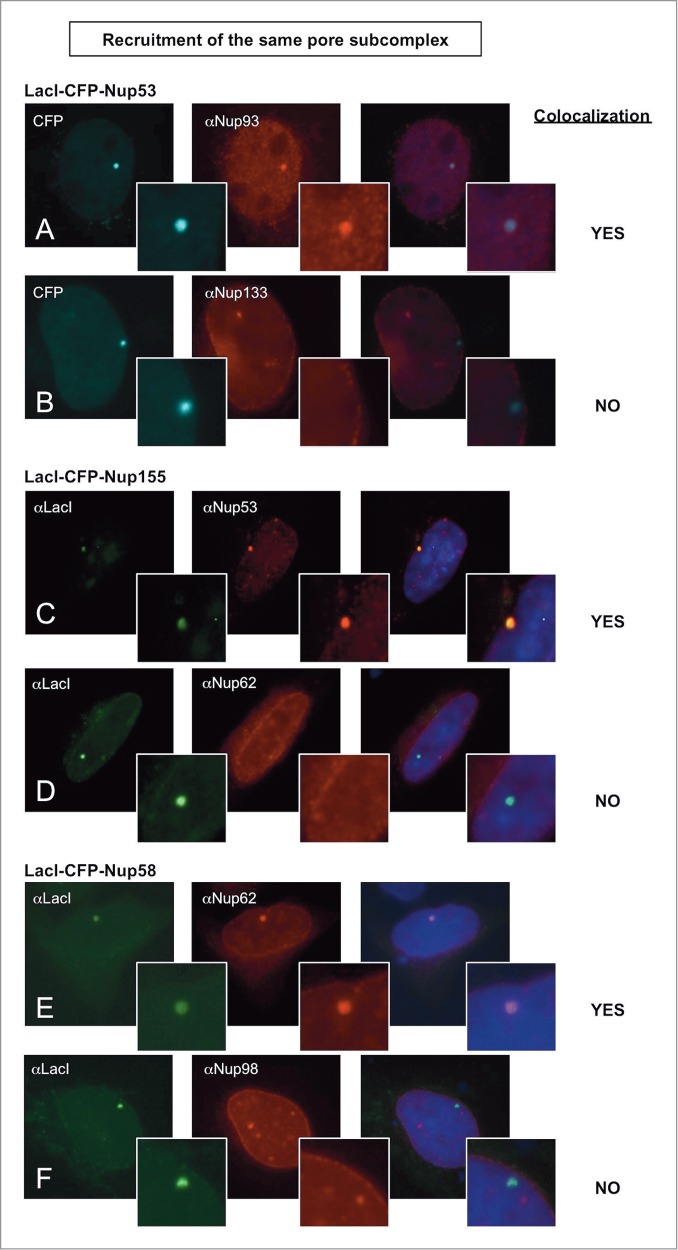
Nucleoporins anchored to the LacO array recruit other nucleoporins from the same subcomplex. Immunofluorescence microscopy of U2OS 2–6–3 cells transiently transfected with either LacI-CFP-Nup53 (A, B), LacI-CFP-Nup155 (C, D), or LacI-CFP-Nup58 (E, F) were stained with the indicated antibodies. Chromatin is visualized using DAPI stain. In panels (A) and (B), LacI-CFP-Nup53 was detected by CFP fluorescence. The right hand panels show a merged image of the 3 stains. The smaller insets show a magnified image of the LacO array in each image. Colocalization of the untagged endogenous Nup in question (middle panels) with the transfected LacI-CFP-Nup being tested (left panels) is summarized at the right.
Table 1.
Summary of nucleoporin interactions and LacO rim targeting. The nucleoporins tested by IF are shown below. The percentages show the amount of colocalization of an endogenous Nup with the LacI-Nup/LacO array, calculated as the percent of transfected cells showing colocalization (triplicate experiments, minimum N = 100 cells per condition). Targeting of the LacO array to the nuclear rim is also shown (last column), calculated as percent of transfected cells with the LacO focus at the rim. The negative control, LacI-CFP, recruited no endogenous nucleoporins to the LacO array and showed no rim targeting, indicating high specificity of the LacI-Nup/LacO system.
| LacI-tagged | Intra-complex interactions | Inter-complex interactions | Low or not recruited | Rim targeting | ||
|---|---|---|---|---|---|---|
| Nup133 | Nup160 (44%) | ELYS (62%) | Nup214 (23%) | Nup62 (5%) | Yes (57%) | |
| Nup85 (86%) | Nup53 (20%) | Nup98 (70%) | ||||
| Nup93 (21%) | Pom121(45%) | |||||
| |
|
mAb414 (18%) |
|
|
|
|
| Nup107 | Nup133 (94%) | ELYS (72%) | Nup62 (24%) | Sometimes (25%) | ||
| Nup160 (39%) | Nup93 (9%) | Pom121 (26%) | ||||
| |
Nup85 (88%) |
mAb414 (20%) |
|
|
|
|
| Nup153 | N/A | Nup133 (35%) | Nup93 (23%) | Yes (76%) | ||
| Nup160 (33%) | Nup214 (6%) | |||||
| Nup85 (58%) | Nup62 (56%) | |||||
| |
|
ELYS (65%) |
Nup98 (11%) |
|
|
|
| Nup53 (14%) | Pom121(31%) | |||||
| Nup50 | N/A | Nup133 (11%) | Nup62 (43%) | Nup160 (4%) | Sometimes (30%) | |
| Nup85 (32%) | mAb414(46%) | |||||
| ELYS (37%) | Nup98 (11%) | |||||
| Nup53 (6%) | Pom121(24%) | |||||
| |
|
Nup93 (28%) |
|
|
|
|
| Nup214 | Not tested | Nup133 (35%) | Nup62 (97%) | Nup53 (0%) | No (0%) | |
| ELYS (40%) | Nup98 (90%) | |||||
| |
|
mAb414 (100%) |
|
|
|
|
| Nup53 | Nup93 (67%) | ELYS (34%) | Nup133 (3%) | Nup62 (2%) | Sometimes (27%) | |
| |
|
|
|
mAb414 (3%) |
Pom121 (0%) |
|
| Nup155 | Nup53 (46%) | Pom121 (56%) | Nup133 (3%) | Nup62 (3%) | Sometimes (16%) | |
| Nup160 (5%) | Nup214 (5%) | |||||
| |
|
|
|
Nup93 (2%) |
Nup98 (2%) |
|
| sPom121 | N/A | Nup133 (8%) | Nup62 (89%) | Nup160 (0%) | No (1%) | |
| Nup85 (7%) | mAb414 (20% | Nup53 (2%) | ||||
| ELYS (14%) | Nup98 (72%) | |||||
| |
|
Nup93 (68%) |
|
|
|
|
| Nup58 | Nup62 (100%) | ELYS (13%) | Nup133 (3%) | Nup21 (1%) | No (1%) | |
| Pom121 (14%) | Nup160 (5%) | Nup98 (5%) | ||||
| |
|
|
|
Nup85 (2%) |
|
|
| mNup85 | ELYS (9%) | Nup133 (0%) | Nup98 (0%) | No (1%) | ||
| Nup160 (2%) | Pom121 (4%) | |||||
| |
|
|
|
Nup53 (0%) |
|
|
| Nup98 | Not tested | ELYS (16%) | Nup133 (2%) | mAb414 (4%) | No (1%) | |
| |
|
Nup62 (8%) |
|
Nup53 (0%) |
Nup214 (1%) |
|
| CFP | Nup133 (0%) | mAb414 (0%) | No (1%) | |||
| Nup160 (0%) | Nup62 (0%) | |||||
| Nup85 (0%) | Nup214 (0%) | |||||
| ELYS (0%) | Nup98 (0%) | |||||
| Nup53 (0%) | Pom121 (0%) | |||||
| Nup93 (0%) | Nup155 (0%) | |||||
Similarly, when a LacI-tagged nucleoporin from the Nup62 central transporter subcomplex,7,65,66 Nup58, (Fig. 1A, middle gold complex) was transfected, it recruited Nup62 to the LacO array in 100% of the transfected cells (Fig. 2E; Table 1). Indeed, the ability to recruit nucleoporins from the same subcomplex was widespread, including LacI-Nup133 and Nup107 recruitment of other Y complex Nups (Supp. Figs. 5A–B; Table 1).
What about the ability to recruit other subcomplexes? One subset of Nups did not extensively recruit other Nup subcomplexes. Two members of the central scaffold complex, Nup53 and Nup15561 were both inefficient in recruiting nucleoporins from the Y complex (≤5% of cells) or recruiting Nup62 from the central transporter region (2–3% of cells) (Figs. 2B and D, respectively; Table 1). Interestingly, Nup53 was seen to unexpectedly recruit the nucleoporin ELYS in 34% of cells, identifying a novel interaction (Table 1). Overall, Nup53 and Nup155 were limited in their ability to recruit other nucleoporin subcomplexes (Table 1).
