Abstract
Mrhl RNA is a nuclear lncRNA encoded in the mouse genome and negatively regulates Wnt signaling in spermatogonial cells through p68/Ddx5 RNA helicase. Mrhl RNA is present in the chromatin fraction of mouse spermatogonial Gc1-Spg cells and genome wide chromatin occupancy of mrhl RNA by ChOP (Chromatin oligo affinity precipitation) technique identified 1370 statistically significant genomic loci. Among these, genes at 37 genomic loci also showed altered expression pattern upon mrhl RNA down regulation which are referred to as GRPAM (Genes Regulated by Physical Association of Mrhl RNA). p68 interacted with mrhl RNA in chromatin at these GRPAM loci. p68 silencing drastically reduced mrhl RNA occupancy at 27 GRPAM loci and also perturbed the expression of GRPAM suggesting a role for p68 mediated mrhl RNA occupancy in regulating GRPAM expression. Wnt3a ligand treatment of Gc1-Spg cells down regulated mrhl RNA expression and also perturbed expression of these 27 GRPAM genes that included genes regulating Wnt signaling pathway and spermatogenesis, one of them being Sox8, a developmentally important transcription factor. We also identified interacting proteins of mrhl RNA associated chromatin fraction which included Pc4, a chromatin organizer protein and hnRNP A/B and hnRNP A2/B1 which have been shown to be associated with lincRNA-Cox2 function in gene regulation. Our findings in the Gc1-Spg cell line also correlate with the results from analysis of mouse testicular tissue which further highlights the in vivo physiological significance of mrhl RNA in the context of gene regulation during mammalian spermatogenesis.
Keywords: Chromatin occupancy, gene regulation, GRPAM, mrhl RNA, p68
Abbreviations
- mrhl
meiotic recombination hotspot locus
- lnc
long non coding
- GRPAM
Genes regulated by physical association of mrhl RNA
- ChOP
Chromatin Oligo Affinity Precipitation
Introduction
The mammalian genome is pervasively transcribed to produce a huge repertoire of RNA molecules. Over recent years, high throughput analyses of the transcriptome have revealed a substantial number of transcripts that lack protein coding capacity and are called non coding RNAs (ncRNAs). In addition to the housekeeping ncRNAs (tRNAs and rRNAs), these ncRNAs also comprise of regulatory small ncRNAs like microRNAs (miRNAs), small interfering RNAs (siRNAs) and piwi-interacting RNAs (piRNAs).1,2 Long ncRNAs (lncRNAs) are arbitrarily classified as ncRNAs longer than 200 nucleotides and have been described in human,3,4 mouse,5-7 fruitfly,8 nematode9 and zebrafish.10 Many studies have demonstrated the regulatory role of lncRNAs in diverse processes like dosage compensation (Xist and roX),11,12 genomic imprinting (Air and Kcnq1ot1),13,14 pluripotency (Evx1as and Hoxb5/6as),15 nuclear architecture (NEAT1)16 etc. LncRNAs employ diverse mechanisms for their biological function(s) which include translation inhibition (lincRNA-p21),17 mRNA degradation (1/2-sbs RNAs),18 RNA decoys (Gas5),19 recruitment of chromatin modifiers (Mistral, HOTTIP)20,21 etc. Many lncRNAs require interaction with regulatory protein or protein complexes which include RNA binding proteins and chromatin modifying or chromatin associated proteins for their functional manifestations.22 Some lncRNAs also recruit specific protein complexes at chromatin loci thereby functioning as elements responsive to developmental cues or signaling pathways.23-25
Recently, our laboratory has discovered mrhl RNA, a 2.4-kb mouse lncRNA which is transcribed from the 14th intron of the Phkb gene located in a meiotic recombination hotspot locus (mrhl) on chromosome 8.26 It is a polyadenylated and unspliced RNA transcript expressed in liver, kidney, spleen and testis. The 2.4 kb mrhl RNA primary transcript is nuclear restricted and gets processed to an 80 nt intermediate RNA by the Drosha machinery.27 Through a series of experiments we have recently demonstrated that mrhl RNA functions as a negative regulator of Wnt signaling in mouse Gc1-Spg cells (derived from mouse B type spermatogonia).28 Furthermore Wnt3a ligand treatment of Gc1-Spg cells resulted in Wnt signaling activation as demonstrated by TOP-FOP luciferase reporter assay. Under the same conditions, mrhl RNA expression was downregulated suggesting a close relationship between mrhl RNA expression and Wnt signaling activation in these Gc1-Spg cells and that negative regulation of Wnt signaling by mrhl RNA is biologically relevant. Mrhl RNA also interacts with p68 (Ddx5 helicase) in the nucleus and down regulation of mrhl RNA resulted in cytoplasmic translocation of p68 that triggered nuclear translocation of β catenin. TCF4 is a key transcription factor of the Wnt signaling pathway and is activated by the nuclear translocated β catenin.29 The β catenin-TCF4 complex binds at the Wnt Responsive Elements (WRE) in the target gene promoters modulating their expression by recruiting co-activators or co-repressors. Thus, p68 plays an important role in the regulation of Wnt signaling by mrhl RNA. In our previous study we observed that upon siRNA mediated down regulation of mrhl RNA, many genes belonging to various functional categories are perturbed in addition to Wnt signaling pathway associated genes (GSE19355).28 Our initial experiments also showed that mrhl RNA is associated with the chromatin fraction of Gc1-Spg cells which prompted us to identify the genome wide chromatin occupancy of mrhl RNA to understand the broader role of mrhl RNA in gene regulation at the chromatin level in spermatogonial cells. For this purpose, we have now carried out a detailed analysis of genome wide occupancy of mrhl RNA by the Chromatin Oligo affinity precipitation (ChOP) technique23 and have also queried whether p68 is necessary for its interaction with the chromatin. Among a total of 1370 statistically significant genomic loci we identified a subset of 37 GRPAM genes as target loci of mrhl RNA mediated gene regulation out of which expression of 27 genes was also dependent on p68 protein. We have also analyzed the proteins associated with mrhl RNA bound chromatin fraction by mass spectrometry.
Results
Mrhl RNA and p68 are associated with chromatin of Gc1-Spg cells
Mrhl RNA is a nuclear restricted RNA and interacts with the p68/Ddx5 helicase protein. In order to determine the localization of mrhl RNA and p68 within the nucleus, Gc1-Spg cell nuclei were fractionated into nucleoplasm and chromatin fractions and scored for the presence of mrhl RNA and p68 protein by RT-PCR and Western blot analysis respectively. Both the mrhl RNA and p68 protein could be detected in the nucleoplasm and chromatin fractions (Figs. 1A and 1B). We have used histone H4 and laminin, a nucleoplasmic marker to authenticate the purity of chromatin and nucleoplasm fraction respectively.30 Further, we also examined the association of mrhl RNA and p68 with the nucleosomal chromatin by performing chromatin immunoprecipitation of histone H3 (H3ChIP) and scoring for p68 and mrhl RNA to be co-precipitated with histone H3. It is clear that both p68 and mrhl RNA co-precipitated with histone H3 as monitored by Western blot and RT-PCR analysis respectively (Fig. 1C), demonstrating their association with oligo nucleosomal chromatin of Gc1-Spg cells.
Figure 1.
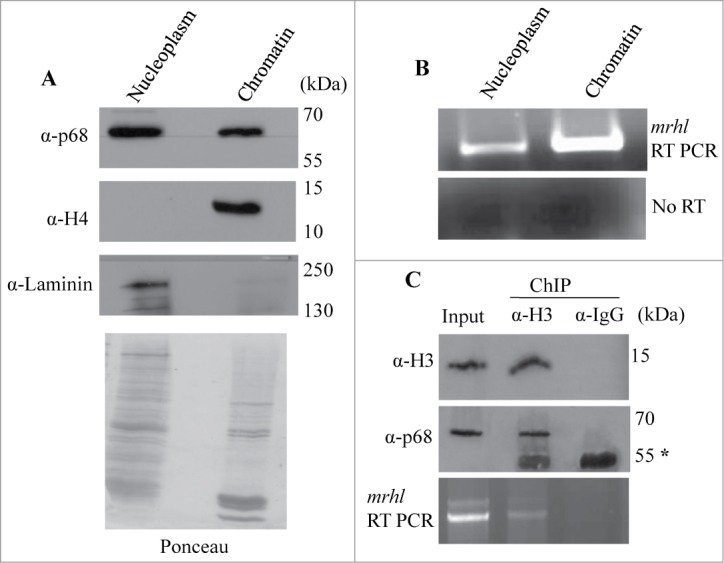
Association of mrhl RNA and p68 (Ddx5) helicase with spermatogonial cell (Gc1-Spg) chromatin. (A) Western blot showing the presence of p68 in the nucleoplasm and chromatin. Histone H4 and Laminin confirmed the purity of nucleoplasm and chromatin fractions respectively. The Ponceau staining pattern of the total proteins in these 2 fractions is given below the Western blot. (B) RT-PCR analysis showing the presence of mrhl RNA in both nucleoplasm and chromatin fractions. (C) Histone H3 ChIP showing co-immunoprecipitation of p68 with histone H3 (Western blot) and association of mrhl RNA with histone H3 ChIP chromatin (RT-PCR), thus demonstrating the presence of p68 and mrhl RNA in the oligo nucleosomal chromatin. Asterik (*) denotes the band corresponding to antibody heavy chain at 55kDa.
