Abstract
In eukaryotes and viruses that infect them, the 5′ end of mRNA molecules, and also many other functionally important RNAs, are modified to form a so-called cap structure that is important for interactions of these RNAs with many nuclear and cytoplasmic proteins. The RNA cap has multiple roles in gene expression, including enhancement of RNA stability, splicing, nucleocytoplasmic transport, and translation initiation. Apart from guanosine addition to the 5′ end in the most typical cap structure common to transcripts produced by RNA polymerase II (in particular mRNA), essentially all cap modifications are due to methylation. The complexity of the cap structure and its formation can range from just a single methylation of the unprocessed 5′ end of the primary transcript, as in mammalian U6 and 7SK, mouse B2, and plant U3 RNAs, to an elaborate m7Gpppm6,6AmpAmpCmpm3Um structure at the 5′ end of processed RNA in trypanosomes, which are formed by as many as 8 methylation reactions. While all enzymes responsible for methylation of the cap structure characterized to date were found to belong to the same evolutionarily related and structurally similar Rossmann Fold Methyltransferase superfamily, that uses the same methyl group donor, S-adenosylmethionine; the enzymes also exhibit interesting differences that are responsible for their distinct functions. This review focuses on the evolutionary classification of enzymes responsible for cap methylation in RNA, with a focus on the sequence relationships and structural similarities and dissimilarities that provide the basis for understanding the mechanism of biosynthesis of different caps in cellular and viral RNAs. Particular attention is paid to the similarities and differences between methyltransferases from human cells and from human pathogens that may be helpful in the development of antiviral and antiparasitic drugs.
Keywords: antiviral drugs, cap, crystallography, methylation, modified nucleotides, mRNA, post-transcriptional modification, RNA maturation, RNA modification, trypanosomes
Introduction
Nascent transcripts produced by RNA polymerases universally carry a 5′ triphosphate (5′ppp). Processed RNA molecules, such as rRNAs and tRNAs, generated from precursors whose 5′ segments were removed by nucleolytic cleavage, carry a 5′ monophosphate (5′p). In several types of cellular and viral RNAs, the 5′ end is further modified enzymatically, by a variety of modification enzymes, to introduce various chemical structures that are collectively dubbed as the “5′ caps”. This cap is absent in bacterial and archaeal transcripts.
The most typical and widely studied cap modification comprises the addition of an N7-methylguanosine (m7G) linked via an inverted 5′–5′ triphosphate bridge to the 5′-terminal nucleoside of the transcript.1 This structure termed cap0 is a characteristic feature of transcripts that are produced by RNA polymerase II, such as messenger RNAs (mRNAs) of all eukaryotic organisms and many viral RNAs. It is typically introduced in sequential steps: (1) hydrolysis of 5′ γ-phosphate of a nascent pre-mRNA to generate a 5′ diphosphate mRNA end; (2) transfer of a guanine monophosphate nucleoside; and (3) methylation of the guanine at the N7 position. The cap0 structure was shown to be essential for cell growth of Saccharomyces cerevisiae2 and survival of mammalian cells;3 it is critical for mRNA interactions with many nuclear and cytoplasmic proteins and has multiple important roles in gene expression, including enhancement of RNA stability, splicing, nucleocytoplasmic transport, and translation initiation.4,5 Enzymes responsible for cap0 formation have been well characterized in many organisms and viruses.
In many instances, m7G-capped RNAs are modified further, in particular by additional methylation steps at the cap0 guanosine or methylation of the first few transcribed nucleoside residues. For instance, the cap0 guanosine is modified by addition of 2 methyl groups at the N2 position, yielding a trimethylguanosine (m2,2,7G or TMG) cap, in some small nuclear RNAs (snRNAs) and nucleolar RNAs (snoRNAs) required for pre-mRNA splicing (e.g., U1, U2, U4, and U5), pre-rRNA processing (U3 and U8), and telomere addition (telomerase RNA), as well as in several selenoprotein mRNAs.6
In higher eukaryotes, the 5′ ends of mRNA and snRNA are modified further by ribose 2′-O-methylation on the first and second transcribed nucleosides, yielding cap1 and cap2 modifications, respectively.7 In humans, cap0 and cap1 methylations are found on all mRNA molecules, while about half of the capped and polyadenylated RNA molecules contain a 2′-O-methylated residue at the second transcribed position.8 The U1, U2, U4, and U5 snRNAs are methylated at both the first 2 positions.9 Cap1 and cap2 methylations in U2 snRNA are required for the formation of spliceosomal E-complex and, as a consequence, for efficient pre-mRNA splicing.10 In some organisms, such as in Trypanosomes, as many as 4 first residues of the nascent transcript undergo ribose methylation, to generate the cap4 structure.11 These additional methylation steps are often important for RNA processing, translation and stability, although their role has not been fully elucidated.
Alternative capping pathways have been invented by certain viruses. For instance, in Alphaviruses, the precursor of cap0 is first methylated to a m7G triphosphate and only then connected to the 5′ end of the RNA.12 Nonsegmented negative-sense (nsNS) RNA viruses have evolved a different mechanism for mRNA cap formation in that the guanylyltransferase transfers GDP rather than GMP onto the 5′ end of the RNA and the resulting cap structure is first monomethylated on the ribose of the first transcribed residue (yielding GpppAm structure), and only later the guanosine is methylated to m7G.13-15
In addition to ribose 2′-O-methylation, base moieties of the first transcribed nucleosides may be methylated, thereby increasing the catalog of 5′ cap structures. In particular, the first adenine nucleoside of the transcript is often methylated at the N6 position.16 In Trypanosomes, the fourth uridine residue is also methylated at the N3 position. The role of these base methylations is unclear, and the enzymes responsible for these modifications remain to be characterized.
Some small RNAs, including mammalian U6 and 7SK, mouse B2, and plant U3, present a completely different 5′ cap structure, which is chemically minimalistic compared to the elaborate guanosine cap. This alternative cap is generated by methylation of a γ-phosphate oxygen at the unprocessed 5′ end of the primary transcript.17
It is clear that apart from guanosine addition to the 5′ end, essentially all cap modifications are due to methylations. The cap structure of mRNAs in trypanosomes, m7Gpppm6,6AmpAmpCmpm3Um, is formed with as many as 8 methylation steps. In all cases that have been experimentally characterized to date, methylations of caps in all organisms and viruses are catalyzed by S-adenosyl-l-methionine (SAM)-dependent methyltransferases (Table 1). For the most common types of methylation reactions implicated in cap modification, the crystal structures of the representative proteins have been determined (Table 2). All cap methyltransferases characterized structurally belong to the Rossmann Fold Methyltransferase (RFM) superfamily.18 The topology of the RFM fold is very similar to the typical Rossmann fold (↓6‑↓5‑↓4‑↓1‑↓2‑↓3), with an additional, 7th ß‑strand inserted into the sheet in an antiparallel manner (↓6‑↑7‑↓5‑↓4‑↓1‑↓2‑↓3) (Fig. 1).18,19 The methyl group donor (SAM) binding site is formed by loops following strands 1, 2, and 3, while the substrate to be methylated is typically bound by loops following strands 4, 5, and 6. Various families of RFM enzymes exhibit fusions with other domains, extensions of termini, and insertions within the conserved RFM domain, in particular following strand 5. These elaborations of the common fold are often involved in substrate binding or in oligomerization.20,21
Table 1.
Representative cellular and viral cap methyltransferases with experimentally characterized RNA cap methyltransferase activities. The enzymes, for which crystal structures were determined, are shown in bold.
| Methylation position | base |
2′-O-ribose |
other |
||||
|---|---|---|---|---|---|---|---|
| Species | cap0 | TMG | cap1 | cap2 | cap3/cap4 | ɣ-phosphate | |
| cellular enzymes | Homo sapiens | RNMT7,26 | TGS169,70 | CMTr138 | CMTr246 | BCDIN382 | |
| Saccharomyces cerevisiae | Abd12 | Tgs164 | |||||
| Encephalitozoon cuniculi | Ecm125 | ||||||
| Giardia lamblia | Tgs165, Tgs268 | ||||||
| Trypanosoma brucei | TbCmt127, TbCgm183 | TbTgs184 | TbMTr141,42 | TbMTr227 | TbMTr347 | ||
| viral enzymes | Vaccinia virus | D1/D1228,29 | VP3985 | ||||
| Flavivirus | NS560 | NS560 | |||||
| Vesicular stomatitis virus | L protein52 | L protein52 | |||||
| Reovirus | lambda 249 | lambda 249 | |||||
| Bluetongue virus | VP451 | VP451 | |||||
| SARS-Coronavirus | nsp1434 | nsp16/nsp1043 | |||||
Table 2.
