Abstract
MicroRNAs (miRNAs) are endogenous, small non-coding RNA molecules that mediate post-transcriptional gene suppression by incomplete matches with their host mRNAs. In the central nervous system, miRNAs that functionally interact with their target genes constitute a flexible, robust and buffered regulatory network, exerting diverse roles in brain evolution and development. However, distinct variation either in hub miRNA expression levels or patterns may initiate and/or progress various adult-onset nerve-related diseases. In this review, we will summarize the current knowledge about the general hallmarks of brain miRNAs that act as vital determinants in increasingly complicated neural activities. We endeavor to provide a constructive insight into the neuroscience research in the quest to comprehend molecular underpinnings of physiological functions and pathological disorders in central nervous system.
Keywords: central nervous system, development, evolution, small non-coding RNA
Introduction
miRNAs constitute a large class of short (∼22nt) endogenous noncoding RNA molecules, which were initially discovered in Caenorhabditis elegans and perceived as the products of heterochronic genes.1 To data, more than 28000 miRNAs have been preserved in the latest miRNA registry2 (Release 21.0, http://www.mirbase.org). The genomic sources of novel miRNA genes, for example, exons, introns, transposable elements and repeats, have been summarized in greater detail by others, yet the appearance and expansion of miRNAs in animal genomes is still in flux.3-9 Exploring the ultimate origin may contribute enormously to understanding the relationship between evolutionary adaptability of miRNAs and biological complexity. The formation and functional mechanism of mature miRNA have been extensively reported3,10-14 (Fig. 1). Generally speaking, miRNAs are originated from long primary transcripts (pri-miRNA) into ∼60–90 nt hairpin-loop precursors (pre-miRNAs) by Drosha-DGCR8 complex in the nucleus, exported by exportin-5 (Exp-5) and further cleaved by the ribonuclease III family member Dicer into mature sequences in the cytoplasm.10-13 The mature miRNA is then incorporated into the miRNA-induced silencing complex (miRISC) that typically guide it to mRNA targets, negatively regulating gene expression at the post-transcriptional level by imperfect matches with the 3-end untranslated regions (3′UTRs) of target mRNAs.3,14
Figure 1.
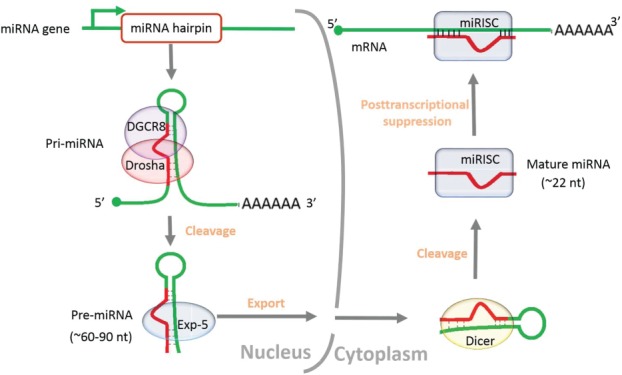
Overview of the generation and functional mechanism of mature miRNA. The sequence of the entire process is ordered by the arrow.
In the last decade, miRNAs have been confirmed to be major gene regulators in animal genomes, functioning by fine-tuning a broad range of biological processes and regulating the expression of at least one-third of all human genes.15-17 Furthermore, miRNAs have been reported to be ubiquitously expressed in midbrain, hippocampus, frontal cortex and cerebellum and ever increasing evidence shows that miRNAs exert diverse functions in the central nervous system18–21 (Fig. 2). These include neural genesis, differentiation and development as well as the process of synapse formation. Previous studies have also revealed that miRNAs gradually increases or decreases at the expression level during brain growth, highlighting a role of miRNA in brain morphogenesis.16,22-24 Considering the pervasiveness and functionality of brain miRNAs, it is likely that distinct variation either in related miRNA expression levels or patterns may initiate and/or progress various adult-onset nerve-related diseases.25-27
Figure 2.
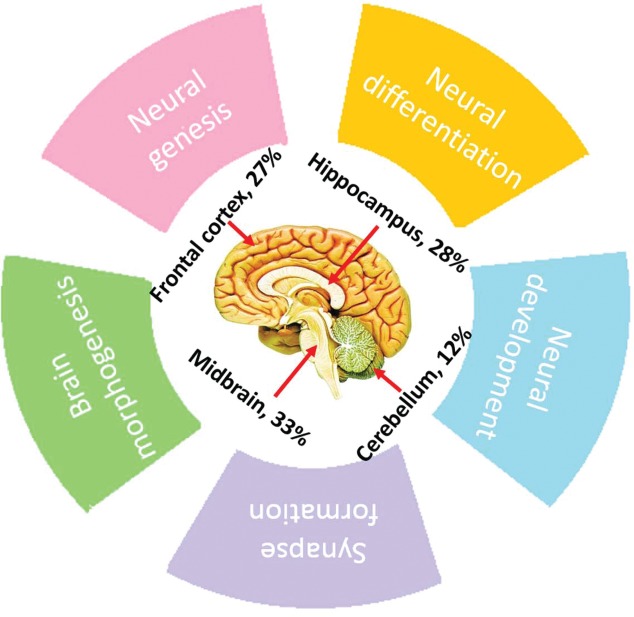
Percentage of miRNAs expressed in different areas of the brain and the related neuro-physiological functions.