Mrhl RNA associates with several genomic loci on the chromatin
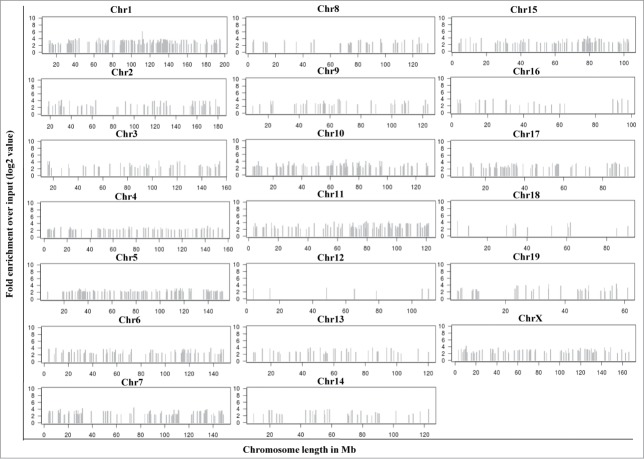
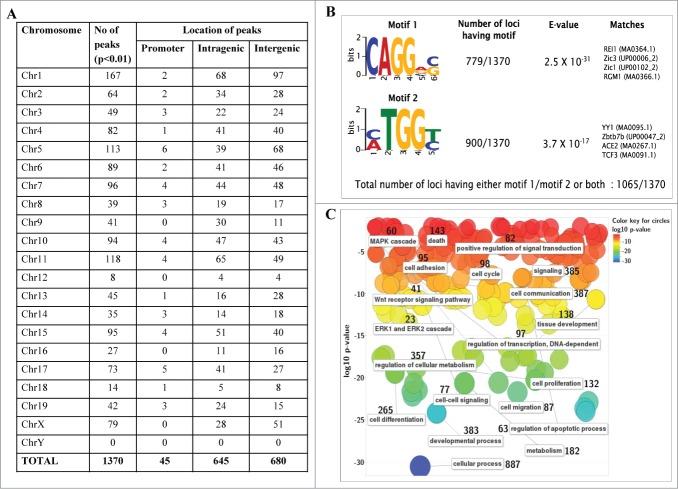
From the above data it is clear that mrhl RNA interacts with the oligo nucleosomal chromatin. We were then interested in identifying and characterizing the genomic sites occupied by mrhl RNA on chromatin. For this purpose, we used the now well established technique of chromatin oligo-affinity precipitation assay (ChOP).23 In this technique, mrhl RNA associated chromatin was affinity purified using biotin labeled oligonucleotides complementary to mrhl RNA. The process of ChOP assay is schematically depicted in Fig. S1A. The 3 different regions of mrhl RNA to which the antisense biotinylated oligonucleotides were designed are shown in Fig. S1B. Biotin labeled scrambled oligonucleotides were used as a control (Detailed procedure is given in Materials and Methods). Deep sequencing of the ChOP DNA showed that mrhl RNA occupies thousands of loci on the chromatin. The highly significant sequence reads were selected on the basis of fold enrichment over the input DNA (Fold enrichment more than 4) and P value (p < 0.01). This exercise resulted in a total of 1370 genomic loci that were associated with mrhl RNA (Table S1). The chromosome wise distribution of the sequence reads enriched in mrhl RNA ChOP as compared to input DNA is shown in Figure 2. The absolute values of fold enrichment of the ChOP sequence reads were in the range of 4 to 66 fold. It is interesting to note that these loci are distributed on all the mouse chromosomes except the Y-chromosome. We analyzed the locations of the sequence reads on the chromosomes and examined whether the reads lie in the promoter of a particular gene (promoter bound) or within the body of a gene (intragenic) or between 2 genes (intergenic). Such topological classification of the 1370 sequence reads revealed that 45, 645 and 680 genomic loci belonged to promoter, intragenic and intergenic classes respectively (Fig. 3A). We also carried out a motif search analysis of the entire 1370 ChOP reads. Two high scoring motifs were obtained with motif 1 being represented in 779 genomic loci and motif 2 in 900 genomic loci with very high statistical significance (Fig. 3B). The total number of loci having either motif 1 or 2 or both was 1065 out of a total of 1370 genomic loci. We also carried out gene ontology analysis for all the genes corresponding to the 1370 sequence reads which is represented graphically (Fig. 3C). Assuming that mrhl RNA occupied chromatin loci might regulate the nearby genes by long range interactions and looping mechanism, we took the genes that are the nearest neighbor (either upstream or downstream) corresponding to the intergenic reads for gene ontology analysis. The genes were classified into a broad range of categories like transcription (97), cell cycle (98), development and differentiation (383), Wnt signaling (41), apoptosis (63), metabolism (357) etc.
Figure 2.
Chromosome wise distribution of 1370 mrhl RNA ChOP sequence reads. Each read shows at least fold4- enrichment over the input DNA with a statistical significance of P < 0.01. The sequences were found to be located across the length of most of the mouse chromosomes except the Y-chromosome.
Figure 3.
Genomic topological distribution of mrhl RNA ChOP sequence reads and gene ontology analysis. (A) The 1370 sequence reads were classified on the basis of their position in the mouse genome with respect to a particular gene as follows 1) Promoter reads (upto 1.5 kb upstream of a gene) 2) Intragenic reads (within the body of the gene) 3) Intergenic reads (1.5 kb–2.5 Mb Upstream or downstream to a particular gene). (B) Motif analysis of mrhl RNA bound ChOP sequences. Two enriched motifs were identified by the DREME software relative to random sequence control. E value measures the significance threshold for motifs. E < 0.05 indicates statistical significance. Both the motifs show very high statistical significance. (C) Gene Ontology (GO) analysis of the genes associated with the ChOP sequence reads using the REViGO software. Data is represented as a 2D-scatterplot of biological process. Circles are color coded based on GO p-value. The number besides each functional term denotes the number of genes involved in that particular function (pathway).
Regulatory targets of mrhl RNA
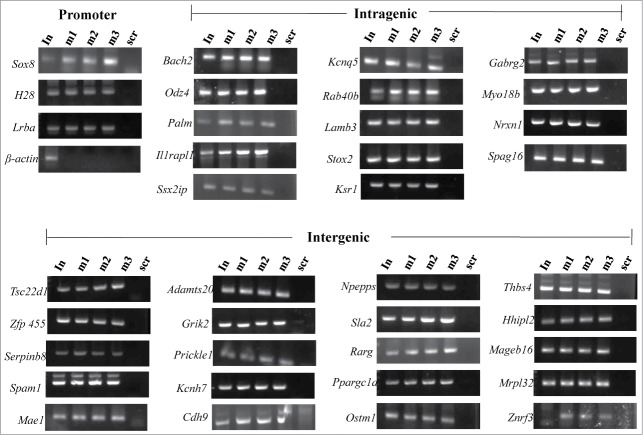
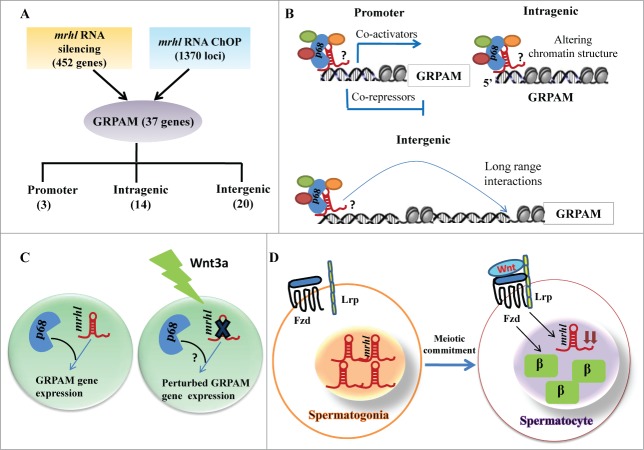
The results presented above clearly identified 1370 genomic loci associated with mrhl RNA with high statistical significance. We next asked the question whether mrhl RNA regulates the expression of genes which are physically associated or in physical proximity with mrhl RNA in cis at these loci. For this purpose we compared only the annotated genes in the 2 data sets namely a. Gene loci occupied by mrhl RNA (1220 genes in ChOP dataset, from the present study) and b. Genes perturbed upon mrhl RNA down regulation (435 genes, GSE19355). The genes common to these 2 data sets represent the ones that are bound by mrhl RNA and also perturbed upon mrhl RNA down regulation. Hence, these genes might represent the most probable regulatory targets of mrhl RNA in the Gc1-Spg cells. This exercise resulted in a subset of 37 genes and we termed them as Genes Regulated by Physical Association of Mrhl RNA (GRPAM). This overlap set of 37 genes is highly significant as revealed by hyper geometric test performed using the entire mouse genome as the background set (p=0.0004). Based on the location of binding of mrhl RNA, we classified these GRPAMs into promoter bound, intragenic and intergenic categories. The list of GRPAMs along with their gene ontology has been summarized in Table 1. The occupancy of mrhl RNA at these 37 GRPAM loci was further experimentally validated by performing ChOP, independently using 3 oligonucleotides binding to 3 different regions of mrhl RNA. Subsequent ChOP-PCR using sequence specific primers and ChOP DNA as the template showed amplification for all the 37 loci when oligonucleotides complementary to mrhl RNA were used for ChOP experiment but not in the scrambled oligonucleotide ChOP experiment (Fig. 4). Mrhl RNA occupancy at the 37 GRPAM loci was also verified by quantitative ChOP-PCR (Fig. S2).
Table 1.
List of GRPAM genes and their ontology
| Gene | Ontology Wnt Signaling | Location of ChOP read w.r.t gene | Gene regulation on mrhl RNA silencing |
|---|---|---|---|
| Odz4 | Wnt signaling, Embryonic Development, differentiation, | Genic | Downregulated |
| Lamb3 | Wnt signaling, differentiation | Genic | Downregulated |
| Tsc22d1 | Wnt signaling, Tgf β signaling, | 51.5 kb upstream | Downregulated |
| Prickle1** | Wnt signaling, differentiation, development | 63.4 kb upstream | Upregulated |
| Znrf3 | Negative regulation of Wnt signaling, stem cell proliferation, tumor supressor | 132.3 kb upstream | Upregulated |
| Ostm1* | Activator of Wnt signaling, differentiation. | 22.2 kb upstream | Downregulated |
| Spermatogenesis | |||
| Sox8 | Sertoli cell function, germ cell differentiation (Also in Wnt signaling) | Promoter | Upregulated |
| Rarg | Retinoic acid signaling, spermatogenesis (Also in Wnt signaling) | 11.1 kb upstream | Upregulated |
| Spag16 | Spermatogenesis | Genic | Upregulated |
| Mael | Spermatogenesis, piRNA pathway, differentiation | 33 kb downstream | Upregulated |
| Spam1 | Spermatogenesis, sperm maturation | 27 kb upstream | Downregulated |
| Mageb16 | testis specific (adult), regulates differentiation in mouse ESCs | 122.5 kb upstream | Downregulated |
| Npepps*,** | Spermatogenesis, antigen processing. | 14.7 kb downstream | Upregulated |
| Cell Adhesion and Transport | |||
| Lrba | Oncogenesis, endosomal transport, | Promoter | Upregulated |
| Gabrg2 | Ion channel transport, post embryonic development | Genic | Upregulated |
| Kcnq5 | Ion transport | Genic | Downregulated |
| Grik2 | Ion transport, apoptosis | 440 kb upstream | Downregulated |
| Kcnh7 | Ion transport | 276 kb upstream | Downregulated |
| Cdh9** | Cell adhesion | 180.2 kb upstream | Upregulated |
| Nrxn1 | Cell adhesion, angiogenesis | Genic | Downregulated |
| Signaling | |||
| Palm | cAMP signaling, cytoskeleton | Genic | Upregulated |
| Rab40b** | Signaling, Protein transport, | Genic | Downregulated |
| Ksr1 | Ras/MAPK signaling, TNF signaling | Genic | Upregulated |
| Ssx2ip | Regulation of Rac signaling, Cell adhesion, | Genic | Downregulated |
| Adamts20 | Integrin signaling, Proteolysis, apoptosis, | 696 kb downstream | Downregulated |
| Sla2 | Calcium signaling, transcription, endocytosis | 0.54 kb downstream | Downregulated |
| Thbs4 | PDGF and PI3K/Akt signaling, Focal adhesion, | 25.5 kb downstream | Downregulated |
| Il1rapl1* | JNK pathway activation, RhoA signaling, differentiation, ion transport | Genic | Downregulated |
| Ppargc1a* | Signaling, differentiation | 183.8 kb downstream | Downregulated |
| Other Functions | |||
| Serpinb8 | Regulation of proteolysis | 0.7 Mb upstream | Downregulated |
| Bach2 | Cell cycle control, transcriptional repressor, differentiation | Genic | Upregulated |
| Zfp455* | Metal ion binding | 24.7 kb downstream | Upregulated |
| Mrpl32** | Ribosomal, translation | 75.75 kb downstream | Downregulated |
| Hhipl2 | Carbohydrate metabolism | 581 kb downstream | Upregulated |
| Myo18b* | Vasculogenesis, cardiac development | Genic | Downregulated |
| Stox2 | Not Annotated | Genic | Upregulated |
| H28 | Interferon induced | Promoter | Downregulated |
Genes that are physically associated with ChOP sequence reads and also showing perturbation of their expression following mrhl RNA down regulation in Gc1-Spg cells (GRPAM). These genes were identified by comparing the genes associated with the ChOP sequence reads (present study) and the transcriptome data from GSE19355. The physical position of occupancy of mrhl RNA with each of the GRPAM (total 37) and its functions are also given. Asterik (*) indicates the GRPAM loci which are not bound by p68. ** indicates the GRPAM loci where mrhl RNA occupancy is not perturbed upon p68 silencing.
