Key Points
Combination of ISS and the EMC92 gene classifier is a novel clinically applicable risk classification for survival in multiple myeloma.
ISS has clear independent additive prognostic value in combination with GEP classifiers or FISH markers.
Abstract
Patients with multiple myeloma have variable survival and require reliable prognostic and predictive scoring systems. Currently, clinical and biological risk markers are used independently. Here, International Staging System (ISS), fluorescence in situ hybridization (FISH) markers, and gene expression (GEP) classifiers were combined to identify novel risk classifications in a discovery/validation setting. We used the datasets of the Dutch-Belgium Hemato-Oncology Group and German-speaking Myeloma Multicenter Group (HO65/GMMG-HD4), University of Arkansas for Medical Sciences-TT2 (UAMS-TT2), UAMS-TT3, Medical Research Council-IX, Assessment of Proteasome Inhibition for Extending Remissions, and Intergroupe Francophone du Myelome (IFM-G) (total number of patients: 4750). Twenty risk markers were evaluated, including t(4;14) and deletion of 17p (FISH), EMC92, and UAMS70 (GEP classifiers), and ISS. The novel risk classifications demonstrated that ISS is a valuable partner to GEP classifiers and FISH. Ranking all novel and existing risk classifications showed that the EMC92-ISS combination is the strongest predictor for overall survival, resulting in a 4-group risk classification. The median survival was 24 months for the highest risk group, 47 and 61 months for the intermediate risk groups, and the median was not reached after 96 months for the lowest risk group. The EMC92-ISS classification is a novel prognostic tool, based on biological and clinical parameters, which is superior to current markers and offers a robust, clinically relevant 4-group model.
Introduction
In multiple myeloma (MM) patients, malignant plasma cells accumulate in the bone marrow, leading to a wide range of clinical symptoms, which include bone disease, hypercalcemia, renal impairment, and anemia.1 The prognosis is variable, with survival for newly diagnosed patients ranging from less than 2 to more than 20 years.2 Adequate prognostication of disease outcome is important in order to make treatment choices and to allocate high-risk patients to alternative treatment options. Clinical trials that address specific treatment of high-risk patients include Total Therapy 4 (NCT00734877), Total Therapy 5 (NCT02128230), and Myeloma United Kingdom 9 (Myeloma UK Clinical Trial Network).
Heterogeneous treatment outcome can in part be explained by different biological subgroups in MM, which are characterized by primary translocations involving genes such as MMSET (t(4;14)), and c-MAF (t(14;16)).3,4 These subgroups can be identified using gene expression profiling.5,6 In addition, gene expression profiling has been used to establish classifiers for prognostication. EMC92 is a robust risk marker for the identification of high-risk MM and was validated in independent clinical trials showing a solid and independent performance in comparison with other MM gene expression profile (GEP) classifiers such as University of Arkansas for Medical Sciences 70 (UAMS70).7-13 Clinical prognostic systems for MM are primarily based on β2-microglobulin (β2M), albumin, lactate dehydrogenase, C-reactive protein, calcium, and creatinine.14,15 The International Staging System (ISS) is based on β2M and albumin, with stage I representing limited disease, stage II intermediate, and stage III the most unfavorable disease.16 Today, it is used as the standard clinical risk classification for MM.
Fluorescence in situ hybridization (FISH)-based cytogenetics and gene expression profiling are biology-based prognostic markers.17 ISS was combined with high-risk cytogenetic markers t(4;14) and deletion of 17p (del17p) to establish novel prognostic risk classifications as proposed by Neben18 and Avet-Loiseau.19 Recently, serum lactate dehydrogenase was added as a component to this marker combination.20 Other prognostic systems include combinations of cytogenetic markers, such as the combination of del17p, translocation t(4;14), and gain of 1q (gain1q).21
The goal of this study was to evaluate all published risk markers used in MM and to compare combinations of FISH, ISS, and GEP based prognostic systems. By applying a study design with independent discovery and validation sets, we demonstrated that ISS can be combined with gene expression signatures into powerful classifiers for MM.
Methods
Clinical data
The clinical data from the Dutch-Belgium Hemato-Oncology Group (HOVON) and German-speaking Myeloma Multicenter Group (GMMG) (HO65/HD4), Medical Research Council-IX (MRC-IX), UAMS Total Therapy (UAMS-TT2 and TT3), Intergroupe Francophone du Myelome (IFM-G; all newly diagnosed patients) and Assessment of Proteasome Inhibition for Extending Remissions (APEX; relapse patients) trials were used.7-9,19,22,23 The IFM-G cohort is a clinical database of patients not separately published and was included in the ISS development.16 Treatment regimens of the trials from which these datasets were derived are summarized in Table 1. Overall survival (OS) or progression-free survival (PFS) and at least 1 prognostic marker were available for all patients (Table 1; see supplemental Figure 1 on the Blood Web site). All patients signed an informed consent in accordance with the Declaration of Helsinki, and all protocols were approved by institutional review boards.
Table 1.