Experimentally determined structures of cap-specific methyltransferases. “cap0 + cap1” indicates 2 activities encoded in separate domains in one polypeptide, while “cap0/cap1” indicates 2 activities associated with one domain. a—indicates structures available in the PDB, for which no corresponding articles are available in the literature.
| MTase type | organism / virus | protein | ligand1 | ligand2 | PDB |
|---|---|---|---|---|---|
| cap0 (m7G) | Homo sapiens | RNMT | sinefungin | — | 3eppa |
| cap0 (m7G) | Homo sapiens | RNMT | SAH | — | 3bgva |
| cap0 (m7G) | Encephalitozoon cuniculi | RNMT | sinefungin | — | 2hv986 |
| cap0 (m7G) | Encephalitozoon cuniculi | RNMT | AzoSAM | — | 1z3c87 |
| cap0 (m7G) | Encephalitozoon cuniculi | RNMT | SAH | m7GpppG | 1ri125 |
| cap0 (m7G) | Encephalitozoon cuniculi | RNMT | — | m7GpppG | 1ri225 |
| cap0 (m7G) | Encephalitozoon cuniculi | RNMT | SAH | — | 1ri325 |
| cap0 (m7G) | Encephalitozoon cuniculi | RNMT | SAM | — | 1ri425 |
| cap0 (m7G) | Encephalitozoon cuniculi | RNMT | — | — | 1ri525 |
| cap0 (m7G) | vaccinia virus | D1, D12 | SAH | — | 2vdw33 |
| cap0 (m7G) | vaccinia virus | D1, D12 | SAH | — | 4cke88 |
| cap0 (m7G) | vaccinia virus | D1, D12 | SAH | GTP | 4ckb88 |
| cap0 (m7G) | vaccinia virus | D1, D12 | SAH | — | 4ckc88 |
| cap1 (XpppNm) | Homo sapiens | CMTr1 | SAM | m7GpppGAUC | 4n4839 |
| cap1 (XpppNm) | Homo sapiens | CMTr1 | SAM | m7GpppG | 4n4939 |
| cap1 (XpppNm) | Homo sapiens | CMTr1 | — | — | 4n4a39 |
| cap1 (XpppNm) | vaccinia virus | VP39 | SAH | m7GpppGAAAAA | 1av6 36 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | m3Ade | 3mag89 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | m1Ade | 1b4289 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | m3Cyt | 3mct89 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | m1Cyt | 1bky89 |
| cap1 (XpppNm) | vaccinia virus | VP39-D182A | — | m7G | 4dcg89 |
| cap1 (XpppNm) | vaccinia virus | VP39-E233Q | — | m7G | 1eqa89 |
| cap1 (XpppNm) | vaccinia virus | VP39-E233A | — | — | 1eam89 |
| cap1 (XpppNm) | vaccinia virus | VP39 | — | — | 1vp3 90 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | — | 1vp9 90 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | m7GpppG | 1v39 90 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | m7GpppG | 1p39 90 |
| cap1 (XpppNm) | vaccinia virus | VP39-dC26 | — | m7GDP | 2vp3 90 |
| cap1 (XpppNm) | vaccinia virus | VP39 | SAM | — | 1vpt 35 |
| cap1 (XpppNm) | vaccinia virus | VP39 | SAH | m7,9G | 1jsz91 |
| cap1 (XpppNm) | vaccinia virus | VP39 | SAH | — | 1jte91 |
| cap1 (XpppNm) | vaccinia virus | VP39 | SAH | m7GpppG | 1jtf91 |
| cap1 (XpppNm) | SARS virus | ns10-ns16 | SAM | — | 3r2443 |
| cap0 + cap1 | reovirus | lambda2 | — | — | 1ej649 |
| cap0 + cap1 | bluetongue virus | VP4 | — | GpppG | 2jha51 |
| cap0 + cap1 | bluetongue virus | VP4 | SAH | — | 2jhp51 |
| cap0 + cap1 | bluetongue virus | VP4 | — | m7GDP | 2jh851 |
| cap0 + cap1 | bluetongue virus | VP4 | — | GTP | 2jh951 |
| cap0 + cap1 | bluetongue virus | VP4 | — | — | 2jhc51 |
| cap0/cap1 | West Nile virus | NS5 | SAH | — | 2oy060 |
| cap0/cap1 | Wesselsbron virus | wv-MTase | SAM | m7GpppG | 3emb61 |
| cap0/cap1 | Wesselsbron virus | wv-MTase | SAM | GpppG | 3elw61 |
| cap0/cap1 | Wesselsbron virus | wv-MTase | SAM | — | 3elu61 |
| cap0/cap1 | Wesselsbron virus | wv-MTase | SAH | — | 3ely61 |
| cap0/cap1 | Wesselsbron virus | wv-MTase | sinefungin | — | 3eld61 |
| cap0/cap1 | Wesselsbron virus | wv-MTase | sinefungin | m7GpppG | 3emd61 |
| cap0/cap1 | Meaban virus | mvMTase | SAH | — | 2oxt62 |
| cap0/cap1 | Murray Valley enc. virus | NS5 | SAH | — | 2px263 |
| cap0/cap1 | Murray Valley enc. virus | NS5 | SAH | — | 2px463 |
| cap0/cap1 | Murray Valley enc. virus | NS5 | SAH | — | 2px563 |
| cap0/cap1 | Murray Valley enc. virus | NS5 | SAH | m7GTP | 2px863 |
| cap0/cap1 | Murray Valley enc. virus | NS5 | SAH | GpppG | 2pxa63 |
| cap0/cap1 | Murray Valley enc. virus | NS5 | SAM | GpppA | 2pxc63 |
| cap0/cap1 | Dengue virus | NS5 | — | m7GpppA | 2p3o59 |
| cap0/cap1 | Dengue virus | NS5 | — | m7GpppG | 2p4059 |
| cap0/cap1 | Dengue virus | NS5 | — | m7GpppGm | 2p4159 |
| cap0/cap1 | Dengue virus | NS5 | — | GpppA | 2p3l59 |
| cap0/cap1 | Dengue virus | NS5 | — | GpppG | 2p3q59 |
| cap0/cap1 | Dengue virus type 2 | NS5 | SAH | — | 1l9k57 |
| cap0/cap1 | Dengue virus type 2 | NS5 | SAH | GMP | 2p1d57 |
| cap0/cap1 | Dengue virus type 2 | NS5 | SAH | ribavirin | 1r6a92 |
| TMG (m2,2,7G) | Homo sapiens | TGS1 | SAH | m7GpppG | 3gdh69 |
| TMG (m2,2,7G) | Homo sapiens | TGS1 | — | m7GpppA | 3egi70 |
| mpppN | Homo sapiens | BCDIN3 | SAM | — | 3g07a |
Figure 1.
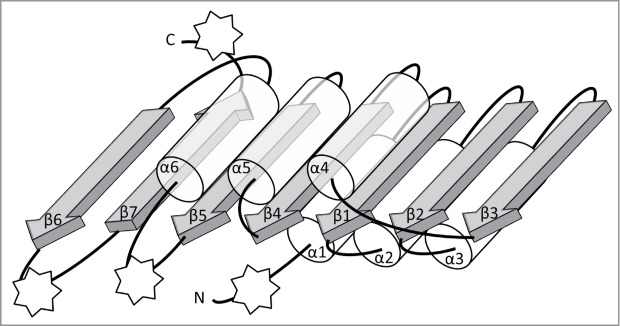
Schematic representation of the conserved core of Rossmann-fold Methyltransferase (RFM) catalytic domains. The β-sheet is composed of 7 β-strands (gray arrows) surrounded by 6 α-helices (semi-transparent tubes) forms the fold that is typical for SAM-dependent methyltransferases. All secondary structure elements of the conserved core are labeled as α1, β1, etc. The stars indicate points of most frequent insertions and terminal fusions with other domains.