Figure 4.
Validation of mrhl RNA occupancy at the GRPAM loci by ChOP-PCR. ChOP DNA isolated from chromatin fraction by using 3 different ChOP oligonucleotides that are complementary to 3 different regions of mrhl RNA (Fig. S1B). Each locus was scored for enrichment in the ChOP DNA by PCR analysis using the primers specific for each of the 37 sequence reads (Primer sequences are given in Table S2). Positive PCR amplification signal was observed in all the 37 GRPAM loci. Promoter region of β-actin gene was used as a negative control. In: Input; m1, m2, m3: complimentary mrhl oligos 1, 2, 3 ; scr: scrambled oligo.
p68 interacts with mrhl RNA in the chromatin and is necessary for mrhl RNA occupancy at GRPAM loci
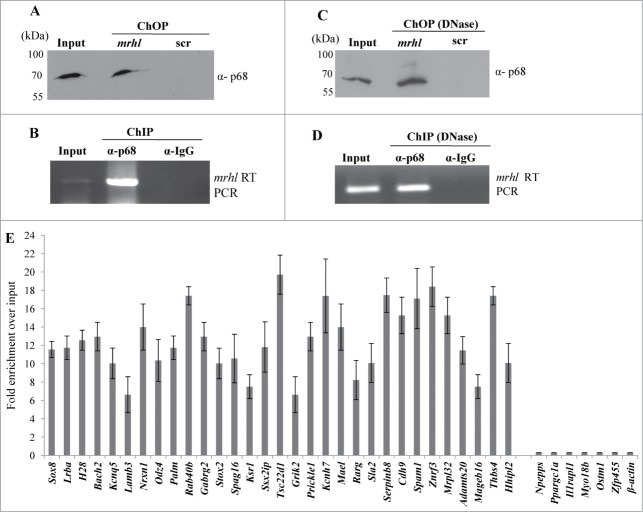
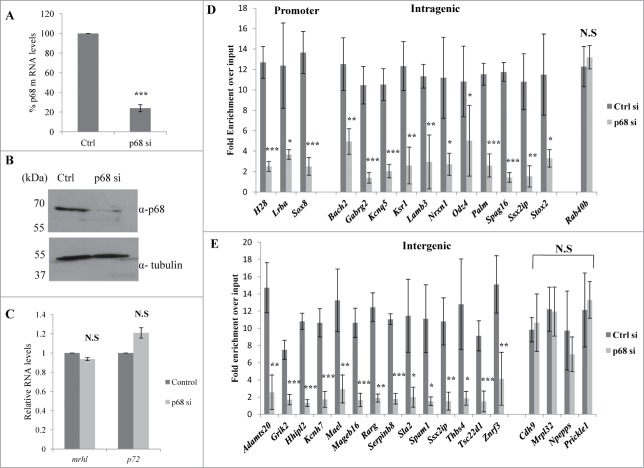
We have shown above that both mrhl RNA and p68 are present in the nucleoplasm and chromatin fraction. We next sought out to examine first whether mrhl RNA and p68 interact with each other in the chromatin context. For this purpose, mrhl RNA was pulled down from the chromatin by the ChOP procedure using biotinylated oligonucleotides. The associated proteins were resolved on SDS-PAGE and the presence of p68 was checked by Western blot analysis. p68 is present both in the input and also in the mrhl ChOP fraction but not in the ChOP chromatin fraction isolated by using biotinylated scrambled oligonucleotides (Fig. 5A). We also carried out the reverse experiment to validate the interaction of mrhl RNA and p68. Chromatin fraction was immunoprecipitated with anti-p68 antibody and the total RNA was extracted from the immunoprecipitate. The presence of mrhl RNA was scored by RT-PCR. Mrhl RNA could be scored in both the input as well as the p68 pull down chromatin fraction but not in the pre immune IgG chromatin IP fraction (Fig. 5B). These results show that p68 and mrhl RNA do interact with the same chromatin fragments. In order to determine whether there is a physical interaction between mrhl RNA and p68, the chromatin fraction was treated with DNase and then the pull down experiments were performed. Western blot upon mrhl RNA pull down with ChOP oligonucleotides showed the presence of p68 (Fig. 5C) while RT-PCR analysis showed the presence of mrhl RNA upon immune pull down of p68 (Fig. 5D) which confirmed the physical interaction between mrhl RNA and p68 on the chromatin template. However, we cannot conclude from this experiment whether p68 and mrhl RNA complex interacts directly with the chromatin or the interaction is mediated through a complex or a bridge protein. We subsequently asked the question whether p68 has a role in the mrhl RNA occupancy at GRPAM loci. To begin with, we first checked whether p68 also binds at these GRPAM chromatin loci. For this purpose we performed quantitative p68 ChIP-PCR which shows that p68 binds to 31 out of 37 GRPAM loci (Fig. 5E). p68 does not bind to Il1rapl1 and Myo18b (intragenic) and Zfp455, Npepps, Ppargc1a and Ostm1 (intergenic) genomic loci which are represented with * in Table 1. For p68 ChIP-PCR we used the same set of primers that were used for mrhl RNA ChOP-PCR. This experiment clearly demonstrated that p68 binds at the same 31 chromatin loci which are bound by the mrhl RNA. We then carried out p68 silencing experiment and checked for the mrhl RNA occupancy at these 31 GRPAM loci. We knocked down p68 in Gc1-Spg cells by using siRNAs targeting 4 different regions of p68 mRNA. All 4 siRNAs were used together for the silencing of p68 gene expression. The down regulation of p68 was checked by qRT PCR (Fig. 6A) as well as by Western blot (Fig. 6B). The levels of mrhl RNA and p72 (which is homologous to p68) do not show any significant change under these experimental conditions (Fig. 6C). The chromatin occupancy of mrhl RNA at the GRPAM loci was then scored by ChOP-PCR and the results were expressed as fold enrichment over input under control scrambled siRNA and p68 siRNA treated conditions (Fig. 6D, 6E). It is very clear from these results that p68 is necessary for the occupancy of mrhl RNA at majority of these GRPAM loci with the exception of Rab40b (intragenic GRPAM) and Cdh9, Mrpl32, Npepps and Prickel1 (intergenic GRPAM) which are represented with ** in Table 1. Thus we conclude that 27 GRPAM loci genes show p68 dependent occupancy of mrhl RNA in Gc1-Spg cells.
Figure 5.
p68-mrhl RNA interaction on chromatin and occupancy of p68 at GRPAM loci (A) Western blot analysis with anti p68 antibody probed on the ChOP chromatin fraction. p68 signal was observed only with the chromatin fraction isolated from mrhl ChOP oligonucleotides but not scrambled oligonucleotides. (B) RT-PCR analysis showing the association of mrhl RNA with the p68-ChIP chromatin fraction isolated by using anti p68 antibody. No signal was observed with pre-immune IgG. (C) Western blot analysis with anti p68 antibody probed on DNase treated ChOP chromatin fraction. p68 signal was observed only with the chromatin fraction isolated from mrhl ChOP oligonucleotides but not scrambled oligonucleotides. (D) RT-PCR analysis showing the association of mrhl RNA with the DNase treated p68-ChIP chromatin fraction isolated by using anti p68 antibody. No signal was observed with pre-immune IgG. (E) Quantitative ChIP-PCR analysis to examine the occupancy of p68 on the 37 GRPAM loci. p68 occupies 31 of the 37 GRPAM loci bound by mrhl RNA. Promoter region of β-actin gene was used as a negative control. Data is an average of 4 independent biological replicates and error bars represent standard deviation.
Figure 6.
p68 is required for the association of mrhl RNA with GRPAM loci. (A) Real Time PCR showing about 80% down regulation of p68 mRNA in siRNA transfected cells. (B) Western blot showing drastic reduction of p68 protein levels upon p68 silencing by siRNA. (C) Real Time PCR showing no significant change in the expression of mrhl RNA and p72 mRNA upon p68 silencing. (D&E) Real Time PCR analysis scoring for the occupancy of mrhl RNA at the GRPAM loci (promoter, intragenic, intergenic) upon p68 silencing. mrhl RNA occupancy is drastically reduced on most of the GRPAM loci upon p68 silencing. Each data is an average of 4 independent biological replicates. Error bars in Figure 6A, C, D, and E represent standard deviation. *** (P ≤ 0.0005), ** P ≤ 0.005), * P ≤ 0.05) (t test). N.S (not significant).
