Abstract
Background
Intestinal microbiota influences the progression of colitis-associated colorectal cancer (CAC). With diet being a key determinant of the gut microbial ecology, dietary interventions are an attractive avenue for the prevention of CAC. Curcumin is the most active constituent of the ground rhizome of the Curcuma Longa plant, which has been demonstrated to have anti-inflammatory, anti-oxidative and anti-proliferative properties.
Methods
Il10−/− mice on 129/SvEv background were used as a model of CAC. Starting at 10 weeks of age, WT or Il10−/− mice received six weekly i.p. injections of azoxymethane (AOM) or saline, and were started on either a control or curcumin-supplemented diet. Stools were collected every 4 weeks for microbial community analysis. Mice were sacrificed at 30 weeks of age.
Results
Curcumin-supplemented diet increased survival, decreased colon weight/length ratio, and at 0.5%, entirely eliminated tumor burden. Although colonic histology indicated improvement with curcumin, no effects of mucosal immune responses have been observed in PBS/Il10−/− mice, and limited effects were seen in AOM/Il10−/− mice. In WT and in Il10−/− mice, curcumin increased bacterial richness, prevented age-related decrease in alpha diversity, increased the relative abundance of Lactobacillales, and decreased Coriobacterales order. Taxonomic profile of AOM/Il10−/− mice receiving curcumin was more similar to those of wild-type mice than those fed control diet.
Conclusions
In AOM/Il10−/− model, curcumin reduced or eliminated colonic tumor burden with limited effects on mucosal immune responses. The beneficial effect of curcumin on tumorigenesis was associated with the maintenance of a more diverse colonic microbial ecology.
Keywords: Colitis-associated colon cancer, Microbiota, Curcumin, Diet, 16S rRNA
INTRODUCTION
Colorectal cancer (CRC) is the third highest cause of cancer-related mortality in the U.S.A. It is estimated that 93,090 people will be diagnosed with colon cancer, 39,610 with rectal cancer, and 49,700 will die from CRC in 2015 in the US alone. 1 Worldwide, CRC is the fourth most common cancer cause of death, accounting for roughly 1.2 million new cases and 600,000 deaths per year.2 Genetic predispositions underlie the pathogenesis of less than 5% of CRC cases, mostly in Lynch syndrome [mutations in the mismatch repair pathway (MMR)], in hereditary non-polyposis colorectal cancer (HNPCC), familial adenomatous polyposis (FAP), and MYH-associated polyposis (MAP). The vast majority of CRC cases, estimated at over 95%, are sporadic cancers occurring in individuals without inheritable predisposition to the disease.3 Environmental factors, inflammation, and gut microbiota have been implicated in the pathogenesis of cancer initiation and the progression from a hyper-proliferative state, through adenoma formation, to progression to carcinoma.
The current concept that the inflammatory microenvironment represents a risk factor for cancer development dates back to the early observations of inflammatory infiltrates within neoplastic lesions by Rudolf Virchov.4 In the case of CRC, the effects of chronic inflammation on cancer are well exemplified in Inflammatory Bowel Diseases (IBD). Crohn’s disease (CD) and ulcerative colitis (UC) are both associated with increased cancer risk 5, with duration and severity of chronic colitis conferring significant risk factors for colitis-associated colon cancer (CAC).6–8 Although the etiology of IBD still remains incompletely understood, animal model-based studies indicate that the host intestinal microbiota triggers and/or perpetuates an immune response required for the onset and progression of the disease. Next generation sequencing approaches have elucidated profound changes in gut microbial ecology in IBD patients and in mouse models, although demonstrating the causality and the mechanisms involved still remain a challenge.9 Intestinal microbiota is also a target of inflammation that affects the development and progression of CAC.10 Recent evidence from a gnotobiotic Il10−/− mouse model indicates that colitis is not sufficient to promote CAC and requires the presence or expansion of a more specific pathobiont, i.e. a symbiont that is able to promote pathology only when host’s specific genetic or environmental conditions are altered.11
Currently, three theories are proposed to explain the microbial influence on colon cancer development: a model in which the pathogenesis is driven by selected pathobiont(s); a model in which global changes to the microbial ecology support carcinogenesis; and a model which combines pathobiont(s) interacting with a changing microbial community.12 T-bet−/−xRag−/− Ulcerative Colitis (TRUC) mice13 provide an example, which supports the latter model. In TRUC mice, the combination of Klebsiella pneumoniae and Proteus mirabilis potently induced colitis in conventionally housed, but not in germ-free animals. This suggested a dependence on the interaction of these two bacterial species with the commensal microbiota community to induce disease.13 The interplay of one or more specific bacterial species with the entire commensal bacterial community to drive disease development highlights the importance global changes in microbial gut ecology. It also suggests that therapeutic approaches with the potential to restore its diversity and normalize the composition of the colonic microbial community would be highly desirable.
Curcumin (diferuloyl-methane), a bioactive component derived from a rhizome of the Curcuma longa plant, has been demonstrated to function as an antiseptic, analgesic, anti-inflammatory, antioxidant, chemopreventive, chemo- and radio-sensitizing agent14–16. Our laboratory has extensively studied the anti-inflammatory benefits of curcumin in murine IBD models.17–19 More recently, we also demonstrated that curcumin inhibits colon cancer cell migration through physical interaction with and activation of PTPN1 tyrosine phosphatase to reduce the abnormally hyperphosphorylated and hyperactive form of cortactin, a protein implicated in cancer motility and tumor invasiveness. 20 We hypothesized that due to its poor bioavailability, orally administered curcumin as a dietary supplement targets and impacts primarily epithelial cells and the gut microbiota, and that curcumin may modulate colonic microbial ecology and prevent the progression of chronic colitis to CAC. Although preventive treatment with curcumin in AOM/DSS model of CAC was shown to be effective,21 chemopreventive effects of curcumin in established immune-mediated colitis and its association with colonic microbiota has not been previously investigated. In this report, we demonstrate that in a model of AOM-induced colon cancer in conventionally housed Il10−/− mice, dietary curcumin dose-dependently reduced or entirely prevented colon cancer development. These chemopreventive effects appeared unrelated to the reduction of inflammation, but rather to the normalizing effects of curcumin on colonic microbial ecology, thus suggesting that curcumin functions as an effective agent for restoring healthy gut homeostasis and microbial-host relationship.
METHODS
Experimental animals
Specific pathogen-free wild type (WT) 129/SvEv mice and germ-free Il10−/− mice on the same genetic background were originally obtained from the National Gnotobiotic Rodent Resource Center at the University of North Carolina, Chapel Hill, re-derived through embryo transfer, and transferred to a conventional animal facility at the University of Arizona Health Sciences Center. Sentinel mice were routinely monitored and determined as free from common murine pathogens (MHV, MPV, MVM, TMEV, Mycoplasma pulmonis, Sendai, EDIM, MNV, ecto- and endoparasites). All animal protocols and procedures were approved by the University of Arizona Animal Care and Use Committee.
Diets
98.05% pure curcumin, free of contaminating curcuminoids (demethoxy-curcumin and bis-demethoxy-curcumin) was obtained from ChromaDex (Irvine, CA). Curcumin was incorporated into NIH-31 modified open formula (base composition listed under the following Link) at 0.05, 0.1, 0.5 and 1% and pelleted by Harlan Teklad (Madison, WI). Based on the average chow intake, the calculated human equivalency dose (HED) of curcumin ranged from 8 mg/kg/day – 162 mg/kg/day, for 0.05% and 1% diets, respectively, and were within the range of well tolerated doses reported by phase I human trials.22, 23 Control diet without curcumin supplementation was processed in parallel. Diets were vacuum-packed in small portions and kept at −20°C until needed.