The human brain, with an estimated 10000 different cell types, contains about 100–200 billion neurons that constitute a sophisticated neural network.26,28 The complexity of information processed exceeds by far any other human system. Understanding the general principles of brain miRNAs related to the constant neural adaptation to environmental cues may be particularly meaningful for illuminating how the brain works through the intricate regulation of miRNAs. In earlier studies, research on miRNA functional features has been hampered due to the limitations of massively parallel detection technology. However, with the rapid development of microarray and high-throughput sequencing technologies, it is possible for investigators to simultaneously detect the sequence and expression change of all the miRNAs in a given sample under a particular condition. In the current review, we will systematically summarize the general features of miRNAs in the central nervous system, in order to shed light on the versatile roles of these small non-coding RNA molecules in brain evolution, development and even disorders.
miRNAs are continuously being added into the genome during brain evolution
Differential gene expressions among species evolutionary lineages have been suggested to be vital for phenotypic differences such as human-specific features, e.g., language and tool-making.29 Protein-coding RNAs, mutations in regulatory elements proximal to genes, changes in expression or sequence of distal regulators and transcription factors have all been reported for differences in cognitive functions.30-34 These unambiguous genetic variations are not enough to explain the differences in brain morphological complexity across Metazoa. However, some noncoding components such as miRNAs may be a supplementary explanation.32,35
miRNAs are continuously being added into metazoan genomes. Several waves of miRNA repertoire expansion have been observed to coincide with the advent of bilaterians, vertebrates, and placental mammals.5,36-38 Meunier et al., based on high-throughput RNA sequencing data, have confirmed increased synthesis and diversity of miRNAs in mammals than in birds and nearly the same number of miRNAs in closely related species through systematic comparative analysis among 5 major organs, including brain and cerebellum, from 6 species.38 Massively parallel sequencing of brain miRNAs between human and chimpanzee or rhesus macaque highlights a cluster of human brain-specific miRNAs.39,40 However, ˜30% of the total human miRNA pool has no detectable expression, implying their recent origin.4,41 It seems that recent miRNA molecules have few conserved targets and low expression and will not be preserved by purifying selection over long evolutionary time periods.42,43 Thus, it appears that human brain-specific miRNA genes are not necessarily associated with human special cognitive function. Dramatically, a recent report found a human brain-specific miRNA termed miR-941 whose target genes are involved in neurotransmitter signaling.44 Another primate-specific and human brain-enriched miRNA is miR-1202, which has be reported to be associated with the pathophysiology of depression.45 The main biological pathways related to neural activities of the 2 miRNAs above are shown in Figure 3. Based on these discoveries, we can speculate that newborn miRNAs in the central nervous system have important implications in the evolution and increased complexity of animal brain and an increasing number of important new miRNAs will be discovered with the rapid development of better miRNA detecting systems and molecular biotechnology.
Figure 3.
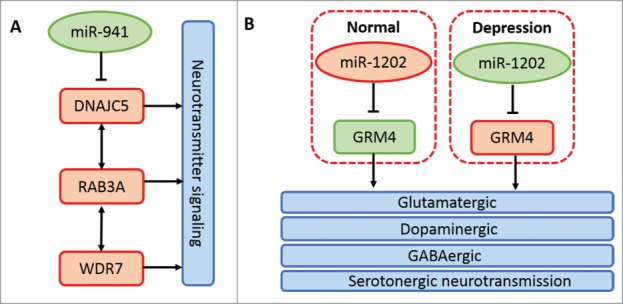
Principal mechanisms of miR-941 and miR-1202 in neural activities. (A) miR-941 inhibits the expression of its host gene DNAJC5. DNAJC5, coupled with its directly interacted gene RAB3A and indirectly interacted gene WDR7, is associated with neurotransmitter release.44 (B) miR-1202 expression is decreased in the donor brain with depression compared with the normal control. While the host gene expression of miR-1202, GRM4, is increased in the donor brain with depression. GRM4 is expressed throughout the brain and is associated with glutamatergic, dopaminergic, GABAergic and serotonergic neurotransmission.45
Non-conservatively expressed miRNAs are involved in brain evolution
The primary sequence of the mature miRNA is highly conserved and rarely mutates across species.36 Comparing the homologous hairpins of the miRNAs cloned from human brain among 17 animal species, 75% of known human brain miRNAs are found to be conserved in vertebrates and mammals, 14% are conserved in invertebrates, and the rest are species-specific.40 Sequence conserved miRNAs are usually more broadly and robustly expressed than non-conserved ones.46,47 Nevertheless, it appears that miRNA sequence conservation does not imply expression conservation. Although 325 miRNAs expressed in the prefrontal cortex are found to have high sequence conservation between human and chimpanzee, 11% are expressed at significantly different levels in these 2 species.39 Comparisons of brain miRNAs among different primates reveal 19 developmentally regulated miRNAs that are 24-fold more divergent in human than in chimpanzee prefrontal cortex, which are found to be evolving far more rapidly than other classes of miRNA genes, indicating the crucial role of miRNAs in metazoan evolution.17 Human-specific expressed miRNAs (miR-184, miR-487a, miR-383, miR-34c-5p and miR-299-3p) located in neurons are found to be inversely related with their target gene expressions.39 Interestingly, the 5 miRNAs, with their host mRNAs, are found to be conserved at the primary sequence level. Most of the target genes of the 5 miRNAs are significantly enriched in neural functions associated with learning and memory pathways (Table 1). Thus, the expression divergence of the 5 miRNAs between human and chimpanzee prefrontal cortex may have a crucial role in human lineage evolution. By fine tuning gene expression, non-conservatively expressed miRNAs, including brain species-specific expressed miRNAs, may to a large extent, be involved in species specific phenotypic adaptations. However, the direct evidence remains to be established.