Expression analysis of GRPAM genes upon p68 silencing in Gc1-Spg cells
Since the occupancy of mrhl RNA on GRPAM loci was drastically reduced upon p68 silencing, the next step was to find out whether this reduced mrhl RNA occupancy caused any perturbation of the expression of GRPAM genes. The expression levels of the 27 GRPAM genes upon p68 silencing were then compared with their perturbation in expression observed upon mrhl RNA silencing (data from GSE19355) (Fig. 7A and 7B). All the 27 GRPAM genes showed perturbation of their expression upon p68 silencing; suggesting that p68 mediated recruitment of mrhl RNA at these GRPAM loci regulates their expression. Interestingly, we do find opposite trend of perturbation of expression (up or down regulation) of some of the genes upon p68 or mrhl RNA down regulation. These genes are Ksr1, Lamb3, Ssx2ip, Stox2 (intragenic class) and Mageb16, Thbs4 (intergenic). The remaining 21 genes showed unidirectional perturbation of expression upon p68 or mrhl RNA down regulation.
Figure 7.
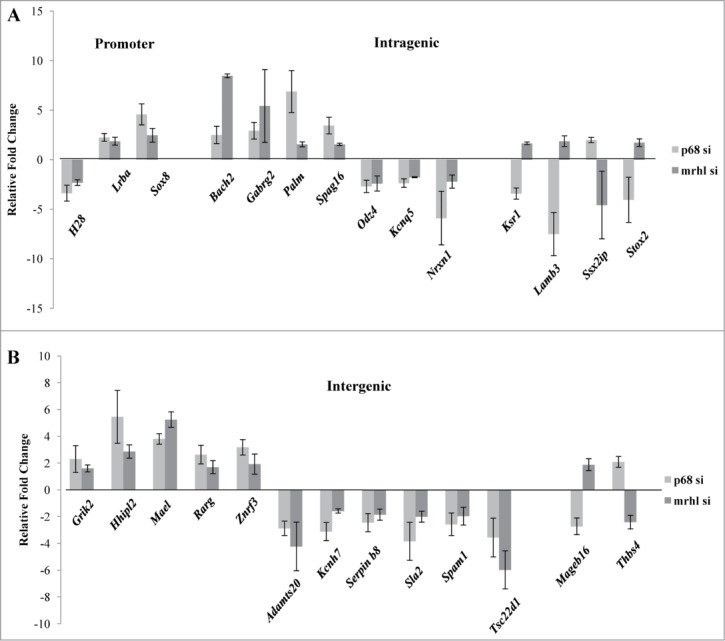
Expression analysis of the GRPAM genes upon silencing of p68 and its comparison with expression levels observed upon down regulation of mrhl RNA (data taken from microarray data GSE 19355). (A) Expression analysis of promoter and intragenic class of GRPAM genes (B) Expression analysis of intergenic class of GRPAM genes. Each data point in 7A and 7B is an average of 4 independent biological replicates and the error bar represents standard deviation.
Expression analysis of GRPAM genes in Gc1-Spg cells upon Wnt3a treatment
We have previously shown that Wnt3a treatment of Gc1-Spg cells down regulates mrhl RNA expression.28 Wnt signaling is an important pathway implicated in the regulation of spermatogenesis.31 Therefore we examined the expression levels of the 27 GRPAM genes in Gc1-Spg cells upon Wnt3a treatment and compared it with that upon mrhl RNA silencing (Figs. 8A and B). Among these, 21 GRPAM genes showed the same kind of perturbation in expression (up or down) upon Wnt 3a treatment as compared to that upon mrhl RNA silencing while 6 GRPAM genes showed the opposite trend (Intragenic: Kcnq5, Nrxn1, Odz4, Ssx2ip and intergenic: Kcnh7, Tsc22d1).
Figure 8.
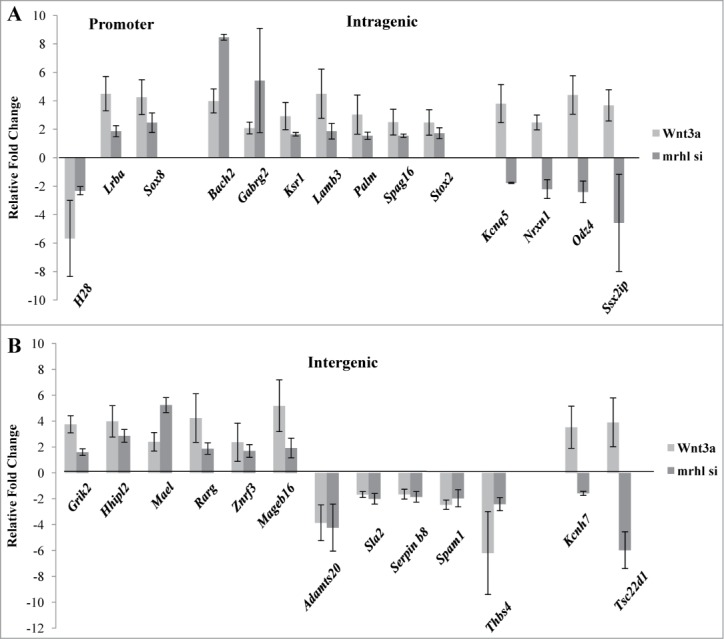
Expression analysis of the GRPAM genes in Gc1-Spg cells upon Wnt3a treatment in comparison with their expression levels observed upon mrhl RNA down regulation (GSE19355) (A) Expression analysis of promoter and intragenic class of GRPAM genes (B) Expression analysis of intergenic class of GRPAM genes. Each data point in 8A and 8B is an average of 4 independent biological replicates and the error bar represents standard deviation.
Proteins associated with mrhl RNA in the chromatin
Our previous study and all the results presented here do indicate that p68/Ddx5 helicase is one of the mrhl RNA interacting protein partners which is necessary for the regulatory function of mrhl RNA in spermatogonial cells. There is an increasing list of long non coding RNAs that carry out chromatin templated functions through different protein complexes.22 Therefore we were curious to find out the nature of the proteins that are present in the mrhl RNA associated chromatin fraction. For this purpose we pulled down mrhl RNA associated chromatin fraction using biotinylated oligonucleotides and resolved the proteins by SDS-PAGE. The coomassie stained gel pattern shows numerous protein bands present in mrhl RNA pull down fraction from the chromatin but not in the scrambled oligonuclotide ChOP-chromatin fraction (Fig. 9A). Protein elutes from the mrhl RNA pull down fraction were subjected to mass spectrometry (nano LC-MS). We could identify many proteins in this analysis and the list of proteins, their molecular masses and identified peptide sequences along with their known biological function(s) are given in Table 2. Among these we further validated the presence of 7 proteins in the mrhl RNA associated chromatin fraction (Pc4, Hmgb2, Rbm3, hnRNPA1, hnRNP A2/B1, hnRNP A/B. p68 was used as a positive control.) by Western blot analysis with the corresponding protein specific antibodies (Fig. 9B). Histone chaperone Npm1 was used as a negative control in this analysis. hnRNP proteins are involved in various aspects of RNA function like transcription and RNA processing.32-34 In this list, it is also comforting to note that all the 4 nucleosomal core histones are also present thus serving as an internal control for the chromatin fraction.
Figure 9.
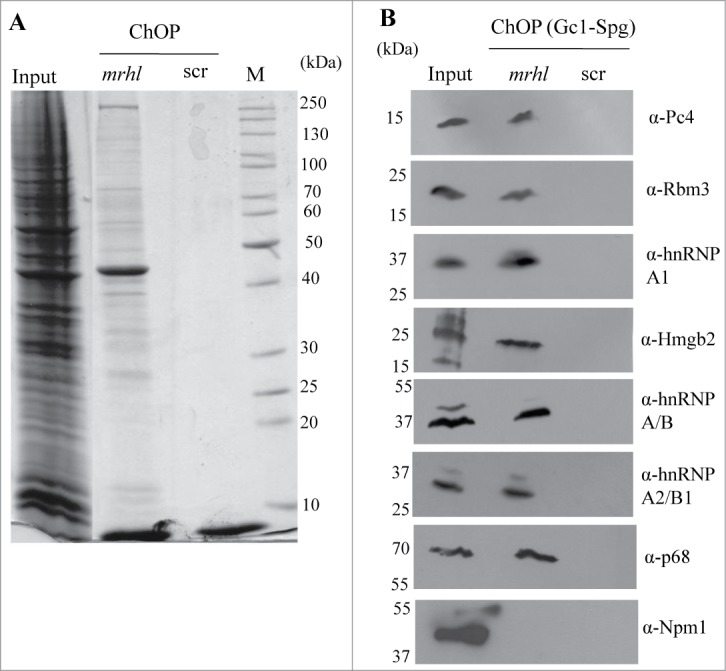
Validation of proteins identified by Mass Spectrometry in the ChOP chromatin by Western blot analysis. (A) Coomassie staining showing the differential proteins associated with mrhl RNA from the chromatin fraction. (B) Validation of association of 7 proteins (Pc4, Rbm3, hnRNP A1, Hmgb2, hnRNP A/B, hnRNP A2/B1 and p68) with mrhl RNA after pull down of mrhl RNA from the chromatin fraction by Western blot analysis. Histone Chaperone Npm1 was used as a negative control.
Table 2.
