Abstract
Microbially produced methane, a versatile, cleaner-burning alternative energy resource to fossil fuels, is sourced from a variety of natural and engineered ecosystems, including marine sediments, anaerobic digesters, shales, and coalbeds. There is a prevailing interest in developing environmental biotechnologies to enhance methane production. Here, we use small-subunit rRNA gene sequencing and metagenomics to better describe the interplay between coalbed methane (CBM) well conditions and microbial communities in the Alberta Basin. Our results show that CBM microbial community structures display patterns of endemism and habitat selection across the Alberta Basin, consistent with observations from other geographical locations. While some phylum-level taxonomic patterns were observed, relative abundances of specific taxonomic groups were localized to discrete wells, likely shaped by local environmental conditions, such as coal rank and depth-dependent physicochemical conditions. To better resolve functional potential within the CBM milieu, a metagenome from a deep volatile-bituminous coal sample was generated. This sample was dominated by Rhodobacteraceae genotypes, resolving a near-complete population genome bin related to Celeribacter sp. that encoded metabolic pathways for the degradation of a wide range of aromatic compounds and the production of methanogenic substrates via acidogenic fermentation. Genomic comparisons between the Celeribacter sp. population genome and related organisms isolated from different environments reflected habitat-specific selection pressures that included nitrogen availability and the ability to utilize diverse carbon substrates. Taken together, our observations reveal that both endemism and metabolic specialization should be considered in the development of biostimulation strategies for nonproductive wells or for those with declining productivity.
INTRODUCTION
The deposition of plant-derived organic matter over geological time has resulted in the formation of stratified hydrocarbon resource environments known as coalbeds. This prevalent energy resource has fueled human development for thousands of years with concomitant environmental impacts, including landscape alteration, waste production, and greenhouse gas emissions (1, 2). In contrast to solid fuel, cleaner-burning coalbed methane (CBM) has become an increasingly attractive global energy resource. While some fraction of CBM is produced under thermogenic conditions, recent studies indicate that microbial communities inhabiting coalbed ecosystems contribute substantially to methane production (3). This has sparked both scientific and biotechnological interest in coalbed microbial communities to enhance CBM production from wells with low methane content or declining productivity (4–6).
Microbial diversity surveys using small-subunit rRNA (SSU; or 16S rRNA) gene sequencing have been conducted across geographically distinct coalbed ecosystems (7–11) and enrichment cultures (6, 12). Taxonomic groups, including Firmicutes, Spirochetes, Bacteroidetes, Proteobacteria (Alpha-, Beta-, Gamma-, and Deltaproteobacteria) and Euryarchaeota, with the potential to mediate syntrophic interactions driving CBM production have been observed in many coalbed ecosystems. While several of these groups, including methanogenic Euryarchaeota, are directly implicated in CBM production, microbial community interactions and metabolic pathways mediating coal conversion into methane remain poorly defined. Moreover, limited information exists on how microbial community structure and function are shaped by in situ well conditions, partially due to restricted access to sampling.
The Western Canadian Sedimentary (Alberta) Basin encompasses a range of CBM ecosystems. Currently, the majority of the Alberta Basin CBM is accessed from the Horseshoe Canyon, Mannville Group, and Ardley (Scollard) formations, where coal rank varies from subbituminous to anthracite. Since 2001 more than 17,000 wells have been drilled across these formations for CBM production, creating an opportunity for accessing subsurface samples for microbial ecology studies (8, 13). The Mannville Group tends to run much deeper than the other formations, with concomitant increases in salinity, temperature, and pressure. Moreover, Mannville coals are “wet,” and waters must be produced from the formation to lower the reservoir pressure prior to well completion. Understanding structural and functional differences between microbial communities inhabiting these formations is necessary for engineering strategies to enhance CBM production, particularly from low-productivity wells penetrating deeper formations. However, previous studies have been limited to investigations of shallow low-rank coals (6) or summaries of community structure and function across hydrocarbon resource environments (14).
Here, we use SSU rRNA gene amplicon sequences recovered from 10 Alberta Basin CBM wells to explore microbial community structure among and between different coal ranks and well conditions. Hierarchical clustering and indicator analysis were used to define microbial consortia unique to each well site with the potential to mediate biogenic CBM production. This information was combined with metagenomic sequencing from a deep coalbed to identify ecosystem-specific selection pressures shaping specific community functions. Resulting data sets reveal patterns of endemism and habitat selection within coalbed ecosystems relevant to design and operation of microbially enhanced CBM production scenarios.
MATERIALS AND METHODS
Sample collection.