Distribution of risk markers and treatments per dataset
| HO65/HD4 | MRC-IX | TT2 | TT3 | APEX | IFM-G | Pooled | ||
|---|---|---|---|---|---|---|---|---|
| Intensive | Nonintensive | |||||||
| N | 827 | 701 | 491 | 351 | 238 | 264 | 1878 | 4750* |
| Age, median (IQR), y | 57 (51-61) | 58 (54-63) | 74 (70-77) | 57 (49-64) | 60 (53-66) | 61 (54-67) | 57 (51-61) | 57 (51-62) |
| Treatment, n | PAD (413) | CTD (351) | CTDa (257) | VTD (175) | VTD (238) | BOR (188) | VD (740) | BOR (1579)/ THAL (783) |
| Control, n | VAD (414) | CVAD (350) | MP (234) | VMD (176) | No controls | DEX (76) | VAD (1138) | BOR (1628)/ THAL (760) |
| High-dose alkylator | Yes | Yes | No | Yes | Yes | Yes | Yes | |
| EMC92, n (% positive) | † | 138 (17) | 109 (24) | 345 (19) | 238 (15) | 264 (16) | 1094 (18) | |
| UAMS17, n (% positive) | 327 (12) | 138 (9) | 109 (16) | † | 238 (14) | 264 (12) | 1076 (12) | |
| UAMS70, n (% positive) | 327 (9) | 138 (7) | 109 (10) | † | 238 (12) | 264 (8) | 1076 (9) | |
| UAMS80, n (% positive) | 327 (8) | 138 (8) | 109 (9) | 345 (9) | † | 264 (7) | 1183 (8) | |
| MRCIX6, n (% positive) | 327 (5) | † | † | 345 (7) | 238 (5) | 264 (3) | 1174 (5) | |
| IFM15, n (% positive) | 327 (25) | 138 (25) | 109 (28) | 345 (24) | 238 (24) | 1157 (25) | ||
| HM19, n (low/medium/high %) | 327 (43/49/8) | 138 (45/48/7) | 109 (39/53/8) | 345 (50/47/8) | 238 (47/47/7) | 264 (41/50/8) | 1420 (44/48/8) | |
| GPI50, n (low/medium/high %) | 327 (34/51/15) | 138 (52/41/7) | 109 (52/38/10) | 345 (63/31/7) | 238 (58/34/8) | 1159 (51/39/10) | ||
| ISS, n (1/2/3%) | 756 (38/37/25) | 636 (25/39/36) | 449 (13/41/45) | 351 (54/25/21) | 208 (50/28 21) | 202 (34/33/33) | 1475 (34/39/28) | 4074 (34/37/30) |
| t(4;14), n (% positive) | 492 (12) | 619 (12) | 434 (10) | 1635 (14) | 3180 (13) | |||
| t(11;14), n (% positive) | 437 (16) | 617 (15) | 434 (12) | 1488 (15) | ||||
| t(14;16), n (% positive) | 360 (2) | 612 (3) | 434 (3) | 456 (4) | 1862 (3) | |||
| t(14;20), n (% positive) | 255 (0) | 612 (2) | 429 (1) | 1296 (1) | ||||
| IgH split, n (% positive) | 372 (48) | 609 (44) | 429 (40) | 1410 (44) | ||||
| gain1q, n (% positive) | 344 (32) | 531 (37) | 371 (41) | 248 (47) | 891 (37) | 2385 (38) | ||
| del13q, n (% positive) | 686 (41) | 612 (46) | 428 (43) | 1807 (48) | 3533 (46) | |||
| del17p, n (% positive) | 351 (11) | 591 (8) | 423 (9) | 1651 (15) | 3016 (12) | |||
| gain9, n (% positive) | 454 (57) | 480 (60) | 351 (66) | 1285 (60) | ||||
| HR.FISH.A, n (% positive) | 354 (46) | 535 (48) | 368 (48) | 116 (100)‡ | 1022 (64) | 2395 (57) | ||
| HR.FISH.B/ISS, n (low/medium/high %) | 334 (60/22/18) | † | † | 516 (55/29/17)† | 850 (57/26/17) | |||
The numbers of patients per data set are given, with the number or percentage of positive patients according to the markers’ risk classification.
Intersection of patients with available data between datasets is shown in supplemental Figure 1.
Training set for these markers. Only the proportion and number that are not used for building the marker, if any, are shown.
The HR.FISH.A compound risk classification is based on a patient having either del17p, t(4;14), or gain of 1q. If only gain of 1q is known (in TT2 patients), these are the only patients classified with certainty as high-risk. The remaining patients cannot be classified because the status of t(4;14) and del17p are unknown. If the missing bias is strong enough (see Methods), that marker is excluded from the combination analyses.
BOR, bortezomib; CTD, cyclophosphamide, thalidomide, dexamethasone; CTDa, attenuated CTD; CVAD, cyclophosphamide, vincristine, doxorubicin, dexamethasone; IgH, immunoglobulin H; IQR, interquartile range; MP, melphalan, prednisone; THAL, thalidomide; PAD, bortezomib, doxorubicin, dexamethasone; VAD, vincristine, doxorubicin, dexamethasone; VD, vincristine, dexamethasone; VMD, bortezomib, melphalan, dexamethasone; VTD, bortezomib, thalidomide, dexamethasone.
GEP
All GEP data are Affymetrix HG U133 Plus 2.0 platform-based, except for the APEX study (Affymetrix U133 A/B platform). HO65/HD4 GEP was performed in our laboratory, as described previously (n = 327; Gene Expression Omnibus (GEO) series GSE19784).6,7,21 Other GEP sets were: TT2 (n = 345; GSE24080),8 TT3 (n = 238; ArrayExpress and GEO accession numbers E-TABM-1138 and GSE24080),24 MRC-IX (n = 247; GSE15695),22 and APEX (n = 264; GSE9782).23 Because of unavailable survival data, the Heidelberg-Montpellier (HM) dataset (n = 206; E-MTAB-362), was used only to determine the probe set means and variances for the training set of the HM19 classifier.12
Standard prognostic markers
Availability of risk markers and patients per dataset is shown in Table 1 and supplemental Figure 1. The ISS was determined by combining serum levels of β2M and albumin.16 Cytogenetics by FISH was used with a 10% cutoff level except for a 20% cutoff used for numerical abnormalities in the MRC-IX trial.19,25-27 Gain of chromosome 9 (gain9), 1 of the hyperdiploid chromosomes and most frequently available marker for this purpose, was used as a proxy for hyperdiploidy.28 FISH probes used in MRC-IX and HO65/HD4 were described previously.25,29 Cytogenetic data obtained by methods other than FISH were excluded. High-risk FISH was defined as having either del17p or t(4;14) or gain1q, denoted here as HR.FISH.A.21 The risk classification described by Avet-Loiseau et al19 is denoted here as HR.FISH.B/ISS. This risk classification distinguishes grade I (ISS = 1 or 2 with FISH markers t(4;14) and del17p both negative), grade 2 (not grades 1 or 3), and grade 3 (ISS = 2 or 3 with FISH markers t(4;14) or del17p-positive). In case of an arbitrary situation because of missing data for 1 of the markers, the observation was excluded.
Gene expression classifiers
The following MM gene expression classifiers were used: EMC92,7 UAMS17,8 UAMS70,8 UAMS80,9 IFM15,10 and MRCIX613 (all 2-risk group classifiers) and HM1912 and GPI5011 (both 3-risk group classifiers). Normalization and cutoffs were calculated as described previously (see supplemental Methods for a brief description).