In this review, we discuss cellular and viral methyltransferases involved in 5′ cap RNA biosynthesis, with emphasis on the sequence structure relationships in the light of the experimentally determined structures of enzymes complexed with their ligands. We focus on comparison of enzymes with similar activities that generate products with chemically similar structures.
Throughout the article, we follow the nomenclature of cap modifying enzymes and their products commonly used in the literature. We use terms “capX methylation” and “capX methyltransferase” (where X is a number) to refer to some enzymatic activity or its product at a particular position X. On the other hand, the term “capX structure” is used to refer to a fully modified cap structure. For instance “cap2 structure” indicates a cap methylated on the inverted guanosine and the first 2 ribose sugars in the nucleotide sequence; i.e., m7GpppN1mN2m. It should be emphasized that some of the cap methyltranserases discussed here have not yet been fully characterized and it cannot be ruled out that they act at multiple positions.
Cap-specific m7G methyltransferases
Cellular and viral RNA cap guanine-N7-methyltrasferases methylate RNA with the GpppN 5′ terminus to form an m7GpppN (cap0) structure. Eukaryotic enzymes catalyze this reaction in the nucleus. Many viruses, however, replicate in the cytoplasm of their eukaryotic host, and the cellular capping machinery is not accessible for their RNAs; hence, these viruses have evolved their own capping enzymes to form a cap structure that can be recognized by the cellular translation machinery for gene expression. Examples include Flaviviridae, Nidovirales, Mononegavirales and Poxviridae (reviewed in ref. 12). While the cellular and viral mRNA capping apparatus is functionally similar, the enzyme organization differs greatly across evolution.
The Abd1 protein from Saccharomyces cerevisiae is a monofunctional cap0 methyltransferase, and biochemically has been one of the best studied methyltransferases involved in cap structure biosynthesis.2,22,23 Its enzymatic activity is critical for yeast cell growth and the gene ABD1 that encodes the Abd1 protein is essential. Abd1 is a founding member of a protein family that is strongly conserved in eukaryotes as well as in viruses.24 The crystal structure of S. cerevisiae Abd1 itself could not be determined, but eventually it was solved for its homolog, the Ecm1 protein from E. cuniculi (Fig. 2).25 Purified Ecm1 is a monomeric protein that catalyzes methyl transfer to GpppRNA to form cap0, but also to free mononucleotides GTP, GDP or dGTP (deoxy-GTP). The methyltransferase domain in Ecm1, and by inference also in other homologous cap0 methyltransferases, exhibits the RFM fold with a characteristic insertion that forms a characteristic β-meander structure involved in the formation of the cap-binding site. This insertion is common between methyltransferases that methylate G to m7G in the RNA cap, and methyltransferases that N-methylate the amino acid glycine. This relationship between 2 different types of methyltransferases, as well as the cap0 methyltransferase structure, were correctly predicted using bioinformatics24 before the first structure of the cap0 methyltransferase was determined.
Figure 2.
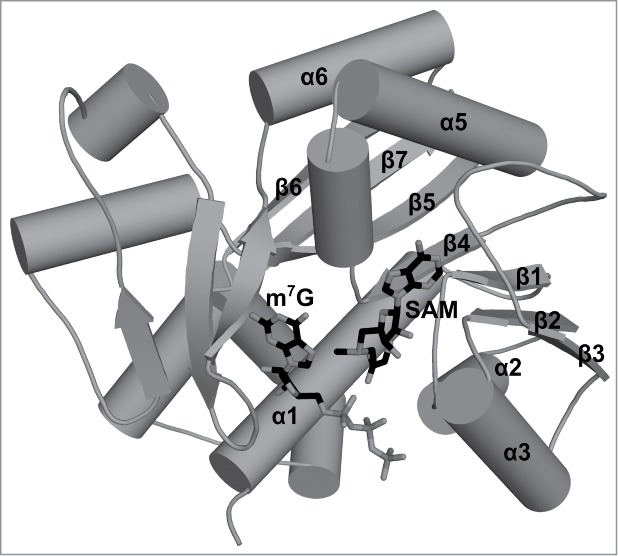
Crystal structure of the cap0 methyltransferase from Encephalitozoon cuniculi. A stick representation of the ligands bound to cap0 methyltransferase. The guanosine cap analog position was defined based on the structure deposited as 1RI2 in the PDB, and the methyl group donor position was depicted based on the structure deposited as 1RI4 in the PDB. Secondary structure elements that correspond to elements of the conserved RFM core are labeled. Secondary structure elements outside of the conserved core are not labeled.
In the human capping system, the cap0 methyltransferase (RNMT) consists of a catalytic subunit related to Abd1 and an obligate activating subunit, RAM (RNMT-activating miniprotein).26 The C-terminal catalytic domain of RNMT has essentially the same structure as Abd1. RNMT also has an N-terminal domain that is conserved in mammals, but not required for catalytic activity. However, it contains 2 nuclear localization signal motifs and the nuclear localization of RNMT is essential for cell viability. The cap0 methyltransferases, members of the above-mentioned family, were also identified and chartacterized in other eukaryotes, including TbCmt1 in Trypanosoma brucei, for example.27
As mentioned above, the viral cap0 methyltransferases possess a catalytic domain that is closely related to the eukaryotic cap0 methyltransferases, but it often functions in the context of other domains. For instance, the vaccinia virus possesses an enzyme that is composed of D1 and D12 polypeptides that execute all 3 steps in cap0 biosynthesis. The D1 subunit contains triphosphatase and guanylyltransferase activities in the N-terminal domain, and a cap0 methyltransferase domain that forms a heterodimer with the D12 subunit.28,29 The methyltransferase active site is located entirely in the D1 subunit and has a weak cap0 modification activity that is stimulated allosterically by D12.30-32 Interestingly, the D12 structure resembles a degenerate cap 2′-O-ribose methyltransferase domain (see below), but it lacks a proper SAM binding site and does not show any methyltransferase activity on its own.33
In the SARS-coronavirus, a nonstructural protein 14 (nsp14) was initially identified as an exoribonuclease (and termed ExoN). Later, it was shown that it also exhibits cap0 methyltransferase activity. Analysis of protein variants with substitutions of conserved residues in the ExoN (N-terminal) and methyltransferase (C-terminal) domains revealed that both active sites are functionally distinct; however, the integrity of the ExoN domain turned out to be essential for the function of the cap0 methyltransferase domain.34 Nsp14 shows little sequence similarity to known methyltransferases; however, its structure has not been determined experimentally, hence its phylogenetic relationships to other enzymes remain unclear.
Cap-specific 2′-O-ribose methyltransferases
A poxvirus cap1-forming enzyme (VP39 protein from vaccinia virus), was the first methyltransferase involved in the cap structure formation, for which a crystal structure was determined35 and also the first one for which a structure of a ternary complex of an enzyme with the cofactor and RNA substrate was determined.36 It has become one of the best studied members of a large family of methyltransferases that act on the 2′-OH-ribose group in RNA, which includes also enzymes such as RrmJ and fibrillarin.37 Although they share little sequence identity with each other, these 2′-O-ribose methyltransferases are characterized by the presence of a conserved tertiary fold characteristic for all RFM enzymes and a conserved K-D-K catalytic triad between the methyl group donor binding site, and the cap binding site. VP39 is a single-domain protein with additional structural elements at both the N- and C-termini, which wrap around the RFM core and form a binding pocket for the cap. In the ternary complex, the m7G base of the cap is bound sandwiched between 2 aromatic side chains, and oriented in such a way that the Hoogsteen edge modified by addition of the methyl group on N7 faces the protein, thus explaining the ability of VP39 to sense the methylation status of the substrate, which is the basis of its preference for substrates that already have an N7-methylated cap.