Table 1.
Functional enrichment analysis of human brain-specific expressed miRNAs
| miRNA ID | KEGG pathway | Biological process |
|---|---|---|
| has-miR-299-3p | Axon guidance | |
| Neurodegenerative Diseases | Synaptic transmission | |
| has-miR-184 | Long-term potentiation | |
| Phosphatidylinositol signaling system | Signal transduction | |
| Melanogenesis | Cell surface receptor mediated signal transduction | |
| has-miR-487a | Adherens junction | Neuronal activities |
| Tight junction | mRNA transcription | |
| has-miR-34c-5p | Colorectal cancer | Other mRNA transcription |
| Prostate cancer | ||
| has-miR-383 | N.A. |
Note: KEGG pathway is determined by each target gene set; biological process is determined by the union set of all the target genes. N.A. represents unknown.
miRNAs constitute more flexible, robust and buffered brain transcriptional regulatory networks
The brain is the most complicated organ system.26,28 It is thought that some characteristics of brain miRNAs; their abundant expression, co-expressed as clusters and more target sites, promote more flexibility, robustness or buffering of miRNA-mRNA regulatory networks. The result is increased combinatorial control of the evolution of brain morphological and functional complexity. But the abnormality of hub miRNAs in the regulatory network may lead to vulnerable regulatory circuits that will cause dysfunctional brain processes.
Brain-enriched miRNAs
It has been reported that approximately 70% of all miRNAs are expressed in brain regions.48,49 This is not surprising given the cellular and transcriptional complexity of the central nervous system. According to previous studies, 65 miRNAs are found to be abundantly expressed in mouse brain based on massively parallel sequencing or chip techniques followed with traditional miRNA detection methods.50-53 We used the miRWalk2.0 database54 (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) Prediction Programs (miRanda, miRDB, miRWalk, RNA22 and TargetScan) to predict target genes and obtained 423 putative host mRNAs. Specifically, based on a functional analysis using DAVID functional analysis tool55 (Release 6.7, http://david.abcc.ncifcrf.gov/), the targets of brain-enriched miRNAs are significantly enriched in various developmental and other basic biological processes (Table 2). Some brain-enriched miRNAs have been reported with important roles in normal brain development and aberrant expressions of these miRNAs are related to cancer or nerve-related diseases. For example, let-7 family members are found highly expressed in the developing mammalian brain.23,56 As a spatiotemporal code for cell fate, let-7 is associated with embryonic and adult neurogenesis.57,58 In addition, let-7 itself has no effect on the proliferation of normal human astrocytes but exerts an anti-tumorigenic effect on glioblastoma cells, abnormal expression of which will cause glioblastoma or neurocytoma.59,60 miR-9, highly expressed in hippocampus, controls not only the timing of neurogenesis, but dendritic development and microglial activation as well as neural apoptosis.52,61-65 miR-9 can also attenuate Aβ-induced synaptotoxicity, appearing to play an important role in the development of Alzheimer's disease.66 Another brain-enriched miRNA is miR-124 which has been confirmed as a neuronal fate determinant, controlling the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression.67,68 Downregulation of miR-124 may be linked to malignant tumor progression and poor prognosis in patients with gliomas.69 Taken together, paying more attention to the potential role of brain-enriched miRNAs may contribute to further unveiling the potential physiological and pathological mechanisms of neuronal regulation.
Table 2.
Significantly enriched biological processes of mouse brain enriched miRNAs
| Biological process | Gene numbers | P-value | Benjamini |
|---|---|---|---|
| nervous system development | 46 | 6.00E-07 | 1.10E-03 |
| negative regulation of biological process | 59 | 5.60E-06 | 5.10E-03 |
| embryonic organ development | 20 | 6.20E-06 | 3.80E-03 |
| negative regulation of cellular process | 54 | 9.50E-06 | 4.30E-03 |
| anatomical structure morphogenesis | 51 | 1.40E-05 | 5.00E-03 |
| localization | 97 | 1.40E-05 | 4.30E-03 |
| blood vessel development | 19 | 2.70E-05 | 7.00E-03 |
| cellular process | 259 | 2.70E-05 | 6.30E-03 |
| embryonic development | 35 | 3.40E-05 | 6.90E-03 |
| vasculature development | 19 | 3.70E-05 | 6.80E-03 |
| protein transport | 34 | 4.60E-05 | 7.70E-03 |
| establishment of protein localization | 34 | 5.40E-05 | 8.20E-03 |
| regulation of cell differentiation | 24 | 6.40E-05 | 9.00E-03 |
| protein localization | 37 | 6.90E-05 | 9.00E-03 |
| system development | 76 | 7.60E-05 | 9.30E-03 |
Note: Fisher's exact test, p < 0.001; Benjamini-Hochberg corrected, p < 0.01.