Interacting proteins of mrhl RNA from chromatin
| No. | Protein | UniProtK B | Mol wt (kDa) | Total peptides | Peptides | Protein Function |
|---|---|---|---|---|---|---|
| 1 | hnRNP A/B | Q99020 | 30.8 | 9 | (K)IFVGGLNPEATEKK(I), (K)EVYQQQQYGSGGR(G), (K)FGEVVDCTIK(M), (K)MFVGGLSWDTSKK(D), | Regulation of transcription, lncRNA function, splicing |
| 2 | Pc4 | P11031 | 14.4 | 6 | (R)DDNMFQIGK(M), (K)EQISDIDDAVR(K), (K)QSSSSRDDNMFQIGK(M), (R)EYWMDSEGEMKPGR(K), (R)EYWMDSEGEMKPGRK(G) | Transcription coactivator |
| 3 | Actg1 | P63260 | 41.7 | 16 | (K)SYELPDGQVITIGNER(F), (K)EITALAPSTMK(I), (R)HQGVMVGMGQK(D), (R)AVFPSIVGRPR(H), (R)GILTLK(Y) | Cell Migration, SignalingTranscription elongation |
| 4 | Histone H4 | P62806 | 11.8 | 4 | (R)ISGLIYEETR(G), (R)TLYGFGG(-), (R)DNIQGITKPAIR(R), (K)VFLENVIR(D) | Nucleosomal component |
| 5 | Rbm3 | O89086 | 16.6 | 2 | (R)YSGGNYRDNYDN(-), (R)GFGFITFTNPEHASDAMR(A) | Regulation of translationProduction of miRNAs |
| 6 | H2B1F | P10853 | 14 | 2 | (R)LLLPGELAK(H), (K)ESYSVYVYK(V) | Nucleosomal component |
| 7 | hnRNP-DL | Q9Z130 | 33.5 | 7 | (R)FGEVVDCTIK(T), (R)FGEVVDCTIK(T), (K)DLTEYLSR(F), (K)DAASVDKVLELK(E), | Regulation of transcription |
| 8 | H2A3 | Q8BFU2 | 2 | (R)AGLQFPVGR(V), (Q)FPVGR(V) | Nucleosomal component | |
| 9 | Annexin A1 | P10107 | 38.7 | 12 | (K)ILVALCGGN(-),(R)FLENQEQEYVQAVK(S), (K)TPAQFDADELRGAMK(G), (K)TPAQFDADELR(G) | Apoptosis, cell cycleCell proliferation |
| 10 | Srsf2 | Q62093 | 25.4 | 4 | (R)DAEDAMDAMDGAVLDGR(E), (R)GFAFVR(F), (R)VGDVYIPR(D), (F)AFVR(F) | RNA spilicngmRNA processing |
| 11 | hnRNP A1 | P49312 | 34.1 | 4 | (K)IEVIEIMTDR(G), (R)SSGPYGGGGQYFAKPR(N), (R)GGGFGGNDNFGR(G), (R)SHFEQWGTLTDCVVMR(D) | RNA spilicngmRNA processing |
| 12 | Histone H3.2 | P84228 | 15.4 | 6 | (K)STELLIR(K), (R)YRPGTVALR(E), (K)RVTIMPK(D), (K)STELLIR(K) | Nucleosomal component |
| 13 | Nucleolin | P09405 | 76.7 | 18 | K)GFGFVDFNSEEDAK(A), (R)SVSLYYTGEK(G), (K)GIAYIEFK(S), (K)NDLAVVDVR(T), | Transcription, p53 pathwayregulation of growth |
| 14 | hnRNP A2 B1 | O88569 | 37.4 | 4 | (R)GGNFGFGDSR(G), (R)GGGGNFGPGPGSNFR(G), (K)TLETVPLER(K), (K)IDTIEIITDR(Q) | Splicing, lncRNA function, Transcription, differentiation |
| 15 | Hmgb1 | P63158 | 24.9 | 5 | (K)YEKDIAAYR(A), (K)GKPDAAKK(G), (K)KHPDASVNFSEFSK(K), (K)IKGEHPGLSIGDVAK(K) | Transcription, DNA repair, apoptosis, differentiation |
| 16 | Myl6 | Q60605 | 16.9 | 6 | (R)HVLVTLGEK(M), (K)ILYSQCGDVMR(A), (R)ALGQNPTNAEVLK(V), (K)EAFQLFDR(T) | Adhesion, migration, signaling |
| 17 | Annexin A2 | P07356 | 38.6 | 9 | (R)TNQELQEINR(V), (K)LMVALAK(G), (R)QDIAFAYQR(R), (R)SEVDMLK(I) | Protein phosphorylationPoly(A) RNA binding |
| 18 | hnRNP D0 | Q60668 | 38.3 | 6 | (K)IFVGGLSPDTPEEK(I),(K)FGEVVDCTLKLDPITGR(S), (R)EYFGGFGEVESIELPMDNK(T), (K)EQYQQQQQWGSR(G) | mRNA processing, mRNA stability |
| 19 | Alyref2 | Q9JJW6–2 | 23.6 | 3 | (R)SLGTADVHFER(R), (K)QQLSAEELDAQLDAYNAR(M), (K)QQLSAEELDAQLDAYNAR(M) | RNA splicing |
| 20 | Hmgb2 | P30681 | 24.1 | 11 | (K)YEKDIAAYR(A), (K)SKFEDLAK(S), (K)IKIEHPGLSIGDTAK(K), (K)KLGEMWSEQSAK(D) | Spermatogenesis, Wnt signaling, Nucelosomal histone binding |
List of interacting proteins of mrhl RNA from ChOP chromatin fraction identified by Mass Spectrometry (nano LC/MS). The peptide sequences obtained for each of the protein are listed. The peptides with high score are also listed for each of the protein where more than 4 peptides were identified. The biological function of each of the protein is given in the last column.
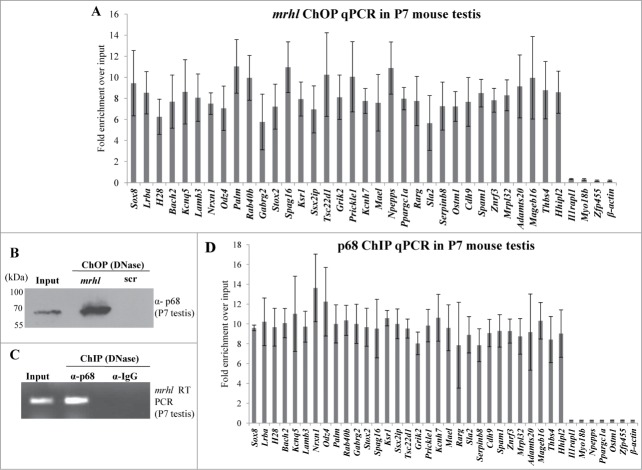
In vivo occupancy of GRPAM loci by mrhl RNA and p68 in mouse testicular chromatin and inverse correlation of nuclear β catenin and mrhl RNA expression
The findings presented so far demonstrate the role of mrhl RNA in gene regulation in a cell culture system. In order to substantiate the physiological relevance of our work, we performed certain key experiments in the mouse testicular tissue. Since Gc1-Spg is a spermatogonial cell line, we selected testis of 7 day old (P7) mice for our further experiments because of the predominance of spermatogonia at that age. We performed mrhl ChOP in the testicular chromatin of P7 mice and checked for the occupancy of mrhl RNA at the GRPAM loci. Similar to Gc1-Spg cells, we could also score for the occupancy of mrhl RNA at the GRPAM loci (34 out of 37) by quantitative ChOP-PCR (Fig. 10A). The physical interaction of mrhl RNA and p68 also holds true in the P7 mouse testicular chromatin (Fig. 10B and 10C) and also the p68 occupancy at 31(out of 37) GRPAM loci as seen in Gc1-Spg cells (Fig. 10C). We also confirmed the association of chromatin proteins with mrhl RNA since we could validate the association of the 7 proteins (Pc4, Hmgb2, hnRNP A/B, hnRNP A2/B1, hnRNP A1, Rbm3 and p68) by Western blot upon mrhl RNA ChOP in P7 mouse testicular chromatin (Fig. 11A). Earlier, we reported the down regulation of mrhl RNA expression upon Wnt signaling activation.28 We further validated this finding in the mouse testicular tissue. P21 mouse testis predominantly contains spermatocytes as opposed to P7 mouse testis which have mostly spermatogonia. Wnt signaling is activated in spermatocyte population and P21 mouse testis as demonstrated by nuclear localization of β catenin.35,36 We compared the expression pattern of mrhl RNA in P7 and P21 testis and there was a drastic reduction of mrhl RNA expression in P21 testis compared to P7 testis (Fig. 11B). The Western blot with nuclear extracts of P7 and P21 testis show nuclear β catenin only in P21 testis (Fig. 11C) confirming the Wnt signaling activation status in the P21 testis.
Figure 10.
(A) Quantitative ChOP-PCR for examining the occupancy of mrhl RNA at 37 GRPAM loci in P7 mouse testis chromatin. mrhl RNA occupies 34 out of 37 GRPAM loci. (B) Western blot analysis with anti p68 antibody probed on DNase treated ChOP chromatin fraction of P7 mouse testis. (C) RT-PCR analysis showing the association of mrhl RNA with the DNase treated p68- ChIP chromatin fraction of P7 mouse testis isolated by using anti p68 antibody. (D) Quantitative ChIP-PCR analysis to examine the occupancy of p68 on the 37 GRPAM loci in P7 mouse testis chromatin. p68 occupies 31 of the 34 GRPAM loci bound by mrhl RNA. Each data point in 10A and 10D is an average of 4 independent biological replicates and the error bar represents standard deviation.
Figure 11.
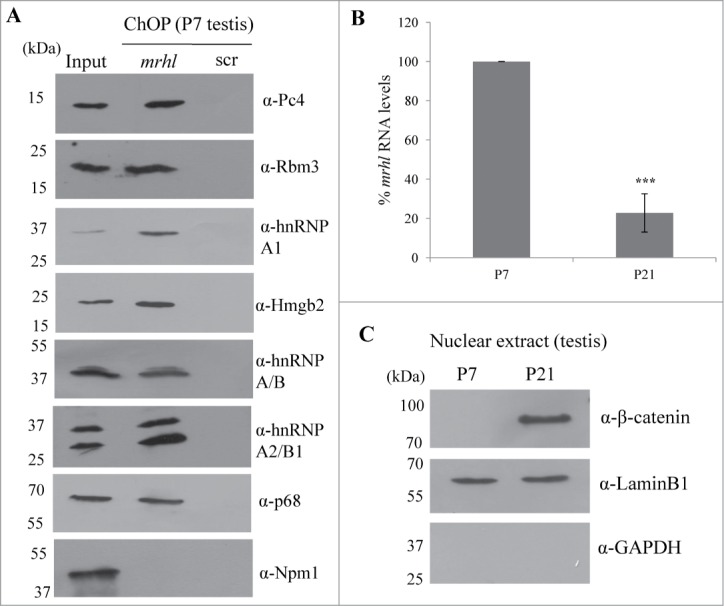
(A) Validation of association of 7 proteins (Pc4, Rbm3, hnRNP A1, Hmgb2, hnRNP A/B, hnRNP A2/B1 and p68) with mrhl RNA after pull down of mrhl RNA from the chromatin fraction of P7 mouse testis. Npm1 was used as a negative control. (B) Real Time PCR analysis showing significant reduction in mrhl RNA expression in P21 mouse testis compared to P7. (C) Western blot analysis showing the presence of β catenin in the nuclear extract of P21 mouse testis but not P7. Lamin and GAPDH Western blots show the purity of the nuclear extracts. *** P ≤ 0.0005) (t-test).