AOM/Il10−/− model of CAC and experimental groups
We utilized a well-documented model to study the development of colitis-associated colon cancers in IL-10-deficient mice. 24, 25 This protocol is based on the use of the mutagenic agent, AOM, which exerts colonotropic carcinogenicity. WT and Il10−/− mice on 129/SvEv genetic background reared and housed in the same room were allowed to age to 10 weeks by which point the Il10−/− mice develop spontaneous and progressive colitis.26, 27 Il10−/− mice entering the study were prescreened by ELISA for systemic marker of inflammation, serum amyloid A to ensure established colitis. 60 WT and 60 Il10−/− mice were assigned into PBS vs. AOM groups, and into respective dietary groups (6 mice per treatment/diet group). Six intraperitoneal injections of AOM (10mg/kg) were administered weekly to initiate tumorigenesis. Mice were monitored for another 14 weeks, and sacrificed at the age of 30 weeks (Fig. 1A) at which time tissues were harvested for histopathological and biochemical analyses.
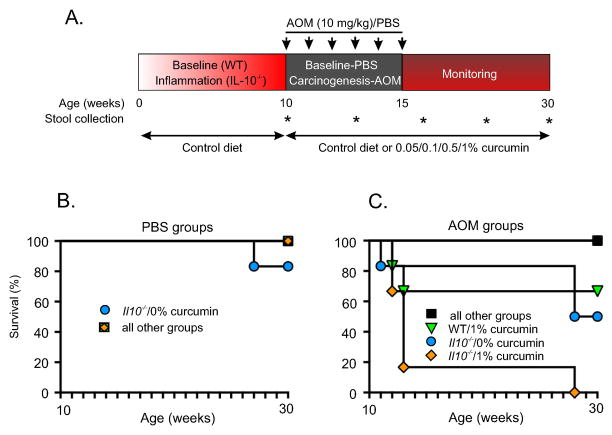
Figure 1. Experimental setup and survival.
(A) Il10−/− mice and wild-type (WT) control mice were maintained until 10 weeks of age to allow development of colitis in the Il10−/− mice. Mice were then randomized into groups of 6 for each curcumin diet dose: 0%, 0.05%, 0.1%, 0.5%, and 1%. Starting on week 10, mice received 6 weekly injections of PBS or AOM and were monitored till 30th week of life. Assessment of survival in PBS- (B) or AOM-injected groups (C). 16% of PBS-injected Il10−/− mice died in the 0% curcumin control diet group. All other PBS-injected mice in every group had 100% survival (A). 50% of the AOM-injected Il10−/− mice died in the 0% curcumin control diet group. Significant toxicity of 1% curcumin diet was documented in AOM-injected mice. All other AOM-injected mice had 100% survival (B). Kaplan-Meier curves were generated using the GraphPad Prism software (v. 5.0a).
Histology and scoring
Colons from WT and Il10−/− mice on different curcumin diets were harvested, Swiss-rolled, and fixed in 10% neutral buffered formalin (Fisher Scientific, Tustin, CA). Fixed tissues were then embedded in paraffin, and 5-μm-thick tissue cuts were stained with hematoxylin and eosin (H&E) for light microscopic examination. Sections were graded by a veterinary pathologist blinded to the study design according to previously published criteria28.
Immunohistochemistry
Immunohistochemical labeling (IHC) of β-catenin was performed with a rabbit monoclonal antibody (clone E247; AbCam). Ki67 IHC was performed using a rabbit polyclonal antibody (#NCL-Ki67p, Leica Biosystems GmbH, Nussloch, Germany). Tissue sections were stained with a Discovery XT Automated Immunostainer (VMSI; Ventana Medical Systems, Inc., Tucson, AZ; VMSI) using VMSI-validated reagents for deparaffinization, cell conditioning (antigen retrieval with a borate-EDTA buffer), primary antibody staining, detection and amplification using a biotinylated-streptavidin-HRP and diaminobenzidine (DAB) system and hematoxylin counterstaining. Following staining, slides were dehydrated through graded alcohols to xylene and coverslipped with mounting medium.
Images were captured using an Zeiss Axioplan microscope (Carl Zeiss MicroImaging, Thornwood, NY) using Nikon Digital Sight DS-Fi1 camera and NIS-Element software (Nikon Instruments, Melville, NY). All images were standardized for light intensity and white balance. Expression of Ki67 and β-catenin in the colonic epithelial cells was evaluated by two independent and unbiased investigators. Ki67-positive cells were counted per 3–5 high-powered fields and expressed as mean +/− SEM in respective groups. β-catenin was evaluated in a similar manner and was expressed as percentage of epithelial cells with exclusively basolateral membrane staining or mixed cytoplasmic/nuclear staining.
Real-time RT-PCR
Real-time RT-PCR was used to evaluate mucosal expression of TNF-α, IFNγ, IL-6, IL-1β, IL-17A, IL-12p40, IL-23 mRNA. Total RNA was isolated from mouse distal colon using TRIzol reagent (Invitrogen, Carlsbad, CA). 250ng of total RNA was reverse-transcribed using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Subsequently, 20 μl of the PCR reactions were set up in 96-well plates containing 10 μl 2x IQ Supermix (Bio-Rad), 1 μl TaqMan® primer/probe set (ABI, Foster City, CA), 2 μl of the cDNA synthesis reaction (10% of RT reaction) and 7 μl of nuclease-free water. Reactions were run and analyzed on a Bio-Rad iCycler iQ real–time PCR detection system. Data were analyzed by using the comparative Ct method as means of relative quantification, normalized to an endogenous reference (TATA Box Bonding Protein, TBP or glyceraldehyde 3-phosphate dehydrogenase, GAPDH) and relative to a calibrator (normalized Ct value obtained from control mice) and expressed as 2−ΔΔCt (Applied Biosystems User Bulletin #2: Rev B “Relative Quantification of Gene Expression”).
Bacterial DNA processing
Fecal samples were collected fresh once per month from individual mice directly into sterile Eppendorf tubes and stored at −80°C until processing. For processing, frozen stool samples were individually placed into an Eppendorf tube containing autoclaved zirconium beads, lysis buffer (100mM NaCl, 10mM Tris, 100mM EDTA and 0.2mg/ml Proteinase K) and a solution of 20% DNAse-free SDS (Sodium dodecyl sulfate) and incubated for 2 hours. The samples were then mixed with 24:24:1 Phenol:Chloroform:IAA (pH 8.0) and placed into an automatic bead beater machine on high settings for 2 min. Samples were centrifuged (8,000 rpm for 3 min.) and the aqueous phase was collected and mixed again with 1:1 Phenol:Chloroform and centrifuged (13,000 rpm, 3 min). The aqueous phase was collected, mixed with −20°C isopropanol and 3M sodium acetate (1:10 ratio), and incubated at 4°C for 20 min. DNA was precipitated by centrifugation (13,000 rpm for 20 min. at 4°C) and excess supernatant was discarded. The remaining pellets were washed with cold (4°C) 100% molecular grade ethanol and centrifuged (13,000 rpm for 5 min. at 4°C). Excess supernatant was discarded and the remaining pelleted material was allowed to air-dry and was then re-suspended in TE (Tris-EDTA, pH 8.0) buffer.
Generation of 16S amplicon library, MiSeq sequencing, and QIIME analysis of microbial gut ecology
The V4 hypervariable region of the 16S rRNA gene was amplified using barcoded PCR with 515F and 806R primers following the Earth Microbiome Project protocol29 and quantified using Nanodrop. Sequencing was performed at Argonne National Laboratories on an Illumina MiSeq (Serial # M02149) using the MCS (MiSeq Control Software) version 2.2.0. 540μL of 6.75pM 16S amplicon library with 597 pooled samples were mixed with 6.75pM PhiX control DNA (10% final), and sequenced in one run, with 17,278,176 total sequences (5′ reads) generated. The raw sequence data was de-multiplexed using QIIME 1.8.0 30 and assigned to sample IDs using QIIME’s default quality filtering process.31 After quality filtering 10,244,883 reads remained. The remaining reads had a median length of 151 bases.