Co-expressed as clusters
Previous studies suggest that miRNAs are separated by intervals as short as a few nucleotides in the genome. These are defined as miRNA clusters which tend to be co-expressed presumably due to sharing common cis-regulatory elements or being derived from polycistronic precursors.53,70 Hence, it seems that miRNAs from the same cluster show a similar expression level and nearly the same host mRNAs and the similar functionalities. A recent study shows that miR-17-92 cluster expressed in the distal axons of primary cortical neurons can enhance axonal outgrowth by local modulation of PTEN protein levels.71 In the developing neocortex, miR-17-92 cluster can facilitate neural stem cells proliferation and differentiation.72 Two members of miR-17-92 cluster, miR-17 and miR-20a, have been validated to act as a vital role in the self-renewal of postnatal neural stem cells by targeting Trp53inp1—a downstream component of the p53 pathway.73 Aberrant expression of miR-17-92 cluster members, such as miR-92, miR-19a, and miR-20, is involved in medulloblastoma and retinoblastoma progressions.74–76 Another interesting miRNA cluster is miR-183-96-182, which exerts pleiotropic effects on cell survival, proliferation and migration in medulloblastoma.77,78 Increased expression of miR-183-96-182 cluster members, miR-183, 96 and 182, are implicated in glioma carcinogenesis.79 Members of the miR-29a/b-1 cluster are brain-enriched non-coding RNA molecules, loss of which may contribute to increased BACE1/β-secretase expression in sporadic Alzheimer's disease and locomotor impairment and ataxia.80,81 miR-132-212 cluster, whose transcriptional activation depends on the neuronal activity-induced transcription factor, CREB, has been confirmed to be critical for the formation of ocular dominance in 2 studies.82-84 In addition, mature miR-212 and miR-132 transcripts upregulate simultaneously in response to the induction of long-term potentiation in the dentate gyrus of adult rats, suggesting the role in synaptic plasticity modulation.85 To sum up, miRNA clusters are closely related to the occurrence and progression of brain tumors or neurological disorders. Thus, they are vital for uncovering the pathogenesis of these challenging lesions and have the prospect of being effective therapeutic targets.
More target sites for miRNAs in brain
The 3′UTRs of mRNAs contain numerous functional motifs, and nearly one-half are miRNA recognition sites.86 It is likely that the length of 3′UTR is correlated with the number of miRNA families that potentially target the mRNA, in other words, it is a vital factor in miRNA-target co-evolution.87-90 With the rapid development of RNA-seq technology followed by bioinformatics analysis and experimental verification, the length of 3′UTRs has been confirmed to possess an unexpected diversity and expression specificity.91 It has been observed that proliferating cells like murine CD4+ T lymphocytes and tumor cells express mRNAs with shortened 3′UTRs and fewer miRNA target sites.92,93 On the contrary, mRNAs expressed in neural tissues, especially in hippocampus, have evidently biased usage of longer 3′UTRs and more miRNA target sites.94,95 This phenomenon together with the 2 hallmarks of brain miRNAs described above—enrichment in brain tissues and co-expressed as clusters—indicate additional possibilities for miRNA mediated regulation; promotion of a more flexible, robust and buffered functional miRNA-mRNA regulatory network in the central nervous system. Furthermore, the sequence of 3′UTR, under much less evolutionary pressure, is not conserved. Changing evolvability of miRNA targets implies that the miRNA-mRNA regulatory network may exert a far-reaching influence on the evolution of organismal diversity.96 The physiological and pathological roles of long 3′UTRs in neural tissues have been previously reviewed.91 Whether mutations in 3′UTRs in specific neural tissues have an impact on the stability of the miRNA-mRNA regulatory network and further cause certain nerve-related diseases is still unclear.
miRNA acts as a spatiotemporal code for brain development
The nervous system enables animals to adapt to environmental stimulus, which relies on a well-conserved toolkit of different molecular mechanisms.97,98 The compensatory responses to a changing world need exquisite spatiotemporal regulation throughout a protracted period of time (Fig. 4). Previous studies based on large numbers of samples consisting of different brain regions or developmental time points describe the spatiotemporal dynamics of gene expression during brain development.22-24,52,58,99-103 That miRNA system controls, in part, both levels and translation of mRNA, shapes the deployment and robust regulation of gene networks during the construction and the remodeling of the brain.97 Rapid growth of sequencing and chip techniques make it conceivable to detect all expressed miRNAs in a given brain region with a wide quantitative range, and highlights that miRNA acts as a spatiotemporal code for brain morphogenesis and neuronal fate.104
Figure 4.
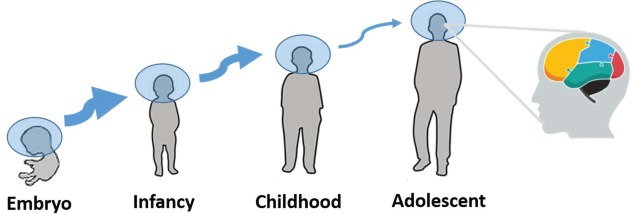
Sketch map of the spatiotemporal regulation pattern of brain miRNAs. The thickness of the arrow is positively correlated with the number of differentially expressed miRNAs during the transition from one period to another. The rightmost diagram with different color areas represents a region-specific expression pattern of brain miRNAs.