However, multiple LacI-tagged Nups could recruit other subcomplexes. For example, LacI-tagged Nup214, a protein of the cytoplasmic ring of the NPC,9 recruited endogenous Nup133 (Fig. 3A) and ELYS in 35% and 45% of cells, respectively (Table 1).
Figure 3.
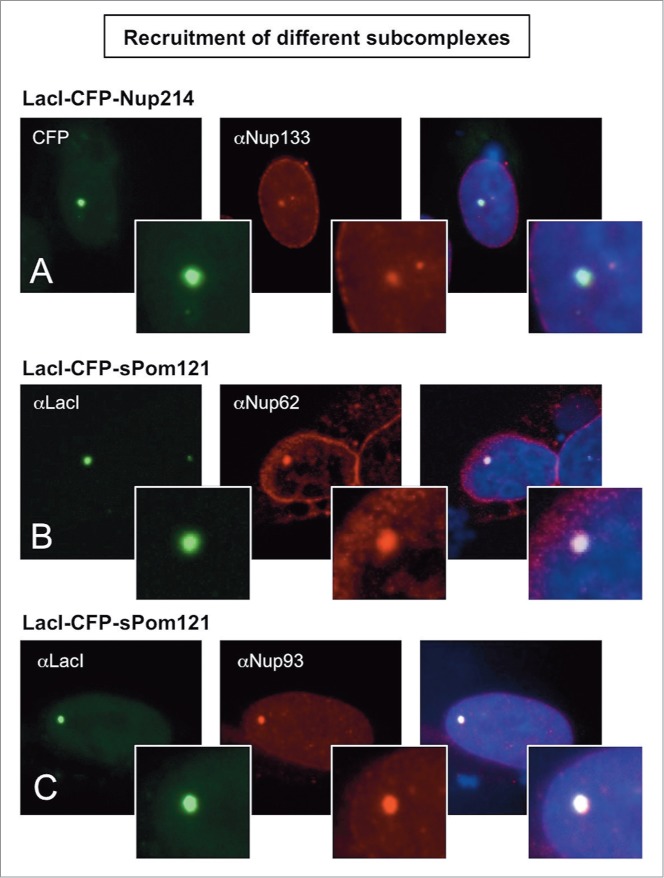
Recruitment of nucleoporins from other NPC subcomplexes to the LacO array. Immunofluorescence microscopy of U2OS 2–6–3 cells transiently transfected with either LacI-CFP-Nup214 (A) or LacI-CFP-Pom121 (B, C), then stained with antibody against LacI or visualized by CFP fluorescence (left panel). The endogenous nucleoporins Nup133 (A), Nup62 (B), or Nup93 (C) were detected with specific antibodies (middle panel). Chromatin is visualized by DAPI staining. The smaller insets show a magnified image of the LacO array in each image. The right hand panels are a merge of the previous 2 images with DAPI staining. All three cases show examples of positive recruitment of an endogenous nucleoporin from a different subcomplex than that of the LacI-CFP-NupX. Full results are summarized in Table 1.
Using the LacI/LacO system, we were also able to study the interaction of the normally integral membrane pore protein, Pom121,64, 67-70 with other Nups and Nup subcomplexes – and to do so away from its normal membrane environs. We observed that LacI-tagged soluble Pom121 expressed in U2OS 2–6–3 cells efficiently recruited Nups from different subcomplexes to the LacO array, including recruitment of Nup93, Nup98, and Nup62 (71, 74, and 89% of cells, respectively) and, with lower efficiency, recruitment of the Y complex and Nup53 (2–14%) (Figs. 3B-C and Table 1).
We conclude that the LacI/LacO system can be used to confirm known interactions within the nuclear pore as well as to propose novel Nup-Nup interactions in an in vivo situation. Moreover, this system allows one to do so without the need for immunoprecipitation or pulldown. Future studies can assess whether such novel interactions are direct or indirect, i.e., whether they occur via an intermediary subcomplex.
Both the Y complex and Nup153/Nup50 nuclear pore basket complex can recruit a near full complement of nucleoporins
Strikingly, LacI-tagged Nup133 of the Y complex was able to efficiently recruit members of all the endogenous nucleoporin subcomplexes that were tested. This included the Y complex nucleoporins Nup160 and Nup85, ELYS, the central scaffold proteins Nup53 and Nup93, the cytoplasmic ring protein Nup214, the central transporter region protein Nup98, and Pom121 (Figs. 4A-G and Table 1; 44, 86, 62, 20, 21, 23, 70, and 45%, respectively). Lower recruitment of the central transporter protein Nup62 was also observed (Table 1, 5% of cells). When a second member of the Y complex was examined, LacI-tagged Nup107, it also efficiently recruited all the endogenous nucleoporins we tested, which included Nup133, Nup160, Nup85, ELYS, Nup62, Pom121, and Nup93 (Supp. Figs. 5A–D; Table 1; 94, 39, 88, 72, 24, 26, and 9%, respectively).
Figure 4.
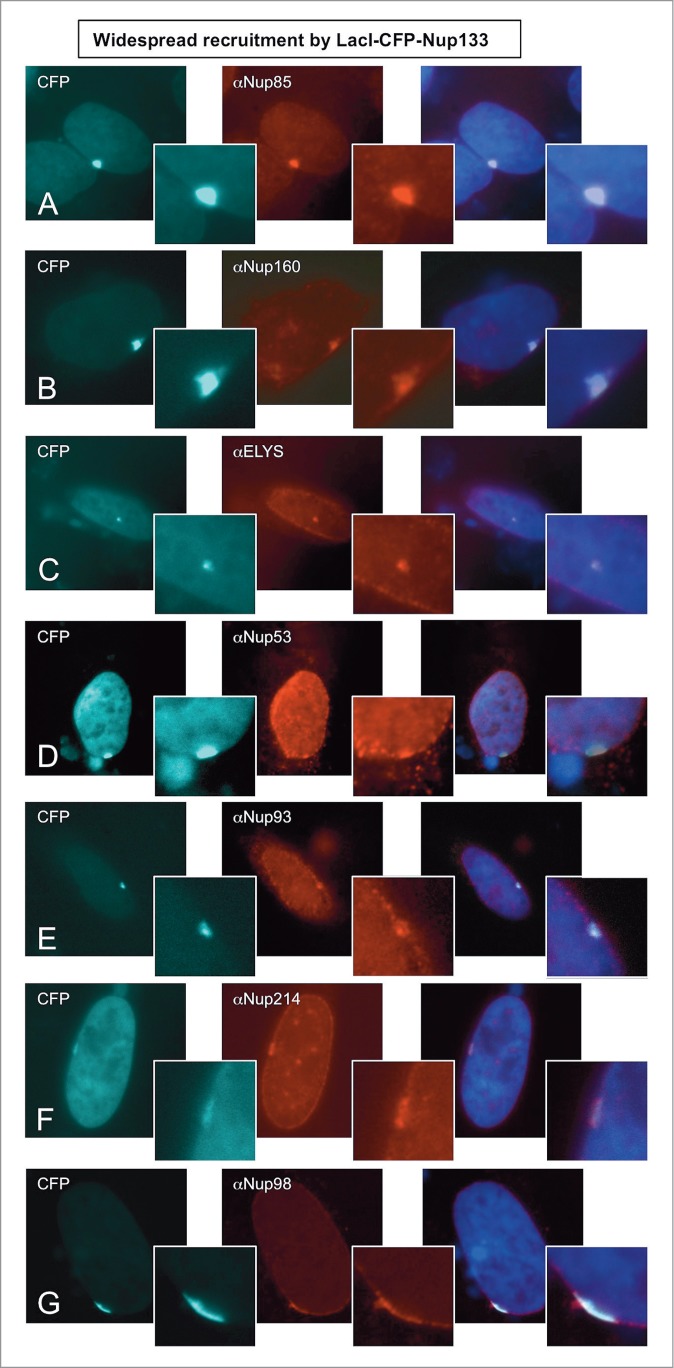
Nup133 is able to recruit an extensive array of nucleoporins to the LacO array. Immunofluorescence microscopy was performed on U2OS 2–6–3 cells transiently transfected with LacI-CFP-Nup133. The location of LacI-CFP-Nup133 was visualized by CFP fluorescence (left panel). To assay for recruitment of endogenous nucleoporins, the transfected cells were stained with antibody against: (A) Nup85, (B) Nup160, (C) ELYS, (D) Nup53, (E) Nup93, (F) Nup214, and (G) Nup98, followed by a Texas Red-labeled secondary antibody (middle panel). DNA was visualized by DAPI staining. The smaller panels show a magnified image of the LacO array. The right hand panels are a merge of the previous 2 images together with a DAPI-stained image. We note that when Nup133 was used as the LacI-CFP-tagged fusion protein, a large percentage of the transfected cells showed rim localization of the LacO array. Quantitation is shown in Table 1.