Multiple industrial partners collected coal cores and associated drilling fluids, cuttings, and production waters during drilling operations at 10 different well sites within the Alberta Basin (Fig. 1; Table 1). Unique identifiers were given to each sample according to its collection method (cores, CR; cuttings, CT; drilling fluids, DF; production waters, PW), well identification (ID) number, coal rank (high-volatile bituminous, HB; volatile bituminous, VB; subbituminous, SB), and depth (see Table S1 in the supplemental material). Drilling crews collected all samples. While it was technically difficult to ensure aseptic collection, efforts were made to prevent sample contamination by providing instructions to crew members on how to minimize contamination risk and by providing sterile collection containers. Core samples were cut into approximately 15-cm lengths and sealed into sterile vacuum bags at the well site prior to laboratory transport. Cuttings were collected into sterile polyvinyl chloride containers and sealed immediately upon reaching the surface. Produced waters were sampled directly into sterile 4-liter fuel cans. All samples were transported and frozen at −80°C within 24 h of collection. Physicochemical parameters for the samples, such as in situ pressures, temperatures, and salinities, were provided by each company, which took the measurements using standard oil and gas operating procedures.
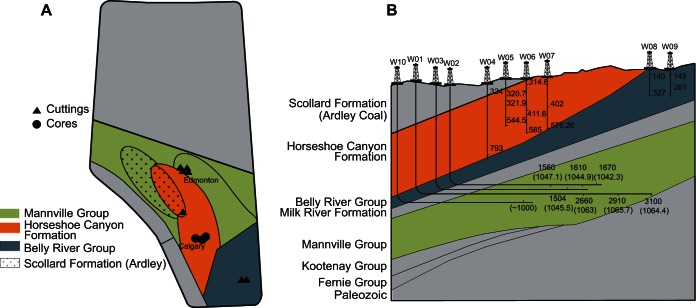
FIG 1.
(A) Plan view outlining well locations sampled across the Alberta Basin. Colors indicate different coal ranks; shapes indicate method used to collect coal samples. (B) Section view outlining well and sample depths for cores and cuttings. Colors indicate major coal formations; wells are indicated by identification number (e.g., W01). Depth was measured as true vertical depth, except for the horizontal wells, which show the measured depth (length of the borehole). The true vertical depths for these samples are shown in parentheses. (The maps were adapted with permission from the Alberta Geological Survey [http://www.ags.gov.ab.ca/energy/cbm/coal_and_cbm_intro.html#coal_occurrences]).
TABLE 1.
Well information
| Well no. | Well IDa | Latitudeb | Longitudeb | Industrial partnerc | Coal formation | Coal rank | Collection method | Date collected |
|---|---|---|---|---|---|---|---|---|
| 1 | 13-5-61-6W5 | 54.2521 | −114.87 | Nexen | Upper Mannville | Volatile bituminous A or B | Cuttings | April 2009 |
| 2 | 8-5-60-5W5 | 54.1575 | −114.705 | Trident | Upper Mannville | Volatile bituminous A or B | Cuttings | June 2009 |
| 3 | 14-11-62-07W5 | 54.3549 | −114.942 | Trident | Upper Mannville | Volatile bituminous A or B | Cuttings | November 2010 |
| 4 | 102-6-4-35-27W4 | 51.973 | −113.8068 | Quicksilver | Scollard (Ardley)/Horseshoe Canyon | Subbituminous/high-volatile bituminous C | Cuttings | July 2009 |
| 5 | 8-30-28-24W4 | 51.4204 | −113.3608 | Encana | Horseshoe Canyon | High-volatile bituminous C | Cores | December 2008 |
| 6 | 5-29-28-24W4 | 51.4204 | −113.3524 | Encana | Horseshoe Canyon | High-volatile bituminous C | Cores | December 2008 |
| 7 | 102/7-29-28-24W4 | 51.4213 | −113.3442 | Encana | Horseshoe Canyon | High-volatile bituminous C | Cores | December 2008 |
| 8 | 100/03-12-018-15W4 | 50.5011 | −111.943 | ARC | Belly River | Subbituminous | Cuttings | October 2010 |
| 9 | 100/03-36-018-15W4 | 50.5594 | −111.943 | ARC | Belly River | Subbituminous | Cuttings | October 2010 |
| 10 | 100/03-28-060-05W5 | 54.2131 | −114.6917 | Trident | Upper Mannville | Volatile bituminous A or B | Produced water | May 2009 |
The well identification (ID) is defined by the Alberta Township Survey legal land description. For example, 13-5-61-6W5 refers to quarter section 13, township 5, range 61, west of the 5th meridian.