Differential miRNA expressions in different brain regions are found. For example, Juhila et al. systematically compared the miRNA expression profiles between mouse frontal cortex and hippocampus and unveiled 39 miRNAs overexpressed in frontal cortex and 40 miRNAs overexpressed in hippocampus.102 Bak et al. dissected 13 different areas of the male Balb/c mouse brain and found 63 differentially expressed miRNAs within the central nervous system, among which 44 miRNAs showed more than threefold enrichment in specific regions.52 Across developmental time within human brain regions, 75 differentially expressed miRNAs were discovered.24 With the exception of spatio-specific expression pattern, miRNA also exerts a temporal-specific expression pattern. The marked change of miRNA expression, for human beings, occurs during the transition from infancy to early childhood and, for mice, occurs within the first postnatal month, suggesting roles in early development.22-24 Only a few differentially expressed miRNAs are found in later developing stages ranging from juvenile to adult.19 Nevertheless, the number of differentially expressed miRNAs between brain regions increases over developmental time; this is in contrast to mRNA expression which is globally similar between brain regions over development.24,100 The spatiotemporal restricted expression pattern indicates the regionalization and dynamism of miRNA regulation. It is likely that understanding the biological roles of the differentially expressed miRNAs in specific regions or times may help to uncover the underlying mechanisms of the proper specialization and interconnectivity among different brain areas or developmental time points.
Conclusion
As post-transcriptional regulators of gene expression, miRNAs in the central nervous system have been regarded as vital triggers for brain development and neurological or psychiatric disease. Dramatic expansions of the brain miRNA repertoire also seems to be related to the emergence of mental and behavioral variation in closely related species during animal evolution. A recent study shows that human-specific glutamate dehydrogenase 2 can promote growth of IDH1R132H glioma, which is considered as an inevitable cost of human evolution and development.105 Ever-increasing miRNA repertoires, in some degree, are conducive to the complexity of the nervous system; on the other hand, it is possible to result in higher animal's susceptibility to nerve-related disorders. It appears that the non-conservatively expressed miRNAs, the robust miRNA-mRNA regulatory network and the particular spatiotemporal expression pattern can guarantee that the supreme commander, brain, can execute the intricate and fabulously complicated behaviors. The challenge is to explore what consequences may result from aberrant happenings to these hallmarks? It is these miRNAs and corresponding host mRNAs with general features that will be the hotspots of neuroscience research in the quest to comprehend the molecular underpinnings of physiological function and pathological disorder in the central nervous system.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
In particular, we thank Dr. Robert A. Haas (California Department of Public Health, USA) for helpful discussions and editing of the manuscript.
Funding
We gratefully thank the support provided by PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (333210140149).
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843–54; PMID:8252621; http://dx.doi.org/ 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014; 42:D68-73; PMID:24275495; http://dx.doi.org/ 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 4.Berezikov E.. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 2011; 12:846-60; PMID:22094948; http://dx.doi.org/ 10.1038/nrg3079 [DOI] [PubMed] [Google Scholar]
- 5.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF; Students of Bioinformatics Computer Labs 2004 and 2005. The expansion of the metazoan microRNA repertoire. BMC Genomics 2006; 7:25; PMID:16480513; http://dx.doi.org/ 10.1186/1471-2164-7-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC. The evolution and functional diversification of animal microRNA genes. Cell Res 2008; 18:985-996; PMID:18711447; http://dx.doi.org/ 10.1038/cr.2008.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 2007; 8:93-103; PMID:17230196; http://dx.doi.org/ 10.1038/nrg1990 [DOI] [PubMed] [Google Scholar]
- 8.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 2008; 23:578-87; PMID:18715673; http://dx.doi.org/ 10.1016/j.tree.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roux J, Gonzalez-Porta M, Robinson-Rechavi M. Comparative analysis of human and mouse expression data illuminates tissue-specific evolutionary patterns of miRNAs. Nucleic Acids Res 2012; 40:5890-900; PMID:22457063; http://dx.doi.org/ 10.1093/nar/gks279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 2005; 24:138-148; PMID:15565168; http://dx.doi.org/ 10.1038/sj.emboj.7600491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi R1, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003; 17:3011-6; PMID:14681208; http://dx.doi.org/ 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001; 409:363-6; PMID:11201747; http://dx.doi.org/ 10.1038/35053110 [DOI] [PubMed] [Google Scholar]
- 13.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005; 6:376-385; PMID:15852042; http://dx.doi.org/ 10.1038/nrm1644 [DOI] [PubMed] [Google Scholar]