Surprisingly, Nup153 of the pore basket also recruited a large set of nucleoporins. LacI-tagged Nup153 recruited to the LacO array endogenous Nup160, Nup133, and Nup85 of the Y complex, ELYS, Nup53, Nup93, Nup62, Nup98, and Pom121 (Figs. 5A-D and Table 1; 33, 35, 58, 65, 14, 23, 56, 11, and 31%, respectively). LacI-tagged Nup50, a partner of Nup153 in the nuclear basket, also showed recruitment of Nup160, Nup133, Nup85, ELYS, Nup53, Nup93, Nup62, Nup98, and Pom121 (Table 1; 4, 11, 32, 37, 6, 28, 43, 11, 24%, respectively).
Figure 5.
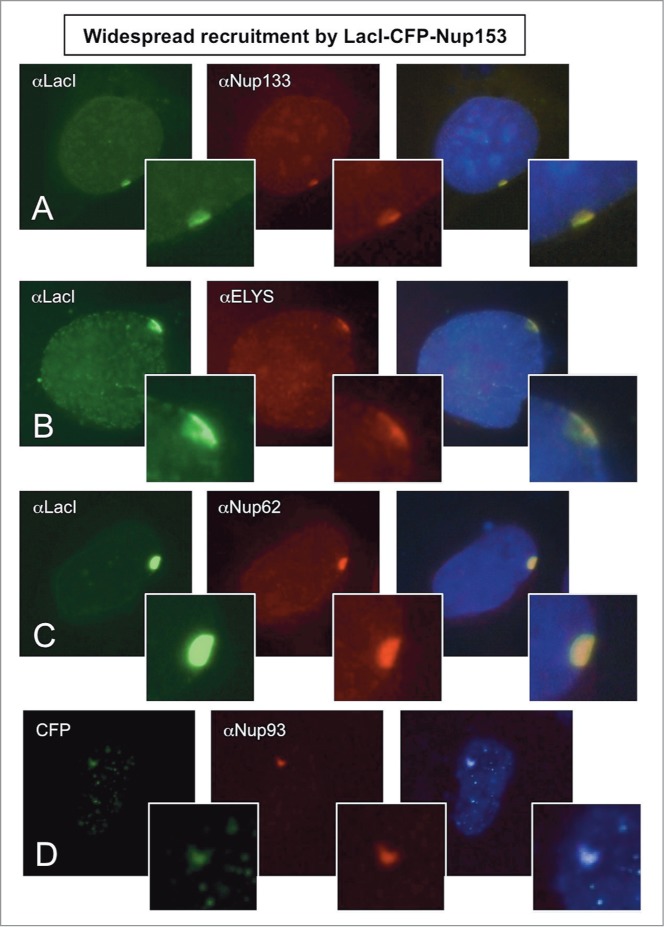
Widespread recruitment of nucleoporins to the LacO array by the LacI-CFP-Nup153 fusion protein. Immunofluorescence microscopy of U2OS 2–6–3 cells transiently transfected with a LacI-CFP-Nup153 construct. The location of LacI-CFP-Nup153 was visualized by CFP fluorescence or staining with anti-LacI antibody (left panel). Endogenous nucleoporin recruitment was assessed with antibody against: (A) Nup133, (B) ELYS, (C) Nup62, or (D) Nup93 (middle panel). Chromatin was visualized by DAPI staining. The smaller panel shows a magnified image of the LacO array for each image; the right hand panel is a merge of the previous 2 images with the DAPI DNA stain. Data for additional nucleoporins tested as well as the quantitation for colocalization and rim localization for all tested nucleoporins are shown in Table 1.
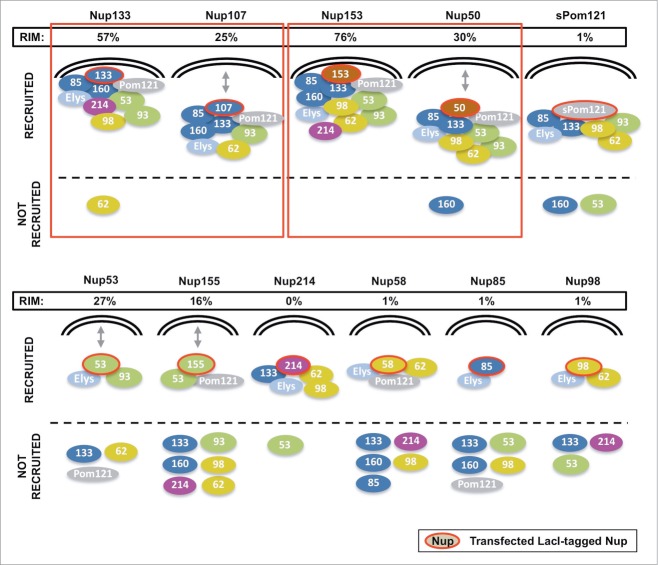
These results indicate that Nup133 and Nup107 of the Y complex and Nup153 and Nup50 of the nuclear pore basket, when anchored to chromatin, are each able to seed the formation of a structure that includes almost all of the nucleoporins we tested.
LacI-tagged Nup133 and -Nup153 efficiently target the LacO chromatin array with recruited nucleoporins to the nuclear rim
For many LacI-tagged Nups, the LacO array in the U2OS 2–6–3 cells appeared as a single spot in the nuclear interior. Such an internal localization is identical to what is seen for the negative control fusion construct, LacI-CFP, which contains no nucleoporin sequence (Fig. 1E, green; Table 1). We observed internal localization for the LacI-tagged fusions of Nup214 (Fig. 3A), Nup98, Nup58 (Figs. 2E–F), Nup85, and soluble Pom121 (Figs. 3B–C) (see Table 1 for quantitation).
Strikingly, LacI-tagged Nup133 and LacI-tagged Nup153 targeted the LacO array and their extensive set of recruited nucleoporins to the nuclear envelope in 57% and 76% of the cells, respectively (Figs. 4 and 5; Table 1). Their respective partners, Nup107 of the Y complex, and Nup50, the partner of Nup153, also targeted the LacO array to the nuclear envelope, albeit in a lesser fraction of the cells (25–30%; see, for example, Supp. Fig. 5D; Table 1).
Interestingly, targeting of the LacO array to the nuclear periphery was also seen with a third subcomplex of Nups: LacI-tagged Nup53 and its partner Nup155, where 27% and 16% of cells, respectively, targeted to the rim (Table 1). This rim targeting indicates that these latter nucleoporins, while each able to recruit only one other nucleoporin subcomplex tested, are nevertheless able to either directly interact with the nuclear membrane, or interact with an intermediating factor that can target them and the LacO array to the nuclear membrane. Nup53 does have the ability to interact with membranes,62 but Nup155 has not been described to have such activity. The LacI/LacO system could prove valuable for future analysis of what the intermediating factor might be.
Together, the above results demonstrate that both Nup133 and Nup153, when targeted to an ectopic site such as the LacO array, are able to recruit an extensive number of nucleoporins to a structure and, importantly, to efficiently localize the structure formed to the nuclear envelope, suggesting that these 2 nucleoporins are able to initiate nuclear pore complex assembly at the correct location.
Combining domain analysis with the LacI/LacO system allows dissection of the role of specific domains in Nup-Nup interaction and nuclear envelope targeting
We reasoned that the LacI/LacO system should also allow us to dissect – in vivo – the role of different structural domains of a given nucleoporin in binding to other nucleoporins. Toward this goal, we examined Nup133, the nucleoporin that shows the most extensive subcomplex recruitment in our study as well as having a high ability to target the resulting Nup/LacO array to the nuclear envelope. Nup133 is known to map to the base of the Y complex37 (Fig. 1B). Nup133 has 2 major structural features: a C-terminal α-helical domain that contains the binding site for Nup107 (aa 934–980) that mediates interaction of Nup133 with the rest of the Y-complex, and a conserved N-terminal β-propeller domain, a type of domain thought to mediate protein-protein interaction.71,72 Indeed, the S. cerevisiae β-propeller domain of Nup133 has been shown to interact with Nup120, the yeast Nup160 homolog.73 In addition, the Nup133 N-terminus contains a putative membrane curvature-sensing motif, termed the ALPS motif, which when mutant decreases localization of Nup133 to the nuclear pore.19,74
To test the role of the N- and C-terminal domains in recruiting nucleoporins to the LacO array, fusion LacI-CFP constructs of these domains, termed Nt-Nup133 and Ct-Nup133 were made and transfected separately into U2OS 2–6–3 cells (Fig. 6A). We found that the Ct-Nup133 fragment (aa 917–1141), which contains the Nup107 binding site, was very efficient at recruiting certain nucleoporins recruited by full length Nup133, i.e., Nup160, ELYS, Nup98 and Pom121 (52–79%, Figs. 6C and D). The Ct domain did not, however, significantly recruit Nup214 or Nup53 (2–3%; Fig. 6D), while the full length LacI-tagged Nup133 had recruited them at 20–23%. When the Nt fragment of Nup133 (aa 1–805) was tested, it failed to recruit any of these nucleoporins except Pom121 (14%; Fig. 6B and D). We conclude that the binding site for Nup107 on Nup133 Ct is critical for Nup133 to recruit other nucleoporins to the LacO array. This further suggests that the nucleoporin recruitment by LacI-tagged full-length Nup133 that we previously observed (Fig. 4) is occurring through a Y complex-initiated mechanism.
Figure 6.