Discussion
Long non coding RNAs (lncRNAs) are emerging as key players in the regulation of diverse cellular functions in the context of development and disease. LncRNAs are also shown to be responsive to signaling pathways as well as function as their regulators. Mrhl RNA is one such RNA that interacts with p68/Ddx5 helicase in the nucleus and negatively regulates Wnt signaling in Gc1-Spg cells (derived from B type spermatogonia). Mrhl RNA is the first lncRNA shown to regulate Wnt signaling. Wnt signaling plays a very important role in the regulation of mammalian spermatogenesis.31 Yeh et al. have shown that Wnt signaling is suppressed in spermatogonial stem cells and undifferentiated spermatogonia while it is activated in differentiated meiotic spermatocytes.35 Thus, an in depth understanding of the mechanism of mrhl RNA function in spermatogonial cells is crucial to elucidate the biological function of this lncRNA in the broader context of spermatogenesis and in more particular the differentiation of spermatogonial cells into meiotic spermatocytes. Toward this direction, we embarked upon mapping the genome wide chromatin occupancy of mrhl RNA on Gc1-Spg cell chromatin to get an insight into the role of mrhl RNA in global gene regulation. There are very few reports in the literature that document the genome wide occupancy of lncRNAs.37,38 The key observations of our present study can be summarized as follows: 1) Occupancy of 1370 genomic loci by mrhl RNA in mouse spermatogonial cell chromatin. 2) Among these, 37 GRPAM loci represent the most probable regulatory targets of mrhl RNA. 3) Most GRPAM loci are occupied by mrhl RNA in a p68 dependent manner. 4) Regulation of GRPAM expression is physiologically relevant in the context of spermatogenesis and Wnt signaling.
One of the significant observations made in the present study is the identification of 27 genes among the 37 GRPAM loci where mrhl RNA occupancy is dependent on p68 and whose expression is perturbed following mrhl RNA down regulation. We consider these genes to be biologically important genomic loci (GRPAM loci) whose regulation is modulated by the occupancy of mrhl RNA in the context of cellular biology of spermatogonial cells. We had shown earlier that down regulation of mrhl RNA resulted in cytoplasmic translocation of phosphorylated form of p68 triggering the stabilization of β catenin in the cytoplasm and the subsequent activation of TCF4 mediated gene expression in the nucleus. From these results, we had proposed that mrhl RNA negatively regulates Wnt signaling by altering the dynamics of p68 in the nucleus and the cytoplasm which is intimately associated with the β catenin/TCF4 mediated regulation of Wnt target genes. The results presented in this communication have shown further that mrhl RNA does play a direct positive role in modulating gene expression by directly associating with specific chromatin loci which is again dependent on p68 protein. Among the 27 GRPAM genes (Table 1), several genes belonged to Wnt pathway and spermatogenesis categories. Sox8 is a transcription factor required for mammalian Sertoli cell function and germ cell differentiation.39 Sox proteins are also known to function as regulators of Wnt signaling.40 Rarg (retinoic acid receptor gamma) protein is necessary for retinoic acid signaling and differentiation of spermatogonia into meiotic spermatocytes.41,42 Mael protein (maelstrom homolog) is essential for spermatogenesis and transposon repression during meiosis.43 Mageb16 is a testis specific gene,44 while Spag16 is a male germ cell nuclear speckle protein important during spermatogenesis.45 Among the genes related to Wnt signaling, recent studies have shown the role of Odz4 in induction of embryonic mesoderm by activation of Wnt signaling and Lamb3 is shown to be downregulated by Wnt5A signaling in breast cancer cells.46,47 Znrf3 is an Wnt antagonist as it promotes Wnt receptor turnover, while Tsc22d1 is an enhancer of Tgf β signaling but is also induced upon Wnt signaling activation in human fibroblasts.48,49 Among the 27 GRPAM genes, 3 genes belonged to promoter class which includes Sox8, Lrba and H28. Lrba is a lipopolysachharide inducible gene and has been shown to be part of the nuclear interactome of the Notch1 ligand.50 Homologues of Lrba in Drosophila and C. elegans are also shown to be involved in Notch signaling pathway.51,52 H28 or Ifi441 protein is a minor histocompatibility antigen and is induced in response to interferon. While it may be easier to conceptualize the mrhl RNA mediated regulation of gene expression of the promoter class of GRPAM genes, the mechanism of gene regulation of intragenic and intergenic classes may be much more complex. It is quite likely that mrhl RNA occupancy at these loci may alter the chromatin structure to either activate or repress the expression of these genes, as in the case of Xist RNA53 or it may involve long range DNA interactions through looping of DNA.38 The biological relevance of regulation of 27 GRPAM genes is further supported from the data obtained from Wnt3a ligand treated Gc1-Spg cells. Wnt3a is a natural ligand of spermatogonial cells and it does bring about the down regulation of mrhl RNA, in turn its occupancy at GRPAM loci and consequent perturbation of gene expression. A careful examination of the nature of perturbation of GRPAM genes reveals that it is not uni-directional, but some genes are induced and some genes are repressed following p68 silencing and Wnt3a treatment. Such a biphasic nature of gene expression regulation indicates a complex mode of regulation at these chromatin loci and possibly depends on the recruitment of either co-activators or co-repressors.54
Our analysis of the 1370 ChOP sequence reads also resulted in the identification of 2 motifs within them (Fig. 4B). However, the actual mechanism by which mrhl RNA interacts with the genomic chromatin can be any one of the following. a. Direct Watson Crick base pairing55 b. DNA-DNA-RNA triple helix formation (For example DHFR lncRNA).56 c. Facilitate interaction with specific loci through protein complexes.19, 57 Since lncRNAs can regulate gene expression by virtue of the proteins that they associate with, we tried to identify the proteins that are co-purified in the mrhl ChOP fraction. We observed proteins that are known to be associated with chromatin templated transcription functions. hnRNP A1 and Srsf2 have been reported to regulate splicing and ultimately gene expression in association with p68.58 hnRNP A/B and hnRNP A2/B1 have recently been shown to be important for function of lincRNA-Cox2 resulting in either gene activation or repression.59 hnRNP class of proteins are also important for chromatin mediated function of lncRNA. For example, hnRNP U is shown to be essential for chromosomal localization of Xist RNA and lncRNA-p21 along with hnRNP K function in p53 dependent gene repression in response to DNA damage.32,33 In addition to hnRNP proteins, chromatin organizer proteins like PRC2 complex and CoREST have also been shown to associate with lncRNA function.60 In this context, we do find chromatin organizer protein Pc4 to be associated with the mrhl associated chromatin fraction. Pc4 protein is a non histone protein involved in chromatin organization and plays a dual role (activation or repression) in gene regulation.61,62 Recently, Pc4 has also been shown to activate Luteinizing Hormone (LH) gene transcription by binding to its promoter.63 Among the other proteins identified by our mass spectrometry analysis, Hmgb2 regulates transcription and formation of nucleoprotein complexes through chromatin and is required for spermatogenesis as well as regulation of Wnt signaling.64,65 We would like to mention here that the protein analysis of mrhl RNA associated chromatin fraction probably reflects an average picture of the protein components of all the 1370 genomic loci that we have identified. However, identifying the protein complexes associated more specifically at the 27 GRPAM loci would decipher the regulation of GRPAM expression.
Mrhl RNA and p68 physically interact with each other and co-occupy most of the GRPAM loci even in the spermatogonial cells of mouse testicular tissue. This in vivo correlation along with the inverse correlation between mrhl RNA expression and nuclear β catenin possibly suggests that mrhl RNA mediated regulation of Wnt signaling is an important biological feature of differentiation of spermatogonial cells into meiotic spermatocytes. The major findings from this study have been summarized schematically in Figure 12. Although we have addressed only 37 GRPAM loci in present study, it is likely that the remaining mrhl RNA bound genomic loci may have Wnt signaling independent function in a context dependent manner during mammalian spermatogenesis.
Figure 12.
Summary of the relationship between chromatin occupancy of mrhl RNA and regulation of gene expression in the context of Wnt signaling in spermatogenesis. (A) Overlap set of mrhl RNA silencing (microarray data) and mrhl RNA ChOP leading to 37 GRPAM genes and their classification based on location of ChOP sequence reads. Number in brackets indicates the number of GRPAM of that particular category. (B) Hypothetical mechanisms of gene regulation by mrhl RNA-p68 complex at GRPAM loci. Oval circles indicate different proteins associated with mrhl RNA which can include co-activators or co-repressors or chromatin modifiers. ? indicates that the nature of interaction of p68-mrhl RNA complex with chromatin template is not clear at present. Regulation of promoter class of GRPAM can be through recruitment of co-activators or co-repressors while intragenic and intergenic GRPAM can be regulated through alteration of chromatin structure and long range interactions respectively. (C) Down regulation of mrhl RNA and perturbation of GRPAM gene expression in Gc1-Spg spermatogonial cell line upon Wnt3a treatment. ? indicates that the status of p68 genome wide occupancy and expression of p68 upon Wnt3a treatment is not known. (D) Inverse correlation between Wnt activated state (as demonstrated by nuclear β catenin) and mrhl RNA expression levels between spermatogonia (Repressed Wnt signaling, higher expression of mrhl RNA) and differentiated spermatocytes (Wnt activation and decreased levels of mrhl RNA ). β: β catenin, Fzd: Frizzled, Lrp: Low-density lipoprotein receptor-related protein.
Materials and Methods
Cell line, Antibodies, siRNA, other chemicals
Gc1-Spg cell line (type B spermatogonia) was obtained from the American Type Culture Collection (ATCC) and was cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen) and 100units/ml penicillin-streptomycin solution (Sigma) at 37oC in a humidified 5% CO2 atmosphere.
Anti p68 antibody was raised in rabbit against full length recombinant p68 protein. Anti Pc4 and anti Npm1 antibodies were kindly provided by Prof Tapas Kundu, JNCASR. Following antibodies were obtained from Abcam (catalog number in brackets): Anti Rbm3 (ab134946), Anti hnRNP A/B (ab94593), Anti hnRNP A1 (ab5832), Anti hnRNP A2/B1 (ab31645), Anti Histone H3 (ab46765), Anti Histone H4 (ab31830), Anti Hmgb2 (ab67282).
Four siRNAs against p68 mRNA were obtained from Sigma. The siRNA ID are: SASI_Mm01_00096788, SASI_Mm01_00096789, SASI_Mm01_00096792 and SASI_Mm01_00096793.
All chemical reagents were of AR grade and were purchased from Sigma. Protein A agarose beads (15918–014) and streptavidin agarose beads (15942–050) were purchased from Invitrogen. DNAseI (MO303) was purchased from New England Biolabs.