- 14.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000; 404: 293-6; PMID:10749213; http://dx.doi.org/ 10.1038/35005107 [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15-20; PMID:15652477; http://dx.doi.org/ 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 16.Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, et al.. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res 2010; 20:1207-18; PMID:20647238; http://dx.doi.org/ 10.1101/gr.106849.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Zhang D, Li G. Insights into the regulation of human CNV-miRNAs from the view of their target genes. BMC Genomics 2012; 13:707; PMID:23244579; http://dx.doi.org/ 10.1186/1471-2164-13-707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiore R, Schratt G. MicroRNAs in synapse development: tiny molecules to remember. Expert Opin Biol Ther 2007; 7:1823-1831; PMID:18034648; http://dx.doi.org/ 10.1517/14712598.7.12.1823 [DOI] [PubMed] [Google Scholar]
- 19.Fiore R, Schratt G. MicroRNAs in vertebrate synapse development. ScientificWorldJournal 2007; 7:167-77; PMID:17982590; http://dx.doi.org/ 10.1100/tsw.2007.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry 2014; 19:410-6; PMID:24468823; http://dx.doi.org/ 10.1038/mp.2013.196 [DOI] [PubMed] [Google Scholar]
- 21.Shinde S, Arora N, Bhadra U. A Complex Network of MicroRNAs Expressed in Brain and Genes Associated with Amyotrophic Lateral Sclerosis. Int J Genomics 2013; 2013:383024; PMID:23936767; http://dx.doi.org/ 10.1155/2013/383024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eda A, Takahashi M, Fukushima T, Hohjoh H. Alteration of microRNA expression in the process of mouse brain growth. Gene 2011; 485:46-52; PMID:21718763; http://dx.doi.org/ 10.1016/j.gene.2011.05.034 [DOI] [PubMed] [Google Scholar]
- 23.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol 2004; 5:R68; PMID:15345052; http://dx.doi.org/ 10.1186/gb-2004-5-9-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry 2014; 19:848-52; PMID:23917947; http://dx.doi.org/ 10.1038/mp.2013.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci 2012; 35:325-34; PMID:22436491; http://dx.doi.org/ 10.1016/j.tins.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Carroll D, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 2013; 38:39-54; PMID:22669168; http://dx.doi.org/ 10.1038/npp.2012.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci 2012; 69:89-102; PMID:21833581; http://dx.doi.org/ 10.1007/s00018-011-0788-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature 2006; 441:1087-1093; PMID:16810244; http://dx.doi.org/ 10.1038/nature04959 [DOI] [PubMed] [Google Scholar]
- 29.Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat Rev Genet 2006; 7:693-702; PMID:16921347; http://dx.doi.org/ 10.1038/nrg1940 [DOI] [PubMed] [Google Scholar]
- 30.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al.. Intra- and interspecific variation in primate gene expression patterns. Science 2002; 296:340-343; PMID:11951044; http://dx.doi.org/ 10.1126/science.1068996 [DOI] [PubMed] [Google Scholar]
- 31.Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A 2003; 100:13030-5; PMID:14557539; http://dx.doi.org/ 10.1073/pnas.2135499100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babbitt CC, Fedrigo O, Pfefferle AD, Boyle AP, Horvath JE, Furey TS, Wray GA. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol Evol 2010; 2:67-79; PMID:20333225; http://dx.doi.org/ 10.1093/gbe/evq002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci U S A 2009; 106:22358-63; PMID:20007773; http://dx.doi.org/ 10.1073/pnas.0911376106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al.. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007; 317:86-94; PMID:17615350; http://dx.doi.org/ 10.1126/science.1139158 [DOI] [PubMed] [Google Scholar]
- 35.Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet 2005; 13:894-7; PMID:15970949; http://dx.doi.org/ 10.1038/sj.ejhg.5201459 [DOI] [PubMed] [Google Scholar]
- 36.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A 2008; 105:2946-50; PMID:18287013; http://dx.doi.org/ 10.1073/pnas.0712259105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays 2009; 31:736-47; PMID:19472371; http://dx.doi.org/ 10.1002/bies.200900033 [DOI] [PubMed] [Google Scholar]
- 38.Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H. Birth and expression evolution of mammalian microRNA genes. Genome Res 2013; 23:34-45; PMID:23034410; http://dx.doi.org/ 10.1101/gr.140269.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu HY, Guo S, Xi J, Yan Z, Fu N, Zhang X, Menzel C, Liang H, Yang H, Zhao M, et al.. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet 2011; 7:e1002327; PMID:22022286; http://dx.doi.org/ 10.1371/journal.pgen.1002327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat Genet 2006; 38:1375-1377; PMID:17072315; http://dx.doi.org/ 10.1038/ng1914 [DOI] [PubMed] [Google Scholar]
- 41.Liang H, Li WH. Lowly expressed human microRNA genes evolve rapidly. Mol Biol Evol 2009; 26:1195-8; PMID:19299536; http://dx.doi.org/ 10.1093/molbev/msp053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res 2007; 17:1865-79; PMID:17989255; http://dx.doi.org/ 10.1101/gr.6593807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nozawa M, Miura S, Nei M. Origins and evolution of microRNA genes in Drosophila species. Genome Biol Evol 2010; 2:180-9; PMID:20624724; http://dx.doi.org/ 10.1093/gbe/evq009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, Taylor MS, Tang L, Li J, Liu J, et al.. Evolution of the human-specific microRNA miR-941. Nat Commun 2012; 3:1145; PMID:23093182; http://dx.doi.org/ 10.1038/ncomms2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, Maussion G, Yang JP, Yerko V, Vigneault E, et al.. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med 2014; 20:764-8; PMID:24908571; http://dx.doi.org/ 10.1038/nm.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 2007; 17:1850-64; PMID:17989254; http://dx.doi.org/ 10.1101/gr.6597907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat Genet 2008; 40:351-55; PMID:18278047; http://dx.doi.org/ 10.1038/ng.73 [DOI] [PubMed] [Google Scholar]