Statistical analyses
In Figure 1, a flowchart of the analyses is given. The association of risk markers with survival was assessed using a Cox survival model (R ‘survival’ package, version 2.38-1).30-32 To account for heterogeneous survival between studies, models were stratified per trial cohort.
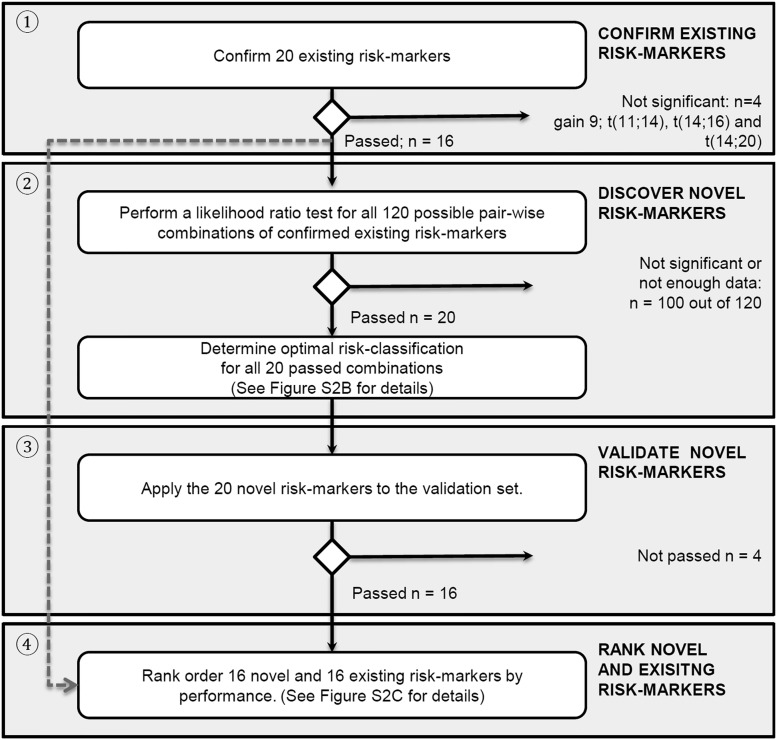
Figure 1.
Flowchart of analyses. The analyses are organized as follows: (1) confirmation of existing risk markers, (2) systematically finding novel risk markers with improved prognostic strength by combining existing risk markers, (3) validating them, and (4) ranking of confirmed existing and validated novel risk markers. See supplemental Figure 2A-C for more details.
The trial cohorts were HO65/HD4, MRC-IX intensive, MRC-IX nonintensive, UAMS-TT2, UAMS-TT3, IFM-G, and APEX. Datasets used for generating risk markers were systematically excluded in validation analyses to avoid training bias. For instance, HO65/HD4 patients were excluded in analyses involving the EMC92 classifier (Table 1).
The method for finding novel combination markers (compound markers) is illustrated in supplemental Figure 2B and extensively described in the supplemental Methods. Briefly, because missing data may confound the analyses, combinations with increased risk for confounding were excluded (supplemental Table 1; supplemental Methods). Subsequently, the data were randomly split into a discovery and validation set. The discovery set was used for finding meaningful combinations of markers as well as the most optimal way to split patients into subgroups using these combinations. Stringent validation was performed in the designated validation set to confirm their prognostic strength. Finally, all new combinations and existing markers were ranked, with a low rank score indicating a high-performing risk marker.
Results
Confirmation of existing risk markers
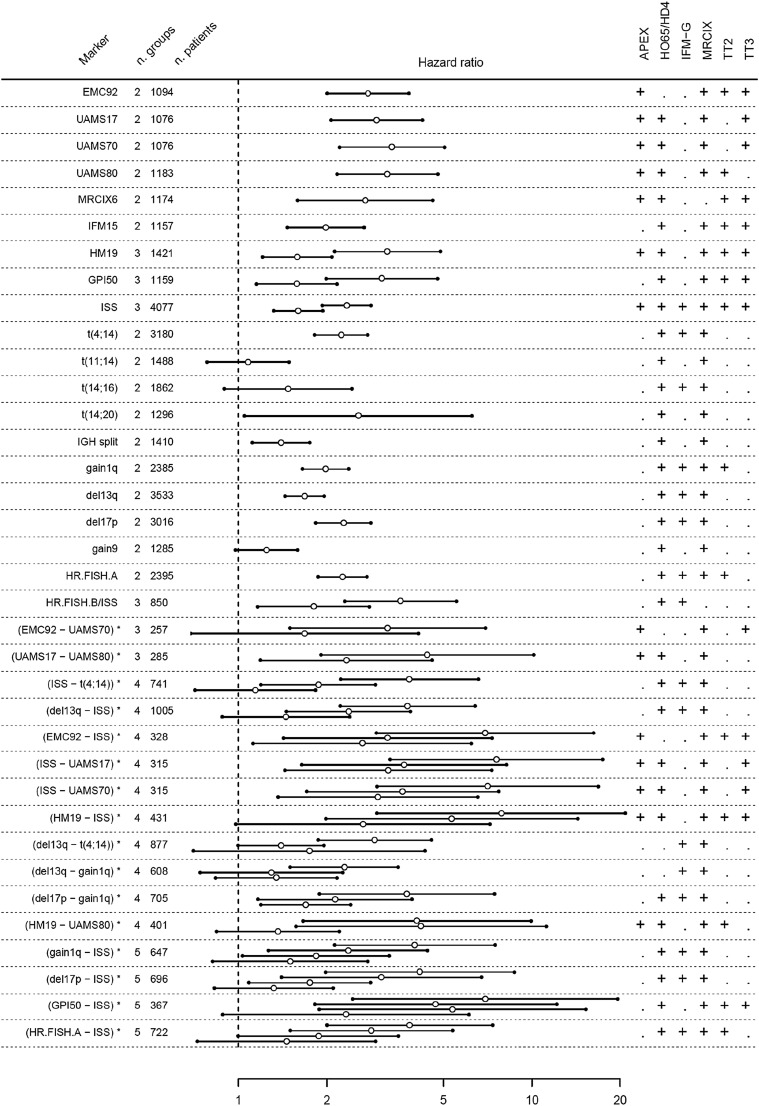
The value of 20 existing risk markers was evaluated in a data set of 4750 patients. The markers and used cohorts are given in Table 1. The prognostic value was evaluated correcting for the differences in survival between cohorts (Figure 2; supplemental Figures 3-5; supplemental Table 2). For all markers, at least 2 cohorts were available. All GEP classifiers demonstrated a highly significant performance for OS. Hazard ratios for GEP classifiers ranged from 2.0 (95% confidence interval [CI], 1.6-2.4; IFM15) up to 3.3 (2.6-4.3; UAMS70). Furthermore, hazard ratios for GEP classifiers were consistently higher than any of the other risk markers, including all FISH markers and ISS. This suggests better risk separation for GEP classifiers compared with FISH markers. GEP classifiers generally performed better for OS than for PFS (supplemental Figures 3A-B, 4, and 5; Table 2) with PFS HRs between 1.8 (95% CI, 1.5-2.1; IFM15) and 2.3 (95% CI, 1.9-2.7; EMC92). The percentage of high-risk patients varied between classifiers: 18% (EMC92), 12% (UAMS17), 10% (GPI50), 9% (UAMS70), and 8% (UAMS80 and HM19; Table 1).