The list of genes perturbed under mrhl RNA down regulation was available from the GEO dataset (GSE19355).28
Chromatin fractionation
Nuclei of Gc1-Spg cells were fractionated into nucleoplasm and chromatin as described before with some minor modifications.66 Gc1-Spg cell pellet (grown to confluence) was resuspended in 5–6 volumes of buffer N250 (15 mM Tris HCl, pH 7.5,15 mM NaCl, 60 mM KCl, 10 mM MgCl2, 1 mM CaCl2, 250 mM sucrose, 1mM DTT, 0.5 mM PMSF and 75 units/ml RNasin) and incubated on ice for 10 minutes. Equal volume of buffer N250 containing 0.6%NP-40 was then added to cells, suspension was gently mixed and further incubated on ice for 20 minutes. Nuclear pellet was separated from the cytoplasm by centrifugation at 2400 X g for 10 min at 4oC. Nuclei were washed once in buffer N250 and then lysed in Pipes-EDTA buffer (10mM Pipes buffer, pH 6.5, 10 mM EDTA, pH 8, 0.5 mM PMSF and 75 units/ml RNasin). Nucleoplasm was obtained in the supernatant and chromatin in the pellet after centrifugation at 6400 X g for 20 min at 4oC. Chromatin pellet was resuspended in sonication buffer (20mM Tris HCl, pH 7.5, 150 mM NaCl, 3mM MgCl2, 0.5 mM PMSF and 75 units/ml RNasin), sonicated for 5 min and supernatant chromatin was obtained by centrifugation at 18,000 X g for 10 minutes.
For RNA isolation, the nucleoplasm and chromatin fractions were treated with DNaseI (10 units/ml) for 45 min and then Proteinase K (80 µg/ml) treatment for 30 min followed by phenol-chloroform extraction and ethanol precipitation. 1µg RNA was used for cDNA synthesis reaction.
Chromatin Oligo Affinity precipitation (ChOP) and DNA sequencing analysis
The ChOP assay was performed as described before with minor modifications.23 Gc1-Spg cells were cross linked using 1% formaldehyde for 10 min at room temperature. Glycine was used at final concentration of 0.125 M for quenching. Cells were washed with PBS, resuspended in 1ml of buffer A (3 mM MgCl2, 10 mM Tris HCl, pH 7.4, 10 mM NaCl, 0.5%v/v NP-40, 0.5 mM PMSF and 75 units/ml RNasin) and incubated on ice for 15 minutes. Nuclei were harvested by centrifugation and resuspended in 800 µl of buffer B (50 mM Tris HCl, pH 7.4, 10 mM EDTA, 0.5% Triton X-100, 0.1%SDS, 0.5 mM PMSF and 75 units/ml RNasin) and incubated on ice for 30 minutes. An equal volume of buffer C (15 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM PMSF and 75 units/ml RNasin) was then added and incubated on ice for 10 minutes. Samples were then sonicated using a Bioruptor sonicator (Diagenode). Sonicated DNA was found to be enriched in the range of 200–500 bp. After centrifugation the chromatin was pre cleared for 30 min using streptavidin-agarose beads. 1 µg of mrhl complementary oligo or 1µg of control scrambled oligo was added to the chromatin solution along with yeast tRNA (100 µg/ml), salmon sperm DNA (100 µg/ml) and BSA (500 µg/ml) and incubated overnight at 4oC (For identification of the mrhl RNA associated proteins, BSA was excluded and all the 3 mrhl complementary oligonucleotides were added). Samples were then incubated with streptavidin agarose beads for 2.5 hrs followed by one wash with Low salt buffer (20 mM Tris HCl, pH7.9,150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 0.5 mM PMSF and 75 units/ml RNasin) and one wash with high salt buffer (20 mM Tris HCl, pH7.9, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 0.5 mM PMSF and 75 units/ml RNasin). The beads were then incubated with elution buffer (1%SDS, 100 mM NaHCO3) for 30 min with intermittent mixing for 30 min followed by Proteinase K (80 µg/ml) digestion and phenol chloroform extraction method for DNA isolation. ChOP-DNA sequencing was performed using Illumina GAII analyzer (single end sequencing, 37 bases reads). Sequence reads were aligned individually to each chromosome using BOWTIE 0.9.8.1 while peak finding was performed using ERANGE 3.1. Total 1370 sequence reads were considered as significant based on the cut off values of fold enrichment over input DNA (>4 ) and p value (p < 0.01). For ChOP-Seq validation, PCR was done using sequence specific primers under standard conditions for 30 cycles. Primers used for ChOP PCR are listed in Table S2.
For ChOP experiment performed with DNase treatment, the sonicated chromatin was diluted fold5- with buffer A to reduce the EDTA concentration to 1mM and then DNaseI was used at concentration of 100 units/ml and incubated for 1.5 hrs at 37°C followed by inactivation of DNaseI by increasing EDTA concentration to 10 mM. Complete DNA degradation was monitored by agarose gel electrophoresis.
For identification of mrhl RNA associated proteins, the protein complex from the Streptavidin agarose beads was eluted in a buffer containing 15 mM Hepes-KOH, pH7.4, 3 mM MgCl2 and 15 mM KCl with intermittent mixing at 55°C.
Chromatin immunoprecipitation (ChIP), p68 silencing and qRT-PCR
ChIP was performed using the same method as described previously.28 6 µg of p68 antibody or 6 µg pre immune IgG was used for p68 ChIP while 4 µg of histone H3 antibody or 4 µg pre immune IgG was used for H3 ChIP. p68 ChIP with DNase treatment was performed by same way as described in the ChOP method section. p68 silencing using siRNA and qRT-PCR were carried out using the same method as described previously.28
In-silicoMotif discovery
Mrhl RNA bound 1370 genomic sequences were obtained from the UCSC table browser and were used for de novo motif identification using DREME v4.9.1 from MEME suite of tools.67,68 TOMTOM motif comparison tool was used to identify nearest matches of motifs identified from DREME using JASPAR CORE database.69 Control data sets of 1370 random sequences, from mouse were extracted from RSAT tools and conducted motif analysis to validate the motifs predicted from ChOP sequence data.70
Gene ontology analysis of ChOP data
Gene ontology enrichment analysis of the genes associated with mrhl RNA bound 1370 genomic loci was performed using GO term finder.71 Terms were considered significant at a p-value <0.01. The GO terms were passed on to REVIGO web server to reduce the redundancy and the functional classification was visualized as scatter plot.72
Liquid chromatography–mass spectrometry analysis
The proteins associated with mrhl RNA in the chromatin fraction were processed for mass spectrometric analysis. Tryptic peptides generated were analyzed by nano LC–MSE using a Nano Acquity UPLC system (Waters Corporation) coupled to a Q-TOF, SYNAPT-HDMS (Waters Corporation). The nano-LC separation was performed using a BEH- C18 reversed phase column (1.7 μm particle size) with an internal diameter of 75 μm and length of 150 mm (Waters Corporation). The binary solvent system used comprised 99.9% water and 0.1% formic acid (mobile phase A) and 99.9% acetonitrile and 0.1% formic acid (mobile phase B). Peptides were initially pre concentrated and desalted online at a flow rate of 5 μL/min using a Symmetry C18 trapping column (internal diameter 180 μm, length 20 mm) (Waters Corporation) with a 0.1% B. After each injection, peptides were eluted into the Nano Lock Spray ion source at a flow rate of 300 nL/min using a gradient of 2–40% B for 35 min, then the column was washed and equilibrated. For lockmass calibrant peptide standard, 600 fmol/μL glu-fibrinopeptide B, was infused into the Nano Lock Spray ion source at a flow rate of 300 nL/min and was analyzed at 30 s intervals. The mass spectrometer was operated in V-mode at a resolution of approx 9000 (FWHM) with an alternating 1 s scans of low (4 V) or high (20−40 V) collision energies are used to generate either intact peptide ions (low energy) or peptide product ions (high energy). For peptide mass analysis, PLGS and MASCOT softwares were used.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to acknowledge Dr. Mahesh Kulkarni and Sneha Bansode from National Chemical Laboratory (NCL, India) for mass-spectrometry. ChOP sequencing was performed by Genotypic Technology (India). We acknowledge Shrinivas Nivrutti Dighe (JNCASR) for his assistance in experiments with mouse testis.