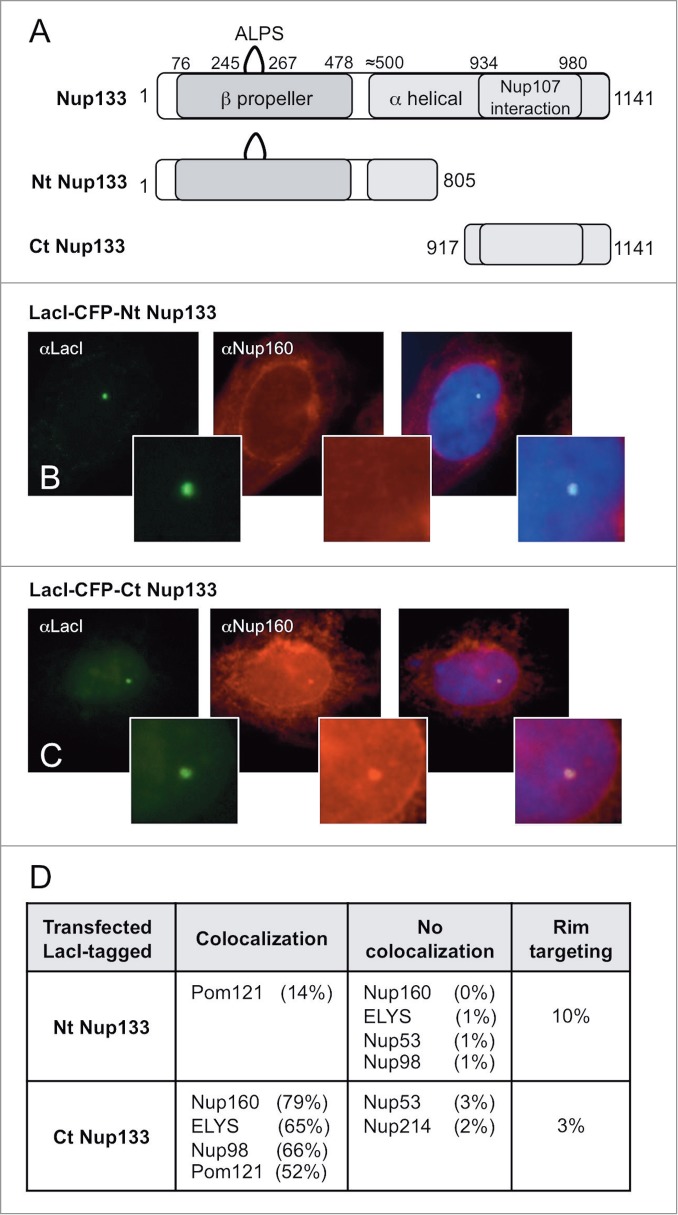
Nup133 N- and C-terminal protein fragments bind nucleoporins differentially. (A) Full length human Nup133 (1141 aa) and specific mapped domains are shown. The N-terminal, designated Nt-Nup133, consisted of 805 aa and contained the β-propeller domain, the ALPS motif, and a portion of the α-helical domain. The C-terminal Ct-Nup133 construct consisted of aa 917–1141 and contained the site for Nup107 interaction (aa 934–980). (B–C) U20S 2–6–3 cells were transfected with the designated Nup133 fragment. The extent of nucleoporin recruitment was then tested by staining with anti-LacI antibody together with antibodies to Nup160 (B-C) or Pom121, ELYS, Nup53, Nup98, and Nup214 (D). The percentage of transfected cells showing rim localization is shown in last column of (D). The comparative data for full length Nup133 is presented in Table 1.
Interestingly, neither the Nt-Nup133 nor the Ct-Nup133 was observed to target the LacO array to the nuclear rim to any large extent (10% and 3% of cells, respectively; Fig. 6D), in clear contrast to the efficient nuclear rim-targeting of the full length LacI-tagged Nup133 construct, seen in 57% of cells (Table 1). We conclude from these results that both the Nt and Ct domains in full length Nup133 must contribute to promoting efficient nuclear rim targeting of the LacO array.
Analysis of defects in Nup recruitment and rim localization caused by the human cardiac arrhythmia mutation Nup155 R391H
A predicted advantage of the LacI-Nup/LacO system is its potential to dissect the mechanism by which a specific nucleoporin mutation disrupts function, by analyzing the effect of the disease mutation on Nup-Nup interaction and nuclear envelope targeting. We focused on the human Nup155 point mutation, R391H, which has been found to lead to atrial fibrillation and early sudden cardiac death in both mice and humans.75
Nup155, similar to Nup133, is predicted to contain 2 distinct regions based on the structure of its homologues in yeast Nup157/170: an N terminal β-propeller, which has been shown to interact with other nucleoporins such as Pom121 and Nup53, and a C-terminal α-solenoid.64, 76–79 The human R391H mutation, located in the predicted β-propeller, has been shown to adversely affect the ability of Nup155 R391H to localize to the nuclear pore in vivo and, furthermore, functionally blocks the nucleocytoplasmic transport of both hsc70 mRNA and hsc70 protein.75 The molecular basis for the mutant's lack of binding to the nuclear pore has been unknown.
Above we observed that a wild type LacI-tagged Nup155 strongly interacts with Nup53 and Pom121 (Fig. 2C; Fig. 7A; Table 1), consistent with the previously published Nup155-interacting nucleoporins.8,64 To ask whether introduction of the R391H disease mutation impaired the ability of Nup155 to interact with these Nup partners, LacI-tagged Nup155 R391H was constructed. When the mutant Nup155 was transfected into U2OS 2–6–3 cells, no colocalization of Nup53 was observed at the LacO array (Figs. 7B and C). Colocalization of Pom121 with the Nup155 R391H/LacO array was also greatly reduced (∼8% of cells as compared to 56% colocalization with wild type LacI-Nup155) (Fig. 7C).
Figure 7.
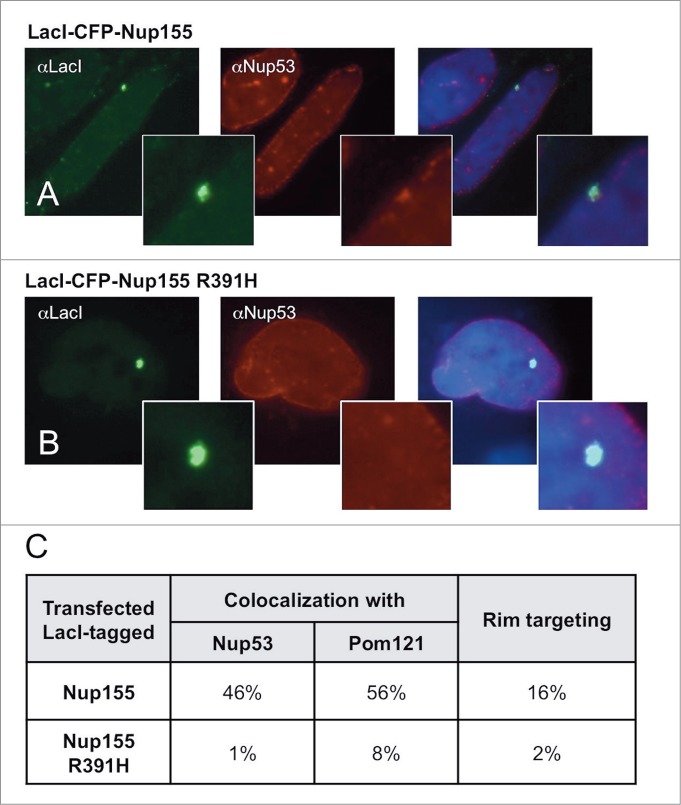
The Nup155 R391H mutation that causes human cardiac arrhythmia abrogates binding of its normal nucleoporin partners and rim localization. U2OS 2–6–3 cells were transfected with a LacI-CFP construct of full-length wild type human Nup155 and the R391H mutant version of the same construct. The transfected cells were tested by immunofluorescence for colocalization of Nup53 or Pom121 with the wild type or mutant Nup155. Note that Nup53 and Pom121 were the only tested nucleoporins that had been seen to be recruited to wild type LacI-CFP-Nup155 (see Table 1). Here Nup53 recruitment can be observed with wild type LacI-CFP-Nup155 (A), but not with the R391H mutant Nup155 (B). (C) Quantitation for the level of Nup53 and Pom121 colocalization and rim targeting is shown for the wild type and mutant Nup155 constructs.
Moreover, the presence of the Nup155 R391H point mutation almost completely prevented nuclear rim localization of the LacO array (2% instead of 16% with wild type LacI-Nup155; Fig. 7C). We conclude that: (1) the cardiac arrhythmia mutation R391H disrupts the ability of Nup155 to interact with Nup53 and Pom121, and (2) this lack of interaction is likely to be the cause for lack of localization to the nuclear rim.