Sequences were assigned to clusters of 97% similarity using QIIME’s uclust-based32 open-reference operational taxonomic unit (OTU) picking protocol33 against the Greengenes 13_5 reference sequence set34. The centroid of each OTU was chosen as the representative sequence for the OTU. The representative sequences were aligned with PyNast35 and the trees were constructed with FastTree36 for phylogenetic calculations. The average number of sequences per sample was 17,160.6 ± 5,944.6 (mean ± SD). After quality filtering and open reference picking, the minimum number of sequences in a given sample in order to retain a sample in the study was set to 6,000. This resulted in the exclusion of 18 samples that contained fewer than 6000 reads.
Beta-diversity calculations were performed using weighted and unweighted-unifrac metrics37 with exactly 6,000 randomly selected sequences per sample using rarefaction as implemented in QIIME. Alpha diversity metrics were calculated using phylogenetic diversity38,39 (a phylogenetic richeness metric) as implemented in QIIME and using Margalef’s diversity index in Excel.
Statistical significance for differences in alpha diversity was determined by nonparametric analysis of variance (ANOVA) tests comparing the alpha diversity results across grouped samples. Statistical significance for differences in beta diversity across sample groups was calculated using ANOSIM (Analysis of Similarity) with 1000 permutations. The Bonferroni-corrected Kroskal-Wallis test was used for to test for differentially abundant OTUs across sample groups.
Prediction of metagenome functional content from the 16S rRNA marker gene was done using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt),40 using PICRUSt’s Kyoto Encyclopedia of Genes and Genomes (KEGG) gene annotation database. This was done by first picking OTUs against 13 August 2013 Greengenes database for compatibility with PICRUSt. The resulting BIOM file was uploaded into the online Galaxy PICRUSt server (http://huttenhower.sph.harvard.edu/galaxy/) for metagenome prediction. The predicted metagenome BIOM file was further analyzed using Statistical Analysis of Metagenomic Profiles (STAMP) software package41, using ANOVA and Tukey-Kramer post-hoc test.
All sequence data generated in this study will be available in Qiita (formerly called the QIIME DB) upon acceptance of the manuscript.
Statistical analysis of non-microbiome data
Normality of data distribution was tested by Shapiro-Wilk test. If normality was confirmed, statistical significance was determined by the factorial analysis of variance (ANOVA) with three main effects (diet, genotype, and/or PBS/AOM groups) followed by the Fisher PLSD post-hoc test with StatView software package v.4.53 (SAS Institute, Cary, NC). In instances where normality test failed (selected groups in histological scoring analysis), Kruskal-Wallis One Way ANOVA on Ranks was employed followed by Dunn’s T test. Where appropriate, unpaired two-tailed T test was used using Excel software. Data were expressed as mean +/− standard error of mean.
RESULTS
Curcumin (0.5%) prevents mortality and improves body weight gain in AOM/Il10−/− mice
We first determined the effect of curcumin on survival and body weight of AOM/Il10−/− mice following a treatment intervention (Fig. 1A). No mortality was observed in WT mice fed curcumin-supplemented diets. One of six Il10−/− mice injected with PBS and fed control diet died before the conclusion of the study. No mortality was observed in chronically inflamed Il10−/− mice injected with PBS and fed curcumin-supplemented diets (Fig. 1B). While 50% mortality was observed in AOM/Il10−/− mice fed control diet, 100% survival was noticed in AOM/Il10−/− mice exposed to 0.5% curcumin diet (Fig. 1C). However, 1% curcumin resulted in increased mortality in both AOM/WT and AOM/Il10−/− mice, therefore these groups were removed from the study. In PBS-treated WT mice, 0.5% curcumin diet tended to result in transient body weight decrease (Fig. S1A), consistent with the previously reported lower food consumption.17 While PBS-injected WT mice on control diet steadily gained weight during the study, similarly treated Il10−/− mice failed to accrue body weight (Fig. S1A). Interestingly, in PBS-treated Il10−/− mice, 0.5% curcumin not only did not improve, but led to a gradual and significant loss of body weight (Fig. S1A). However, in AOM-treated groups, curcumin entirely prevented body weight loss in Il10−/− mice, which was indistinguishable from AOM-treated WT mice on control diet (Fig. S1B).
Curcumin reduced hyperplasia in AOM-treated Il10−/− mice
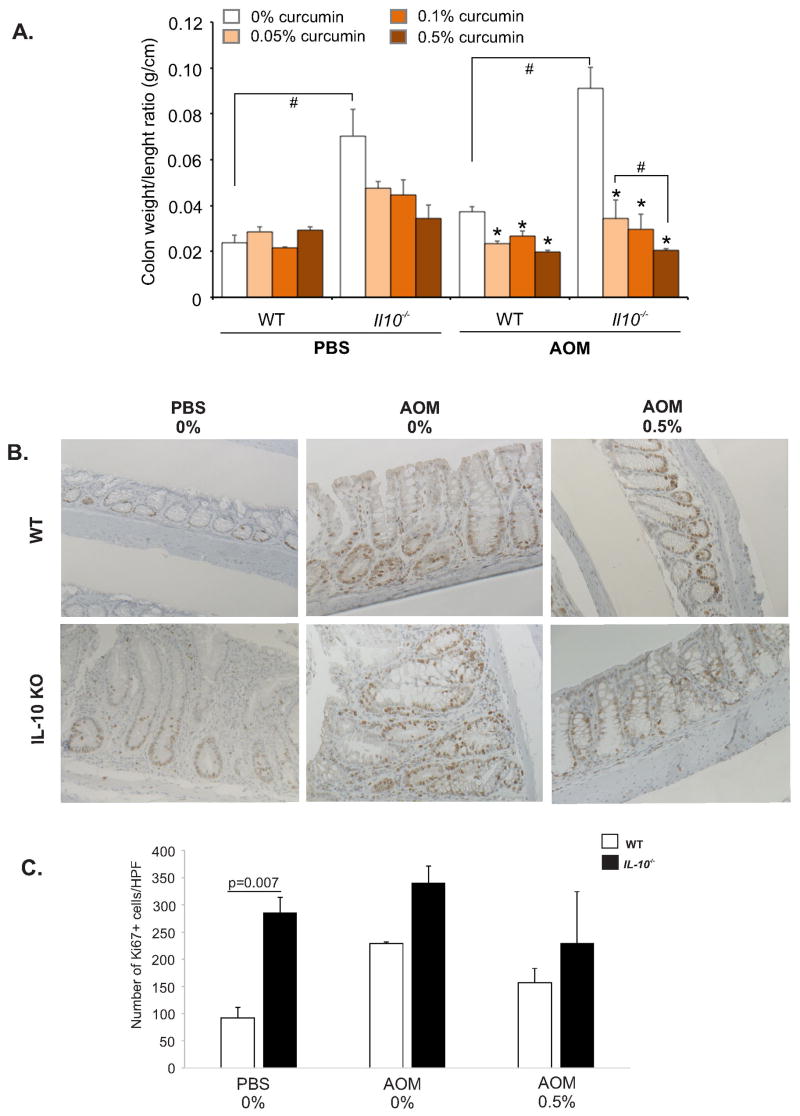
Colon weight/length ratio is a widely used marker of mucosal hyperplasia in chronic colitis, which corresponds well to histological scoring.42 Dietary curcumin dose-dependently reduced weight/length ratio in both Il10−/−/PBS (without reaching statistical significance; ANOVA p=0.15), in WT/AOM, and especially in AOM/Il10−/− groups (p<0.05; Fig. 2A). Analysis of Ki67+ proliferating epithelial cells in the distal colon (primary site of AOM-induced tumorigenesis) was assessed in areas free of frank and histologically confirmed tumors. It showed a modest trend in curcumin-mediated reduction in the numbers of actively proliferating epithelial cells (Fig. 2B,C). This finding implies that in our model, the effects of curcumin on the colonic weight/length ratio was likely secondary to decreased edema and reduced thickness of the smooth muscle layers rather than direct generalized effect on epithelial cell proliferation.
Figure 2. The effects of dietary curcumin on colonic hyperplasia (weight/length ratio) and proliferation in Il10−/−/AOM mice.