In humans, cap1 formation is catalyzed by the CMTr1 enzyme.38 It is composed of several domains, including the N-terminal catalytic RMF domain with a conserved K-D-K triad characteristic for 2′-O-ribose methyltransferases and a guanylyltransferase-like domain that lacks catalytic residues.39 The N-terminal domain of CMTr1 shares a global architecture with the VP39 protein and is sufficient for cap1 activity in vitro. Interestingly, while the cofactor-binding sites, active sites, and the sites of binding of the nascent RNA chain exhibits similarities with the VP39 and CMTr1 enzymes (and likewise the conformations of the respective ligands), their cap-binding sites exhibit large differences in the shape of the m7G-binding pocket. As a result, CMTr1 binds m7G in a different way, in which the sugar edge of the cap guanosine faces the protein, and the methyl group on N7 faces the solvent (Fig. 3). These structural differences explain why CMTr1 is relatively insensitive to the absence of cap0 methylation and therefore is able to act, at least in vitro, on substrates with unmethylated guanosine.
Figure 3.
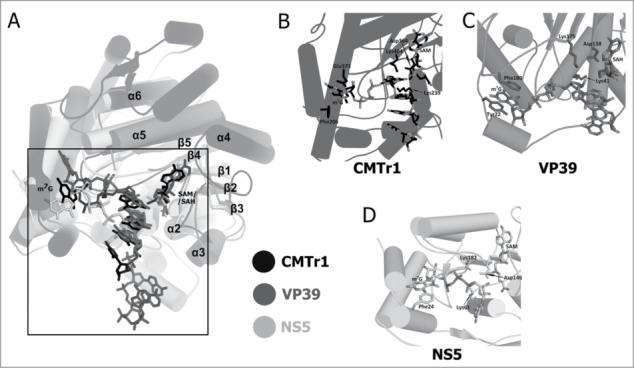
Comparison of the crystal structures of 2′-O-ribose methyltransferases. (A) Superimposition of the catalytic domain of human CMTr1 methyltransferase (colored black; PDB ID: 4N48), VP39 methyltransferase from the vaccinia virus (colored dark gray; PDB ID: 1AV6) and the NS5 protein from the Wesselsbron virus (colored bright gray; PDB ID: 3EMB). The ligands are shown in stick representation and they are colored corresponding to the hue used for protein molecules representation. Secondary structure elements that correspond to elements of the conserved RFM core are labeled (α1, β6, and β7 are hidden behind other elements and their labels have been omitted). Secondary structure elements outside of the conserved core are not labeled. (B) The capped oligoribonucleotide (m7GpppGAUC) located in its binding pocket on the surface of human CMTr1 MTase is shown in stick representation. The side chains of Phe206 and Glu373 that correspond to stacking residues in viral methyltransferases and the 3 catalytic residues are also displayed. (C) The crystal structure of the VP39 methyltransferase from vaccinia virus in complex with m7GpppGAAAAA (shown in stick representation). The methylated guanine ring is stacked by 2 aromatic rings of Tyr22 and Phe180. (D) A stick representation of the cap0 structure analog—m7GpppG bound by NS5 flaviviral 2′-O-ribose methyltransferase.
Proteins with cap1 methyltransferase activities were also characterized in the alfalfa looper moth Autographa californica nucleopolyhedrovirus (orf6940) and in T. brucei (TbMTr141,42). Both of these enzymes are relatively closely related to the human CMTr1 enzyme.
In the SARS virus, cap1 methylation is catalyzed by a complex comprised of 2 partners: the nsp16 protein that is clearly related to the above-mentioned cap1 methyltransferases, but is inactive on its own, and a small regulatory protein nsp10 that is required for nsp16 to bind both the SAM methyl group donor and the RNA substrate. The crystal structure of the snp10-nsp16 complex showed that, in nsp16, the SAM-binding region is partially degenerated compared to “partner-independent” ribose methyltransferases, and nsp10 stabilizes the SAM binding pocket and extends the RNA-binding groove of nsp16.43
Apart from the enzymes responsible for cap1 methylation, methyltransferases have been characterized that act on additional residues in the nascent RNA chain. Many eukaryotic organisms possess a 2′-O-ribose methyltransferase that methylates the 2nd residue in mRNA and in other RNA molecules. The cap2 methyltransferase has been characterized in T. brucei (TbMTr244,45) and in humans (CMTr246). Interestingly, while CMTr2 appears to be closely related to its human paralog CMTr1 as well as to TbMTr1, TbMTr2 is more closely related to the vaccinia virus cap1 methyltransferase.44 In trypanosomes, a third 2′-O-ribose cap methyltransferase was identified and termed TbMTr3, which is responsible for the methylation of the third residue of the cap and is required for the methylation of the fourth residue.47,48 TbMTr3 is a close relative of TbMTr2 and of VP39, and is only remotely related to other eukaryotic cap 2′-O-ribose methyltransferases, which suggests that trypanosomes acquired enzymes for “additional” methylation by adapting proteins from viruses. A phylogenetic study of 2′-O-ribose methyltransferases revealed that the relationships between cellular and viral enzymes are quite complex, and that these proteins can vary greatly in number even in closely-related organisms. Furthermore, alveolate species were identified that possessed as many as 4 2′-O-ribose methyltransferases, suggesting that certain enzymes of this group may act with different substrate specificities or that new cap structures with additional methylation sites remain to be discovered.46
Proteins with cap0 and cap1 methyltransferase activities
A number of viral proteins were reported to possess both cap0 and cap1 methyltransferase activities. In most of them, this is due to the presence of multiple domains. For instance, in the human reovirus (a virus with a dsRNA genome), the cap structure formation is catalyzed by a large multidomain protein lambda 2, which in turn is a part of the reovirus core: an assembly with a relative molecular mass of 52 MDa that synthesizes, modifies and exports viral mRNA. The structure of the human reovirus core has been solved at low resolution, revealing a series of domains that include a putative guanylyltransferase domain and 2 putative methyltransferase (RFM) domains.49 It has been suggested that the order of the domains in the lambda 2 protein corresponds to the order of the capping reactions: guanosine transfer followed by cap0 and cap1 methylation. However, comparison of domain structures suggested that the functional assignments may be different, as the RFM domain 1 shared a putative active site with the corresponding structurally characterized 2′-O-ribose methyltransferases, including the cap1 methyltransferase, whereas the RFM domain 2 exhibited structural similarity to the cap0 methyltransferases.50 It should be noted that the putative cap1 methyltransferase domain of reovirus exhibits a similar cap-binding platform formed by N- and C-terminal extensions, as in VP39 and human CMTr1 enzymes; however, its putative m7G-binding site is more open.
In bluetongue virus, another member of the reoviruses, the structure of the VP4 protein revealed a multi-domain protein with an N-terminal guanylyltransferase domain and 2 RFM domains, of which one was inserted into another. The inserted RFM domain exhibited clear similarities to the cap1 methyltransferases and in 3 crystal forms had GpppG, m7GDP, or GTP bound in the position of the cap-binding site, while the other RFM domain exhibited low but significant similarity to known cap0 methyltransferase structures.51
In the non-segmented, negative-sense single-stranded RNA viruses [order Mononegavirales (MNV)] that include pathogens such as respiratory syncytial virus, measles, mumps, rabies, parainfluenza, vesicular stomatitis virus (VSV), and Marburg and Ebola viruses, one of the common components of the viral ribonucleoprotein core is the large (L) protein, which encodes multiple functions such as the RNA-dependent RNA polymerase and activities responsible for mRNA capping, cap0 and cap1 methylation, poly(A) polymerase and protein kinase.52,53 Using bioinformatics methods, we and others predicted that the C-terminal region of that protein (conserved region VI) encodes a domain homologous to 2′-O-ribose methyltransferases and is likely to function as a cap1 methyltransferase.54,55 Later it was found that, in VSV, this region is essential not only for cap1, but also for cap0 methyltransferase activity and that the same SAM-binding site and part of the K-D-K triad is used for both reactions.15,56 The structural basis of this phenomenon remains to be determined.