- 48.De Pietri Tonelli D, Clovis YM, Huttner WB. Detection and monitoring of microRNA expression in developing mouse brain and fixed brain cryosections. Methods Mol Biol 2014; 1092:31-42; PMID:24318812 [DOI] [PubMed] [Google Scholar]
- 49.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci 2006; 7:911-20; PMID:17115073; http://dx.doi.org/ 10.1038/nrn2037 [DOI] [PubMed] [Google Scholar]
- 50.Ling KH, Brautigan PJ, Hahn CN, Daish T, Rayner JR, Cheah PS, Raison JM, Piltz S, Mann JR, Mattiske DM, et al.. Deep sequencing analysis of the developing mouse brain reveals a novel microRNA. BMC Genomics 2011; 12:176; PMID:21466694; http://dx.doi.org/ 10.1186/1471-2164-12-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al.. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A 2004; 101:9740-4; PMID:15210942; http://dx.doi.org/ 10.1073/pnas.0403293101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA 2008; 14:432-44; PMID:18230762; http://dx.doi.org/ 10.1261/rna.783108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004; 5:R13; PMID:15003116; http://dx.doi.org/ 10.1186/gb-2004-5-3-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 2011; 44:839-47; PMID:21605702; http://dx.doi.org/ 10.1016/j.jbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 55.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/ 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 56.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002; 12:735-9; PMID:12007417; http://dx.doi.org/ 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- 57.Rehfeld F, Rohde AM, Nguyen DT, Wulczyn FG. Lin28 and let-7: ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res 2015; 359:145-60; PMID:24825413; http://dx.doi.org/ 10.1007/s00441-014-1872-2 [DOI] [PubMed] [Google Scholar]
- 58.Kucherenko MM, Barth J, Fiala A, Shcherbata HR. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J 2012; 31:4511-23; PMID:23160410; http://dx.doi.org/ 10.1038/emboj.2012.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee ST, Chu K, Oh HJ, Im WS, Lim JY, Kim SK, Park CK, Jung KH, Lee SK, Kim M, et al.. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J Neurooncol 2011; 102:19-24; PMID:20607356; http://dx.doi.org/ 10.1007/s11060-010-0286-6 [DOI] [PubMed] [Google Scholar]
- 60.Buechner J, Tomte E, Haug BH, Henriksen JR, Lokke C, Flægstad T, Einvik C. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer 2011; 105:296-303; PMID:21654684; http://dx.doi.org/ 10.1038/bjc.2011.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coolen M, Thieffry D, Drivenes O, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev Cell 2012; 22:1052-64; PMID:22595676; http://dx.doi.org/ 10.1016/j.devcel.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 62.Davila JL, Goff LA, Ricupero CL, Camarillo C, Oni EN, Swerdel MR, Toro-Ramos AJ, Li J, Hart RP. A positive feedback mechanism that regulates expression of miR-9 during neurogenesis. PLoS One 2014; 9:e94348; PMID:24714615; http://dx.doi.org/ 10.1371/journal.pone.0094348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giusti SA, Vogl AM, Brockmann MM, Vercelli CA, Rein ML, Trümbach D, Wurst W, Cazalla D, Stein V, Deussing JM, et al.. MicroRNA-9 controls dendritic development by targeting REST. Elife 3 2014; 3:e02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, Kook Y, Niu F, Liao K, Fu M, et al.. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun 2014; 5:4386; PMID:25019481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, Peng T, Li L, Wang X, Duan R, Gao H, Guan W, Lu J, Teng J, Jia Y. MicroRNA-9 regulates neural apoptosis in methylmalonic acidemia via targeting BCL2L11. Int J Dev Neurosci 2014; 36:19-24; PMID:24798023; http://dx.doi.org/ 10.1016/j.ijdevneu.2014.04.005 [DOI] [PubMed] [Google Scholar]
- 66.Chang F, Zhang LH, Xu WP, Jing P, Zhan PY. microRNA-9 attenuates amyloidbeta-induced synaptotoxicity by targeting calcium/calmodulin-dependent protein kinase kinase 2. Mol Med Rep 2014; 9:1917-22; PMID:24603903 [DOI] [PubMed] [Google Scholar]
- 67.Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci 2012;32:8879-8889; PMID:22745489; http://dx.doi.org/ 10.1523/JNEUROSCI.0558-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neo WH, Yap K, Lee SH, Looi LS, Khandelia P, Neo SX, Makeyev EV, Su IH. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J Biol Chem 2014; 289:20788-801; PMID:24878960; http://dx.doi.org/ 10.1074/jbc.M113.525493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen T, Wang XY, Li C, Xu SJ. Downregulation of microRNA-124 predicts poor prognosis in glioma patients. Neurol Sci 2015; 36:131-5; PMID:25112530; http://dx.doi.org/ 10.1007/s10072-014-1895-1 [DOI] [PubMed] [Google Scholar]
- 70.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005; 11:241-7; PMID:15701730; http://dx.doi.org/ 10.1261/rna.7240905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Ueno Y, Liu XS, Buller B, Wang X, Chopp M, Zhang ZG. The MicroRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J Neurosci 2013; 33:6885-94; PMID:23595747; http://dx.doi.org/ 10.1523/JNEUROSCI.5180-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, Garg V, Sun T. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep 2013; 3:1398-406; PMID:23623502; http://dx.doi.org/ 10.1016/j.celrep.2013.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garg N, Po A, Miele E, Campese AF, Begalli F, Silvano M, Infante P, Capalbo C, De Smaele E, Canettieri G, et al.. microRNA-17-92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J 2013; 32:2819-32; PMID:24076654; http://dx.doi.org/ 10.1038/emboj.2013.