(A) Upon euthanasia, colons were removed from the ileocecal junction to the anal verge at the end of the study and the ratio of the weight (mg) to length (cm) was measured. * indicates a significant (p≤0.05) difference from 0% curcumin dietary group within respective treatment group (bar cluster; ANOVA followed by Fisher LSD test). # indicates a significant (p≤0.05) difference between bars indicated by brackets (unpaired Student t-test). (B) Non-tumor tissue sections were labeled with anti-Ki67 antibody. (A) Representative Ki67 staining in WT and Il10−/− mice treated with PBS or AOM and fed control (0%) or 0.5% curcumin-supplemented diet (Magn. 200x). (C) Summary of the numbers of Ki67-positive cells assessed by two investigators blinded to sample group assignment. P=0.007 indicates statistically significant difference between WT and Il10−/− mice on control diet (unpaired t-test).
Curcumin reduced tumor burden in AOM-treated Il10−/− mice
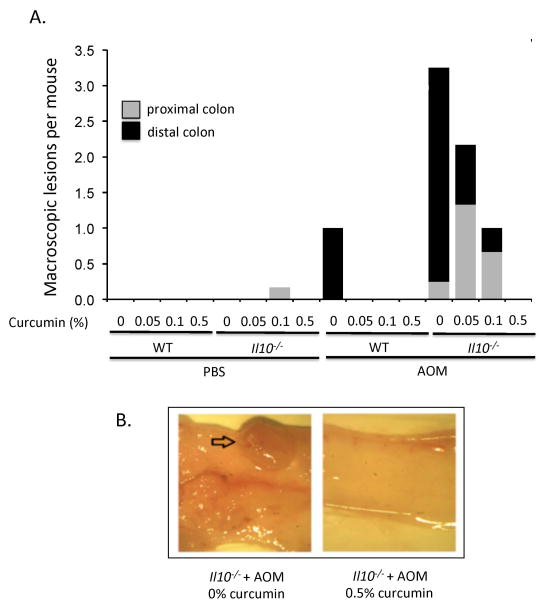
We assessed the occurrence of colonic tumors in PBS- and AOM-treated mice and whether dietary curcumin affects CAC development. PBS-treated WT mice fed control or supplemented diets had no spontaneous macro- or microscopic tumors. AOM-treated Il10−/− mice showed more than 3-fold higher number of macroscopic lesions predominantly localized in the distal colon, characteristic of the model (Fig. 3A).43 Curcumin dose-dependently reduced tumor burden in AOM/Il10−/− mice, with the 0.5% dosage entirely eliminating all macroscopic lesions (Fig. 3A). Microscopic evaluation of tissue sections closely correlated with the results presented in Fig. 3A (not shown). Figure 3B depicts representative photograph of distal colon of AOM-treated Il10−/− mice on control or 0.5% curcumin diet. No lesions were observed in AOM-treated WT mice exposed to curcumin at any of the three doses used.
Figure 3. Curcumin dose-dependently reduces tumor burden in AOM-treated Il10−/− mice.
(A) Average numbers of macroscopic tumors per mouse in the proximal and distal colon were determined at the time of euthanasia with a stereomicroscope. Macroscopic tumors (shown in B) were confirmed by histological analysis by an unbiased pathologist (not shown). The average number of macroscopic tumors per mouse for the AOM/Il10−/− mice is underestimated, since two mice in this group had a high tumor burden classified as “too many to count” (above 10 tumors, numerous and small) and were assigned an arbitrary number of “10” for the purposes of calculation.
Curcumin reduced aberrant localization of β-catenin in AOM-treated Il10−/− mice
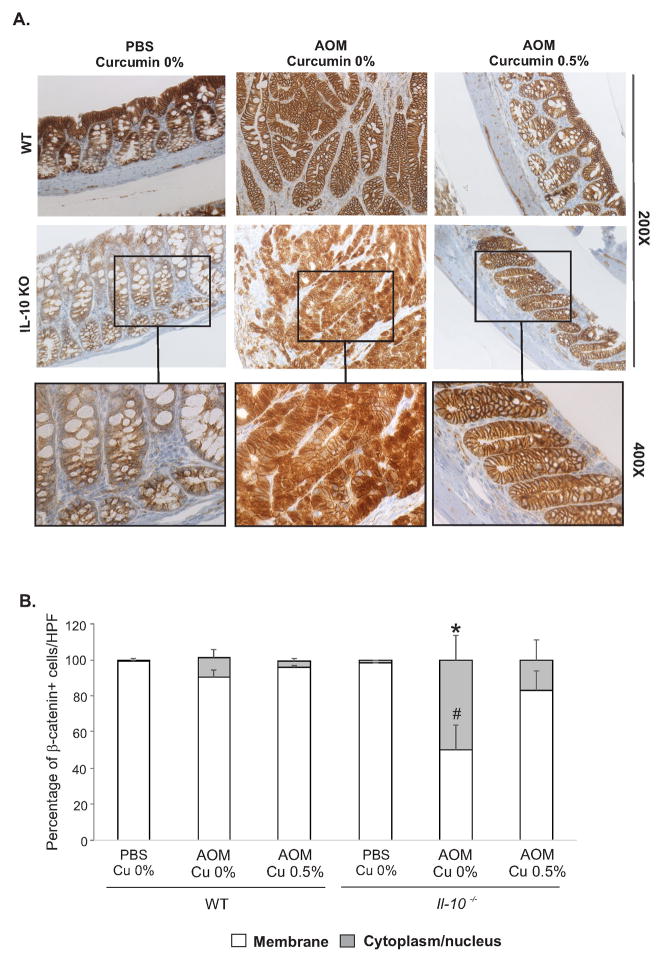
We next evaluated the general expression and localization of β-catenin, a protein involved in cell-cell adhesion, gene transcription, cellular proliferation and differentiation, and which is commonly mutated in colon cancer.44 In PBS-treated WT and Il10−/− mice, β-catenin was almost exclusively localized to the plasma membrane, and it was not affected by curcumin-supplemented diet (Fig. 4A). In AOM-treated WT mice on control diet, we observed mucin-depleted crypt foci with aberrant cytoplasmic and nuclear localization of β-catenin (Fig. 4B). 0.5% curcumin restored β-catenin expression pattern to that observed in PBS-treated WT mice on control diet (Fig. 4B). In AOM-treated Il10−/− mice, analysis of mucosal section outside of frank tumors (absent in 0.5% curcumin-treated mice) showed inflammatory lesions with disorganized crypts with mixed focal mucin-depleted lesions with predominantly cytoplasmic and nuclear localization of β-catenin (Fig. 4B). Striking normalization of β-catenin staining was observed in AOM-treated Il10−/− mice fed 0.5% curcumin diet (Fig. 4B).
Figure 4. Curcumin normalizes β-catenin localization in colonic tissue.
(A) Representative images of β-catenin expression and cellular localization as analyzed by immunohistochemistry. (B) Summary of β-catenin expression with consideration to the number of cells per high-powered field of vision (HPF) with membrane-associated, or evident cytoplasmic/nuclear localization of β-catenin. * indicates statistically significant increase in cytoplasmic/nuclear-positive cells, while # indicates a significant decrease in membrane-bound-positive cells in AOM/Il10−/−/0.5% group compared to all other experimental groups (ANOVA followed by Fisher LSD test).