Figure 2.
Risk markers in relation to overall survival. Both existing markers and validated novel combinations are shown. For novel combinations, the results shown represent the validation. For confirmation of existing markers, no discovery/validation split is required and results shown are based on all available data. In the left panel, existing markers and novel combinations (denoted by an asterisk) are listed. For each marker, the number of risk groups (n. groups) and number of available patients are given (number [n.] of patients). Markers are sorted by the number of risk groups. In the center panel, the hazard ratios are shown (open circles), with Bonferroni adjusted 95% CIs (indicated by 2 lines and closed circles). For coherent notation, hazard ratios are expressed relative to the lowest risk group. Every additional risk group results in an extra hazard ratio. For instance, for the novel combination EMC92-ISS, 4 risk groups result in 3 hazard ratios, as indicated in the text and supplemental Table 2A (intermediate-low risk relative to low risk: hazard ratio, 2.6; 95% CI, 1.6-4.5); intermediate-high risk relative to low risk: hazard ratio, 3.2 (95% CI, 1.9-5.4); and high risk relative to low risk: hazard ratio, 6.9 [95% CI, 4.1-12]). In the right panel, a plus sign indicates whether a data set could be used for the analysis of a specific marker or combination (for details of available data, see Table 1 and supplemental Figure 1). For the EMC92-ISS combination, the following datasets could be used: APEX, MRC-IX, TT2, and TT3.
FISH markers with prognostic strength can be distinguished from markers with no or disputable value. For OS, markers t(4;14), del17p, gain1q, and del13q performed well, with hazard ratios ranging between 1.7 (95% CI, 1.5-1.8; del13q) and 2.3 (95% CI, 2.0-2.6; del17p). The markers gain9, t(11;14), t(14;16), and t(14;20) were clearly not significant or had high variance from a lack of predictive value or a small number of positive cases. These markers were excluded from further analyses. A similar pattern was found for PFS, but the strength of the markers was generally lower with PFS hazard ratios ranging from 1.4 (95% CI, 1.3-1.5; del13q) to 1.8 (95% CI, 1.6-2.0; t(4;14)).
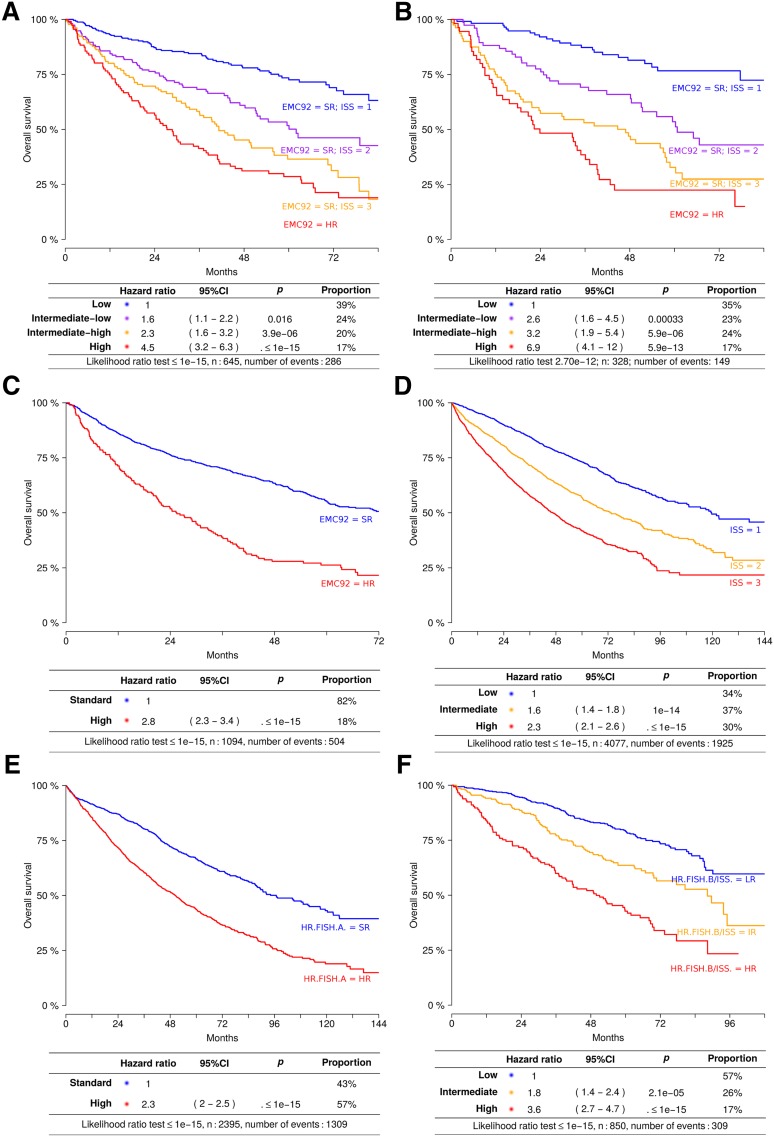
ISS was confirmed as a valuable and highly significant prognostic marker. Hazard ratios of 1.6 (95% CI, 1.4-1.8; ISS = 2) and 2.3 (95% CI, 2.1-2.6; ISS = 3) were found for OS and 1.4 (95% CI, 1.3-1.6; ISS = 2) and 1.7 (95% CI, 1.6-1.9; ISS = 3) for PFS.