In Flaviviruses (positive-sense, single-stranded RNA viruses), an RFM domain with a similar dual methyltransferase function was identified. In a non-structural protein 5, the N-terminus was first unambiguously characterized as a cap1 (2′-O-ribose) methyltransferase. Later, it was shown that this domain takes part also in cap0 (m7G) methylation using the same SAM-binding site during cap synthesis.57,58 Interestingly, in these viruses, the order of methylation is different than in Mononegavirales, as cap0 methylation precedes cap1 methylation. Several structures were determined for the flavivirus cap methyltransferases known or predicted to be bifunctional, including Dengue,59 West Nile,60 Wesselbron,61 Meaban,62 and Murray Valley encephalitis63 viruses and they all revealed high similarity to the cap1 methyltransferases, and little if any similarity to the classical cap0 methyltransferases. It should be noted that these methyltransferases share a similar cap-binding platform structure with VP39 and human CMTr1 enzymes (a platform formed by N- and C-terminal extensions); however, the orientation of the bound guanosine residue suggests that their mode of cap-recognition is different from both poxvirus and human enzymes (Fig. 3).
Tgs1/Tgs2
The enzyme responsible for the trimethylguanosine (m2,2,7G, TMG) synthesis was first identified in yeast and named yTgs1.64 The Tgs enzymes of budding and fission yeast and Giardia are relatively small polypeptides (239–315 amino acids) consisting of little more than an RFM methyltransferase catalytic domain (Fig. 4), whereas metazoan Tgs1 proteins are much larger, because they include an N-terminal extension not found in lower eukaryotes.65,66 Tgs1 activity is strictly dependent on prior cap0 (m7G) methylation, thereby restricting its activity to RNAs that were already methylated by cap0 methyltransferase.67 Similar substrate requirements are characteristic for the Giardia Tgs2 enzyme.68 Interestingly, in contrast to Tgs1 methyltransferases able to catalyze 2 sequential N2 methylation steps leading to TMG cap formation, Tgs2 activity is apparently limited to a single round of N2 methylation, resulting in the synthesis of a 2,7-dimethylguanosine (m2,7G) product. Bioinformatics analyses predicted that the Tgs enzymes are related to a large group of RFM enzymes that act on exocyclic amine groups in nucleic acid bases, including m6A, m4C, and m2G and have a characteristic NPPY-like motif at the active site.66 The crystal structure of the active C-terminal methyltransferase domain of the human TGS enzyme bound to a minimal substrate m7GTP as well as the reaction product SAH has been reported, confirming these predictions and revealing the atomic details of these protein-ligand interactions.69,70
Figure 4.
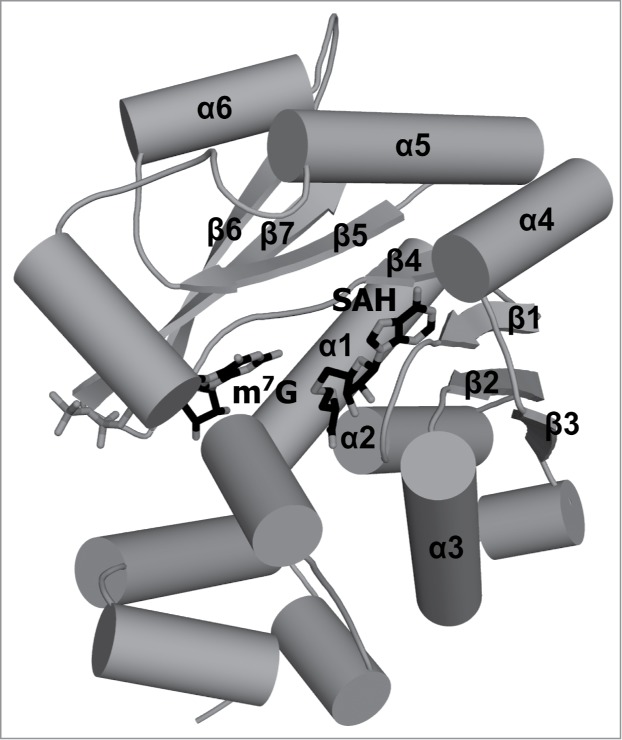
Crystal structure of the human TGS1 protein. Trimethylguanosine synthase catalyzes hypermethylation of cap0 structure. In a 2-step reaction, 2 methyl groups are transferred to the amine group of m7G and, as a result, the m2,2,7G structure is formed. The crystal structure of human TGS1 methyltransferase in complex with m7Gppp and SAH (shown in stick representation) is deposited in the PDB as 3GDH. Secondary structure elements that correspond to elements of the conserved RFM core are labeled. Secondary structure elements outside of the conserved core are not labeled.
Other methyltransferases involved in cap-specific base modifications
Studies of cap composition of human mRNAs conducted in mid-70s revealed that when the first nucleotide of the transcript is an adenosine, this base can be methylated to m6A.8,71 The enzyme that catalyzes the conversion of m7GpppAm ends of mRNA to m7Gpppm6Am has been isolated from a cytoplasmic fraction of HeLa cells. The isolated enzyme showed no activity toward internal adenosines.72 Recently, Schwartz and coworkers studied the m6A mRNA methylome following depletion of multiprotein methyltransferase complex components METTL3, METTL14, KIAA1429, and WTAP, and implicated the involvement of the METTL3, METTL14, and KIAA1429 proteins in m6A formation at the internal sites but not at the 5′ sites.73 The full characterization of the cap-specific m6A methyltransferase activity requires further studies in vitro.
In trypanosomes, the first adenine of the hypermethylated cap4 structure is not only methylated at the ribose, but also dimethylated at the N6 position, to form m6,6Am. The methyltransferase responsible for the latter reaction remains unknown.11 Further, the fourth uracil in that structure is modified to m3U, and the enzyme responsible for this modification also remains unknown.
Bin3/γ–methyltransferases
The γ-methylphosphate cap structure is unique in that it is an alternative to the guanosine-containing cap. It is formed by a single methyltransfer reaction to a γ -phosphate oxygen at the 5′ end of the primary transcripts of certain small RNA molecules such as mammalian U6 and 7SK, mouse B2 and plant U3.74 The enzyme responsible for this reaction, Bicoid-interacting protein 3 (Bin3), is a methyltransferase conserved in eukaryotes. It is, however, absent from S. cerevisiae.75,76 A structure of the human Bin3 homolog (BCDIN3) was determined, revealing a conserved RFM core (Fig. 5). An enzyme-substrate complex is not yet available, and the details of protein-RNA recognition and the mechanism of discrimination between Bin3 substrates and non-substrates remain to be determined.
Figure 5.
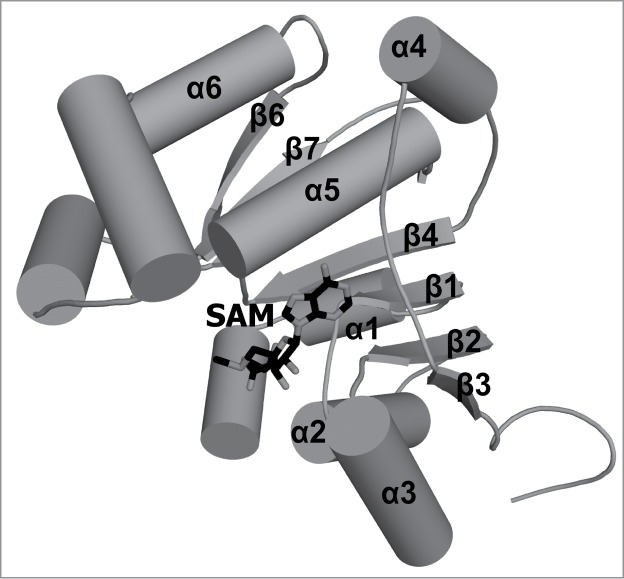
Crystal structure of human BCDIN3 γ-methyltransferase. A stick representation of SAM as a donor of the methyl group which is transferred by the BCDIN3 (PDB ID: 3G07) enzyme on the 5′ γ-phosphate group of the 7SK snRNA molecule. Secondary structure elements that correspond to elements of the conserved RFM core are labeled (β5 is hidden behind α5 and therefore its label has been omitted). Secondary structure elements outside of the conserved core are not labeled.