Curcumin has limited anti-inflammatory effects in PBS- or AOM-injected Il10−/− with established colitis
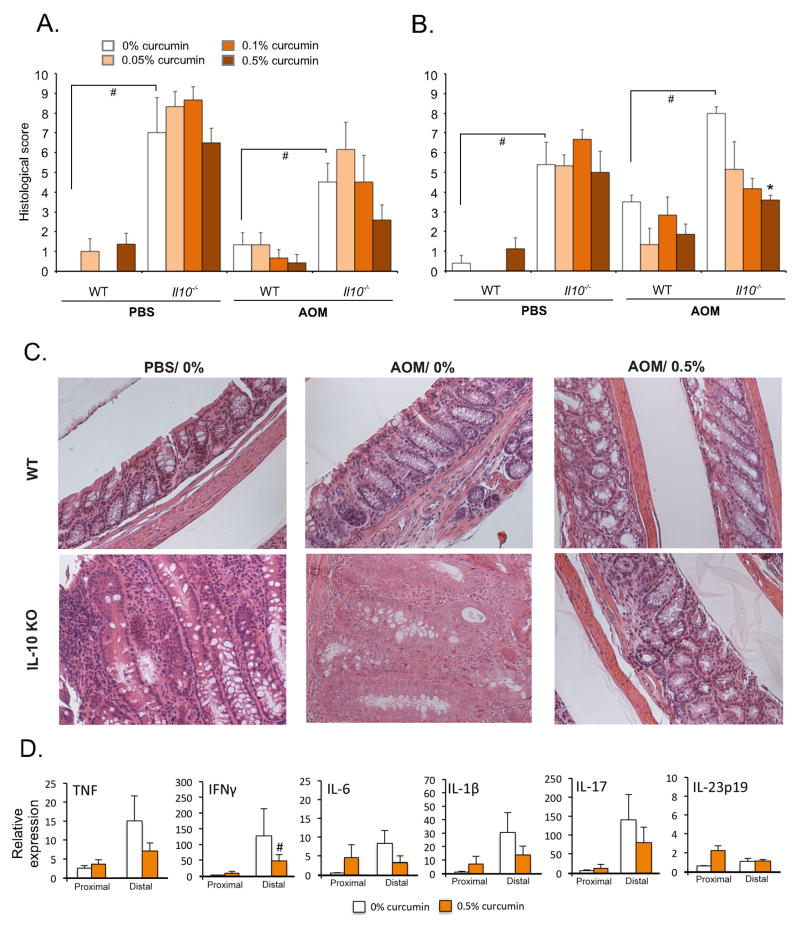
Inflammation is an important risk factor for CRC development and we next investigated whether curcumin-mediated decreased in carcinogenesis was linked to the inflammatory status of AOM/Il10−/− mice. In mice fed control diets, histological scores in the proximal and distal colon were significantly higher in both PBS-injected and in AOM-injected Il10−/− mice as compared to WT mice (Fig. 5). Curcumin did not significantly impact colonic inflammation in either segment in PBS-injected Il10−/− mice, thus suggesting that in the settings of established colitis, curcumin has limited anti-inflammatory potential in any of the three doses analyzed (Fig. 5A,B). In curcumin-treated AOM-injected WT and Il10−/− mice, we observed a trend toward improvement in inflammatory score, albeit without reaching statistical significance (Fig. 5A). In the distal colon of AOM-treated Il10−/− mice, 0.5% curcumin significantly decreased histological score (Fig. 5B). Representative images of H&E-stained tissues of AOM-treated mice are depicted in Figure 5C. This improvement in histological inflammation appeared to be largely driven by decreased hyperplasia, reduced submucosal edema and smooth muscle thickness, and not by suppression of mucosal immune responses. Curcumin treatment was not associated with statistically significant decreases in mucosal cytokine expression in the proximal colon of either PBS- or AOM-treated Il10−/− mice (Fig. 5D, Suppl. Fig. S2). Although in the distal segment, curcumin reduced the mucosal expression of IL-6, IL-1β, IL-17A, and IL-23p19 in PBS-injected Il10−/− mice, it remained without statistically significant effect in AOM-injected Il10−/− mice (Fig. 5D, Suppl. Fig. S2). These findings suggested that despite the overall improvement in the colonic mucosal architecture, curcumin-mediated strong decreased carcinogenesis was not primarily due to a significant decrease in mucosal immune responses in AOM/Il10−/− mice.
Figure 5. The effects of dietary curcumin on colonic histology and mucosal cytokine expression.
(A) Histological scoring of colonic sections from the proximal and (B) distal segments. * indicates a significant (p≤0.05) difference from 0% curcumin dietary group within respective treatment group (bar cluster; ANOVA followed by Fisher LSD test). # indicates a significant (p≤0.05) difference between bars indicated by brackets (unpaired Student t-test). (C) Representative H&E images from the distal colons of WT and Il10−/− mice treated with PBS or AOM and fed control (0%) or 0.5% curcumin-supplemented diet (Magn. 200x). (D) qPCR analysis of mucosal cytokine expression. Each sample was normalized to GAPDH expression and analyzed relative to PBS-injected WT mice on control diet using 2−ΔΔCt method. Bars represent means and SEM. # indicates a significant (p≤0.05) difference from 0% curcumin dietary group within respective treatment group (bar cluster; unpaired t-test). All others are not statistically significant. More detailed analysis which includes all experimental groups is presented in Suppl. Fig. S2.
Microbial analysis of the effect of curcumin-supplemented diet
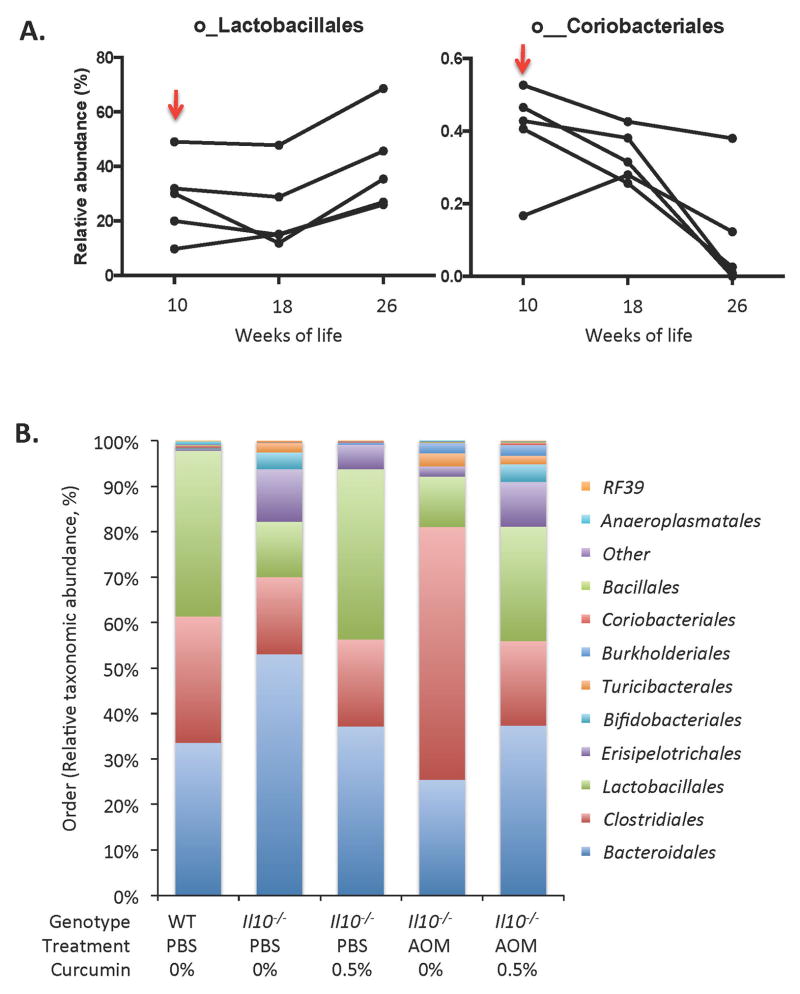
A non-parametric ANOSIM (Analysis of Similarity) method showed that among single variables, mouse gender or diet (all doses considered) were not significantly different based on a unifrac beta diversity metric (Supplemental Table 1). However, because of the striking reduction of tumor burden in mice receiving 0.5% curcumin diet and modest effect on inflammation, we further investigated whether curcumin primary effect was associated with a change in microbial diversity. We first assessed whether curcumin at the most efficacious dietary concentration (0.5%) had a long-term effect on colonic luminal microbial composition in healthy mice. WT mice fed control diet showed age-dependent reduction in species richness between 10th and 30th week of age (810.3 ± 20.9 vs. 652.8 ± 116.1; p<0.05) and in Margalef index (93.0 ± 1.2 vs. 74.9 ± 13.3; p<0.05). Interestingly, WT/PBS mice switched to 0.5% curcumin-supplemented diet at 10 weeks of age maintained high diversity until 30 weeks of life, expressed both as the number of observed species (735.9 ± 28.7 and 738.2 ± 63.2, 10 weeks vs. 30 weeks, respectively; p=0.97) or as the Margalef index (84.48 ± 3.2 and 84.74 ± 7.27, 10 weeks vs. 30 weeks, respectively; p=0.97). At the taxonomic level, to increase the statistical power, we used repeated-measures non-parametric Friedman test to compare fecal microbial ecology in WT PBS-injected control mice at the time of diet switch (10 weeks), and 8 weeks and 16 weeks into the period of 0.5% curcumin diet (18 and 26 weeks of age). As a control for age-related changes, we analyzed WT/PBS mice fed control diet throughout this period. The taxonomic analysis (relative abundances) at the order level was performed with 16 orders comprising >99.9% of OTU’s at this level. We observed gradual increase in the relative abundance of Lactobacillales (represented mainly by genus Lactobacillus), gradual loss of Coriobacteriales (Actinobacteria phylum) (Fig. 6A) and a transient increase (at 18 weeks of age) in the putative order YS2 (Cyanobacteria phylum) (not shown). Two other changes were observed in WT mice fed curcumin-supplemented diets: gradual decrease in Bifidobacteriales and a transient increase in Verrucomicrobiales at week 18. These appeared largely age-related as WT mice fed control diets showed similar trends, albeit without reaching statistical significance (data not shown).