Other previously published compound risk markers, denoted here as HR.FISH.A21 (either t(4;14) or del17p or gain1q) and a combined FISH/ISS marker (HR.FISH.B/ISS19) showed good performance. The hazard ratio was 2.3 (95% CI, 2.0-2. 5; HR.FISH.A). For the 3 group HR.FISH.B/ISS risk classifications, hazard ratios of 1.8 (95% CI, 1.4-2.4; intermediate risk) and 3.6 (95% CI, 2.7-4.7; high risk) were found.
To correct for heterogeneity between studies, all analyses were corrected for the survival differences between trials as a result of differences in treatment, disease stage, and patient populations. To evaluate the effect of this correction, all analyses were repeated per cohort and highly similar results were obtained, suggesting that these risk markers perform similarly across different cohorts (supplemental Results).
Pairwise combinations of risk markers
The next analysis was performed to explore combinations of risk markers. As indicated previously, 16 of 20 evaluated markers had significant associations with OS and/or PFS. Based on these 16, all possible pairwise combinations were generated. Twenty combinations were significant in the discovery set, of which 16 remained significant in the independent validation set (Figure 2; supplemental Figure 8A-B; supplemental Tables 2 and 3). In 10 of 16 combinations, ISS was combined with either GEP classifiers (n = 5) or FISH markers (n = 5), illustrating the strong additive power of ISS to these markers. Combinations of GEP (n = 3) and FISH markers were observed (n = 3), but not combinations of FISH with GEP. Two combinations divided patients into 3 groups, 10 into 4 groups, and 4 into 5 groups.
Ranking of existing and novel markers
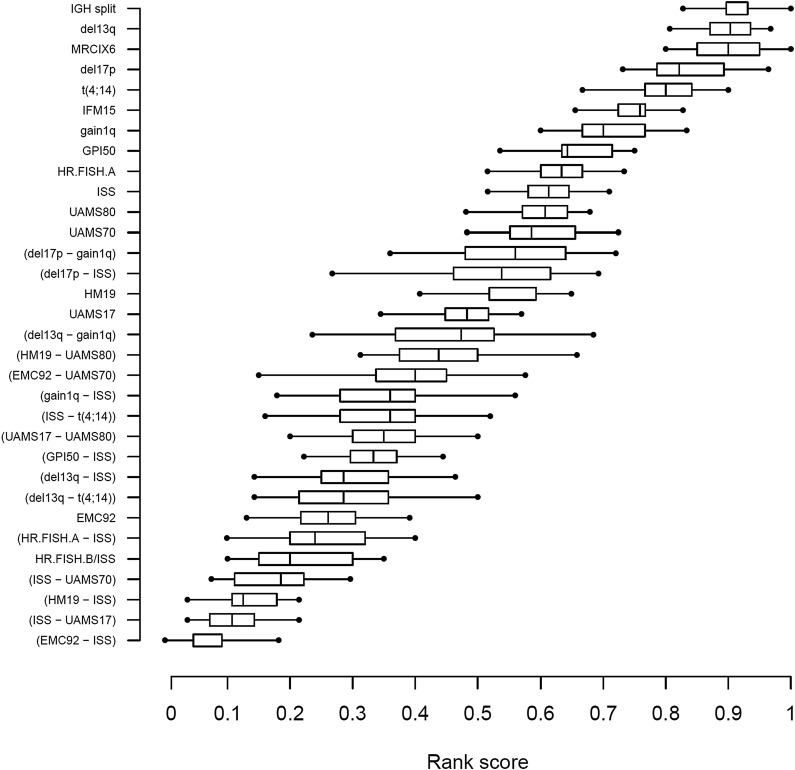
The markers described previously (ie, 16 existing plus 16 validated new risk markers) were ranked on the basis of performance, as described in the supplemental Methods.
ISS-GEP combinations consistently ranked at the top, with the EMC92-ISS compound risk marker having the best median rank score (RS) (Figure 3; RS = 0.05). Other high-scoring markers included ISS-UAMS17 (RS = 0.11), ISS-HM19 (RS = 0.13), and ISS-UAMS70 (RS = 0.19). The HR.FISH.B/ISS compound marker ranked in fifth place (RS = 0.20) and ISS ranked in 23rd place (of 32; RS = 0.61). In general, compound markers tended to score better than single markers. The best single marker was EMC92 in seventh position (RS = 0.26).
Figure 3.
Ranking of confirmed existing risk markers and validated novel risk markers in relation to overall survival on the validation data. The markers are vertically ordered by rank score, which reflects the observed proportion of risk markers with a better performance. Each box shows the interquartile range of the rank score per marker.
EMC92-ISS classifies patients into 4 groups with proportions of 38%, 24%, 22%, and 17% for the lowest to the highest risk group, respectively (Figure 4A-B). The hazard ratios relative to the lowest risk group were 2.6 (95% CI, 1.6-4.5; intermediate-low), 3.2 (95% CI, 1.9-5.4; intermediate-high), and 6.9 (95% CI, 4.1-11.7; high). Median survival times were 24 (high), 47 (intermediate-high), and 61 months (intermediate-low) for the 3 highest risk groups, with median survival not reached after 96 months for the lowest risk group. To gain insight into the performance of this marker over time, we determined the proportions of surviving patients in each risk group and analyzed the EMC92-ISS at different time points. This marker is clearly applicable to younger as well as older and relapsed patients and holds its value during follow-up (Table 2; supplemental Figure 10).
Figure 4.
Survival analysis of EMC92-ISS, FISH, and ISS. Kaplan-Meier plots and Cox regression model data are given. Kaplan-Meier plots are not stratified; Cox regression results are stratified (ie, corrected for differences in survival in different cohorts). (A) EMC92-ISS in the discovery set, (B) EMC92-ISS in the validation set, (C) EMC92-ISS in all data, (D) ISS in all data, (E) HR.FISH.A in all data, and (F) HR.FISH.B/ISS in all data. In order of increasing risk: low (blue); intermediate-low (purple); intermediate-high (orange); high (red). HR, high-risk; SR, standard-risk. Below the Kaplan-Meier curves, results of the stratified Cox model are found. Hazard, hazard ratio relative to the lowest risk group; P, P value relative to the lowest risk group; % positive, percentage of patients within the specified risk group. The bottom line shows the result of the likelihood ratio goodness-of-fit test.