Discussion
In this study we adapted a highly efficient LacI-LacO system52,53 for the study of nucleoporin-nucleoporin interactions and of initiation of steps in nuclear pore complex assembly. Our system, where nucleoporins are anchored to a specific ectopic site on chromatin, enables us to study assembly in the nuclear environment and to examine the ability of different nucleoporins to induce localization of the forming structure to the nuclear envelope. We focused on 11 different nucleoporins derived from multiple distinct NPC subcomplexes. We found that most of the tested nucleoporins recruited Nups from their own subcomplex to the LacO array. A number also recruited neighboring subcomplexes (Table 1; Intra- and Inter-complexes). Strikingly, certain Nups were capable of initiating much more inclusive assembly as well as rim localization, as summarized in Figure 8. The system in addition allowed targeted analysis of the role of individual Nup domains in promoting assembly and rim localization, as well as providing a unique platform for dissecting the functional consequences of disease mutations in human cells.
Figure 8.
A schematic summary of the recruitment and rim localization results for each LacI-CFP-Nup tested. The individual LacI-tagged Nup construct transfected (outlined in red) is indicated above each drawing. Endogenous nucleoporins tested that were recruited to the LacI-Nup/LacO array are shown above the dotted line, while the non-recruited Nups are shown below the dotted line. Nups belonging to the same subcomplex are in the same color (and correlate with the subcomplexes in Fig. 1A). Rim localization of the LacI-Nup/LacO array (% of cells) is denoted by close proximity of the recruited nucleoporins to the double nuclear membrane lines, and also by the % of cells that showed rim localization of the array (RIM: %). Double headed arrows indicate the Nups where the LacI-Nup/LacO array was found adjacent to the membranes, but in lesser percentages. A number of LacI-Nup/LacO arrays (sPom121, Nup214, Nup58, Nup85, and Nup98) showed essentially no rim localization. Red squares denote the members of the 2 subcomplexes that are able to initiate both extensive Nup recruitment to the array and rim localization of the array.
The Y complex and Nup153/Nup50 were each found to initiate extensive Nup recruitment and nuclear rim targeting
Significantly, when the Y complex protein Nup133 was tagged with LacI-CFP, it recruited a near full complement of nuclear pore subcomplexes (Fig. 8, Table 1). In addition, the LacO/Nup133 assemblage became targeted to the nuclear rim in a majority of transfected cells (57%, Table 1, right column). Nup107, another member of the Y complex, when targeted to the LacO array by LacI, was similarly able to recruit multiple nucleoporin subcomplexes. However, the Nup107-initiated structure localized to the nuclear rim in only 25% of cells. Nup133, as the base of the Y complex, might be concluded from this data to be more ideally suited to initiate recruitment and to be able to do so in a more hierarchical, ordered manner.
Unexpectedly, the basket nucleoporin Nup153 was also able to efficiently recruit a near full complement of nucleoporin subcomplexes and target the assemblage to the nuclear envelope (76% of cells) (Fig. 8, Table 1). In an in vivo time course done previously in HeLa cells, a fraction of GFP-Nup153 was seen to be recruited to telophase chromosomes very early in the formation of nuclear pores.30,80 Recruiting Nup153 onto chromatin in our study might either substitute for the need for ELYS to initiate nuclear pore assembly or, alternately, recruit the Y complex with bound ELYS, and in that way seed nucleoporin assembly. In the HeLa study, recruitment of a small fraction of Nup50, partner to Nup153, was also seen early in mitotic NPC assembly,30 which suggested that Nup50 too might be part of an early nuclear pore intermediate. Indeed, we found that LacI-tagged Nup50 was able to recruit many of the subcomplexes recruited by Nup153 (Fig. 8), although to a lesser extent particularly in case of the Y complex (Table 1). It is interesting to point out that all 3 putative “seeds” found to have the capacity to initiate pore assembly, i.e., ELYS,22,23,26,27 the Y complex, and Nup153/50, are found on the nuclear face of the NPC.
The central scaffold proteins, Nup53 and Nup155, recruit few nucleoporins but can induce rim localization of the LacO array
Prior to this study, we might have predicted that targeting of the LacO array to the nuclear rim would require a large and complex set of pore proteins, as seen above. However, we found that LacI-tagged Nup155 and Nup53, members of the central scaffold subcomplex (Fig. 1A), were able to target the LacO array to the nuclear rim in 16 and 27% of cells. This was surprising to us, especially given our initial prediction, because we found that neither of these Nups recruited more than one nucleoporin, at least of those we tested, other than from their own subcomplex (Fig. 8). In the case of Nup155, the explanation could lie in the fact that the only nucleoporin from a different subcomplex that it efficiently recruited was the endogenous transmembrane nucleoporin Pom121 (56% of cells), previously shown biochemically to directly bind to Nup155.64 Nup53, on the other hand, showed no Pom121 recruitment (0%), but Nup53 itself has been shown to have membrane-binding regions and also to interact with lamin B, which could more readily explain its nuclear membrane/rim targeting in the face of very few other nucleoporins.8,61,62 In sum, because of their very limited ability to recruit the majority of nucleoporins, we think that – with respect to nuclear pore assembly – Nup155 and Nup53s rim-targeting may simply represent dead-end pathways, i.e., Nup155 and Nup53 cannot act as “seeds” for further assembly.
Soluble Pom121 cannot act as an efficient seed in the LacI/LacO system
Pom121 is a critical, transmembrane FG-nucleoporin essential for interphase NPC assembly in vivo.5,19,20,64,67,69 LacI-CFP-sPom121, a soluble form of Pom121, when immobilized to the LacO array in our study recruited multiple nucleoporins, including the FG Nup, Nup62 (89% of cells), which was previously observed to be recruited in a study where Pom121 was inserted into the mitochondrial membrane of HeLa and 3T3 cells.68 LacI-tagged sPom121 also efficiently recruited the FG nucleoporin Nup98 (72% of cells) and the central scaffold protein Nup93 (68% of cells). However, soluble Pom121 did not substantially recruit Nup53 (2% of cells; a partner of Nup93), or proteins of the Y complex or ELYS (0–14% of cells). These results indicate that in our system the soluble form of Pom121 cannot act as a major initiator of Nup recruitment.
Stochastic, hierarchical, or seeded assembly?
The different mechanisms of assembly of multi-subunit structures have been addressed and categorized in a recent review, and fall into 3 major types of assembly: stochastic, hierarchical, or seeded.56 For a stochastic assembly process, theoretically any subunit can initiate the recruitment and assembly of all the other subunits into a final full structure.56 Stochastic assembly has been proposed for Cajal bodies in a previous study that used the LacI/LacO system in U2OS 2–6–3 cells,54 although certain caveats still remain.56 For an ordered, hierarchical assembly mechanism, a very defined pathway of assembly is followed, with each subunit being recruited in a specific order. Lastly, for seeded assembly, one or, at most, a few different subunits have the ability to initiate or “seed” assembly of the majority of the structure, while all the others do not. However, with this model, after the initial “seeding event," the remainder of assembly could theoretically be stochastic (any downstream pathway is OK) or ordered (a specific downstream order of assembly must be followed).
Given the bulk of LacI-tagged nucleoporins that we tested were unable to initiate the recruitment of most of the other subcomplexes, this argues against a stochastic mechanism of assembly for nuclear pore structure.
Two nucleoporins, Nup85 of the Y complex and Nup98, had the lowest ability to recruit other nucleoporins. One possibility is that the LacI-tagging of their N-termini disrupted their structure or their ability to interact with other Nups. LacI-tagged Nup85 did indeed show low ability to incorporate into endogenous nuclear pores of U2OS cells (Supp. Fig. 6E), potentially indicating a defective Nup85. However, LacI-Nup98 efficiently incorporated into endogenous nuclear pores of U2OS cells (Supp. Fig. 6G), which indicates that the LacI-Nup98 maintains its ability to interact with nucleoporins. We thus believe that LacI-Nup98s observed recruitment to the LacO array of only Nup62 (8% of cells) and ELYS (16% of cells) (Table 1) further contributes to the argument against a stochastic model for nuclear pore assembly.
Importantly, the finding that proteins of 2 different complexes, Nup133 (the base of the Y complex) and Nup153 (critical for formation of the nuclear pore basket), could initiate recruitment of most nucleoporin subcomplexes points toward a seeding model with 2 different molecular seeds possible. In the case of the Y complex, our observations using the LacI/LacO system, are consistent with the critical roles proposed for the Y complex and its chromatin adaptor ELYS in studies. ELYS, the AT-rich DNA-binding protein identified as a “seed” in post-mitotic studies and important to the targeting of the forming complex to chromatin,19,22,23,26,27 would thus map upstream of the 2 seeds that act in our ectopic studies, or be a parallel seed. The two subcomplexes identified as being able to act as seeds in the LacI/LacO system, where they are directly tethered to chromatin, could be predicted to be recruited under normal cellular conditions after the DNA-binding NPC initiator ELYS.