Figure 6. Taxonomic analysis of the effect of 0.5% curcumin-supplemented diet.
(A) Repeated-measures non-parametric Friedman test was used to compare fecal microbial ecology in WT PBS-injected control mice at the time of diet switch (10 weeks), and 8 weeks and 16 weeks into the period of 0.5% curcumin diet (18 and 26 weeks of age). The taxonomic analysis (relevant abundances) at the order level was performed with 16 orders comprising >99.9% of OTU’s at this level. (B) Taxonomic analysis of the effect of 0.5% curcumin-supplemented diet in PBS- and AOM-treated Il10−/− mice. Relative order level microbial abundance in control WT mice (baseline) is visually compared with PBS- and AOM-injected IL10−/− mice fed control (0%) or 0.5% curcumin-supplemented diets. Data derived from the 30th week of life (at euthanasia).
The effects of dietary curcumin in AOM-induced CRC in Il10−/− mice
AOM administration to Il10−/− mice between 10th and 30th weeks of life did not significantly affect alpha diversity expressed either as the number of observed species (495.5 ± 39.6 and 564.5 ± 81.1, PBS vs. AOM, respectively) or as the Margalef index (56.8 ± 4.6 and 64.7 ± 9.3, PBS vs. AOM, respectively). Taxonomic analysis of microbial composition clearly indicated preservative effects (pattern resembling WT/PBS mice) of curcumin on major taxa, especially visible at the order level in Fig. 6B. This normalization pattern in response to curcumin was observed for both PBS-injected and in AOM-injected Il10−/− mice, and particularly for Lactobacillales and Clostridiales orders. Among the AOM/II10−/− groups, curcumin significantly reduced the relative abundance of Clostridiales while increasing the abundance of Lactobacillales, Bifidobacteriales, Erisipelotrichales, Coriobacteriales, and the putative order YS2 from the Cyanobacteria phylum (Fig. 6B).
At the genus level however, the effects were less clear, an observation, which may explain the lack of clear beta diversity clustering patterns (not shown). The most significant effects of curcumin at this taxonomic level were observed for Lactobacillus, and several low-abundance genera from the Firmicutes phylum (data not shown).
We next used PICRUSt to predict the differential metabolic functions represented by each microbial cluster. Imputed metagenomic content based on microbial profiling has been shown to provide a good representation of functional changes in complex, vertebrate-associated microbial communities.40 To simplify the presentation, we compared the following three experimental groups: PBS/WT/0% (baseline), AOM/Il10−/−/0%, and AOM/Il10−/−/0.5% at the time of sacrifice (week 30). 38 KEGG categories were significantly different by ANOVA (p≤0.05). Among those, we selected 19, which in post-hoc comparison (Tukey-Kramer test) showed no statistically significant difference between PBS/WT/0% and AOM/Il10−/−/0.5%, thus suggesting the preserving or normalizing effects of dietary curcumin. These 19 categories are depicted in Suppl. Fig. S3. These include pathways under-represented in the gut microbiome of AOM/Il10−/−/0% mice (Alzheimer’s disease, β-lactam resistance, primary and secondary bile acid biosynthesis, amino-acid related enzymes, nucleotide excision repair, and ubiquinone and other terpenoid-quinone biosynthesis), as well as categories over-represented in the gut microbiome of AOM/Il10−/−/0% mice (mineral absorption, other transporters, ABC transporters, styrene degradation, arachidonic acid metabolism, basal transcription factors, transcription factors, bacterial chemotaxis, bacterial motility proteins, sporulation).
DISCUSSION
Crohn’s disease and ulcerative colitis are both associated with an increased risk of inflammation-associated CRC, which becomes one of the most important causes for morbidity and mortality in IBD patients. Chemoprevention is focused on induction of remission and on the use of anti-inflammatory drugs targeting pathways known to be involved in sporadic CRC. 45 However, the probability of relapse remains high, and endoscopic evidence of remission does not rule out subclinical mucosal inflammation, which chronically may contribute the genomic instability. Moreover, inconsistent clinical data on chemoprevention of IBD-associated CRC suggests that there is a need for novel approaches. In this preclinical study with a mouse model of colitis-associated CRC,25, 46 we demonstrate that dietary curcumin, in a dose-dependent fashion improves survival, reduces tumor burden and hyperplasia, and normalizes β-catenin expression pattern in the colonocytes. In our study, the most effective dietary concentration of curcumin was 0.5%, which entirely eliminated the occurrence of macroscopic tumors. This dose roughly corresponds to HED of ca. 80 mg/kg/day, well within the range of well-tolerated doses reported by phase I human trials.22, 23 Higher dose (1%) proved detrimental in the Il10−/− mouse model, an observation that was anticipated based on our previous studies with Il10−/− model.47
The chemopreventive effects of curcumin did not correspond with a strong suppression of mucosal inflammatory responses. This observation is in line with a previous study showing minimal effect of curcumin on inflammation in Il10−/− mice.47 Although histological score indicated significant improvement in the distal colon of Il10−/−/AOM mice fed 0.5% curcumin, this is most likely the result of diminished hyperplasia, submucosal edema, smooth muscle thickness, and dysplastic and neoplastic changes, which were all integral parts of the scoring system. Mucosal cytokine expression, a more objective measure of mucosal inflammatory tone, with the exception of IFNγ in the distal colon, did not indicate a significant reduction of inflammatory response in AOM/Il10−/− mice fed control of curcumin-supplemented diet (Fig. 5, Fig. S2). Therefore, we concluded that contrary to the results obtained from the ApcMin/+ model,48 the observed striking chemopreventive effects of curcumin did not correlate with significant suppression of inflammatory resposes.
Numerous in vitro studies with curcumin have demonstrated the inhibitory effects on intrinsic cellular mechanisms associated with neoplastic transformation, proliferation and apoptosis, and cell migration and invasion.16, 49 These intrinsic mechanisms likely contributed to the chemopreventive effects observed in our study. We investigated colonic microbiome as an extrinsic factor with a critical role in the pathogenesis of colitis-associated CRC.12, 46, 50 To this end, we determined that dietary curcumin maintained high microbial diversity, which otherwise declined with age between 10 and 30 weeks of age. Interestingly, this was associated with the expansion of Lactobacillales (represented mainly by genus Lactobacillus). Moreover, the relative abundance of the Lactobacillales order was decreased in colitic Il10−/− and in AOM/Il10−/− mice, and in both cases, dietary curcumin restored this order to control levels. Lactobacillus strains have been used successfully in the CRC prevention in animal models51–54, have been shown to exhibit significant antigenotoxicity55, and the Lactobacillus genus was shown to be associated with cell cycle arrest and induction of apoptosis in colon cancer cell lines56,57. Although the results of systematic and well-controlled clinical studies in CRC prevention are not yet available, published reports suggest cautious optimism. 58–60 Although our study design does not address causality, it is plausible that curcumin contributes to chemoprevention via expansion of the native Lactobacilli in the context of overall increase of colonic microbiome richness. Similar effects may be ascribed to increased Bifidobacteria, which have also been shown to reduce aberrant crypt foci in mice receiving dimethylhyrazine.61 Future experiments using gnotobiotic approaches would help establish the role of these microorganisms on colitis-associated CRC.