Table 2.
Proportion of surviving patients at multiple time points per EMC92-ISS risk group in a Kaplan-Meier analysis of the validation data (from top to bottom: 6, 12, 24, and 72 mo, respectively)
| Pooled | <65 y | ≥65 y | Relapse | |
|---|---|---|---|---|
| 6 mo | ||||
| Low risk (%) | 98 | 97 | 96 | 95 |
| Intermediate-low risk (%) | 96 | 95 | 95 | 85 |
| Intermediate-high risk (%) | 86 | 93 | 77 | 79 |
| High risk (%) | 84 | 88 | 75 | 57 |
| Total survival (%) | 92 | 94 | 87 | 83 |
| 12 mo | ||||
| Low risk (%) | 97 | 97 | 96 | 89 |
| Intermediate-low risk (%) | 87 | 93 | 91 | 54 |
| Intermediate-high risk (%) | 74 | 93 | 73 | 42 |
| High risk (%) | 67 | 72 | 56 | 57 |
| Total survival (%) | 84 | 91 | 81 | 60 |
| 24 mo | ||||
| Low risk (%) | 92 | 97 | 92 | 55 |
| Intermediate-low risk (%) | 76 | 88 | 73 | 23 |
| Intermediate-high risk (%) | 57 | 77 | 58 | 24 |
| High risk (%) | 46 | 56 | 31 | 0 |
| Total survival (%) | 72 | 84 | 67 | 30 |
| 72 mo | ||||
| Low risk (%) | 77 | 86 | 69 | — |
| Intermediate-low risk (%) | 43 | 59 | 32 | — |
| Intermediate-high risk (%) | 27 | 39 | 28 | — |
| High risk (%) | 22 | 33 | 0 | — |
| Total survival (%) | 48 | 62 | 36 |
In the left column, patient groups are pooled (n = 328). Subsequent columns show percentages for newly diagnosed patients younger than 65 y (n = 174), newly diagnosed older than 65 y (n = 90), and relapsed patients (n = 64), respectively. The 72-mo time point is not available for the last item.
The composition of the 4 groups in terms of ISS, EMC92, and FISH markers is shown in Table 3. Interestingly, within the EMC92-ISS lowest risk group, 75% of patients—with truly favorable prognosis (supplemental Table 4)—were positive for either t(4;14), del(17p), or gain1q. In the other risk categories 32%, 42%, and 86% of patients were positive (intermediate-low, intermediate-high, and high risk, respectively), indicating that EMC92-ISS and FISH only partly represent overlapping patient sets.
Table 3.
Distribution of markers in each of the 4 EMC92-ISS–based risk groups
| EMC92 | ISS | del17p | del13q | 1q gain | HR.FISH.A | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EMC92-ISS | HR | n | 1 | 2 | 3 | n | Positive | n | Positive | n | Positive | n | HR | n |
| Low | 0% | 365 | 100% | 0% | 0% | 365 | 8% | 39 | 44% | 39 | 34% | 154 | 75% | 76 |
| Intermediate-low | 0% | 231 | 0% | 100% | 0% | 231 | 5% | 60 | 37% | 60 | 34% | 92 | 44% | 70 |
| Intermediate-high | 0% | 211 | 0% | 0% | 100% | 211 | 8% | 66 | 44% | 66 | 41% | 101 | 55% | 84 |
| High | 100% | 166 | 30% | 32% | 39% | 166 | 16% | 38 | 74% | 39 | 76% | 90 | 93% | 76 |
The numbers in the data for which the EMC92-ISS risk classification could be determined. For the classifications based on del13q, 1q gain, and HR.FISH.A, a clear correlation was found to the EMC92-ISS classifications. For instance, 93% of EMC92-ISS high-risk patients are positive for HR.FISH.A compared with 44 to 55% of the intermediate group and 75% of the low group.
HR, the percentage of patients indicated as high-risk according to the specified marker; n, number of patients in the EMC92-ISS–based risk group for which the specified marker was available; positive, the percentage of patients positive for the specified marker.
Biological relevance of GEP classifiers
Genes within GEP classifiers are selected based on association with survival, rather than a direct link to biology. Still, a gene ontology enrichment analysis33 can highlight biological processes important for a poor outcome (supplemental Table 5A-H). All GEP classifiers had enrichment of cell cycle–related genes. When all probe sets in all classifiers were pooled, 191 biological processes were found to be enriched (false discovery rate <0.05). Top processes included “nuclear division,” “mitosis,” and “cell division,” which share the genes BIRC5, BUB1, and UBE2C. Other prominent processes included “DNA metabolic process,” “DNA packaging,” and “DNA replication” (with genes such as TOP2A and MCM2).
Discussion
Important prognostic markers in MM are based on ISS, FISH markers, and GEP classifiers.7-13,16,17 Previously, we showed that combining various GEP classifiers resulted in a stronger prediction of the high-risk population.7 Here, we systematically evaluated additional, new combinations of prognostic markers. We limited the search for new compound risk markers to pairwise combinations of existing markers. This choice is mainly driven by the lack of complete data sets that contain all risk markers (as shown in supplemental Figure 1), which hinders the analyses of more complex risk models. The number of patients positive for specific markers was remarkably stable between cohorts, irrespective of the type of marker. This adds strength to the belief that these markers—and thus decisions based on them—can be reliably replicated.
Three findings are of particular interest: first, ISS has a clear and independent value in combination with either GEP classifiers or FISH markers. GEP classifiers combined with ISS are the strongest risk classifications found here. By combining the EMC92 gene classifier with ISS, patients are effectively stratified into 4 risk groups, including a distinctive low-risk group of 38% and a high-risk group of 17%. This strong additive strength of ISS to GEP has been recognized before in a previous smaller study.34 Also, ISS was integrated with GEP and other factors, but this risk score did not take into account correlations between markers and was generated without using a solid discovery/validation design.35 In contrast, we have opted for a study design in which part of the data was reserved for validation.