What are the recruited entities? - Individual proteins and subcomplexes
A clear question involves the nature of the recruited entities. Nup133, Nup107, and Nup153 all appear to recruit the 3 tested members of the Y complex and ELYS. This argues that the recruited entity in this case is likely the full Y complex itself. On the other hand, LacI-tagged Nup155, a member of the central scaffold complex (Fig. 1A; Nup155/53/93/205/188), when anchored to the LacO array, efficiently recruited Nup53 (46% of cells), but only recruited Nup93 to a low level (2% of cells). This latter result argues that certain Nup subcomplexes are less stable, since we see selective protein recruitment. We conclude that both individual pore proteins and full subcomplexes can be recruited, depending on the stability or steric conformation of their specific molecular interactions.
FG-based interactions may also make a contribution to nucleoporin recruitment to the LacO array. For instance, as described above, the soluble form of Pom121, which contains FG repeats, was observed to recruit 2 other FG-rich nucleoporins, Nup62 and Nup98, to the LacO array. FG repeats are hydrophobic and their interaction is known to be disrupted by the addition of 1,6-hexanediol or other aliphatic alcohols.14,81,82 As such, hexanediol may in future help reveal the contribution of FG-repeats to nucleoporin recruitment.
With respect to the nature of the large structures formed at the nuclear rim, different possibilities can be imagined: a large cluster of fully formed nuclear pores embedded in the nuclear membranes with the LacO DNA array on the inner face of the nuclear envelope, a structure containing most or all of the nucleoporins but not perforating the nuclear membranes, and a structure containing only a small set of subcomplexes, at least one of which has membrane-binding capability (as in Nup155/53-initiated structures). Determining which structure each LacI-tagged Nup initiates will require careful electron microscopic analysis, a study beyond the scope of this present work.
The system allows for detection of highly specific interactions and domain dissection
Beyond addressing large questions in nuclear pore assembly, the LacI-Nup/LacO system proves quite useful for approaching specific molecular questions. By examining different subdomains, we were able to distinguish roles for individual Nup133 domains and compare them to full length Nup133. The efficient binding of Nup160, ELYS, and Nup98 by the C-terminal domain, Ct Nup133 (65–79% of cells), and lack of binding of these by the N-terminal domain, Nt Nup133 (0–1% of cells), highlights the fact that the system can detect Nup-Nup interactions with a very high degree of specificity. With respect to the ALPS domain, prior studies have shown that the ALPS motif of Nup133 on its own is ER-membrane binding and, in the context of full Nup133, mutation of the ALPS motif decreases integration of Nup133 into existing nuclear pores.19 We found that a construct containing the Nt Nup133 domain that includes the ALPS motif did not target the LacO array to the nuclear rim as efficiently (10%) compared to full length Nup133 (57%; Table 1). The ability of the LacI-Nup/LacO system to precisely address such questions will be of use in increasingly detailed studies of protein-protein interaction with respect to nuclear pore structure.
The human cardiac arrhythmia mutation, Nup155 R391H, disrupts Nup recruitment and rim localization
The cardiac arrhythmia mutation Nup155 R391H prevents mutant Nup155 from associating with nuclear pores, creating the overall human disease phenotype of atrial fibrillation and/or early sudden death.75 Our data clearly reveal that the R391H mutation renders Nup155 unable to interact with one of its major partners, Nup53, and greatly reduces its ability to interact with the membrane protein Pom121 (Fig. 7). This data argues that these defects can well be the basis for the lack of association of mutant Nup155 with the nuclear pore that results in human and mouse cardiac disease.
Summary and conclusions
In sum, we took advantage of the LacI-LacO system to study nucleoporin interactions and nuclear pore assembly. Using this extremely versatile tool we demonstrate formation of a partial nuclear pore complex containing a large number of nucleoporins by immobilizing specific single nucleoporins. Importantly, our results show that not every nucleoporin that is essential for assembly (i.e., pore structure) also has the ability to recruit other nucleoporins and initiate assembly. Therefore, there is a distinction between nucleoporins that may be important for maintaining NPC structure and those that function as a platform or “seed” for NPC assembly. Our analysis revealed that Nup133 and Nup153 upon recruitment to chromatin act as seeds sufficient to initiate much of NPC assembly.
We further conclude that the LacI-Nup/LacO system can prove effective in analyzing the modularity of the NPC, both through allowing the study of different nucleoporin domains as compared to full-length proteins, as well as providing a powerful approach to determining the effects of specific Nup mutations important to human health.
Materials and Methods
Plasmids
To generate LacI-CFP-Nup expressing constructs, Nup153, Nup53, Nup98 and Nup58 were PCR amplified from pEGFP-NUP expression plasmids (Euroscarf, Frankfurt, Germany; deposited by Dr. Jan Ellenberg). Nup133 and Nup107 were amplified from pEGFP-C2, and Nup50 L isoform was amplified from pEGFP-N2 (all were the kind gift of Dr. Katharine Ullman, University of Salt Lake City, USA). Nup214 was amplified from pEFGP-N2 (the kind gift of Dr. Maureen Powers, Emory University School of Medicine, Atlanta, USA). Mouse Nup85 was amplified from pET28. Subsequently, all were subcloned into a pSV2-LacI-CFP vector (the kind gift of Dr. David Spector, CSH Labs, USA), using either EcoRI and XhoI, or XhoI and SalI. Pom121 was amplified from a pEXPR-PEF-1alpha-Pom121A-venus clone (the kind gift of Dr. Naoko Imamoto, Cellular Dynamics Laboratory, RIKEN, Wako, Saitama, Japan) and subcloned into the pSV2-LacI-CFP vector using BglII and SalI. All of the above encode human nucleoporins except for the mouse Nup85, which is 92% identical to the human homolog.
Constructs of N-terminal and C-terminal fragments of Nup133 (Fig. 6A) were constructed as follows: pSV2-LacI-CFP-Nup133 was digested with AflII, which cut the plasmid at 2 sites, one immediately before the sequence encoding amino acid residue 806 of hNup133 and at a site 276 nucleotides downstream from the Nup133 ORF. The fragment containing the vector plus the N terminus of Nup133 (aa 1–805) was purified and religated, generating pSV2-LacI-CFP-Nt Nup133. To clone the desired LacI-CFP-tagged C-terminal fragment of Nup133, specific oligos were designed to amplify the region on pSV2-LacI-CFP-Nup133 between amino acid residue 917 and the STOP codon after amino acid 1141. This PCR fragment was subcloned into pSV2-LacI-CFP using XhoI and SalI.
Wild type human Nup155 was amplified from pBlueScriptR-hNup155 (clone Image: 5295664) and subcloned into the pSV2-LacI-CFP vector using SmaI. The point mutant Nup155 R391H was constructed by site-directed mutagenesis PCR from the wild type pSV2-LacI-CFP-hNup155, with primers designed to change the nucleotide G1172 to A. All of the above clones were sequenced to verify correctness.
Antibodies
Antibodies used in this study included rabbit anti-Nup133–555 conjugated,40 anti-Nup160,40 anti-Nup53, anti-Nup85,40 anti-Nup62 (Sigma-Aldrich, SAB1410438), anti-Nup214 (Abcam, ab84357), Anti-Nup98 (Cell Signaling Technology, #2598), anti-Nup93,83 anti-Pom121 (Gene-Tex, GTX102128), anti-ELYS (Bethyl Laboratories, A300–166A), and mouse anti-FG Nup mAb414 (Biolegend, #mms-120p) and anti-LacI (Merck Millipore, 05–503).
Cell culture and transfection
U2OS 2–6–3 cells were kindly provided by Dr. David Spector, Cold Spring Harbor Laboratory, NY, USA. U2OS 2–6–3 cells contain, stably integrated into a single locus on their genome, approximately 200 copies of the expression plasmid, p3216PECMS2β, which is composed of 256 copies of the Lac operator, 96 tetracycline response elements, a minimal CMV promoter, CFP fused to the peroxisomal targeting signal SKL, 24 MS2 translational operators (MS2 repeats), a rabbit β-globin intron/exon module, and a cleavage/polyadenylation signal.52,84
U2OS and U2OS 2–6–3 cells were grown in DMEM media (CellGro, 10–017-CV) supplemented with 10% FBS (fetal bovine serum; Gemini Bio-products, #100–106), 1% glutamine, 1% penicillin/streptomycin, with 500 μg/ml hygromycin (Invitrogen, 10687–010) added in the case of the U2OS 2–6–3 cells. Before transfection, the cells were seeded on cover slips (12 mm) in 12-well plates containing 1 ml of DMEM media (CellGro, 10–017-CV) supplemented with 10% FBS (fetal bovine serum; Gemini Bio-products, #100–106), 1% glutamine, at ∼7.5 × 104 cells per well and grown overnight at 37°C in 5% CO2. Cells were transfected using a JetPEI transfection kit (Polyplus, cat n. 89129–938), as per manufacturer's instructions. The media was removed 24 hours post-transfection and the cells incubated with fresh DMEM medium plus antibiotics for 24 additional hours before proceeding with immunofluorescence.
As a control, to ensure that the fusion proteins were functional with respect to assembly into the existing NPCs on the nuclear envelope, we transfected the above LacI-CFP-Nup fusion plasmids individually into U2OS cells that did not contain the LacO array and determined their ability to localize to the nuclear pore. As shown in Supplemental Figure 6, 10 out of the 11 LacI-CFP-Nup fusion proteins clearly localized to the nuclear rim, showing a typical nuclear pore-staining pattern (i.e., a punctate nuclear rim). This indicated that the LacI-CFP tag does not disrupt the ability of the tagged nucleoporins to assemble into endogenous nuclear pores. LacI-CFP-Nup50 exhibited relatively high nucleoplasmic staining, but one could still see its presence at the nuclear rim (Supp. Fig. 6). We note that Nup98 showed both rim staining of nuclear pores and also the expected staining of Nup98 intranuclear bodies85–89 (Supp. Fig 6E, green). LacI-CFP-Pom121 is a soluble form of the transmembrane nucleoporin, due to absence of a functional ER signal sequence; instead of localizing to the nuclear rim as in the case of wild type POM121, it localized to the nuclear interior20,60 (Supp. Fig. 6K, green).
Immunofluorescence and microscopy
Cells on coverslips were washed with 1x phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 10 minutes, washed again with PBS, then permeabilized in 0.2% Triton X-100/PBS for 10 minutes. After being washed again with PBS, cells were blocked with 5% Fetal Goat Serum/PBS (Gemini Bio-products, #100–109) for 2h at room temperature, and immunolabeled using specific antibodies against different nucleoporins (see Antibodies for a list of the antibodies used) overnight at 4°C. Primary antibodies were either conjugated to a fluorophore (Alexa Fluor 555) or detected by incubation with a fluorescently labeled secondary antibody [Texas Red-labeled goat-anti-rabbit (Jackson Immunoresearch Laboratories Inc.., #111–075–144) or FITC-labeled goat anti-mouse (Jackson Immunoresearch Laboratories Inc.., #115–095–146)], at a 1:500 dilution for 1 hour at room temperature. The cells on coverslips were mounted on slides with 2 ml Vectashield (Vector, H-1200) containing DAPI DNA dye (0.75 mg/ml), and viewed in a Zeiss Axioplan 2 fluorescence microscope (Zeiss) using a 63X oil objective. Images were taken using an AxioCam HRc camera. Alternatively, the slides were viewed with a Leica SP5 II Upright Spectral Confocal using a 63X oil objective. All the experiments were performed at least 3 times and more than a 100 cells were counted overall for each combination of transfected LacI-CFP-tagged nucleoporin and each different antibody.
Immunoblotting
Approximately 106 transfected U2OS 2–6–3 cells were washed twice with cold PBS. They were then resuspended with 100 ml 2x Laemmli buffer (100 mM Tris-HCl pH 6.8, 200 mM DTT, 4% SDS, 20% glycerol, 0.008% bromophenol blue), and boiled for 4 minutes. 70 ml of the transfected U2OS 2–6–3 whole cell extracts were resolved using gradient SDS-PAGE gels, transferred to a nitrocellulose membrane (BioRad, 162–0115), and blocked with 5% milk (Non-fat dry milk, Apex Bioresearch products, #20–241) for 30 min. Following blocking, the LacI-CFP fusion proteins produced were immunodetected using a mouse anti-LacI antibody (Merck Millipore, 05–503) and a secondary goat anti-mouse conjugated to HRP (Invitrogen, 626520). Detection was performed using Western Lightning Plus-ECL (Perkin Elmer, NEL103001EA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. David L. Spector, Dr. Katharine Ullman, Dr. Maureen Powers, Dr. Jan Ellenberg, and Dr. Naoko Imamoto for kindly providing plasmids and cell lines. We also thank all of the members of the Forbes lab for helpful discussions and Nishdallyh Beltran-Raygoza for help with the Nup107 cloning.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by NIH R01- GM033279 grant to DJF and NS047101 grant to the Neuroscience Core Facility, UCSD.
References
- 1. Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett 2008; 582:2004-16; PMID:18328825; http://dx.doi.org/ 10.1016/j.febslet.2008.02.067 [DOI] [PubMed] [Google Scholar]
- 2. D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 2008; 18:11; PMID:18786826; http://dx.doi.org/ 10.1016/j.tcb.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol 2005; 15:6; PMID:15837182; http://dx.doi.org/ 10.1016/j.sbi.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 4. Wente SR. Gatekeepers of the nucleus. Science 2000; 288:1374-7; PMID:10827939; http://dx.doi.org/ 10.1126/science.288.5470.1374 [DOI] [PubMed] [Google Scholar]
- 5. Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 2005; 17:83-92; PMID:15629719; http://dx.doi.org/ 10.1016/j.molcel.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 6. Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andres-Pons A, Glavy JS, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 2013; 155:1233-43; PMID:24315095; http://dx.doi.org/ 10.1016/j.cell.2013.10.055 [DOI] [PubMed] [Google Scholar]
- 7. Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol 1991; 114:169-83; PMID:2050741; http://dx.doi.org/ 10.1083/jcb.114.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hawryluk-Gara L, Platani M, Santarella R, Wozniak RW, Mattaj IW. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell 2008; 19:1753-62; PMID:18256286; http://dx.doi.org/ 10.1091/mbc.E07-08-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci U S A 1994; 91:1519-23; PMID:8108440; http://dx.doi.org/ 10.1073/pnas.91.4.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol 1995; 128:721-36; PMID:7876300; http://dx.doi.org/ 10.1083/jcb.128.5.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. The JBiol Chem 1995; 270:14209-13; PMID:7775481; http://dx.doi.org/ 10.1074/jbc.270.23.14209 [DOI] [PubMed] [Google Scholar]
- 12. Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature 1995; 376:184-8; PMID:7603572; http://dx.doi.org/ 10.1038/376184a0 [DOI] [PubMed] [Google Scholar]
- 13. Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 2006; 314:815-7; PMID:17082456; http://dx.doi.org/ 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- 14. Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 2007; 129:83-96; PMID:17418788; http://dx.doi.org/ 10.1016/j.cell.2007.01.044 [DOI] [PubMed] [Google Scholar]
- 15. Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryotic cell 2009; 8:1814-27; PMID:19801417; http://dx.doi.org/ 10.1128/EC.00225-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imamoto N, Funakoshi T. Nuclear pore dynamics during the cell cycle. Curr Opin Cell Biol 2012; 24:453-9; PMID:22770730; http://dx.doi.org/ 10.1016/j.ceb.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 17. Schooley A, Vollmer B, Antonin W. Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly and chromatin de-condensation. Chromosoma 2012; 121:539-54; PMID:23104094; http://dx.doi.org/ 10.1007/s00412-012-0388-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wandke C, Kutay U. Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell 2013; 152:1222-5; PMID:23498932; http://dx.doi.org/ 10.1016/j.cell.2013.02.046 [DOI] [PubMed] [Google Scholar]
- 19. Doucet C, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 2010; 141:1030-41; PMID:20550937; http://dx.doi.org/ 10.1016/j.cell.2010.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaulov L, Gruber R, Cohen I, Harel A. A dominant-negative form of POM121 binds chromatin and disrupts the two separate modes of nuclear pore assembly. J Cell Sci 2011; 124:3822-34; PMID:22100917; http://dx.doi.org/ 10.1242/jcs.086660 [DOI] [PubMed] [Google Scholar]
- 21. Kimura N, Takizawa M, Okita K, Natori O, Igarashi K, Ueno M, Nakashima K, Nobuhisa I, Taga T. Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells 2002; 7:435-46; PMID:11952839; http://dx.doi.org/ 10.1046/j.1365-2443.2002.00529.x [DOI] [PubMed] [Google Scholar]
- 22. Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA 2006; 103:17801-6; PMID:17098863; http://dx.doi.org/ 10.1073/pnas.0608484103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 2007; 8:165-72; PMID:17235358; http://dx.doi.org/ 10.1038/sj.embor.7400889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gillespie P, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 2007; 17:1657-62; PMID:17825564; http://dx.doi.org/ 10.1016/j.cub.2007.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, Rasala BA, Forbes DJ. Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol Biol Cell 2009; 20:4043-58; PMID:19641022; http://dx.doi.org/ 10.1091/mbc.E09-02-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell 2008; 19:3982-96; PMID:18596237; http://dx.doi.org/ 10.1091/mbc.E08-01-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell 2009; 20:4031-42; PMID:19625448; http://dx.doi.org/ 10.1091/mbc.E09-02-0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bilokapic S, Schwartz TU. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc Natl Acad Sci U S A 2012; 109:15241-6; PMID:22955883; http://dx.doi.org/ 10.1073/pnas.1205151109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bilokapic S, Schwartz TU. Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains. Structure 2013; 21:572-80; PMID:23499022; http://dx.doi.org/ 10.1016/j.str.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol 2008; 180:857-65; PMID:18316408; http://dx.doi.org/ 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fichtman B, Ramos C, Rasala B, Harel A, Forbes DJ. Inner/Outer nuclear membrane fusion in nuclear pore assembly: biochemical demonstration and molecular analysis. Mol Biol Cell 2010; 21:4197-211; PMID:20926687; http://dx.doi.org/ 10.1091/mbc.E10-04-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoelz A, Debler E, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem 2011; 80:613-43; PMID:21495847 [DOI] [PubMed] [Google Scholar]
- 33. Belgareh N, Rabut G, Baï SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina O V, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol 2001; 154:1147-60; PMID:11564755; http://dx.doi.org/ 10.1083/jcb.200101081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 2004; 6:1114-21; PMID:15502822; http://dx.doi.org/ 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- 35. Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol 2001; 155:339-54; PMID:11684705; http://dx.doi.org/ 10.1083/jcb.200108007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure 2009; 17:1156-68; PMID:19748337; http://dx.doi.org/ 10.1016/j.str.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J 2002; 21:387-97; PMID:11823431; http://dx.doi.org/ 10.1093/emboj/21.3.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane coating module of the nuclear pore complex. Nat Struct Mol Biol 2009; 16:782-8; PMID:19503077; http://dx.doi.org/ 10.1038/nsmb.1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boehmer T, Schwartz TU. Purification, crystallization and preliminary X-ray analysis of a Nup107-Nup133 heterodimeric nucleoporin complex. Acta Crystallogr Sect F Struct Biol Cryst Commun 2007; 63:816-8; PMID:17768364; http://dx.doi.org/ 10.1107/S1744309107040523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harel A, Orjalo A V, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 2003; 11:853-64; PMID:12718872; http://dx.doi.org/ 10.1016/S1097-2765(03)00116-3 [DOI] [PubMed] [Google Scholar]
- 41. Walther T, Alves A, Pickersgill H, Loïodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 2003; 113:195-206; PMID:12705868; http://dx.doi.org/ 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- 42. Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A 2003; 100:981-5; PMID:12552102; http://dx.doi.org/ 10.1073/pnas.252749899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szymborska A, de Marco A, Daigle N, Cordes VC, Briggs JAG, Ellenberg J. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 2013; 341:655-8; PMID:23845946; http://dx.doi.org/ 10.1126/science.1240672 [DOI] [PubMed] [Google Scholar]
- 44. Walther T, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J 2001; 20:5703-14; PMID:11598013; http://dx.doi.org/ 10.1093/emboj/20.20.5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mackay D, Ullman KS. Coordinating postmitotic nuclear pore complex assembly with abscission timing. Nucleus 2011; 2:283-8; PMID:21941107; http://dx.doi.org/ 10.4161/nucl.2.4.16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ball JR, Ullman KS. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 2005; 114:319-30; PMID:16133350; http://dx.doi.org/ 10.1007/s00412-005-0019-3 [DOI] [PubMed] [Google Scholar]
- 47. Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, Aebi U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol 2002; 140:254-67; PMID:12490173; http://dx.doi.org/ 10.1016/S1047-8477(02)00524-5 [DOI] [PubMed] [Google Scholar]
- 48. Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol 2001; 154:71-84; PMID:11448991; http://dx.doi.org/ 10.1083/jcb.200101089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. D'Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009; 136:284-95; PMID:19167330; http://dx.doi.org/ 10.1016/j.cell.2008.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fichtman B, Harel A. Stress and aging at the nuclear gateway. Mech Ageing Dev 2014; 135:24-32; PMID:24447784; http://dx.doi.org/ 10.1016/j.mad.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 51. Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, 3rd, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013; 154:971-82; PMID:23993091; http://dx.doi.org/ 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Janicki S, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth K V, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell 2004; 116:683-98; PMID:15006351; http://dx.doi.org/ 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 1996; 135:1685-700; PMID:8991083; http://dx.doi.org/ 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science 2008; 322:1713-7; PMID:18948503; http://dx.doi.org/ 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- 55. Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13:95-101; PMID:21170033; http://dx.doi.org/ 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet 2011; 27:295-306; PMID:21680045; http://dx.doi.org/ 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Griffis E, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell 2003; 14:600-10; PMID:12589057; http://dx.doi.org/ 10.1091/mbc.E02-09-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. The Nup153-Nup50 protein interface and its role in nuclear import. J Biol Chem 2012; 287:38515-22; PMID:23007389; http://dx.doi.org/ 10.1074/jbc.M112.378893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 1995; 81:215-22; PMID:7736573; http://dx.doi.org/ 10.1016/0092-8674(95)90331-3 [DOI] [PubMed] [Google Scholar]
- 60. Funakoshi T, Maeshima K, Yahata K, Sugano S, Imamoto F, Imamoto N. Two distinct human POM121 genes: requirement for the formation of nuclear pore complexes. FEBS Lett 2007; 581:4910-6; PMID:17900573; http://dx.doi.org/ 10.1016/j.febslet.2007.09.021 [DOI] [PubMed] [Google Scholar]
- 61. Hawryluk-Gara L, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell 2005; 2005:2382-94; PMID:15703211; http://dx.doi.org/ 10.1091/mbc.E04-10-0857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B, Antonin W. Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J 2012; 31:4072-84; PMID:22960634; http://dx.doi.org/ 10.1038/emboj.2012.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J 2005; 24:3519-31; PMID:16193066; http://dx.doi.org/ 10.1038/sj.emboj.7600825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mitchell J, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol 2010; 191:505-21; PMID:20974814; http://dx.doi.org/ 10.1083/jcb.201007098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol 1996; 134:589-601; PMID:8707840; http://dx.doi.org/ 10.1083/jcb.134.3.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ulrich A, Partridge JR, Schwartz TU. The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution. Mol Biol Cell 2014; 25:1484-92; PMID:24574455; http://dx.doi.org/ 10.1091/mbc.E13-12-0745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell 2011; 22:1058-69; PMID:21289085; http://dx.doi.org/ 10.1091/mbc.E10-07-0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stavru F, Nautrup-Pedersen G, Cordes VC, Görlich D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J Cell Biol 2006; 173:477-83; PMID:16702234; http://dx.doi.org/ 10.1083/jcb.200601002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Talamas J, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol 2011; 194:27-37; PMID:21727197; http://dx.doi.org/ 10.1083/jcb.201012154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol 1993; 122:513-21; PMID:8335683; http://dx.doi.org/ 10.1083/jcb.122.3.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Berke I, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol 2004; 167:591-7; PMID:15557116; http://dx.doi.org/ 10.1083/jcb.200408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell 2008; 30:721-31; PMID:18570875; http://dx.doi.org/ 10.1016/j.molcel.2008.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci U S A 2009; 106:14281-6; PMID:19706512; http://dx.doi.org/ 10.1073/pnas.0907453106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol 2007; 14:138-46; PMID:17220896; http://dx.doi.org/ 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- 75. Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 2008; 135:1017-27; PMID:19070573; http://dx.doi.org/ 10.1016/j.cell.2008.10.022 [DOI] [PubMed] [Google Scholar]
- 76. Flemming D, Sarges P, Stelter P, Hellwig A, Bottcher B, Hurt E. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J Cell Biol 2009; 185:387-95; PMID:19414606; http://dx.doi.org/ 10.1083/jcb.200810016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Seo HS, Blus BJ, Jankovic NZ, Blobel G. Structure and nucleic acid binding activity of the nucleoporin Nup157. Proc Natl Acad Sci U S A 2013; 110:16450-5; PMID:24062435; http://dx.doi.org/ 10.1073/pnas.1316607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Whittle JR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem 2009; 284:28442-52; PMID:19674973; http://dx.doi.org/ 10.1074/jbc.M109.023580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Busayavalasa K, Chen X, Farrants A-KÖ, Wagner N, Sabri N. The Nup155-mediated organisation of inner nuclear membrane proteins is independent of Nup155 anchoring to the metazoan nuclear pore complex. J Cell Sci 2012; 125:4214-8; PMID:22718353; http://dx.doi.org/ 10.1242/jcs.105809 [DOI] [PubMed] [Google Scholar]
- 80. Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci 1999; 112(Pt 1):2253-64; PMID:10362555 [DOI] [PubMed] [Google Scholar]
- 81. Ribbeck K, Gorlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J 2002; 21:2664-71; PMID:12032079; http://dx.doi.org/ 10.1093/emboj/21.11.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shulga N, Goldfarb DS. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol Cell Biol 2003; 23:534-42; PMID:12509452; http://dx.doi.org/ 10.1128/MCB.23.2.534-542.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miller BR, Forbes DJ. Purification of the vertebrate nuclear pore complex by biochemical criteria. Traffic 2000; 1:941-51; PMID:11208084 [PMC free article] [PubMed] [Google Scholar]
- 84. Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol 2000; 2:871-8; PMID:11146650; http://dx.doi.org/ 10.1038/35046510 [DOI] [PubMed] [Google Scholar]
- 85. Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 2002; 13:1282-97; PMID:11950939; http://dx.doi.org/ 10.1091/mbc.01-11-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell 2004; 15:1991-2002; PMID:14718558; http://dx.doi.org/ 10.1091/mbc.E03-10-0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010; 140:372-83; PMID:20144761; http://dx.doi.org/ 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010; 140:360-71; PMID:20144760; http://dx.doi.org/ 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- 89. Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet 2013; 9:e1003308; PMID:23468646; http://dx.doi.org/ 10.1371/journal.pgen.1003308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.