PICRUSt analysis has been shown to provide a good representation of functional changes in complex communities, and may predict the results of metagenomic analyses.40,62 Analysis with the focus on the most abundant KEGG orthologs, which were affected by curcumin-supplementation in AOM/Il10−/− mice showed reduced abundance of functions involved in transport, transcription factors, bacterial chemotaxis and bacterial motility proteins. 56, 57, 61, 63–67 Curcumin has been observed to inhibit bacterial motility and cytotoxicity of Vibrio vulnificus68, to arrest Helicobacter pylori growth during infections69, as well as inhibit biofilm formation in Pseudomonas aeruginosa70. This suggests that the protective effect of 0.5% curcumin diet shifts the microbial composition to be more stationary, which may contribute to a decrease in microbial invasiveness and a decrease in overall disease severity. Interestingly, curcumin reduced the relative abundance of taxa associated with arachidonic acid (ARA) metabolism. This may represent host-associated response to curcumin, which is known for its ability to inhibit ARA metabolism in experimental CRC,71 or actual changes in the microbial community. While little is know on that topic, reduced ARA derived from neutrophils may lead to decreased numbers of bacteria utilizing ARA for growth, such as Pseudomonas.72
Although the microbial breakdown of AOM to genotoxic metabolites has been initially suggested to represent at least part of the mechanism of CRC induction, recent results from Zhen et al.73 showed that germ-free mice form significantly more and larger tumors compared with specific-pathogen free mice in DSS/AOM model despite the lack of early acute inflammation in response to DSS. This suggests that microbial AOM metabolism is not a prerequisite for the genotoxic effects of AOM. Therefore, it is very unlikely that the striking effects of curcumin on AOM-induced CRC in Il10−/− mice could be explained by altered AOM metabolism as a result of compositional changes in colonic microbiota.
In summary, dietary curcumin may represent an efficacious means of chemoprevention of inflammation-associated CRC irrespective of its direct anti-inflammatory effects. While our study cannot assert causality between shifts in the colonic microbial ecology (and potentially metabolism) and reduced (or entirely eliminated) tumor formation, we demonstrated efficacy of curcumin on inflammation associated colorectal cancer and normalizing effects of colonic microbial ecology. Curcumin may represent a promising approach to chemoprevention in IBD, regardless of its anti-inflammatory potential.
Supplementary Material
Acknowledgments
Grant support: NIH R01DK67286
16S sequencing was generated by IGSB-NGS (Institute for Genomics & Systems Biology-Next Generation Sequencing) Core Facility at Argonne National Laboratory by Sarah Owens. Immunohistochemical data generated by the TACMASS Core (Tissue Acquisition and Cellular/Molecular Analysis Shared Service) is supported by the University of Arizona Cancer Center in Tucson, AZ (NIH CA023074). We would also like to thank Andrea Grantham from the Department of Cellular and Molecular Medicine Histology Service Lab for her technical assistance.
Footnotes
Disclosures: The authors declare no conflict of interest to disclose.
References
- 1.American Cancer Society. Cancer Facts and Figures 2015. 2015 [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Watson AJ, Collins PD. Colon cancer: a civilization disorder. Digestive diseases. 2011;29:222–8. doi: 10.1159/000323926. [DOI] [PubMed] [Google Scholar]
- 4.Heidland A, Klassen A, Rutkowski P, Bahner U. The contribution of Rudolf Virchow to the concept of inflammation: what is still of importance? Journal of nephrology. 2006;19(Suppl 10):S102–9. [PubMed] [Google Scholar]
- 5.Feagins LA, Souza RF, Spechler SJ. Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat Rev Gastroenterol Hepatol. 2009;6:297–305. doi: 10.1038/nrgastro.2009.44. [DOI] [PubMed] [Google Scholar]
- 6.Gupta RB, Harpaz N, Itzkowitz S, Hossain S, Matula S, Kornbluth A, et al. Histologic inflammation is a risk factor for progression to colorectal neoplasia in ulcerative colitis: a cohort study. Gastroenterology. 2007;133:1099–105. doi: 10.1053/j.gastro.2007.08.001. quiz 340–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 8.Rutter M, Saunders B, Wilkinson K, Rumbles S, Schofield G, Kamm M, et al. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126:451–9. doi: 10.1053/j.gastro.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory Bowel Disease as a Model for Translating the Microbiome. Immunity. 2014;40:843–54. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–12. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur JC, Jobin C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes. 2013;4:253–8. doi: 10.4161/gmic.24220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–28. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramamoorthi G, Sivalingam N. Molecular mechanism of TGF-beta signaling pathway in colon carcinogenesis and status of curcumin as chemopreventive strategy. Tumour Biol. 2014 doi: 10.1007/s13277-014-1840-1. [DOI] [PubMed] [Google Scholar]
- 15.Goel A, Aggarwal BB. Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutrition and cancer. 2010;62:919–30. doi: 10.1080/01635581.2010.509835. [DOI] [PubMed] [Google Scholar]
- 16.Shehzad A, Wahid F, Lee YS. Curcumin in cancer chemoprevention: molecular targets, pharmacokinetics, bioavailability, and clinical trials. Archiv der Pharmazie. 2010;343:489–99. doi: 10.1002/ardp.200900319. [DOI] [PubMed] [Google Scholar]
- 17.Billerey-Larmonier C, Uno JK, Larmonier N, Midura AJ, Timmermann B, Ghishan FK, et al. Protective effects of dietary curcumin in mouse model of chemically induced colitis are strain dependent. Inflamm Bowel Dis. 2008;14:780–93. doi: 10.1002/ibd.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larmonier CB, Midura-Kiela MT, Ramalingam R, Laubitz D, Janikashvili N, Larmonier N, et al. Modulation of neutrophil motility by curcumin: implications for inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:503–15. doi: 10.1002/ibd.21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Midura-Kiela MT, Radhakrishnan VM, Larmonier CB, Laubitz D, Ghishan FK, Kiela PR. Curcumin inhibits interferon-gamma signaling in colonic epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2012;302:G85–96. doi: 10.1152/ajpgi.00275.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radhakrishnan VM, Kojs P, Young G, Ramalingam R, Jagadish B, Mash EA, et al. pTyr421 cortactin is overexpressed in colon cancer and is dephosphorylated by curcumin: involvement of non-receptor type 1 protein tyrosine phosphatase (PTPN1) PLoS One. 2014;9:e85796. doi: 10.1371/journal.pone.0085796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villegas I, Sanchez-Fidalgo S, de la Lastra CA. Chemopreventive effect of dietary curcumin on inflammation-induced colorectal carcinogenesis in mice. Molecular nutrition & food research. 2011;55:259–67. doi: 10.1002/mnfr.201000225. [DOI] [PubMed] [Google Scholar]
- 22.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, et al. Phase I clinical trial of oral curcumin: biomarkers of systemic activity and compliance. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:6847–54. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 23.Cheng AL, Hsu CH, Lin JK, Hsu MM, Ho YF, Shen TS, et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer research. 2001;21:2895–900. [PubMed] [Google Scholar]