Second, our study confirmed that FISH markers can be divided into those consistently associated with shorter OS as opposed to inconsistent markers. Consistent FISH markers included t(4;14), gain1q, del17p, and del13q. Combinations of any of these markers with ISS constituted solid prognostic predictors. As reported previously, t(4;14) and del17p are currently regarded as the most important high-risk FISH markers.17 Third, by combining these FISH markers into the previously defined risk classifications HR.FISH.A and HR.FISH.B/ISS, a major improvement of prognostic strength is achieved. Interestingly, patients classified as high risk according to the HR.FISH.A marker, but that actually had favorable survival, were correctly identified as low-risk patients by the EMC92-ISS compound marker.
In addition to validating EMC92-ISS, we have now also validated the HR.FISH.B/ISS risk classification for the first time in independent data by excluding training data from the analyses. Combining FISH and ISS is thus a valid choice for routine clinical practice, including the existing HR-FISH.B/ISS, as proposed by Avet-Loiseau et al.19 Incorporating lactate dehydrogenase and bone imaging was outside the scope of this study because these markers were not consistently available.20
Combining GEP with ISS may become an attractive option for prognostication. The EMC92-ISS classification is independent from therapy choice: EMC92 was shown to function in bortezomib clinical trials as well as in thalidomide and more conventional regimens.7 In contrast, bortezomib and other novel agents may abrogate the unfavorable impact of some FISH markers on PFS.29 EMC92-ISS is useful because it can identify both high- and low-risk MM. This is an advantage over FISH markers, which only seem to identify high-risk patients. Moreover, the technical applicability of GEP and its costs are thought to be comparable to FISH.36
The agreement between GEP classifiers in terms of pathways is of interest. Although the primary force for classifier discovery is association with survival, the genes within classifiers appear to converge on the cell-cycle pathways. Indeed, proliferative capacity, assessed as the plasma cell labeling index or by Ki-67 staining, has long been recognized to be an important prognostic factor.37,38
The clinical applicability of stratification into 4 risk groups will be increasingly relevant in the era of novel treatment modalities being available. First, increased accuracy of prognosis can improve patient counseling.17 Second, and more important, risk stratification may lead to adaptation of treatment according to risk status. This composite risk marker opens the way to better risk stratification in clinical trials and explore novel drugs in different risk groups.39,40 This could effectively be a first step toward a more individual treatment, using patient-specific markers as a directional key.
Based on the current study, we conclude that the combination of EMC92 with ISS is a strong disease-based prognosticator for survival in MM. This risk classification is a good candidate to stratify patients for treatment options in a clinical trial.
Acknowledgments
The authors acknowledge Mathijs Sanders for his contribution.
This work was supported by the Center for Translational Molecular Medicine (BioCHIP), a clinical research grant from the European Hematology Association, an Early-Mid Career Researchers Translational Research Grant, a Federal Ministry of Education and Research of Germany grant from CLIOMMICS (Clinically-applicable, omics-based assessment of survival, side effects, and targets in multiple myeloma; 01ZX1309A), SkylineDx, Janssen Pharmaceuticals, and a Seventh Framework Programme grant from the Myeloma Stem Cell Network (MSCNET;LSHC-Ct-2006-037602).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.K., M.v.D., M.H.v.V., and P.S. designed the research; A.B. collected data; R.K. and M.v.D. analyzed and interpreted the data and wrote the manuscript; M.H.v.V. and E.H.v.B. critically reviewed the article; B.v.d.H. provided the clinical data of HO65/HD4 and critically reviewed the article; L.e.J. performed central data management for the HO65/HD4; G. Mulligan provided the CEL files from the APEX dataset and critically reviewed the article; G. Morgan and W.M.G. provided CEL files and clinical data from the MRC-IX dataset and critically reviewed the article H.A.-L. provided the IFM-G clinical data and critically reviewed the article; H.G. is principal investigator research for the German part of the HO65/HD4 and critically reviewed the article; H.M.L. organized the trial and critically reviewed the article; and P.S. organized the trial, is the principal investigator of the performed research and HO65/HD4; and critically reviewed the article.
Conflict-of-interest disclosure: R.K., A.B., B.v.d.H., L.e.J., and M.v.D. have no disclosures; M.H.v.V. and E.H.v.B. are employees of SkylineDx; W.M.G. received unrestricted educational grants from Novartis, Schering Health Care Ltd, Chugai, Pharmion, Celgene, and Ortho Biotech; G. Morgan, H.A.L., G. Mulligan, H.G., and H.M.L. are members of advisory boards of pharmaceutical companies; and P.S. served on an advisory board of Skyline DX and received honoraria and research funding from Janssen-Cilag, Celgene, Onyx, and research funding from Millennium.
Correspondence: P. Sonneveld, Erasmus MC Cancer Institute, Department of Hematology, Na-822, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: p.sonneveld@erasmusmc.nl.