Conclusions and Future Perspectives
In recent years significant progress has been made in understanding the mechanism of formation of different RNA cap structures. This progress has been driven in particular by the identification and characterization of novel methyltransferases that take part in cap biosynthesis, and by the determination of their crystal structures. This knowledge also has a practical dimension, as the capping process is essential for eukaryotic cells as well as for the life cycle of viruses that infect them. In this context, the difference between the structures of the human enzymes and the enzymes from human pathogens could be exploited to develop new drugs. In particular, viruses that evolved alternative enzymes to synthesize the same cap structures as are synthesized by human cellular machinery are attractive targets for the development of inhibitors that could specifically block viral methyltransferases.
To date, numerous high-resolution structures of viral RNA capping enzymes have been determined, in particular for cap methyltransferases from various flaviviruses, which have been considered an attractive new antiviral target.77 Based on knowledge of structures, efforts have been made toward the identification of specific inhibitors of these enzymes. For instance, a structure-based search for new inhibitors was performed for the dengue virus methyltransferase.78-81 The development of compounds that specifically inhibit viral methyltransferases will be aided by the recent structure determination of the catalytic domain of the human cap1 methyltransferase, which shares the global architecture, but exhibits a different cap-binding site compared to the viral enzymes.39 The human cap1 methyltransferase appears to be essential and cannot be knocked out in human cells (our unpublished data), therefore the development of inhibitors specific against that human enzyme could be also useful as tools to study the cellular function of cap1 methylation.
The study on the process of SL RNA maturation in trypanosomal parasites could benefit from structure analysis of trypanosomal methyltransferases. While the cap0 and cap1 methyltransferases in trypanosomes are relatively closely related to their human counterparts, bioinformatics analyses identified cap2 and cap3/4 methyltransferases as close homologs of the vaccinia virus cap1 methyltransferase. While the analysis of protein-RNA interactions and search for potential regulatory molecules (e.g., inhibitors) could be guided by homology models developed so far,47 experimental determination of high resolution structures for cap methyltransferases in trypanosomes would be definitely useful.
A complete understanding of RNA cap biosynthesis requires not only structure determination of the enzymes that are well characterized biochemically, but also the identification of the genes and proteins that encode the cap methylation machinery. Some of the prominent enzymatic activities known to exist that are still awaiting unequivocal identification of the corresponding proteins include m6A methylation of the first transcribed nucleoside of capped RNAs in humans, and m6,6A and m3U methylation of the first and the fourth residues in the cap4 structure in capped RNAs in trypanosomes. A comprehensive biochemical and structural characterization of these enzymes could further contribute to the possibility of developing new drugs against trypanosomal parasites and new tools to study RNA metabolism in human cells.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This analysis was supported in part by the Foundation for Polish Science (FNP, grant TEAM/2009-4/2 to J.M.B.). M.B. and E.P. were additionally supported by National Science Center (NCN, 2011/03/D/NZ1/03247 to E.P.). M.Ś. was additionally supported by the START Fellowship from the FNP. J.M.B. was additionally supported by the European Research Council (ERC, StG grant RNA+P = 123D) and by the "Ideas for Poland" fellowship from the FNP. Research on structural aspects of protein-RNA interactions in the Bujnicki laboratory has been also supported by grants from the European Union's Seventh Framework Program (REGPOT grant FishMed; grant agreement n° 316125), the Polish Ministry of Science and Higher Education (MNiSW, grant POIG.02.03.00-00-003/09) and by the infrastructure financed by the European Union—the European Regional Development Fund within the Operational Program “Innovative economy” for 2007–2013 (CePT, grant POIG.02.02.00-14-024/08-00).
References
- 1. Muthukrishnan S, Filipowicz W, Sierra JM, Both GW, Shatkin AJ, Ochoa S. mRNA methylation and protein synthesis in extracts from embryos of brine shrimp, Artemia salina. J Biol Chem 1975; 250:9336-41; PMID:1194288 [PubMed] [Google Scholar]
- 2. Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol 1995; 15:4167-74; PMID:7623811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shafer B, Chu C, Shatkin AJ. Human mRNA cap methyltransferase: alternative nuclear localization signal motifs ensure nuclear localization required for viability. Mol Cell Biol 2005; 25:2644-9; PMID:15767670; http://dx.doi.org/ 10.1128/MCB.25.7.2644-2649.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA 2011; 2:277-98; PMID:21957010; http://dx.doi.org/ 10.1002/wrna.52 [DOI] [PubMed] [Google Scholar]
- 5. Hocine S, Singer RH, Grunwald D. RNA processing and export. Cold Spring Harb Perspect Biol 2010; 2:a000752; PMID:20961978; http://dx.doi.org/ 10.1101/cshperspect.a000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wurth L, Gribling-Burrer AS, Verheggen C, Leichter M, Takeuchi A, Baudrey S, Martin F, Krol A, Bertrand E, Allmang C. Hypermethylated-capped selenoprotein mRNAs in mammals. Nucleic Acids Res 2014; 42:8663-77; PMID:25013170; http://dx.doi.org/ 10.1093/nar/gku580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res 2000; 55:135-84; PMID:11050942; http://dx.doi.org/ 10.1016/S0065-3527(00)55003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci U S A 1975; 72:1904-8; PMID:1057180; http://dx.doi.org/ 10.1073/pnas.72.5.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Massenet S, Mougin A, Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. Modification Editing RNA 1998:201-27 [Google Scholar]
- 10. Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. Rna 2004; 10:1925-33; PMID:15525712; http://dx.doi.org/ 10.1261/rna.7186504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem 1992; 267:9805-15; PMID:1349605 [PubMed] [Google Scholar]
- 12. Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 2012; 10:51-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Testa D, Banerjee AK. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J Virol 1977; 24:786-93; PMID:201777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammond DC, Lesnaw JA. The fates of undermethylated mRNA cap structures of vesicular stomatitis virus (New Jersey) during in vitro transcription. Virology 1987; 159:229-36; PMID:3039729; http://dx.doi.org/ 10.1016/0042-6822(87)90459-4 [DOI] [PubMed] [Google Scholar]
- 15. Li J, Wang JT, Whelan SP. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A 2006; 103:8493-8; PMID:16709677; http://dx.doi.org/ 10.1073/pnas.0509821103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei C, Gershowitz A, Moss B. N6, O2'-dimethyladenosine a novel methylated ribonucleoside next to the 5' terminal of animal cell and virus mRNAs. Nature 1975; 257:251-3; PMID:1161029; http://dx.doi.org/ 10.1038/257251a0 [DOI] [PubMed] [Google Scholar]
- 17. Wierzchowski KL, Shugar D. Further studies on the photochemistry of pyrimidines, with special reference to 5-and 6-substituted derivatives in relation to photoreactivation in the T-even bacteriophages. Acta Biochim Pol 1960; 7:63-84; PMID:13844652 [PubMed] [Google Scholar]
- 18. Bujnicki JM. Comparison of protein structures reveals monophyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol 1999; 1:175-82; PMID:11479932 [PubMed] [Google Scholar]
- 19. Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein. Nature 1974; 250:194-9; PMID:4368490; http://dx.doi.org/ 10.1038/250194a0 [DOI] [PubMed] [Google Scholar]