- 24.Kanneganti M, Mino-Kenudson M, Mizoguchi E. Animal models of colitis-associated carcinogenesis. J Biomed Biotechnol. 2011;2011:342637. doi: 10.1155/2011/342637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 27.Gomes-Santos AC, Moreira TG, Castro-Junior AB, Horta BC, Lemos L, Cruz DN, et al. New insights into the immunological changes in IL-10-deficient mice during the course of spontaneous inflammation in the gut mucosa. Clinical & developmental immunology. 2012;2012:560817. doi: 10.1155/2012/560817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiela PR, Midura AJ, Kuscuoglu N, Jolad SD, Solyom AM, Besselsen DG, et al. Effects of Boswellia serrata in mouse models of chemically induced colitis. Am J Physiol Gastrointest Liver Physiol. 2005;288:G798–808. doi: 10.1152/ajpgi.00433.2004. [DOI] [PubMed] [Google Scholar]
- 29.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–9. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. [Google Scholar]
- 39.Chao A. Nonparametric estimation of the number of classes in a population. Scandinavian Journal of Statistics. 1984;11:265–70. [Google Scholar]
- 40.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–4. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostanin DV, Bao J, Koboziev I, Gray L, Robinson-Jackson SA, Kosloski-Davidson M, et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol. 2009;296:G135–46. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nature protocols. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 44.Miyaki M, Iijima T, Kimura J, Yasuno M, Mori T, Hayashi Y, et al. Frequent mutation of beta-catenin and APC genes in primary colorectal tumors from patients with hereditary nonpolyposis colorectal cancer. Cancer research. 1999;59:4506–9. [PubMed] [Google Scholar]
- 45.Foersch S, Neurath MF. Colitis-associated neoplasia: molecular basis and clinical translation. Cellular and molecular life sciences : CMLS. 2014;71:3523–35. doi: 10.1007/s00018-014-1636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–3. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larmonier CB, Uno JK, Lee KM, Karrasch T, Laubitz D, Thurston R, et al. Limited effects of dietary curcumin on Th-1 driven colitis in IL-10 deficient mice suggest an IL-10-dependent mechanism of protection. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1079–91. doi: 10.1152/ajpgi.90365.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy EA, Davis JM, McClellan JL, Gordon BT, Carmichael MD. Curcumin’s effect on intestinal inflammation and tumorigenesis in the ApcMin/+ mouse. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31:219–26. doi: 10.1089/jir.2010.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shehzad A, Lee J, Lee YS. Curcumin in various cancers. BioFactors. 2013;39:56–68. doi: 10.1002/biof.1068. [DOI] [PubMed] [Google Scholar]
- 50.Arthur JC, Jobin C. The struggle within: microbial influences on colorectal cancer. Inflamm Bowel Dis. 2011;17:396–409. doi: 10.1002/ibd.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lightfoot YL, Yang T, Sahay B, Mohamadzadeh M. Targeting aberrant colon cancer-specific DNA methylation with lipoteichoic acid-deficient Lactobacillus acidophilus. Gut Microbes. 2013;4:84–8. doi: 10.4161/gmic.22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asha, Gayathri D. Synergistic impact of Lactobacillus fermentum, Lactobacillus plantarum and vincristine on 1,2-dimethylhydrazine-induced colorectal carcinogenesis in mice. Experimental and therapeutic medicine. 2012;3:1049–54. doi: 10.3892/etm.2012.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khazaie K, Zadeh M, Khan MW, Bere P, Gounari F, Dennis K, et al. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10462–7. doi: 10.1073/pnas.1207230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang JH, Shim YY, Cha SK, Reaney MJ, Chee KM. Effect of Lactobacillus acidophilus KFRI342 on the development of chemically induced precancerous growths in the rat colon. Journal of medical microbiology. 2012;61:361–8. doi: 10.1099/jmm.0.035154-0. [DOI] [PubMed] [Google Scholar]
- 55.Raman M, Ambalam P, Kondepudi KK, Pithva S, Kothari C, Patel AT, et al. Potential of probiotics, prebiotics and synbiotics for management of colorectal cancer. Gut Microbes. 2013;4:181–92. doi: 10.4161/gmic.23919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan Y, Xin Y, Zhang C, Wu D, Ding D, Tang L, et al. Fermentation supernatants of inhibit growth of human colon cancer cells and induce apoptosis through a caspase 3-dependent pathway. Oncol Lett. 2014;7:1738–42. doi: 10.3892/ol.2014.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang SM, Zhang LW, Fan RB, Han X, Yi HX, Zhang LL, et al. Induction of HT-29 cells apoptosis by lactobacilli isolated from fermented products. Res Microbiol. 2014;165:202–14. doi: 10.1016/j.resmic.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Gianotti L, Morelli L, Galbiati F, Rocchetti S, Coppola S, Beneduce A, et al. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World journal of gastroenterology : WJG. 2010;16:167–75. doi: 10.3748/wjg.v16.i2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishikawa H, Akedo I, Otani T, Suzuki T, Nakamura T, Takeyama I, et al. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. International journal of cancer Journal international du cancer. 2005;116:762–7. doi: 10.1002/ijc.21115. [DOI] [PubMed] [Google Scholar]
- 60.Rafter J, Bennett M, Caderni G, Clune Y, Hughes R, Karlsson PC, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. The American journal of clinical nutrition. 2007;85:488–96. doi: 10.1093/ajcn/85.2.488. [DOI] [PubMed] [Google Scholar]
- 61.Liboredo JC, Anastacio LR, do Peluzio MC, Valente FX, Penido LC, Nicoli JR, et al. Effect of probiotics on the development of dimethylhydrazine-induced preneoplastic lesions in the mice colon. Acta Cir Bras. 2013;28:367–72. doi: 10.1590/s0102-86502013000500008. [DOI] [PubMed] [Google Scholar]
- 62.Xu Z, Malmer D, Langille MG, Way SF, Knight R. Which is more important for classifying microbial communities: who’s there or what they can do? ISME J. 2014;8:2357–9. doi: 10.1038/ismej.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest. 2011;121:2126–32. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–24. e1–2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukhopadhya I, Hansen R, Nicholl CE, Alhaidan YA, Thomson JM, Berry SH, et al. A comprehensive evaluation of colonic mucosal isolates of Sutterella wadsworthensis from inflammatory bowel disease. PLoS One. 2011;6:e27076. doi: 10.1371/journal.pone.0027076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Houot L, Chang S, Absalon C, Watnick PI. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect Immun. 2010;78:1482–94. doi: 10.1128/IAI.01356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Na HS, Cha MH, Oh DR, Cho CW, Rhee JH, Kim YR. Protective mechanism of curcumin against Vibrio vulnificus infection. FEMS Immunol Med Microbiol. 2011;63:355–62. doi: 10.1111/j.1574-695X.2011.00855.x. [DOI] [PubMed] [Google Scholar]
- 69.De R, Kundu P, Swarnakar S, Ramamurthy T, Chowdhury A, Nair GB, et al. Antimicrobial activity of curcumin against Helicobacter pylori isolates from India and during infections in mice. Antimicrob Agents Chemother. 2009;53:1592–7. doi: 10.1128/AAC.01242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rudrappa T, Bais HP. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J Agric Food Chem. 2008;56:1955–62. doi: 10.1021/jf072591j. [DOI] [PubMed] [Google Scholar]
- 71.Rao CV, Simi B, Reddy BS. Inhibition by dietary curcumin of azoxymethane-induced ornithine decarboxylase, tyrosine protein kinase, arachidonic acid metabolism and aberrant crypt foci formation in the rat colon. Carcinogenesis. 1993;14:2219–25. doi: 10.1093/carcin/14.11.2219. [DOI] [PubMed] [Google Scholar]
- 72.Sorrell TC, Muller M, Sztelma K. Bacterial metabolism of human polymorphonuclear leukocyte-derived arachidonic acid. Infect Immun. 1992;60:1779–85. doi: 10.1128/iai.60.5.1779-1785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhan Y, Chen PJ, Sadler WD, Wang F, Poe S, Nunez G, et al. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer research. 2013;73:7199–210. doi: 10.1158/0008-5472.CAN-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.