References
- 1.Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15(12):e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Gondos A, Pulte D. Recent major improvement in long-term survival of younger patients with multiple myeloma. Blood. 2008;111(5):2521–2526. doi: 10.1182/blood-2007-08-104984. [DOI] [PubMed] [Google Scholar]
- 3.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci USA. 1996;93(24):13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101(11):4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 5.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broyl A, Hose D, Lokhorst H, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116(14):2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 7.Kuiper R, Broyl A, de Knegt Y, et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012;26(11):2406–2413. doi: 10.1038/leu.2012.127. [DOI] [PubMed] [Google Scholar]
- 8.Shaughnessy JD, Jr, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 9.Shaughnessy JD, Jr, Qu P, Usmani S, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118(13):3512–3524. doi: 10.1182/blood-2010-12-328252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decaux O, Lodé L, Magrangeas F, et al. Intergroupe Francophone du Myélome. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myélome. J Clin Oncol. 2008;26(29):4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 11.Hose D, Rème T, Hielscher T, et al. Proliferation is a central independent prognostic factor and target for personalized and risk-adapted treatment in multiple myeloma. Haematologica. 2011;96(1):87–95. doi: 10.3324/haematol.2010.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rème T, Hose D, Theillet C, Klein B. Modeling risk stratification in human cancer. Bioinformatics. 2013;29(9):1149–1157. doi: 10.1093/bioinformatics/btt124. [DOI] [PubMed] [Google Scholar]
- 13.Dickens NJ, Walker BA, Leone PE, et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin Cancer Res. 2010;16(6):1856–1864. doi: 10.1158/1078-0432.CCR-09-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greipp PR, Lust JA, O’Fallon WM, Katzmann JA, Witzig TE, Kyle RA. Plasma cell labeling index and beta 2-microglobulin predict survival independent of thymidine kinase and C-reactive protein in multiple myeloma. Blood. 1993;81(12):3382–3387. [PubMed] [Google Scholar]
- 15.Kyle RA, Gertz MA, Witzig TE, et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clin Proc. 2003;78(1):21–33. doi: 10.4065/78.1.21. [DOI] [PubMed] [Google Scholar]
- 16.Greipp PR, San Miguel J, Durie BG, et al. International staging system for multiple myeloma [published correction appears in J Clin Oncol. 2005;23(25):6281]. J Clin Oncol. 2005;23(15):3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 17.Chng WJ, Dispenzieri A, Chim CS, et al. International Myeloma Working Group. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28(2):269–277. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 18.Neben K, Jauch A, Bertsch U, et al. Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica. 2010;95(7):1150–1157. doi: 10.3324/haematol.2009.016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avet-Loiseau H, Durie BG, Cavo M, et al. International Myeloma Working Group. Combining fluorescent in situ hybridization data with ISS staging improves risk assessment in myeloma: an International Myeloma Working Group collaborative project. Leukemia. 2013;27(3):711–717. doi: 10.1038/leu.2012.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreau P, Cavo M, Sonneveld P, et al. Combination of international scoring system 3, high lactate dehydrogenase, and t(4;14) and/or del(17p) identifies patients with multiple myeloma (MM) treated with front-line autologous stem-cell transplantation at high risk of early MM progression-related death. J Clin Oncol. 2014;32(20):2173–2180. doi: 10.1200/JCO.2013.53.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broyl A, Kuiper R, van Duin M, et al. Dutch-Belgian HOVON group; German GMMG Group. High cereblon expression is associated with better survival in patients with newly diagnosed multiple myeloma treated with thalidomide maintenance. Blood. 2013;121(4):624–627. doi: 10.1182/blood-2012-06-438101. [DOI] [PubMed] [Google Scholar]
- 22.Morgan GJ, Davies FE, Gregory WM, et al. Long-term follow-up of MRC Myeloma IX trial: Survival outcomes with bisphosphonate and thalidomide treatment. Clin Cancer Res. 2013;19(21):6030–6038. doi: 10.1158/1078-0432.CCR-12-3211. [DOI] [PubMed] [Google Scholar]
- 23.Mulligan G, Mitsiades C, Bryant B, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109(8):3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 24.Nair B, van Rhee F, Shaughnessy JD, Jr, et al. Superior results of Total Therapy 3 (2003-33) in gene expression profiling-defined low-risk multiple myeloma confirmed in subsequent trial 2006-66 with VRD maintenance. Blood. 2010;115(21):4168–4173. doi: 10.1182/blood-2009-11-255620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross FM, Ibrahim AH, Vilain-Holmes A, et al. UK Myeloma Forum. Age has a profound effect on the incidence and significance of chromosome abnormalities in myeloma. Leukemia. 2005;19(9):1634–1642. doi: 10.1038/sj.leu.2403857. [DOI] [PubMed] [Google Scholar]
- 26.Walker BA, Leone PE, Chiecchio L, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116(15):e56–e65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 27.Ross FM, Avet-Loiseau H, Ameye G, et al. European Myeloma Network. Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica. 2012;97(8):1272–1277. doi: 10.3324/haematol.2011.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wuilleme S, Robillard N, Lodé L, et al. Intergroupe Francophone de Myélome. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005;19(2):275–278. doi: 10.1038/sj.leu.2403586. [DOI] [PubMed] [Google Scholar]
- 29.Sonneveld P, Schmidt-Wolf IG, van der Holt B, et al. Bortezomib induction and maintenance treatment in patients with newly diagnosed multiple myeloma: results of the randomized phase III HOVON-65/ GMMG-HD4 trial. J Clin Oncol. 2012;30(24):2946–2955. doi: 10.1200/JCO.2011.39.6820. [DOI] [PubMed] [Google Scholar]
- 30.Cox DR, Oakes D. Analysis of Survival Data. Boca Raton, FL: Taylor & Francis; 1984. [Google Scholar]
- 31.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310(6973):170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waheed S, Shaughnessy JD, van Rhee F, et al. International staging system and metaphase cytogenetic abnormalities in the era of gene expression profiling data in multiple myeloma treated with total therapy 2 and 3 protocols. Cancer. 2011;117(5):1001–1009. doi: 10.1002/cncr.25535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner T, Seckinger A, Rème T, et al. Gene expression profiling in multiple myeloma—reporting of entities, risk, and targets in clinical routine. Clin Cancer Res. 2011;17(23):7240–7247. doi: 10.1158/1078-0432.CCR-11-1628. [DOI] [PubMed] [Google Scholar]
- 36.Hose D, Seckinger A, Jauch A, et al. The role of fluorescence in situ hybridization and gene expression profiling in myeloma risk stratification. Srp Arh Celok Lek. 2011;139(Suppl 2):84–89. doi: 10.2298/sarh11s2084h. [DOI] [PubMed] [Google Scholar]
- 37.Steensma DP, Gertz MA, Greipp PR, et al. A high bone marrow plasma cell labeling index in stable plateau-phase multiple myeloma is a marker for early disease progression and death. Blood. 2001;97(8):2522–2523. doi: 10.1182/blood.v97.8.2522. [DOI] [PubMed] [Google Scholar]
- 38.Gastinne T, Leleu X, Duhamel A, et al. Intergroupe Francophone du Myelome. Plasma cell growth fraction using Ki-67 antigen expression identifies a subgroup of multiple myeloma patients displaying short survival within the ISS stage I. Eur J Haematol. 2007;79(4):297–304. doi: 10.1111/j.1600-0609.2007.00915.x. [DOI] [PubMed] [Google Scholar]
- 39.Ludwig H, Sonneveld P, Davies F, et al. European perspective on multiple myeloma treatment strategies in 2014. Oncologist. 2014;19(8):829–844. doi: 10.1634/theoncologist.2014-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawley C, Iacobelli S, Björkstrand B, Apperley JF, Niederwieser D, Gahrton G. Reduced-intensity conditioning for myeloma: lower nonrelapse mortality but higher relapse rates compared with myeloablative conditioning. Blood. 2007;109(8):3588–3594. doi: 10.1182/blood-2006-07-036848. [DOI] [PubMed] [Google Scholar]