- 20. Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res 2002; 30:1427-64; PMID:11917006; http://dx.doi.org/ 10.1093/nar/30.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct Biol 2005; 5:19; PMID:16225687; http://dx.doi.org/ 10.1186/1472-6807-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mao X, Schwer B, Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol Cell Biol 1996; 16:475-80; PMID:8552073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang SP, Shuman S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J Biol Chem 1997; 272:14683-9; PMID:9169431; http://dx.doi.org/ 10.1074/jbc.272.23.14683 [DOI] [PubMed] [Google Scholar]
- 24. Bujnicki JM, Feder M, Radlinska M, Rychlewski L. mRNA:guanine-N7 cap methyltransferases: identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure-function relationships. BMC Bioinformatics 2001; 2:2; PMID:11472630; http://dx.doi.org/ 10.1186/1471-2105-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell 2004; 13:77-89; PMID:14731396; http://dx.doi.org/ 10.1016/S1097-2765(03)00522-7 [DOI] [PubMed] [Google Scholar]
- 26. Gonatopoulos-Pournatzis T, Dunn S, Bounds R, Cowling VH. RAM/Fam103a1 is required for mRNA cap methylation. Mol Cell 2011; 44:585-96; PMID:22099306; http://dx.doi.org/ 10.1016/j.molcel.2011.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hall MP, Ho CK. Characterization of a Trypanosoma brucei RNA cap (guanine N-7) methyltransferase. Rna 2006; 12:488-97; PMID:16431985; http://dx.doi.org/ 10.1261/rna.2250606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu L, Shuman S. Mutational analysis of the RNA triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol 1996; 70:6162-8; PMID:8709242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Myette JR, Niles EG. Domain structure of the vaccinia virus mRNA capping enzyme. Expression in Escherichia coli of a subdomain possessing the RNA 5'-triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. J Biol Chem 1996; 271:11936-44; PMID:8662635; http://dx.doi.org/ 10.1074/jbc.271.20.11936 [DOI] [PubMed] [Google Scholar]
- 30. Higman MA, Christen LA, Niles EG. The mRNA (guanine-7-)methyltransferase domain of the vaccinia virus mRNA capping enzyme. Expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J Biol Chem 1994; 269:14974-81; PMID:8195132 [PubMed] [Google Scholar]
- 31. Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem 1994; 269:24472-9; PMID:7929111 [PubMed] [Google Scholar]
- 32. Schwer B, Hausmann S, Schneider S, Shuman S. Poxvirus mRNA cap methyltransferase. Bypass of the requirement for the stimulatory subunit by mutations in the catalytic subunit and evidence for intersubunit allostery. J Biol Chem 2006; 281:18953-60; PMID:16707499; http://dx.doi.org/ 10.1074/jbc.M602867200 [DOI] [PubMed] [Google Scholar]
- 33. De la Pena M, Kyrieleis OJ, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. Embo J 2007; 26:4913-25; PMID:17989694; http://dx.doi.org/ 10.1038/sj.emboj.7601912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 2009; 106:3484-9; PMID:19208801; http://dx.doi.org/ 10.1073/pnas.0808790106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hodel AE, Gershon PD, Shi X, Quiocho FA. The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell 1996; 85:247-56; PMID:8612277; http://dx.doi.org/ 10.1016/S0092-8674(00)81101-0 [DOI] [PubMed] [Google Scholar]
- 36. Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5'-capped mRNA by a cap-modifying enzyme. Mol Cell 1998; 1:443-7; PMID:9660928; http://dx.doi.org/ 10.1016/S1097-2765(00)80044-1 [DOI] [PubMed] [Google Scholar]
- 37. Feder M, Pas J, Wyrwicz LS, Bujnicki JM. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2'-O-methyltransferases. Gene 2003; 302:129-38; PMID:12527203; http://dx.doi.org/ 10.1016/S0378-1119(02)01097-1 [DOI] [PubMed] [Google Scholar]
- 38. Belanger F, Stepinski J, Darzynkiewicz E, Pelletier J. Characterization of hMTr1, a human cap1 2'O-ribose methyltransferase. J Biol Chem 2010; 285:33037-44; PMID:20713356; http://dx.doi.org/ 10.1074/jbc.M110.155283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smietanski M, Werner M, Purta E, Kaminska KH, Stepinski J, Darzynkiewicz E, Nowotny M, Bujnicki JM. Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat Commun 2014; 5:3004; PMID:24402442; http://dx.doi.org/ 10.1038/ncomms4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu X, Guarino LA. Autographa californica nucleopolyhedrovirus orf69 encodes an RNA cap (nucleoside-2'-O)-methyltransferase. J Virol 2003; 77:3430-40; PMID:12610118; http://dx.doi.org/ 10.1128/JVI.77.6.3430-3440.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zamudio JR, Mittra B, Foldynova-Trantirkova S, Zeiner GM, Lukes J, Bujnicki JM, Sturm NR, Campbell DA. The 2′-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei. Mol Cell Biol 2007; 27:6084-92; PMID:17606627; http://dx.doi.org/ 10.1128/MCB.00647-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mittra B, Zamudio JR, Bujnicki JM, Stepinski J, Darzynkiewicz E, Campbell DA, Sturm NR. The TbMTr1 spliced leader RNA cap 1 2′-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity. J Biol Chem 2008; 283:3161-72; PMID:18048356; http://dx.doi.org/ 10.1074/jbc.M707367200 [DOI] [PubMed] [Google Scholar]
- 43. Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 2011; 7:e1002294; PMID:22022266; http://dx.doi.org/ 10.1371/journal.ppat.1002294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hall MP, Ho CK. Functional characterization of a 48 kDa Trypanosoma brucei cap 2 RNA methyltransferase. Nucleic Acids Res 2006; 34:5594-602; PMID:17028101; http://dx.doi.org/ 10.1093/nar/gkl573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arhin GK, Ullu E, Tschudi C. 2′-O-methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol Biochem Parasitol 2006; 147:137-9; PMID:16516986; http://dx.doi.org/ 10.1016/j.molbiopara.2006.01.011 [DOI] [PubMed] [Google Scholar]
- 46. Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, Zamudio JR, Sturm NR, Jaworski J, Bujnicki JM. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res 2011; 39:4756-68; PMID:21310715; http://dx.doi.org/ 10.1093/nar/gkr038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zamudio JR, Mittra B, Zeiner GM, Feder M, Bujnicki JM, Sturm NR, Campbell DA. Complete cap 4 formation is not required for viability in Trypanosoma brucei. Eukaryot Cell 2006; 5:905-15; PMID:16757738; http://dx.doi.org/ 10.1128/EC.00080-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arhin GK, Li H, Ullu E, Tschudi C. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei. Rna 2006; 12:53-62; PMID:16301606; http://dx.doi.org/ 10.1261/rna.2223406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature 2000; 404:960-7; PMID:10801118; http://dx.doi.org/ 10.1038/35010041 [DOI] [PubMed] [Google Scholar]
- 50. Bujnicki JM, Rychlewski L. Reassignment of specificities of two cap methyltransferase domains in the reovirus lambda 2 protein. Genome Biol 2001; 2:RESEARCH0038; PMID:11574057; http://dx.doi.org/ 10.1186/gb-2001-2-9-research0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sutton G, Grimes JM, Stuart DI, Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol 2007; 14:449-51; PMID:17417654; http://dx.doi.org/ 10.1038/nsmb1225 [DOI] [PubMed] [Google Scholar]
- 52. Emerson SU, Yu Y. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol 1975; 15:1348-56; PMID:167189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hamaguchi M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus. I. Both L and P proteins are required to constitute an active complex. Virology 1983; 128:105-17; PMID:6683907; http://dx.doi.org/ 10.1016/0042-6822(83)90322-7 [DOI] [PubMed] [Google Scholar]
- 54. Bujnicki JM, Rychlewski L. In silico identification, structure prediction and phylogenetic analysis of the 2'-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng 2002; 15:101-8; PMID:11917146; http://dx.doi.org/ 10.1093/protein/15.2.101 [DOI] [PubMed] [Google Scholar]
- 55. Ferron F, Longhi S, Henrissat B, Canard B. Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem Sci 2002; 27:222-4; PMID:12076527; http://dx.doi.org/ 10.1016/S0968-0004(02)02091-1 [DOI] [PubMed] [Google Scholar]