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, et al.. The miR-17˜92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A 2009; 106:2812-7; PMID:19196975; http://dx.doi.org/ 10.1073/pnas.0809579106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kauppinen S, Roussel MF. Silencing of the miR-17˜92 cluster family inhibits medulloblastoma progression. Cancer Res 2013; 73:7068-78; PMID:24145352; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D. miR-17˜92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev 2011; 25:1734-45; PMID:21816922; http://dx.doi.org/ 10.1101/gad.17027411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, et al.. Pleiotropic effects of miR-183˜96˜182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol 2012; 123:539-52; PMID:22402744; http://dx.doi.org/ 10.1007/s00401-012-0969-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, Li S, Cheng SY. The miR-183 approximately 96 approximately 182 cluster promotes tumorigenesis in a mouse model of medulloblastoma. J Biomed Res 2013; 27:486-94; PMID:24285947; http://dx.doi.org/ 10.7555/JBR.27.20130010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q, Xu G, Wu M, Li G. The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets 2013; 13:221-31; PMID:23252827; http://dx.doi.org/ 10.2174/1568009611313020010 [DOI] [PubMed] [Google Scholar]
- 80.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A 2008; 105:6415-20; PMID:18434550; http://dx.doi.org/ 10.1073/pnas.0710263105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papadopoulou AS, Serneels L, Achsel T, Mandemakers W, Callaerts-Vegh Z, Dooley J, Lau P, Ayoubi T, Radaelli E, Spinazzi M, et al.. Deficiency of the miR-29a/b-1 cluster leads to ataxic features and cerebellar alterations in mice. Neurobiol Dis 2014; 73C:275-88; PMID:25315682 [DOI] [PubMed] [Google Scholar]
- 82.Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 2010; 20:492-8; PMID:19557767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, et al.. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci 2011; 14:1240-2; PMID:21892155; http://dx.doi.org/ 10.1038/nn.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tognini P, Putignano E, Coatti A, Pizzorusso T. Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat Neurosci 2011; 14:1237-9; PMID:21892154; http://dx.doi.org/ 10.1038/nn.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003; 9:1274-1281; PMID:13130141; http://dx.doi.org/ 10.1261/rna.5980303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 2005; 434:338-45; PMID:15735639; http://dx.doi.org/ 10.1038/nature03441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 2005; 310:1817-21; PMID:16308420; http://dx.doi.org/ 10.1126/science.1121158 [DOI] [PubMed] [Google Scholar]
- 88.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific sign atures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A 2006; 103:2746-51; PMID:16477010; http://dx.doi.org/ 10.1073/pnas.0511045103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stark A1, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 2005; 123:1133-46; PMID:16337999; http://dx.doi.org/ 10.1016/j.cell.2005.11.023 [DOI] [PubMed] [Google Scholar]
- 90.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 2011; 12:846-60; PMID:22094948; http://dx.doi.org/ 10.1038/nrg3079 [DOI] [PubMed] [Google Scholar]
- 91.Wang L, Yi R. 3′UTRs take a long shot in the brain. Bioessays 2014; 36:39-45; PMID:24115048; http://dx.doi.org/ 10.1002/bies.201300100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009; 138:673-84; PMID:19703394; http://dx.doi.org/ 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008; 320:1643-7; PMID:18566288; http://dx.doi.org/ 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al.. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 2012; 1:277-89; PMID:22685694; http://dx.doi.org/ 10.1016/j.celrep.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res 2013; 23:812-25; PMID:23520388; http://dx.doi.org/ 10.1101/gr.146886.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu J, Zhang R, Shen Y, Liu G, Lu X, Wu CI. The evolution of evolvability in microRNA target sites in vertebrates. Genome Res 2013; 23:1810-16; PMID:24077390; http://dx.doi.org/ 10.1101/gr.148916.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron 2012; 75:363-379; PMID:22884321; http://dx.doi.org/ 10.1016/j.neuron.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci 2009; 10:724-35; PMID:19763105; http://dx.doi.org/ 10.1038/nrn2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strand AD, Aragaki AK, Baquet ZC, Hodges A, Cunningham P, Holmans P, Jones KR, Jones L, Kooperberg C, Olson JM. Conservation of regional gene expression in mouse and human brain. PLoS Genet 2007; 3:e59; PMID:17447843; http://dx.doi.org/ 10.1371/journal.pgen.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G. Spatio-temporal transcriptome of the human brain. Nature 2011; 478:483-489; PMID:22031440; http://dx.doi.org/ 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al.. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129:1401-14; PMID:17604727; http://dx.doi.org/ 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Juhila J, Sipila T, Icay K, Nicorici D, Ellonen P, Kallio A, Korpelainen E, Greco D, Hovatta I. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PLoS One 2011; 6:e21495; PMID:21731767; http://dx.doi.org/ 10.1371/journal.pone.0021495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith B, Treadwell J, Zhang D, Ly D, McKinnell I, Walker PR, Sikorska M. Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS One 2010; 5:e11109; PMID:20559549; http://dx.doi.org/ 10.1371/journal.pone.0011109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, et al.. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A 2005; 102:10357-62; PMID:16002470; http://dx.doi.org/ 10.1073/pnas.0503357102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen R, Nishimura MC, Kharbanda S, Peale F, Deng Y, Daemen A, Forrest WF, Kwong M, Hedehus M, Hatzivassiliou G, et al.. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci U S A 2014; 111:14217-22; PMID:25225364; http://dx.doi.org/ 10.1073/pnas.1409653111 [DOI] [PMC free article] [PubMed] [Google Scholar]