Decimal degrees.
Nexen, a subsidiary of CNOCC, Ltd.; Trident, Trident Exploration Corp.; Quicksilver, Quicksilver Resources Canada, Inc.; Encana, Encana Corporation; ARC, ARC Resources, Ltd.
DNA extraction.
A subsection from the center of the cores was sampled in an anaerobic chamber (5% H2, 95% N2) and aseptically crushed into fines. Cuttings were used directly. Production waters and drilling fluids were filtered through a 0.22-μm-pore-size filter, and DNA was extracted directly from the filter paper. However, a recent report on the presence of ultrasmall bacteria in groundwater indicates the potential of smaller phyla not being represented in our community analyses (15). A previously developed bead-beating method was used to extract DNA from coal samples (16). Briefly, 0.5 g of each sample was added to tubes containing 0.5 g of both 0.5- and 10-μm-diameter zircon beads, 300 μl of 50 mM phosphate buffer (pH 7.0), 300 μl of lysis buffer (10% SDS, 100 mM NaCl, 500 mM Tris, pH 8.0) and 300 μl of 24:1 chloroform-isoamyl alcohol. Samples were prepared in triplicate. Sample tubes were subsequently shaken on a bead-beater apparatus for 1 min, followed by centrifugation at 13,000 rpm for 5 min. The supernatants were removed to new tubes, and 7 M ammonium acetate was added to each tube to a final concentration of 2.5 M. The tubes were gently mixed, left on their sides for 2 min, vortexed briefly, and finally centrifuged at 13,000 rpm for 7 min. The supernatants were transferred to new tubes, 1 volume of 100% isopropanol was added to each tube, and the tubes were kept at −20°C overnight. The tubes were then centrifuged at 13,000 rpm for 30 min at 4°C, the supernatant was removed, and the DNA pellet was allowed to dry for 2 h before being resuspended on ice for 30 min in 30 μl of sterile nuclease-free water (Ambion).
PCR amplification and pyrosequencing.
The V6-V8 region of the SSU rRNA gene was amplified from DNA templates using the universal primer pair 926F (5′-AAACTYAAAKGAATTGRCGG-3′) and 1392R (5′-ACGGGCGGTGTGTRC3′). Primers were modified to include 454 pyrosequencing adaptors, and reverse primers included a 5-bp bar code according to previously published protocols (17, 18). Sixty samples were multiplexed on a single analysis chip. PCRs (50 μl) were performed in 12 replicates and pooled to minimize PCR bias. Each reaction mixture contained 2 μl of each primer (20 pmol/μl), 25 μl of PCR master mix (containing 0.05 units/μl Taq DNA polymerase, reaction buffer, 4 mM MgCl2, and 0.4 mM each deoxynucleoside triphosphate [dNTP; Fermentas]), 21 μl of nuclease-free water, and 2 μl of DNA template (10 to 100 ng). Negative controls were included with each reaction to ensure that no contamination of DNA had occurred. PCR amplicons were diluted to 5 ng/μl and pooled in equal concentrations prior to sequencing. Emulsion PCR and pyrosequencing were performed at Genome Quebec and McGill University Innovation Centre (Montreal, Canada) on the Roche 454 GS FLX Titanium platform (454 Life Sciences, Branford, CT, USA) according to the manufacturer's instructions.
Analysis of pyrotag sequencing data.
SSU rRNA gene sequences were processed using the software package Quantitative Insights Into Microbial Ecology (QIIME), version 1.4.0 (19). Sequences with less than 150 bases, ambiguous N bases, and homopolymer runs were removed before chimera detection. Chimeric sequences were identified with QIIME via ChimeraSlayer and removed prior to taxonomic assignment. A total of 763,488 nonchimeric SSU rRNA gene sequences were clustered at 97% into operational taxonomic units (OTUs). Representative sequences from each cluster were queried against the SILVA 111 rRNA database (20) using the Basic Local Alignment Search Tool (BLAST) (21) to assign taxonomy. Singleton OTUs (represented by one read only) were omitted from downstream analysis to reduce overprediction of rare OTUs (22). Prior to analysis, data sets were normalized to the total number of reads per sample. Diversity metrics were calculated based on OTU tables rarefied to 1,050 reads (see Table S2 in the supplemental material).
Statistical analysis.
Indicator analysis was performed to identify OTUs specifically associated with microbial communities of similar taxonomic compositions using the package labdsv, version 1.5.0, in R based on previously published methods (23, 24). