Abstract
Insulin-like growth factor binding protein 2 (IGFBP2) is a pleiotropic oncogenic protein that has both extracellular and intracellular functions. Despite a clear causal role in cancer development, the tumor-promoting mechanisms of IGFBP2 are poorly understood. The contributions of intracellular IGFBP2 to tumor development and progression are also unclear. Here we present evidence that both exogenous IGFBP2 treatment and cellular IGFBP2 overexpression lead to aberrant activation of EGFR, which subsequently activates STAT3 signaling. Furthermore, we demonstrate that IGFBP2 augments the nuclear accumulation of EGFR to potentiate STAT3 transactivation activities, via activation of the nuclear EGFR signaling pathway. Nuclear IGFBP2 directly influences the invasive and migratory capacities of human glioblastoma cells, providing a direct link between intracellular (and particularly nuclear) IGFBP2 and cancer hallmarks. These activities are also consistent with the strong association between IGFBP2 and STAT3-activated genes derived from the TCGA database for human glioma. A high level of all 3 proteins (IGFBP2, EGFR and STAT3) was strongly correlated with poorer survival in an independent patient dataset. These results identify a novel tumor-promoting function for IGFBP2 of activating EGFR/STAT3 signaling and facilitating EGFR accumulation in the nucleus, thereby deregulating EGFR signaling by 2 distinct mechanisms. As targeting EGFR in glioma has been relatively unsuccessful, this study suggests that IGFBP2 may be a novel therapeutic target.
Keywords: IGFBP2, EGFR, STAT3, glioma
INTRODUCTION
Secreted proteins such as growth factors and hormones exert their function by binding to the extracellular domain of membrane receptors. Secreted factors can also enter the cell through receptor-mediated endocytosis [1-4]. Once internalized, these proteins can regulate intracellular cytoplasmic signal transduction and transcriptional activity in the nucleus [5-11]. Insulin-like growth factor (IGF) binding protein 2 (IGFBP2) is a secreted protein that was initially characterized as binding and modulating the activity of IGF-I and -II [12-14]. IGFBP2 can also function independently of IGF binding, and its versatility as a secreted or cytoplasmic signaling effector has been widely characterized. IGFBP2 can bind integrins [15-17] and activate PI3K/AKT [18], NFκB [15] and ERK [19]. Recently, a classic nuclear localization signal sequence that is responsible for nuclear entry has been identified in IGFBP2 [20]. However, the functional and clinical significance of nuclear IGFBP2 has not been clearly elucidated [21-24].
In mammals, IGFBP2 is expressed at high levels in embryonic tissues, but the expression is drastically decreased after birth. However, IGFBP2 expression has been observed postnatally in hematopoietic stem cells and in liver and spleen progenitor cell populations [25-30]. IGFBP2 is reactivated during progression of a wide spectrum of cancer types, including glioma and prostate, breast and lung cancers [18, 30-32]. IGFBP2 plays an oncogenic role in tumor initiation and progression to high-grade glioma [33] and is reported as one of the 9 genes in a signature associated with poor clinical outcome in high-grade glioma [34]. IGFBP2 mediates cell expansion and survival of glioma stem cells [35, 36]. Despite the clear role for IGFBP2 in tumorigenesis, the mechanisms underlying nuclear IGFBP2’s contribution to the tumorigenic program remain unknown.
EGFR/IGFBP2 and EGFR/STAT3 [37, 38] are concurrently co-expressed in glioma. EGFR, a cell surface tyrosine kinase receptor, is activated in 30-50% of high-grade gliomas through amplification, overexpression or mutation [39-41]. EGFR signal transduction can be mediated by STAT3. STAT3 interacts with EGFR at 2 autophosphorylation sites in the cytoplasmic domain, tyrosine 1068 or tyrosine 1086 [42], and is activated by phosphorylation at tyrosine 705 (Y705) [43]. In addition to this cytoplasmic interaction, EGFR and STAT3, after translocation into the nucleus, can form a complex to activate transcription of genes such as COX2 [44], iNOS [45] and c-MYC [46]. Nuclear EGFR expression in glioma and other cancers, such as breast carcinoma [47], esophageal squamous cell carcinoma [48] and ovarian cancer [49], is associated with poor survival and linked to an aggressive tumor phenotype [50]. Furthermore, IGFBP2 regulates expression of the VEGF, MMP2, TIMP1, TWIST, BCL2 and HIF1A genes [20, 51], which are known transcriptional targets of STAT3. Recent research implicated nuclear IGFPB2 in angiogenesis through activation of VEGF, a STAT3 target gene [20].These observations suggest that there is a functional connection between IGFBP2, EGFR and STAT3 in glioma. Here we tested this hypothesis and provide evidence that IGFBP2 mediates the tumorigenic program through a tightly linked IGFBP2-EGFR-STAT3 regulatory signaling network.
RESULTS
IGFBP2 activates the STAT3 signaling pathway via an EGFR-dependent mechanism
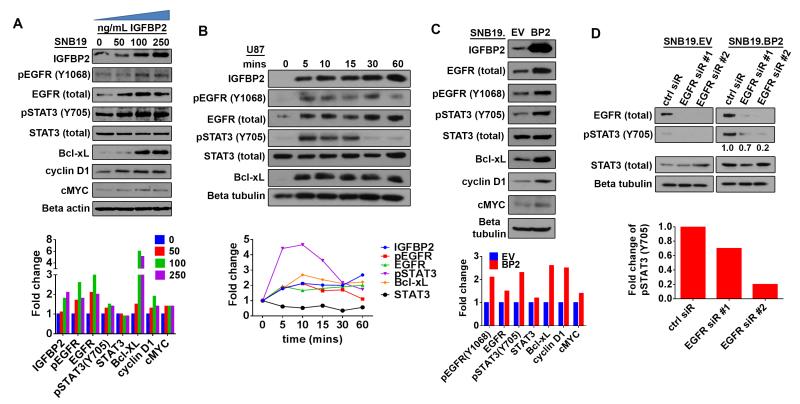
To explore the functional interaction between IGFBP2, EGFR and STAT3, we stimulated SNB19 parental (SNB19.par) glioma cells, which had been serum-starved overnight, with increasing amounts of exogenous IGFBP2 protein. Immunoblotting analysis demonstrated increased expression of both total EGFR and EGFR activated via phosphorylation at tyrosine Y1068, or pEGFR(Y1068), in parallel with IGFBP2 uptake into the cell (Fig. 1A). STAT3 activation via phosphorylation at tyrosine 705, designated pSTAT3(Y705), and expression of STAT3 transcriptional targets Bcl-xL, cyclin D1 and c-MYC also increased in response to IGFBP2 stimulation.
Figure 1. IGFBP2 activates STAT3 through EGFR.
(A) Immunoblot analysis of SNB19 cells starved of serum overnight then stimulated with exogenous IGFBP2 protein at the indicated dosages (0, 50, 100, 250 ng/mL) for 60 minutes. Densitometric analysis shown below the immunoblot indicates fold-change relative to unstimulated control cells (normalized to beta-actin loading control or total protein for phosphorylated proteins). (B) Immunoblot analysis of U87 cells starved of serum overnight then stimulated with exogenous IGFBP2 (100ng/mL) for the indicated time points (0, 5, 10, 15, 30, 60 minutes). Densitometric analysis shown below the immunoblot indicates fold-change relative to unstimulated control cells (normalized to loading control or total protein for phosphorylated proteins). (C) Immunoblot analysis comparing stable SNB19 empty vector cells (SNB19.EV) to SNB19 cells stably overexpressing IGFBP2 (SNB19.BP2). Densitometric analysis shown below the immunoblot indicates fold-change relative to SNB19.EV after normalization to beta-tubulin loading control (or total protein for phosphorylated proteins). (D) Immunoblot analysis comparing SNB19.EV and SNB19.BP2 cells depleted of EGFR via 2 independent pools of EGFR siRNA (EGFR sir#1, EGFR sir#2) to cells transfected with scrambled negative control siRNA (ctrl siR). The intensity of pSTAT3(Y705), quantified by densitometry, is shown below the immunoblot as fold-change relative to control siRNA, normalized to total STAT3.
Next, we performed a time-course experiment in which U87 glioma cells, which lack endogenous IGFBP2 expression, were stimulated with exogenous IGFBP2 after overnight serum-starvation. Immunoblotting analysis revealed induction of EGFR, STAT3 and Bcl-xL expression as early as 5 minutes following addition of exogenous IGFBP2 (Fig. 1B). In comparison with EGF (EGFR ligand) stimulation, IGFBP2 stimulation of EGFR activation was substantially less than that produced by EGF (data not shown). Furthermore, immunoblotting analysis of SNB19 cells stably overexpressing IGFBP2 (SNB19.BP2) demonstrated that, compared to SNB19 cells stably transfected with empty vector (SNB19.EV), IGFBP2 overexpression resulted in increased expression of EGFR and phosphorylated STAT3, along with Bcl-xL, cyclin D1 and c-MYC (Fig. 1C).
To examine the involvement of EGFR in IGFBP2-mediated STAT3 activation, we depleted EGFR by using 2 different pools of small interfering RNA (siRNA) in SNB19.BP2 cells and observed decreases in STAT3 activation (Fig. 1D), supporting the hypothesis that IGFBP2 mediates STAT3 activation through EGFR. To rule out the possibility of off-target effects of EGFR siRNA-mediated knockdown, we knocked down EGFR in SNB19.BP2 cells and stimulated the cells with recombinant IL6. We observed STAT3 phosphorylation in these cells, confirming that EGFR knockdown impairs STAT3 activation by IGFBP2 without compromising alternate STAT3 activation pathways (Supplementary Fig 1).
EGFR can be indirectly activated through transactivation, which involves a disintegrin and metalloproteinases (ADAMs) [52]. To determine whether ADAMs are involved in IGFBP2-mediated EGFR activation, we inhibited ADAMs by treatment with 2 different ADAM inhibitors, TAPI2 and marimastat [53, 54]. U87 cells serum-starved overnight were pretreated with 20 μM TAPI-2 or marimastat, then stimulated with exogenous IGFBP2 for 5 minutes (Supplementary Fig 2A). Immunoblotting analysis demonstrated that exogenous IGFBP2 stimulated EGFR and STAT3 activation despite ADAMs inhibition. Furthermore, because ADAM17 is essential to regulation of EGFR transactivation [52], we knocked down ADAM17 using 2 different pools of siRNA to evaluate whether IGFBP2-mediated EGFR activation involves ADAM17 (Supplementary Fig 2B). Immunoblotting analysis showed that ADAM17 knockdown did not affect EGFR and STAT3 activation in SNB19.BP2 cells. These data demonstrate that ADAMs are not involved in IGFBP2-mediated EGFR/STAT3 activation.
IGFBP2 is significantly correlated with STAT3 pathway activation in glioma
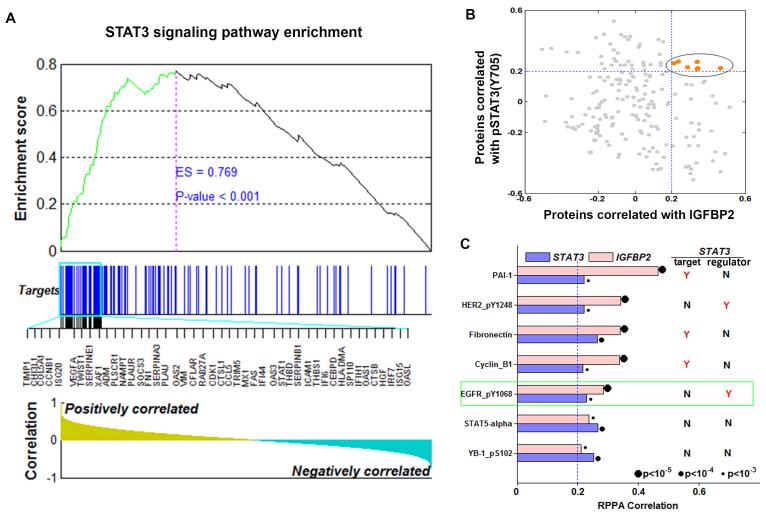
Previous studies showed that IGFBP2 regulates the expression of many STAT3 target genes [20, 51], and our results demonstrate that IGFBP2 can stimulate STAT3 activation through EGFR. To gain a comprehensive view of the relationship between IGFBP2 and STAT3 signaling, we analyzed the whole-genome gene expression profiling data from The Cancer Genome Atlas (TCGA) low-grade glioma (LGG) database. Gene set enrichment analysis (GSEA) revealed that STAT3-activated genes were significantly and positively correlated with IGFBP2 (Fig. 2A), suggesting that IGFBP2 expression is associated with STAT3 activation. To further substantiate the IGFBP2-STAT3 link, we performed hierarchical clustering on the 157 experimentally validated STAT3 target genes across all samples in the REpository for Molecular BRAin Neoplasia DaTa (REMBRANDT) dataset. Two distinct clusters were formed, associated with tumor grade and IGFBP2 and STAT3 expression but not with other transcription factors such as beta-catenin (CTNNB1) or Forkhead box protein M1 (FOXM1) (Supplementary Fig. 3). This finding further validates that expression of IGFBP2 and STAT3 are tightly linked.
Figure 2. IGFBP2 is strongly and significantly correlated with STAT3 pathway genes.
(A) GSEA demonstrated enrichment for STAT3 target genes based on correlation with IGFBP2 expression in the TCGA low-grade glioma database. The top of the panel shows the enrichment score (ES) for genes associated with STAT3 signaling pathway targets. The blue lines indicate where the STAT3 target genes appear in the ranked gene list, and the black lines represent the top 45 highly correlated targets. The bottom of the panel shows the ranking scores (correlation of all genes associated with the STAT3 signaling pathway targets with IGFBP2). (B) Correlation of expression of proteins in the TCGA RPPA data with IGFBP2 (x-axis) and pSTAT3(Y705) (y-axis). Each dot represents a protein. Proteins with correlation coefficients greater than 0.2 are highlighted in orange. (C) Correlation of the 7 proteins with the highest correlation coefficients with both IGFBP2 and STAT3. Also shown is the relationship of each protein with STAT3 (“target” = STAT3 transcriptional target; “regulator” = STAT3 upstream regulator). Y = yes, a known target or upstream regulator of STAT3; N = not a known target or upstream regulator of STAT3.
Next we postulated that the most functionally important of the correlated genes would likely be associated with STAT3 activity (as measured by phosphorylation) in the reverse-phase protein array (RPPA) data of the same TCGA cohort. In this proteomic analysis, we identified the 7 proteins (Fig. 2B, 2C) that were most significantly and strongly correlated with both IGFBP2 and pSTAT3(Y705) (correlation coefficients >0.2). Of these 7 strongly correlated proteins, 5 are closely related to the STAT3 signaling pathway, namely plasminogen activator inhibitor-1 (PAI-1), fibronectin, cyclin B1, pHER2(Y1248), and, notably, pEGFR(Y1068). HER2 is a member of the EGFR family and an upstream regulator of STAT3, however it has not been shown to have clinical significance in glioma [55-59]. Thus these results from patient samples are consistent with the results of our in vitro cell line–based studies, and together these results illustrate the potential importance of the IGFBP2-EGFR-STAT3 signaling axis in glioma.
IGFBP2 co-precipitates and co-localizes with EGFR
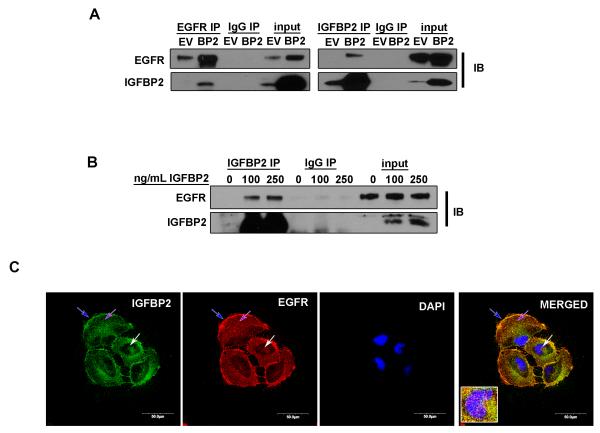
To further evaluate the functional relationship between IGFBP2 and EGFR, we performed reciprocal immunoprecipitation (IP) studies followed by immunoblotting comparing IGFBP2-overexpressing SNB19 cells and empty vector control cells. Co-IP experiments revealed co-precipitation of IGFBP2 and EGFR (Fig. 3A). We next treated U87 cells, which had been serum-starved overnight, with 2 different doses of exogenous IGFBP2 followed by IP analysis and immunoblotting. The results showed a dose-dependent increase of IGFBP2 co-precipitated with EGFR (Fig 3B). Confocal imaging analysis of SNB19.BP2 cells demonstrated clear co-localization of IGFBP2 and EGFR proteins on the cell membrane and in the cytoplasm and nucleus (Fig. 3C). Co-localization of IGFBP2 and EGFR provides further evidence of a complex containing IGFBP2 and EGFR.
Figure 3. IGFBP2 co-precipitates and co-localizes with EGFR.
(A) Co-immunoprecipitation (IP) of IGFBP2 and EGFR in SNB19.EV control cells versus SNB19.BP2 cells analyzed by immunoblot (IB). (B) Immunoprecipitation of IGFBP2 in U87 cells starved of serum overnight then stimulated with 2 different doses of IGFBP2 for 30 minutes, analyzed by immunoblotting. (C) Confocal microscopy images of immunofluorescence staining for IGFBP2 (green), EGFR (red) and DAPI (blue) in SNB19.BP2 cells show IGFBP2 and EGFR co-localization; blue arrow = cell membrane; purple arrow = cytoplasm; white arrow = nucleus.
IGFBP2 facilitates EGFR nuclear accumulation
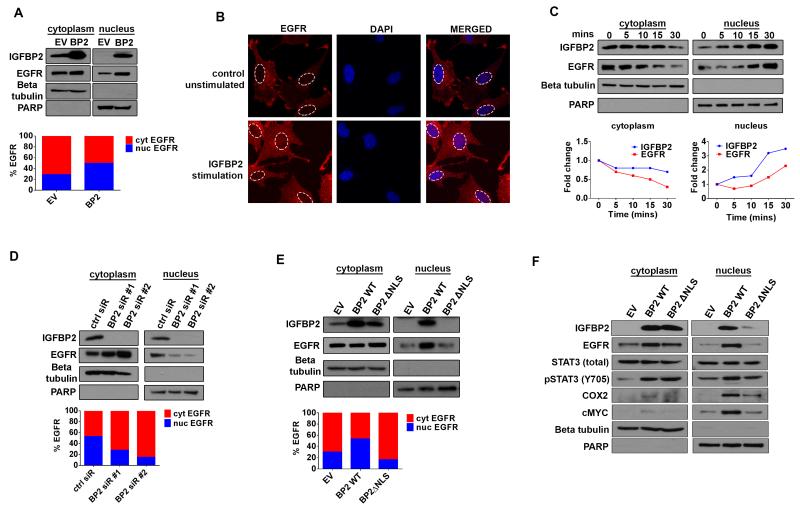
Because we observed IGFBP2 and EGFR co-localization in the cytoplasm and nucleus, we investigated whether nuclear IGFBP2 is closely linked to nuclear EGFR and whether this complex augments STAT3 transcriptional activation. We first fractionated SNB19.BP2 and SNB19.EV cells into cytoplasmic and nuclear fractions and performed immunoblotting to detect IGFBP2, EGFR and STAT3. Our results revealed that a substantial proportion of IGFBP2 and EGFR localized to the nucleus in SNB19.BP2 cells (Fig. 4A). We then determined the ratio of nuclear to cytoplasmic EGFR via densitometric analysis and found that SNB19.BP2 cells had more than twice as much nuclear EGFR as SNB19.EV cells.
Figure 4. IGFBP2 drives EGFR nuclear accumulation.
(A) Immunoblot analysis of cytoplasmic (cyt) and nuclear (nuc) fractions of SNB19.EV and SNB19.BP2 cells. Beta-tubulin represents a loading control for the cytoplasmic fraction, and PARP represents a loading control for the nuclear fraction. Densitometric analysis represented by the bar graph, demonstrates percentage of cytoplasmic or nuclear EGFR. (B) Confocal images of SNB19 parental cells and SNB19 parental cells stimulated with exogenous IGFBP2 protein (250ng/mL for 30 minutes). Cells were stained for EGFR (red) and the nuclei stained with DAPI (blue). (C) Immunoblot analysis of cytoplasmic and nuclear fractions of SNB19 parental cells stimulated with exogenous IGFBP2 (250ng/mL for indicated times). The graph represents fold-change of cytoplasmic or nuclear IGFBP2 and EGFR calculated from densitometric analysis of the immunoblot bands. (D) Immunoblot analysis comparing cytoplasmic and nuclear fractions of SNB19.BP2 cells depleted of IGFBP2 via 2 independent pools of IGFBP2 siRNA (BP2 siR #1, #2) to cells transfected with scrambled negative control siRNA (ctrl siR). Densitometric analysis represented by the bar graph, demonstrates percentage of cytoplasmic or nuclear EGFR. (E) Immunoblot analysis of cytoplasmic and nuclear fractions of transiently transfected SNB19.EV, SNB19.BP2 wild type (BP2 WT) and SNB19 with a mutated IGFBP2 nuclear localization signal (BP2ΔNLS). Densitometric analysis represented by the bar graph, demonstrates percentage of cytoplasmic or nuclear EGFR. (F) Immunoblot analysis of cytoplasmic and nuclear proteins in stable SNB19.EV, SNB19.BP2 WT and SNB19.BP2ΔNLS cells.
To investigate whether IGFBP2 facilitates EGFR nuclear accumulation, we stimulated SNB19.par cells, which had been serum-starved overnight with exogenous IGFBP2 protein and then visualized EGFR protein localization by confocal imaging. IGFBP2 stimulation of SNB19.par cells resulted in EGFR accumulation in the nucleus (Fig. 4B). A time-course study with the same cells demonstrated that IGFBP2 nuclear accumulation paralleled EGFR nuclear accumulation in a time-dependent manner (Fig. 4C). To validate that EGFR nuclear accumulation is mediated through IGFBP2, we knocked down IGFBP2 using 2 different pools of siRNA in SNB19.BP2 cells and performed immunoblotting analysis on the fractionated cells. IGFBP2 depletion led to impaired EGFR nuclear localization with coordinate cytoplasmic accumulation of EGFR, whereas control knockdown did not affect EGFR nuclear accumulation (Fig. 4D). These results suggest that IGFBP2 plays a role in promoting EGFR nuclear accumulation.
Nuclear translocation of IGFBP2 is required for IGFBP2-mediated EGFR nuclear accumulation
To better understand the mechanism of nuclear IGFBP2–mediated EGFR nuclear accumulation, we generated an IGFBP2 construct with a mutant nuclear localization signal [60] (BP2ΔNLS; Supplementary Fig. 4A). Transient transfection of BP2ΔNLS plasmid into SNB19.par cells resulted in the expected compromise of IGFBP2 nuclear entry and also impaired EGFR nuclear accumulation (Fig. 4E). Next, we created a stable BP2ΔNLS-overexpressing cell line (SNB19.BP2ΔNLS). Impaired EGFR nuclear accumulation in fractionated stable SNB19.BP2ΔNLS cells, compared to SNB19.BP2 WT (wild-type IGFBP2), resulted in decreased nuclear expression of COX2 and cMYC, which are known downstream targets of nuclear EGFR/STAT3 complex (Fig. 4F). These results were replicated in another glioma cell line, T98G (Supplementary Fig. 5A, 5B). To determine whether BP2ΔNLS can bind to EGFR, we transiently transfected U87 cells with BP2 WT or BP2ΔNLS plasmid and performed IP followed by immunoblotting (Supplementary Fig. 6). The results showed that mutation of IGFBP2 NLS does not affect binding to EGFR, demonstrating that nuclear translocation of IGFBP2 is important for mediating EGFR nuclear accumulation. Because IGFBP2 is involved in glioma cell migration and invasion [15, 51], we then performed migration and invasion assays using the SNB19.EV, SNB19.BP2 WT, and SNB19.BP2ΔNLS cell lines. Migration and invasion potential were significantly impaired in the SNB19.BP2ΔNLS cells compared to SNB19.BP2 WT (Supplementary Fig. 4B, 4C), indicating that nuclear IGFBP2 is important for the invasive phenotype of glioma cells, plausibly through regulation of nuclear EGFR-STAT3 activity.
Levels of nuclear EGFR, nuclear IGFBP2 and pSTAT3 are significantly correlated in glioma
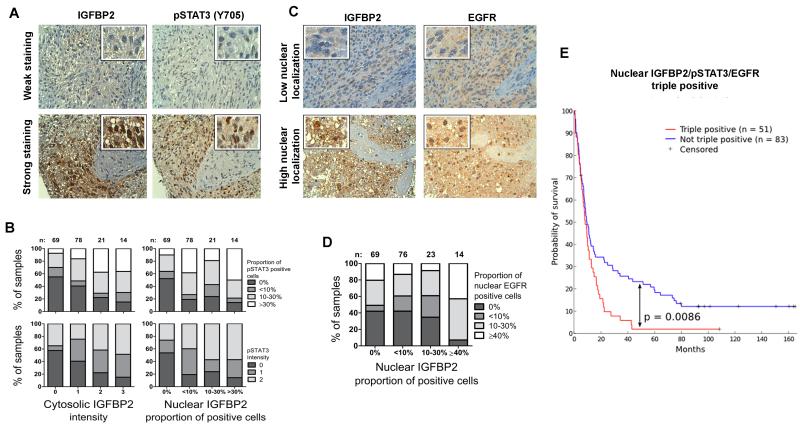
The RPPA LGG data from TCGA revealed a close relationship between IGFBP2, activated EGFR and activated STAT3, but did not provide spatial information. To further investigate localization of these proteins, we performed immunohistochemical analysis to determine the association between IGFBP2, EGFR and pSTAT3(Y705) in a clinical glioma tissue microarray (TMA) comprising 222 samples of grade 2-4 gliomas. We observed both cytosolic and nuclear localization of IGFBP2, both of which were strongly associated with STAT3 phosphorylation in these gliomas (Fig. 5A, 5B, Supplementary Table 1). Both cytosolic and nuclear IGFBP2 expression positively correlated with increased fraction and degree of phosphorylation of STAT3 (p=0.023 and p=0.018, respectively), suggesting a functional link between IGFBP2 expression and STAT3 phosphorylation.
Figure 5. IGFBP2 correlates with STAT3 activation and nuclear EGFR localization in clinical samples.
Expression and localization of IGFBP2, pSTAT3(Y705) and EGFR were detected with immunohistochemistry from a TMA that included 222 human grade 2-4 gliomas. (A) TMA immunostaining images (magnification 40×) representing weak and strong staining of IGFBP2 and pSTAT3(Y705). (B) Cytosolic and nuclear IGFBP2 expression associated with the percentage of cells positive for pSTAT3 and with pSTAT3 staining intensity. Bar graphs illustrate the increasing fractions of pSTAT3-positive cells and pSTAT3 intensity upon increasing IGFBP2 intensity or nuclear accumulation. (C) TMA immunostaining images (magnification 40×) representing low and high nuclear localization of IGFBP2 and EGFR. (D) Nuclear IGFBP2 associated with nuclear EGFR. The bar graph illustrates the fraction of samples with increasing nuclear EGFR localization upon increasing nuclear accumulation of IGFBP2. (E) Nuclear co-localization of IGFBP2, EGFR and phosphorylated STAT3 predicted poor survival among patients with human grade 2-4 glioma. Patients were stratified into 2 cohorts based on the nuclear staining of all 3 proteins: triple positives (≥1% of cells with nuclear expression, n=51, red line) and all other cases (n=83, blue line). Survival rates were visualized by using a Kaplan-Meier survival plot (p=0.0086).
We observed nuclear co-localization of IGFBP2 and EGFR in the clinical samples (Fig. 5C, 5D, and Supplementary Table 1). Cytosolic IGFBP2 did not correlate with nuclear EGFR and nuclear IGFBP2 did not correlate with cytosolic EGFR. However, nuclear IGFBP2 positively associated with nuclear EGFR localization (p=0.011). Furthermore, clinical samples that were triply positive for nuclear accumulation of IGFBP2, phosphorylated STAT3 and EGFR were strongly associated with poor survival (Figure 5E).
DISCUSSION
Conventionally, proteins are categorized as secreted, membrane-bound or intracellular. However, accumulating evidence demonstrates that signaling molecules are spatiotemporally dynamic [61-66]; in addition to cell surface–initiated signaling, upon internalization, EGFR can mediate signaling in the endosome [67-70], nucleus [71] or mitochondria [72, 73]. EGFR crosstalks with STAT3 through 2 levels: tyrosine kinase–mediated activation of STAT3 [42, 43, 74] and nuclear cooperation as transcriptional cofactors [44-46]. Our results significantly expand our understanding of this network by demonstrating that IGFBP2 stimulation or overexpression contributes to activation of EGFR-STAT3 and downstream pathways. IGFBP2 forms a complex containing EGFR and facilitates the nuclear accumulation of EGFR to potentiate nuclear EGFR-STAT3 activity. Thus, IGFBP2 controls 2 fundamental functions of EGFR, cytoplasmic signal transduction and nuclear accumulation.
These results also elucidate newly identified oncogenic functions for nuclear IGFBP2. By facilitating nuclear EGFR accumulation, nuclear IGFBP2 activates an EGFR-STAT3–mediated transcriptional program, which promotes transcription of iNOS, cMYC and COX2 [44-46]. The resulting aberrant activation of these genes leads to uncontrolled cell proliferation, survival and metastasis, and hence poor prognosis. COX2 is upregulated and correlated with poor survival in glioma [75-77], iNOS is critical for glioma stem cell survival and tumorigenesis [78-81] and cMYC is correlated with high-grade glioma and important for glioma stem cell maintenance [82-84]. We also demonstrated a role for nuclear IGFBP2 in promoting cancer cell migration and invasion, a key hallmark of cancer and a major aspect of glioma aggressiveness and treatment response.
The dynamic nature of these oncogenic signaling molecules may contribute to the ineffectiveness of EGFR-targeted therapy in glioma, perhaps because EGFR is being actively translocated into the nucleus by IGFBP2, rendering the cells resistant to therapies targeting membrane EGFR. Thus IGFBP2 may serve as an escape mechanism for glioma cells from EGFR-targeted therapy. EGFR can be activated by 8 different ligands [85, 86], including EGF, transforming growth factor α (TGFα) and heparin-binding EGF (HB-EGF). EGF, TGFα and HB-EGF are constitutively expressed in developing and normal adult brain [87-90]. Therefore, therapeutic targeting of these ligands is not feasible. Conversely, IGFBP2 is highly expressed in fetal brain and gliomas, but not in normal adult brain [26, 27, 30, 91, 92], making it a better therapeutic target than the known EGF ligands. EGFR amplification and activation are among the most common oncogenic events in human cancer [93, 94], and IGFBP2 overexpression is also a frequent event in glioma and other cancers. Our analysis of 3 independent clinical datasets revealed strong correlation between IGFBP2 and EGFR in terms of both gene expression and protein localization in the cell. The development of therapeutic strategies that target both molecules may represent a rational approach for cancer therapeutics.
MATERIALS AND METHODS
Cell culture, treatments, plasmids and transfections
SNB19, U87 and T98G cells were obtained from ATCC (Manassas, VA). Cells were cultured in Dulbecco modified essential/F12 50:50 medium supplemented with 10% fetal bovine serum (FBS) and 5% penicillin/streptomycin in an incubator with 5% CO2 at 37 °C. SNB19.EV (empty vector) and SNB19.BP2 WT (IGFBP2 wild-type) cells were created as previously described [51]. To generate BP2ΔNLS (IGFBP2 mutation at the nuclear localization signal), amino acid residues 179PKKLRPP185 of the IGFBP2 nuclear localization signal were mutated to 179PNNLAPP185 using the Quikchange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s protocol. A stable SNB19.BP2ΔNLS cell line was created by transfection of pcDNA3.1.IGFBP2ΔNLS plasmid via FuGENE HD (Promega, Fitchburg, WI) according to the manufacturer’s protocol, followed by G418 selection for 3 weeks.
IGFBP2 stimulation experiments were performed by using recombinant IGFBP2 (ab63223; Abcam, Cambridge, MA) with cells starved of serum overnight. Depletion of IGFBP2 and EGFR was achieved via transfection of Lipofectamine RNAiMAX (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol with 2 different pools of siRNA from Mission siRNA (Sigma, St Louis, MO) for 48 hours. Some cells were treated with a broad-spectrum ADAM inhibitor, TAPI-2 (#14695; Cayman Chemical, Ann Arbor, MI) or marimatstat (#M2699; Sigma), at 20 μM for 2 hours. Depletion of ADAM17 siRNA was achieved via transfection of Lipofectamine RNAiMAX according to the manufacturer’s protocol with 2 different pools of siRNA from Life Technologies (#s13718 and #s13719).
Gene set enrichment analysis
A total of 268 LGG samples obtained from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/)were subjected to RNA sequencing. The gene expression data were median-centered and then transformed to log2 space. We calculated the correlation of IGFBP2 gene expression with all other genes in the genome and ranked the genes in descending order based on the correlation coefficients. Using the gene expression correlation as the ranking metric, GSEA was then used to calculate the score for the degree of enrichment of the genes with higher correlation coefficients among genes involved in the STAT3 signaling pathway [95].
In a similar manner, the correlation of IGFBP2 or STAT3 protein expression with proteins in the TCGA TMA was calculated for 257 LGG samples for which reverse phase protein array (RPPA) data were available. Proteins that had higher correlation coefficients with both IGFBP2 and STAT3 proteins were considered the most likely candidates to represent molecular mechanisms underlying the association of IGFBP2 and the STAT3 signaling pathway.
Ingenuity Pathway Analysis
The interaction network feature of Ingenuity Pathway Analysis was used to determine direct downstream targets of STAT3. Interactions were filtered on the basis of their confidence level so that only interactions experimentally observed in humans were included in the table of results. Interactions were also filtered by relationship type so that only interactions of type “expression” or “transcription” were included.
Immunoprecipitation, immunoblotting and cellular fractionation
For immunoprecipitation (IP), cells were subjected to lysis in NP-40 buffer with 0.1% phosphatase inhibitor cocktail (Pierce Biotechnology, Thermo Fisher Scientific, Waltham, MA). After preclearing for 1 hour at 4 °C with Protein G agarose beads (SC#2002; Santa Cruz Biotechnology, Santa Cruz, CA) and appropriate species normal IgG, lysates were immunoprecipitated overnight at 4 °C with Protein G agarose beads using antibodies to IGFBP2 (#SC-6001; Santa Cruz Biotechnology; 1:100) and EGFR (#2256; Cell Signaling Technology, Beverly, MA; 1:100). Beads were washed with NP-40 buffer 3 times and boiled in Laemmli buffer. Proteins from the IP experiment or extracted from cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (10%) in running buffer and transferred onto an Immobilon TM-PVDF membrane (Millipore, Billerica, MA) for 1 hour at 100V in transfer buffer (24 mM Tris base, 191 mM glycine and 20% [v/v] methanol). Membranes were blocked for 1 hour at room temperature with 5% (w/v) non-fat milk powder in phosphate-buffered saline solution (PBS) with 0.1% Tween-20 (PBST) and incubated overnight at 4 °C with primary antibody: IGFBP2 (#SC-6001; 1:500); EGFR (#4267; Cell Signaling Technology; 1:1000), EGFR-Y1068 (#3777; Cell Signaling Technology; 1:1000), beta-tubulin (#2128; Cell Signaling Technology; 1:1000), PARP (#9542; Cell Signaling Technology; 1:1000), STAT3 (#9139; Cell Signaling Technology; 1:1000), STAT3-Y705 (#9145; Cell Signaling Technology; 1:1000), Bcl-xL (#2764; Cell Signaling Technology; 1:1000), cyclin D1 (#2978; Cell Signaling Technology; 1:1000), c-MYC (#SC-40; Santa Cruz Biotechnology; 1:1000), COX-2 (#160112; Cayman Chemical; 1:250), or ADAM17 (#T5442; Sigma; 1:500) in blocking solution. After washing in PBST, blots were incubated for 1 hour at room temperature in PBST with secondary antibodies (anti-goat IgG, anti-rabbit IgG, or anti-mouse IgG; Santa Cruz Biotechnology; 1:5000) couple to horseradish peroxidase (HRP). Immunoblots were incubated with enhanced chemiluminescence SuperSignal West Pico or Femto solution (Pierce Biotechnology). Cellular fractionation was performed by using the NE-PER nuclear and cytoplasmic kit (Pierce Biotechnology) according to the manufacturer’s protocol. Densitometric analysis of immunoblot bands were quantified using ImageJ software (U.S. National Institutes of Health, Bethesda, MD).
Confocal imaging
Cell on chamber slides were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and incubated with primary antibody to EGFR (#4267; 1:100) and IGFBP2 (#SC-6001; 1:100) at 4 °C overnight. They were then incubated with secondary antibody (Life Technologies [Alexa Fluor]; 1:500) for 1 hour at room temperature in 1% bovine serum albumin/PBS buffer. They were mounted in Vectashield (Vector Laboratories, Burlingame, CA), and nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride). Immunofluorescence images were acquired by using an Olympus FV1000 Laser Confocal Microscope at 40×/NA 1.3 objective (stacking from basement membrane to apical site at 1-μM intervals).
Tissue microarray construction and immunohistochemical analysis
Tumor samples were collected and the TMA comprising formalin-fixed, paraffin-embedded astrocytoma tissues was processed at Tampere University Hospital as described previously [96]. Briefly, histologically representative tumor regions were selected by a neuropathologist (HH), and samples from these areas were placed in TMA blocks using a custom-built instrument (Beecher Instruments, Sun Prairie, WI). The diameter of the tissue cores in the microarray block was 1 mm. Altogether, 222 diffusely infiltrating astrocytomas (167 glioblastomas, 17 grade 3 astrocytomas, and 38 grade 2 astrocytomas) were included in the immunohistochemical analysis. For staining, 5-μm sections from TMA blocks were deparaffinized in xylene or hexane and rehydrated through an ethanol dilution series. Immunohistochemical staining was performed with goat antibodies against human IGFBP2 (#SC-6001; 1:300), phosphorylated STAT3 (#9145; 1:100), and EGFR (GR-01, Calbiochem, San Diego, CA; 1:50), together with the HRP-diaminobenzidine (DAB)–based Cell and Tissue Staining Kit (R&D Systems, Minneapolis, MN) or the Envision+System HRP-DAB kit (Dako, Carpenteria, CA).
Intensity of cytosolic expression levels of the proteins in tumor cells was manually quantified by using a scoring system from 0 to 3 (0 = no signal, 1 = weak signal, 2 = moderate signal and 3 = strong signal). The proportion of the cells with nuclear protein localization was manually classified into 4 categories: 0%, <10%, 10-30% and ≥30%. Intensity of nuclear expression levels in tumor cells was manually quantified by using a scoring system from 0 to 2 (0 = no signal, 1 = weak signal, 2 = strong signal). The TMA samples were examined and scored by 2 neuropathologists who were blinded to the clinical data. A survival association analysis of the patients from whom these samples were taken compared survival in patients with nuclear co-localization of all 3 proteins—IGFBP2, EGFR and phosphorylated STAT3 (≥1% cells with nuclear staining)—with survival of all the other patients. The survival data were analyzed by the log-rank test and visualized with a Kaplan-Meier plot. Statistical analyses were run with SPSS 20.0 software for Windows (SPSS Inc., Chicago, IL). The statistical significance of associations was evaluated by using the Pearson chi-square test.
Invasion and migration assays
The cell invasion assay was performed in triplicate in Matrigel-coated transwell chambers (8-μm pore size; (BD Biosciences, San Jose, CA). The cells were plated in 500 μL of serum-free medium (4 × 104 cells per transwell) and allowed to invade toward a medium containing 10% FBS for 16 hours. Cells that invaded into the underside of the filter were fixed and stained with HEMA-DIFF solution (Thermo Fisher Scientific). The numbers of invaded cells from 5 randomly chosen fields from each membrane were counted. The cell migration assay was performed the same way as the invasion assay, using transwell chambers (8-μm pore size, BD Biosciences) and the cells were allowed to migrate for 4 hours. The data were expressed as means±s.e.m., and analyzed by student t-test for difference between two groups.
Statistical analysis
Experiments were performed at least three times. GraphPrism 6 (GraphPad, La Jolla, CA, USA) and SPSS 20.0 software for Windows (SPSS Inc., Chicago, IL) were used for statistical analysis and graphing. The Spearman correlation test was used to examine correlation between protein or phosphoprotein expression in the TCGA RPPA data set. The survival data were analyzed by the log-rank test and visualized with a Kaplan-Meier plot. The statistical significance of protein associations in the TMA data set was evaluated by using the Pearson chi-square test. Statistical test on GSEA was estimated as previously described [95]. Student t-tests were used for paired comparisons where variances were estimated to be similar. Except for one-side test for the GSEA analysis [95], all other tests were two-sided with P<0.05 as the threshold for statistical significance in all tests. Indicated annotations correspond to the following P-values: *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Oliver Bogler, Zhimin Lu, Frederick Lang, Paul Chiao and Tapio Visakorpi for their helpful comments and discussions, and Kathryn L. Hale, Department of Scientific Publications at MD Anderson Cancer Center, for editing the manuscript. We thank Ville Kytölä for contributions to the immunohistochemical association analyses. This work was partially supported by grants from the U.S. National Institutes of Health (CA098503, CA141432 and CA143835 to W.Z and G.N.F and U24 CA143835 to W.Z.), by funding for the Cancer Systems Informatics Center from the National Foundation for Cancer Research (W.Z), by NIH/NCI grant P30CA016672 to MD Anderson Cancer Center supporting the Flow Cytometry and Cellular Imaging Core Facility, and by the Finnish Funding Agency for Technology and Innovation Finland Distinguished Professor program and the Academy of Finland (grant 259038 to K.G.). W.K.C. is a Fellow of the National Foundation for Cancer Research.
Footnotes
The authors have no potential conflicts of interest to disclose.
REFERENCES
- 1.Dobrowolski R, De Robertis EM. Endocytic control of growth factor signalling: multivesicular bodies as signalling organelles. Nat Rev Mol Cell Biol. 2012;13(1):53–60. doi: 10.1038/nrm3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorkin A, Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol. 2002;3(8):600–14. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 3.Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10(9):609–22. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–33. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 5.Baxter RC. Insulin-like growth factor binding protein-3 (IGFBP-3): Novel ligands mediate unexpected functions. J Cell Commun Signal. 2013;7(3):179–89. doi: 10.1007/s12079-013-0203-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joy A, et al. Nuclear accumulation of FGF-2 is associated with proliferation of human astrocytes and glioma cells. Oncogene. 1997;14(2):171–83. doi: 10.1038/sj.onc.1200823. [DOI] [PubMed] [Google Scholar]
- 7.Stachowiak MK, et al. Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J Cell Biochem. 2003;90(4):662–91. doi: 10.1002/jcb.10606. [DOI] [PubMed] [Google Scholar]
- 8.Arese M, et al. Nuclear activities of basic fibroblast growth factor: potentiation of low-serum growth mediated by natural or chimeric nuclear localization signals. Mol Biol Cell. 1999;10(5):1429–44. doi: 10.1091/mbc.10.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin SY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3(9):802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 10.Sarfstein R, Werner H. Minireview: nuclear insulin and insulin-like growth factor-1 receptors: a novel paradigm in signal transduction. Endocrinology. 2013;154(5):1672–9. doi: 10.1210/en.2012-2165. [DOI] [PubMed] [Google Scholar]
- 11.Rakowicz-Szulczynska EM, et al. Chromatin binding of epidermal growth factor, nerve growth factor, and platelet-derived growth factor in cells bearing the appropriate surface receptors. Proc Natl Acad Sci U S A. 1986;83(11):3728–32. doi: 10.1073/pnas.83.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baxter RC. Circulating binding proteins for the insulinlike growth factors. Trends Endocrinol Metab. 1993;4(3):91–6. doi: 10.1016/1043-2760(93)90085-s. [DOI] [PubMed] [Google Scholar]
- 13.Clemmons DR. Insulinlike growth factor binding proteins. Trends Endocrinol Metab. 1990;1(8):412–7. doi: 10.1016/1043-2760(90)90102-9. [DOI] [PubMed] [Google Scholar]
- 14.Scharf J, et al. Synthesis of insulinlike growth factor binding proteins and of the acid-labile subunit in primary cultures of rat hepatocytes, of Kupffer cells, and in cocultures: regulation by insulin, insulinlike growth factor, and growth hormone. Hepatology. 1996;23(4):818–27. doi: 10.1053/jhep.1996.v23.pm0008666337. [DOI] [PubMed] [Google Scholar]
- 15.Holmes KM, et al. Insulin-like growth factor-binding protein 2-driven glioma progression is prevented by blocking a clinically significant integrin, integrin-linked kinase, and NF-kappaB network. Proc Natl Acad Sci U S A. 2012;109(9):3475–80. doi: 10.1073/pnas.1120375109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang GK, et al. An interaction between insulin-like growth factor-binding protein 2 (IGFBP2) and integrin alpha5 is essential for IGFBP2-induced cell mobility. J Biol Chem. 2006;281(20):14085–91. doi: 10.1074/jbc.M513686200. [DOI] [PubMed] [Google Scholar]
- 17.Pereira JJ, et al. Bimolecular interaction of insulin-like growth factor (IGF) binding protein-2 with alphavbeta3 negatively modulates IGF-I-mediated migration and tumor growth. Cancer Res. 2004;64(3):977–84. doi: 10.1158/0008-5472.can-03-3056. [DOI] [PubMed] [Google Scholar]
- 18.Mehrian-Shai R, et al. Insulin growth factor-binding protein 2 is a candidate biomarker for PTEN status and PI3K/Akt pathway activation in glioblastoma and prostate cancer. Proc Natl Acad Sci U S A. 2007;104(13):5563–8. doi: 10.1073/pnas.0609139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han S, et al. Exogenous IGFBP-2 promotes proliferation, invasion, and chemoresistance to temozolomide in glioma cells via the integrin beta1-ERK pathway. Br J Cancer. 2014;111(7):1400–9. doi: 10.1038/bjc.2014.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azar WJ, et al. IGFBP-2 enhances VEGF gene promoter activity and consequent promotion of angiogenesis by neuroblastoma cells. Endocrinology. 2011;152(9):3332–42. doi: 10.1210/en.2011-1121. [DOI] [PubMed] [Google Scholar]
- 21.Terrien X, et al. Intracellular colocalization and interaction of IGF-binding protein-2 with the cyclin-dependent kinase inhibitor p21CIP1/WAF1 during growth inhibition. Biochem J. 2005;392(Pt 3):457–65. doi: 10.1042/BJ20050517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyako K, et al. PAPA-1 Is a nuclear binding partner of IGFBP-2 and modulates its growth-promoting actions. Mol Endocrinol. 2009;23(2):169–75. doi: 10.1210/me.2008-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeflich A, et al. Peri/nuclear localization of intact insulin-like growth factor binding protein-2 and a distinct carboxyl-terminal IGFBP-2 fragment in vivo. Biochem Biophys Res Commun. 2004;324(2):705–10. doi: 10.1016/j.bbrc.2004.09.111. [DOI] [PubMed] [Google Scholar]
- 24.Besnard V, et al. Distinct patterns of insulin-like growth factor binding protein (IGFBP)-2 and IGFBP-3 expression in oxidant exposed lung epithelial cells. Biochim Biophys Acta. 2001;1538(1):47–58. doi: 10.1016/s0167-4889(00)00136-1. [DOI] [PubMed] [Google Scholar]
- 25.Gerrard DE, Okamura CS, Grant AL. Expression and location of IGF binding proteins-2, -4, and -5 in developing fetal tissues. J Anim Sci. 1999;77(6):1431–41. doi: 10.2527/1999.7761431x. [DOI] [PubMed] [Google Scholar]
- 26.Green BN, et al. Distinct expression patterns of insulin-like growth factor binding proteins 2 and 5 during fetal and postnatal development. Endocrinology. 1994;134(2):954–62. doi: 10.1210/endo.134.2.7507840. [DOI] [PubMed] [Google Scholar]
- 27.Lee WH, Michels KM, Bondy CA. Localization of insulin-like growth factor binding protein-2 messenger RNA during postnatal brain development: correlation with insulin-like growth factors I and II. Neuroscience. 1993;53(1):251–65. doi: 10.1016/0306-4522(93)90303-w. [DOI] [PubMed] [Google Scholar]
- 28.Wood TL, Streck RD, Pintar JE. Expression of the IGFBP-2 gene in post-implantation rat embryos. Development. 1992;114(1):59–66. doi: 10.1242/dev.114.1.59. [DOI] [PubMed] [Google Scholar]
- 29.Huynh H, et al. IGF binding protein 2 supports the survival and cycling of hematopoietic stem cells. Blood. 2011;118(12):3236–43. doi: 10.1182/blood-2011-01-331876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuller GN, et al. Reactivation of insulin-like growth factor binding protein 2 expression in glioblastoma multiforme: a revelation by parallel gene expression profiling. Cancer Res. 1999;59(17):4228–32. [PubMed] [Google Scholar]
- 31.Busund LT, et al. Significant expression of IGFBP2 in breast cancer compared with benign lesions. J Clin Pathol. 2005;58(4):361–6. doi: 10.1136/jcp.2004.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazawa T, et al. Neuroendocrine cancer-specific up-regulating mechanism of insulin-like growth factor binding protein-2 in small cell lung cancer. Am J Pathol. 2009;175(3):976–87. doi: 10.2353/ajpath.2009.081004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunlap SM, et al. Insulin-like growth factor binding protein 2 promotes glioma development and progression. Proc Natl Acad Sci U S A. 2007;104(28):11736–41. doi: 10.1073/pnas.0703145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colman H, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12(1):49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh D, et al. IGFBP2 promotes glioma tumor stem cell expansion and survival. Biochem Biophys Res Commun. 2010;397(2):367–72. doi: 10.1016/j.bbrc.2010.05.145. [DOI] [PubMed] [Google Scholar]
- 36.Li X, et al. Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene. 2014 doi: 10.1038/onc.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo HW, et al. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res. 2008;14(19):6042–54. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scrideli CA, et al. Prognostic significance of co-overexpression of the EGFR/IGFBP-2/HIF-2A genes in astrocytomas. J Neurooncol. 2007;83(3):233–9. doi: 10.1007/s11060-007-9328-0. [DOI] [PubMed] [Google Scholar]
- 39.Nishikawa R, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91(16):7727–31. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekstrand AJ, et al. Functional characterization of an EGF receptor with a truncated extracellular domain expressed in glioblastomas with EGFR gene amplification. Oncogene. 1994;9(8):2313–20. [PubMed] [Google Scholar]
- 41.Heimberger AB, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11(4):1462–6. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 42.Shao H, et al. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003;63(14):3923–30. [PubMed] [Google Scholar]
- 43.Park OK, Schaefer TS, Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc Natl Acad Sci U S A. 1996;93(24):13704–8. doi: 10.1073/pnas.93.24.13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo HW, et al. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8(2):232–45. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo HW, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7(6):575–89. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Jaganathan S, et al. A functional nuclear epidermal growth factor receptor, SRC and Stat3 heteromeric complex in pancreatic cancer cells. PLoS One. 2011;6(5):e19605. doi: 10.1371/journal.pone.0019605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Lo HW, et al. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65(1):338–48. [PubMed] [Google Scholar]
- 48.Hoshino M, et al. Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74(1):15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- 49.Xia W, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48(7):610–7. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006;94(2):184–8. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, et al. Insulin-like growth factor binding protein 2 enhances glioblastoma invasion by activating invasion-enhancing genes. Cancer Res. 2003;63(15):4315–21. [PubMed] [Google Scholar]
- 52.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6(1):32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 53.Zhou BB, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10(1):39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng X, et al. Inhibition of ADAM17 reduces hypoxia-induced brain tumor cell invasiveness. Cancer Sci. 2007;98(5):674–84. doi: 10.1111/j.1349-7006.2007.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haynik DM, Roma AA, Prayson RA. HER-2/neu expression in glioblastoma multiforme. Appl Immunohistochem Mol Morphol. 2007;15(1):56–8. doi: 10.1097/01.pai.0000213133.09160.da. [DOI] [PubMed] [Google Scholar]
- 56.Waage IS, Vreim I, Torp SH. C-erbB2/HER2 in human gliomas, medulloblastomas, and meningiomas: a minireview. Int J Surg Pathol. 2013;21(6):573–82. doi: 10.1177/1066896913492196. [DOI] [PubMed] [Google Scholar]
- 57.Torp SH, et al. C-erbB-2/HER-2 protein in human intracranial tumours. Eur J Cancer. 1993;29A(11):1604–6. doi: 10.1016/0959-8049(93)90305-y. [DOI] [PubMed] [Google Scholar]
- 58.Tuzi NL, et al. Expression of growth factor receptors in human brain tumours. Br J Cancer. 1991;63(2):227–33. doi: 10.1038/bjc.1991.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haapasalo H, et al. c-erbB-2 in astrocytomas: infrequent overexpression by immunohistochemistry and absence of gene amplification by fluorescence in situ hybridization. Br J Cancer. 1996;73(5):620–3. doi: 10.1038/bjc.1996.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azar WJ, et al. IGFBP-2 nuclear translocation is mediated by a functional NLS sequence and is essential for its pro-tumorigenic actions in cancer cells. Oncogene. 2014;33(5):578–88. doi: 10.1038/onc.2012.630. [DOI] [PubMed] [Google Scholar]
- 61.Smalheiser NR. Proteins in unexpected locations. Mol Biol Cell. 1996;7(7):1003–14. doi: 10.1091/mbc.7.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganesan A, Zhang J. How cells process information: quantification of spatiotemporal signaling dynamics. Protein Sci. 2012;21(7):918–28. doi: 10.1002/pro.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7(3):165–76. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Housden BE, Perrimon N. Spatial and temporal organization of signaling pathways. Trends Biochem Sci. 2014;39(10):457–64. doi: 10.1016/j.tibs.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiley HS. Trafficking of the ErbB receptors and its influence on signaling. Exp Cell Res. 2003;284(1):78–88. doi: 10.1016/s0014-4827(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 66.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, et al. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol Cell Biol. 2002;22(20):7279–90. doi: 10.1128/MCB.22.20.7279-7290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pennock S, Wang Z. Stimulation of cell proliferation by endosomal epidermal growth factor receptor as revealed through two distinct phases of signaling. Mol Cell Biol. 2003;23(16):5803–15. doi: 10.1128/MCB.23.16.5803-5815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miaczynska M, Pelkmans L, Zerial M. Not just a sink: endosomes in control of signal transduction. Curr Opin Cell Biol. 2004;16(4):400–6. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 70.Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell. 2001;12(6):1897–910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brand TM, et al. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov Med. 2011;12(66):419–32. [PMC free article] [PubMed] [Google Scholar]
- 72.Boerner JL, et al. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol. 2004;24(16):7059–71. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Demory ML, et al. Epidermal growth factor receptor translocation to the mitochondria: regulation and effect. J Biol Chem. 2009;284(52):36592–604. doi: 10.1074/jbc.M109.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coffer PJ, Kruijer W. EGF receptor deletions define a region specifically mediating STAT transcription factor activation. Biochem Biophys Res Commun. 1995;210(1):74–81. doi: 10.1006/bbrc.1995.1629. [DOI] [PubMed] [Google Scholar]
- 75.Shono T, et al. Cyclooxygenase-2 expression in human gliomas: prognostic significance and molecular correlations. Cancer Res. 2001;61(11):4375–81. [PubMed] [Google Scholar]
- 76.Joki T, et al. Expression of cyclooxygenase 2 (COX-2) in human glioma and in vitro inhibition by a specific COX-2 inhibitor, NS-398. Cancer Res. 2000;60(17):4926–31. [PubMed] [Google Scholar]
- 77.Xu K, Wang L, Shu HK. COX-2 overexpression increases malignant potential of human glioma cells through Id1. Oncotarget. 2014;5(5):1241–52. doi: 10.18632/oncotarget.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang DI, et al. NO-mediated chemoresistance in C6 glioma cells. Ann N Y Acad Sci. 2002;962:8–17. doi: 10.1111/j.1749-6632.2002.tb04052.x. [DOI] [PubMed] [Google Scholar]
- 79.Giannopoulou E, et al. Expression of HIF-1alpha and iNOS in astrocytic gliomas: a clinicopathological study. In Vivo. 2006;20(3):421–5. [PubMed] [Google Scholar]
- 80.Hara A, Okayasu I. Cyclooxygenase-2 and inducible nitric oxide synthase expression in human astrocytic gliomas: correlation with angiogenesis and prognostic significance. Acta Neuropathol. 2004;108(1):43–8. doi: 10.1007/s00401-004-0860-0. [DOI] [PubMed] [Google Scholar]
- 81.Jahani-Asl A, Bonni A. iNOS: a potential therapeutic target for malignant glioma. Curr Mol Med. 2013;13(8):1241–9. doi: 10.2174/1566524011313080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3(11):e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orian JM, et al. Overexpression of multiple oncogenes related to histological grade of astrocytic glioma. Br J Cancer. 1992;66(1):106–12. doi: 10.1038/bjc.1992.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Herms JW, et al. c-myc oncogene family expression in glioblastoma and survival. Surg Neurol. 1999;51(5):536–42. doi: 10.1016/s0090-3019(98)00028-7. [DOI] [PubMed] [Google Scholar]
- 85.Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. J Cell Physiol. 2009;218(3):460–6. doi: 10.1002/jcp.21635. [DOI] [PubMed] [Google Scholar]
- 86.Swindle CS, et al. Epidermal growth factor (EGF)-like repeats of human tenascin-C as ligands for EGF receptor. J Cell Biol. 2001;154(2):459–68. doi: 10.1083/jcb.200103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaser MR, Lakshmanan J, Fisher DA. Comparison between epidermal growth factor, transforming growth factor-alpha and EGF receptor levels in regions of adult rat brain. Brain Res Mol Brain Res. 1992;16(3-4):316–22. doi: 10.1016/0169-328x(92)90241-3. [DOI] [PubMed] [Google Scholar]
- 88.Kudlow JE, et al. Transforming growth factor-alpha in the mammalian brain. Immunohistochemical detection in neurons and characterization of its mRNA. J Biol Chem. 1989;264(7):3880–3. [PubMed] [Google Scholar]
- 89.Ferrer I, et al. Transforming growth factor-alpha immunoreactivity in the developing and adult brain. Neuroscience. 1995;66(1):189–99. doi: 10.1016/0306-4522(94)00584-r. [DOI] [PubMed] [Google Scholar]
- 90.Lazar LM, Blum M. Regional distribution and developmental expression of epidermal growth factor and transforming growth factor-alpha mRNA in mouse brain by a quantitative nuclease protection assay. J Neurosci. 1992;12(5):1688–97. doi: 10.1523/JNEUROSCI.12-05-01688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Becher OJ, et al. IGFBP2 is overexpressed by pediatric malignant astrocytomas and induces the repair enzyme DNA-PK. J Child Neurol. 2008;23(10):1205–13. doi: 10.1177/0883073808321766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McDonald KL, et al. IQGAP1 and IGFBP2: valuable biomarkers for determining prognosis in glioma patients. J Neuropathol Exp Neurol. 2007;66(5):405–17. doi: 10.1097/nen.0b013e31804567d7. [DOI] [PubMed] [Google Scholar]
- 93.Santarius T, et al. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10(1):59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 94.Salomon DS, et al. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19(3):183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 95.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sallinen SL, et al. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000;60(23):6617–22. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.