- 56. Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. Ribose 2'-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol 2009; 83:11043-50; PMID:19710136; http://dx.doi.org/ 10.1128/JVI.01426-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J 2002; 21:2757-68; PMID:12032088; http://dx.doi.org/ 10.1093/emboj/21.11.2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5'-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 2006; 80:8362-70; PMID:16912287; http://dx.doi.org/ 10.1128/JVI.00814-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol 2007; 372:723-36; PMID:17686489; http://dx.doi.org/ 10.1016/j.jmb.2007.07.005 [DOI] [PubMed] [Google Scholar]
- 60. Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol 2007; 81:3891-903; PMID:17267492; http://dx.doi.org/ 10.1128/JVI.02704-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bollati M, Milani M, Mastrangelo E, Ricagno S, Tedeschi G, Nonnis S, Decroly E, Selisko B, de Lamballerie X, Coutard B, et al. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in Flavivirus. J Mol Biol 2009; 385:140-52; PMID:18976670; http://dx.doi.org/ 10.1016/j.jmb.2008.10.028 [DOI] [PubMed] [Google Scholar]
- 62. Mastrangelo E, Bollati M, Milani M, Selisko B, Peyrane F, Canard B, Grard G, de Lamballerie X, Bolognesi M. Structural bases for substrate recognition and activity in Meaban virus nucleoside-2'-O-methyltransferase. Protein Sci 2007; 16:1133-45; PMID:17473012; http://dx.doi.org/ 10.1110/ps.072758107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Assenberg R, Ren J, Verma A, Walter TS, Alderton D, Hurrelbrink RJ, Fuller SD, Bressanelli S, Owens RJ, Stuart DI, et al. Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J Gen Virol 2007; 88:2228-36; PMID:17622627; http://dx.doi.org/ 10.1099/vir.0.82757-0 [DOI] [PubMed] [Google Scholar]
- 64. Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 2002; 9:891-901; PMID:11983179; http://dx.doi.org/ 10.1016/S1097-2765(02)00484-7 [DOI] [PubMed] [Google Scholar]
- 65. Hausmann S, Ramirez A, Schneider S, Schwer B, Shuman S. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res 2007; 35:1411-20; PMID:17284461; http://dx.doi.org/ 10.1093/nar/gkl1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mouaikel J, Bujnicki JM, Tazi J, Bordonne R. Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res 2003; 31:4899-909; PMID:12907733; http://dx.doi.org/ 10.1093/nar/gkg656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hausmann S, Shuman S. Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1). J Biol Chem 2005; 280:4021-4; PMID:15590684; http://dx.doi.org/ 10.1074/jbc.C400554200 [DOI] [PubMed] [Google Scholar]
- 68. Hausmann S, Shuman S. Giardia lamblia RNA Cap Guanine-N2 Methyltransferase (Tgs2). J Biol Chem 2005; 280:32101; PMID:16046409 [DOI] [PubMed] [Google Scholar]
- 69. Monecke T, Dickmanns A, Ficner R. Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1. Nucleic Acids Res 2009; 37:3865-77; PMID:19386620; http://dx.doi.org/ 10.1093/nar/gkp249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Monecke T, Dickmanns A, Strasser A, Ficner R. Structure analysis of the conserved methyltransferase domain of human trimethylguanosine synthase TGS1. Acta Crystallogr D Biol Crystallogr 2009; 65:332-8; PMID:19307714; http://dx.doi.org/ 10.1107/S0907444909003102 [DOI] [PubMed] [Google Scholar]
- 71. Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell 1975; 4:379-86; PMID:164293; http://dx.doi.org/ 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- 72. Keith JM, Muthukrishnan S, Moss B. Effect of methylation of the N6 position of the penultimate adenosine of capped mRNA on ribosome binding. J Biol Chem 1978; 253:5039-41; PMID:670177 [PubMed] [Google Scholar]
- 73. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Reports 2014; 8:284-96; PMID:24981863; http://dx.doi.org/ 10.1016/j.celrep.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci U S A 1989; 86:8280-3; PMID:2813391; http://dx.doi.org/ 10.1073/pnas.86.21.8280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Forget D, Lacombe AA, Cloutier P, Al-Khoury R, Bouchard A, Lavallee-Adam M, Faubert D, Jeronimo C, Blanchette M, Coulombe B. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics 2010; 9:2827-39; PMID:20855544; http://dx.doi.org/ 10.1074/mcp.M110.003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol 2009; 26:2821-30; PMID:19734296; http://dx.doi.org/ 10.1093/molbev/msp198 [DOI] [PubMed] [Google Scholar]
- 77. Dong H, Zhang B, Shi PY. Flavivirus methyltransferase: A novel antiviral target. Antiviral Res 2008; 80:1-10; PMID:18571739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Podvinec M, Lim SP, Schmidt T, Scarsi M, Wen D, Sonntag LS, Sanschagrin P, Shenkin PS, Schwede T. Novel inhibitors of dengue virus methyltransferase: discovery by in vitro-driven virtual screening on a desktop computer grid. J Med Chem 2010; 53:1483-95; PMID:20108931; http://dx.doi.org/ 10.1021/jm900776m [DOI] [PubMed] [Google Scholar]
- 79. Idrus S, Tambunan US, Zubaidi AA. Designing cyclopentapeptide inhibitor as potential antiviral drug for dengue virus ns5 methyltransferase. Bioinformation 2012; 8:348-52; PMID:22570514; http://dx.doi.org/ 10.6026/97320630008348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lim SV, Rahman MB, Tejo BA. Structure-based and ligand-based virtual screening of novel methyltransferase inhibitors of the dengue virus. BMC Bioinformatics 2011; 12 Suppl 13:S24; PMID:22373153; http://dx.doi.org/ 10.1186/1471-2105-12-S13-S24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B, et al. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem 2011; 286:6233-40; PMID:21147775; http://dx.doi.org/ 10.1074/jbc.M110.179184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 2007; 27:262-74; PMID:17643375; http://dx.doi.org/ 10.1016/j.molcel.2007.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Takagi Y, Sindkar S, Ekonomidis D, Hall MP, Ho CK. Trypanosoma brucei encodes a bifunctional capping enzyme essential for cap 4 formation on the spliced leader RNA. J Biol Chem 2007; 282:15995-6005; PMID:17416901; http://dx.doi.org/ 10.1074/jbc.M701569200 [DOI] [PubMed] [Google Scholar]
- 84. Ruan JP, Ullu E, Tschudi C. Characterization of the Trypanosoma brucei cap hypermethylase Tgs1. Mol Biochem Parasitol 2007; 155:66-9; PMID:17610965; http://dx.doi.org/ 10.1016/j.molbiopara.2007.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Barbosa E, Moss B. mRNA(nucleoside-2'-)-methyltransferase from vaccinia virus. Characteristics and substrate specificity. J Biol Chem 1978; 253:7698-702; PMID:701282 [PubMed] [Google Scholar]
- 86. Zheng S, Hausmann S, Liu Q, Ghosh A, Schwer B, Lima CD, Shuman S. Mutational analysis of Encephalitozoon cuniculi mRNA cap (guanine-N7) methyltransferase, structure of the enzyme bound to sinefungin, and evidence that cap methyltransferase is the target of sinefungin's antifungal activity. J Biol Chem 2006; 281:35904-13; PMID:16971388; http://dx.doi.org/ 10.1074/jbc.M607292200 [DOI] [PubMed] [Google Scholar]
- 87. Hausmann S, Zheng S, Fabrega C, Schneller SW, Lima CD, Shuman S. Encephalitozoon cuniculi mRNA cap (guanine N-7) methyltransferase: methyl acceptor specificity, inhibition BY S-adenosylmethionine analogs, and structure-guided mutational analysis. J Biol Chem 2005; 280:20404-12; PMID:15760890; http://dx.doi.org/ 10.1074/jbc.M501073200 [DOI] [PubMed] [Google Scholar]
- 88. Kyrieleis OJ, Chang J, de la Pena M, Shuman S, Cusack S. Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus. Structure 2014; 22:452-65; PMID:24607143; http://dx.doi.org/ 10.1016/j.str.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hu G, Gershon PD, Hodel AE, Quiocho FA. mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc Natl Acad Sci U S A 1999; 96:7149-54; PMID:10377383; http://dx.doi.org/ 10.1073/pnas.96.13.7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hodel AE, Gershon PD, Shi X, Wang SM, Quiocho FA. Specific protein recognition of an mRNA cap through its alkylated base. Nat Struct Biol 1997; 4:350-4; PMID:9145102; http://dx.doi.org/ 10.1038/nsb0597-350 [DOI] [PubMed] [Google Scholar]
- 91. Hu G, Oguro A, Li C, Gershon PD, Quiocho FA. The “cap-binding slot” of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap. Biochemistry 2002; 41:7677-87; PMID:12056899; http://dx.doi.org/ 10.1021/bi0201926 [DOI] [PubMed] [Google Scholar]
- 92. Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, Canard B. A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate. J Biol Chem 2004; 279:35638-43; PMID:15152003; http://dx.doi.org/ 10.1074/jbc.M400460200 [DOI] [PubMed] [Google Scholar]