Funding
M.R.S Rao thanks Department of Science and Technology for J.C Bose and SERB Distinguished fellowships. Vijay Suresh Akhade is a CSIR- Senior Research Fellow. This work was financially supported by Department of Biotechnology, Govt. of India (Grant Number BT/01/COE/07/09).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Kim VN. Small RNAs: Classification, Biogenesis, and Function. Mol Cells 2005; 19:1-15; PMID:15750334; http://dx.doi.org/10.1016/j.molcel.2005.05.026 [PubMed] [Google Scholar]
- 2.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007; 318:761-64; PMID:17975059; http://dx.doi.org/10.1126/science.1146484 [DOI] [PubMed] [Google Scholar]
- 3.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011; 25:1915-27; PMID:21890647; http://dx.doi.org/10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 2012; 22:1775-89; PMID:22955988; http://dx.doi.org/10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science 2005; 309:1559-63; PMID:16141072; http://dx.doi.org/10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- 6.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458:223-7; PMID:19182780; http://dx.doi.org/10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L,Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature Biotechnol 2010; 28:503-10; http://dx.doi.org/10.1038/nbt.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, Ponting CP. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol 2012; 4:427-42; PMID:22403033; http://dx.doi.org/10.1093/gbe/evs020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam JW. and Bartel D. Long non-coding RNAs in C. elegans. Genome Res 2012; 22:2529-40; PMID:22707570; http://dx.doi.org/10.1101/gr.140475.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011; 147:1537-50; PMID:22196729; http://dx.doi.org/10.1016/j.cell.2011.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert SL, Pehrson JR, Sharp PA. XIST RNA associates with specific regions of the inactive X chromatin. J Biol Chem 2000; 275:36491-4; PMID:11006266; http://dx.doi.org/10.1074/jbc.C000409200 [DOI] [PubMed] [Google Scholar]
- 12.Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 2002; 298:1620-3; PMID:12446910; http://dx.doi.org/10.1126/science.1076686 [DOI] [PubMed] [Google Scholar]
- 13.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008; 32:232-46; PMID:18951091; http://dx.doi.org/10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- 14.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002; 14:68-73 [DOI] [PubMed] [Google Scholar]
- 15.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008; 18:1433-45; PMID:18562676; http://dx.doi.org/10.1101/gr.078378.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemenson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA:NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33:717-26; PMID:19217333; http://dx.doi.org/10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA p21 suppresses target mRNA translation. Mol Cell 2012; 47:648-55; PMID:22841487; http://dx.doi.org/10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong C. and Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 2011; 470:284-8; PMID:21307942; http://dx.doi.org/10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010; 3:ra8; PMID:20124551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 2011; 43:1040-46; PMID:21925392; http://dx.doi.org/10.1016/j.molcel.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472:120-4; PMID:21423168; http://dx.doi.org/10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL. and Chang HY. Genome regulation by Long Noncoding RNAs. Annu Rev Biochem 2012; 81:145-66; PMID:22663078; http://dx.doi.org/10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 2008; 29:499-509; PMID:18313387; http://dx.doi.org/10.1016/j.molcel.2007.12.013 [Google Scholar]
- 24.Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K, Takahashi S, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J 2013; 32:1665-80; PMID:23644382; http://dx.doi.org/10.1038/emboj.2013.99 [Google Scholar]
- 25.Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta 2014; 1843:372-86; PMID:24184209; http://dx.doi.org/10.1016/j.bbamcr.2013.10.016 [Google Scholar]
- 26.Nishant KT, Ravishankar H, Rao MRS. Characterization of a mouse recombination hot spot locus encoding a novel non-protein-coding RNA. Mol Cell Biol 2004; 24:5620-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganesan G. and Rao SM. A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. RNA 2008; 14:1399-1410; PMID:18515546; http://dx.doi.org/10.1261/rna.838308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arun G, Akhade VS, Donakonda S, Rao MR. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol Cell Biol 2012; 32:3140-52; PMID:22665494; http://dx.doi.org/10.1128/MCB.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logan CY. and Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004; 20:781-810; PMID:15473860; http://dx.doi.org/10.1146/annurev.cellbio.20.010403.113126 [DOI] [PubMed] [Google Scholar]
- 30.Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene 2004; 23:4454-65; PMID:15064750; http://dx.doi.org/10.1038/sj.onc.1207579 [Google Scholar]
- 31.Kerr GE, Young JC, Horvay K, Abud HE, Loveland KL. Regulated Wnt/Beta-Catenin signaling sustains adult spermatogenesis in mice. Biol Reprod 2014; 90:1-12; http://dx.doi.org/10.1095/biolreprod.112.105809 [DOI] [PubMed] [Google Scholar]
- 32.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 2009; 106:11667-72; PMID:19571010; http://dx.doi.org/10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142:409-19; PMID:20673990; http://dx.doi.org/10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 2010; 19:469-76; PMID:20833368; http://dx.doi.org/10.1016/j.devcel.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 35.Yeh JR, Zhang X, Nagano MC.. 2011. Wnt 5a is a cell-extrinsic factor that supports self-renewal of mouse spermatogonial stem cells. J Cell Sci 2011; 124:2357-66; PMID:21693582; http://dx.doi.org/10.1242/jcs.080903 [DOI] [PubMed] [Google Scholar]
- 36.Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 2009; 138:151-62; PMID:19419993; http://dx.doi.org/10.1530/REP-08-0510 [DOI] [PubMed] [Google Scholar]
- 37.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011; 44:667-678; PMID:21963238; http://dx.doi.org/10.1016/j.molcel.2011.08.027 [Google Scholar]
- 38.Vance KW. and Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends in Genetics 2014; pii: S0168-9525(14)00086-9. doi: 10.1016/j.tig.2014.06.001; PMID:24974018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrionuevo F. and Scherer G. SOXE genes: SOX9 and SOX8 in mammalian testis development. Int J Biochem Cell Biol 2010; 42:433-6; PMID:19647095; http://dx.doi.org/10.1016/j.biocel.2009.07.015 [DOI] [PubMed] [Google Scholar]
- 40.Kormish JD, Sinner D, Zorn MA. Interactions between SOX factors and Wnt/β-catenin signaling in development and disease. Dev Dyn 2010; 239:56-68; PMID:19655378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gely-Pernot A, Raverdeau M, Célébi C, Dennefeld C, Feret B, Klopfenstein M, Yoshida S, Ghyselinck NB, Mark M. Spermatogonia differentiation requires retinoic acid receptor γ. Endocrinology 2012; 153:438-49; PMID:22045663; http://dx.doi.org/10.1210/en.2011-1102 [DOI] [PubMed] [Google Scholar]
- 42.Raverdeau M, Gely-Pernot A, Féret B, Dennefeld C, Benoit G, Davidson I, Chambon P, Mark M, Ghyselinck NB. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc Natl Acad Sci 2012; 109:16582-7; PMID:23012458; http://dx.doi.org/10.1073/pnas.1214936109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 2008; 15:285-97; PMID:18694567; http://dx.doi.org/10.1016/j.devcel.2008.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Wang M, Jiang S, Lu Y, Tao D, Yang Y, Ma Y, Zhang S. Demethylation of CpG islands in the 5' upstream regions mediates the expression of the human testis-specific gene MAGEB16 and its mouse homolog Mageb16. BMB Rep 2014; 47:86-91; PMID:24219866; http://dx.doi.org/10.5483/BMBRep.2014.47.2.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagarkatti-Gude DR, Jaimez R, Henderson SC, Teves ME, Zhang Z, Strauss JF 3rd. Spag16, an axonemal central apparatus gene, encodes a male germ cell nuclear speckle protein that regulates SPAG16 mRNA expression. PLos One 2011; 6:e20625; PMID:21655194; http://dx.doi.org/10.1371/journal.pone.0020625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura H, Cook RN, Justice MJ. Mouse Tenm4 is required for mesoderm induction. BMC Dev Biol 2013; 13:9; PMID:23521771; http://dx.doi.org/10.1186/1471-213X-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang W, Crossman DK, Mitchell EH, Sohn P, Crowley MR, Serra R. WNT 5A inhibits metastasis and alters splicing of Cd44 in breast cancer cells. PLos One 2013; 8:e58329; PMID:23484019; http://dx.doi.org/10.1371/journal.pone.0058329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012; 485:195-200; PMID:22575959; http://dx.doi.org/10.1038/nature11019 [DOI] [PubMed] [Google Scholar]
- 49.Klapholz-Brown Z, Walmsley GG, Nusse YM, Nusse R, Brown PO. Transcriptional program induced by Wnt protein in human fibroblasts suggests mechanisms for cell cooperativity in defining tissue microenvironments. PLos One 2007; 2:e945; PMID:17895986; http://dx.doi.org/10.1371/journal.pone.0000945 [Google Scholar]
- 50.Yatim A, Benne C, Sobhian B,Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell 2012; 48:445-58; PMID:23022380; http://dx.doi.org/10.1016/j.molcel.2012.08.022 [Google Scholar]
- 51.Han JD, Baker NE, Rubin CS. Molecular characterization of a novel A kinase anchor protein from Drosophila melanogaster. J Biol Chem 1997; 272:26611-9; PMID:9334242 [DOI] [PubMed] [Google Scholar]
- 52.De Souza N, Vallier LG, Fares H, Greenwald I. SEL-2, the C.elegans neurobleachin/LRBA homolg, is a negative regulator of lin-12/Notch activity and affects endosomal traffic in polarized epithelial cells. Development 2007; 134:691-702; PMID:17215302; http://dx.doi.org/10.1242/dev.02767 [DOI] [PubMed] [Google Scholar]
- 53.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 2006; 20:2223-37; PMID:16912274; http://dx.doi.org/10.1101/gad.380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol 2011; 3:a003756; PMID:20573714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buske FA, Mattick JS, Bailey TL. Potential in vivo roles of nucleic acid triple-helices. RNA Biol 2011; 8:427-39; PMID:21525785; http://dx.doi.org/10.4161/rna.8.3.14999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007; 445:666-70; PMID:17237763; http://dx.doi.org/10.1038/nature05519 [DOI] [PubMed] [Google Scholar]
- 57.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011; 43:621-9; PMID:21642992; http://dx.doi.org/10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guil S, Gattoni R, Carrascal M, Abián J, Stévenin J, Bach-Elias M. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol 2003; 23:2927-41; PMID:12665590; http://dx.doi.org/10.1128/MCB.23.8.2927-2941.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013; 341:789-92; PMID:23907535; http://dx.doi.org/10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329:689-93; PMID:20616235; http://dx.doi.org/10.1126/science.1192002 [Google Scholar]
- 61.Das C, Hizume K, Batta K, Kumar BR, Gadad SS, Ganguly S, Lorain S, Verreault A, Sadhale PP, Takeyasu K, et al. Transcriptional coactivator PC4, a chromatin associated protein, induces chromatin condensation. Mol Cell Biol 2006; 26:8303-15; PMID:16982701; http://dx.doi.org/10.1128/MCB.00887-06 [Google Scholar]
- 62.Conesa C. and Acker J. Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA Biol 2010; 7:287-90; PMID:20305379; http://dx.doi.org/10.4161/rna.7.3.11491 [DOI] [PubMed] [Google Scholar]
- 63.Liao M, Zhang Y, Kang JH, Dufau ML. Coactivator function of positive cofactor 4 (PC4) in Sp1-directed luteinizing hormone receptor (LHR) gene transcription. J Biol Chem 2011; 286:7681-91; PMID:21193408; http://dx.doi.org/10.1074/jbc.M110.188532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ronfani L, Ferraguti M, Croci L, Ovitt CE, Schöler HR, Consalez GG, Bianchi ME. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 2001; 128:1265-73; PMID:11262228 [Google Scholar]
- 65.Itou J, Taniguchi N, Oishi I, Kawakami H, Lotz M, Kawakami Y. HMGB factors are required for posterior digit development through integrating signaling pathway activities. Dev Dyn 2011; 240:1151-62; PMID:21384471; http://dx.doi.org/10.1002/dvdy.22598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci 2005; 102:2808-13; PMID:15710904; http://dx.doi.org/10.1073/pnas.0406060102 [Google Scholar]
- 67.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 2004; 32(Database issue):D493-6; PMID:14681465; http://dx.doi.org/10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 2011; 27:1653-59; PMID:21543442; http://dx.doi.org/10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol 2007; 8(2):R24; PMID:17324271; http://dx.doi.org/10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas-Chollier M, Defrance M, Median-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res 2011; 39(webserver issue):W86-91; PMID:21715389; http://dx.doi.org/10.1093/nar/gkr377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlcok G. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004; 20:3710-15; PMID:15297299; http://dx.doi.org/10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011; 6:e21800; PMID:21789182; http://dx.doi.org/10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.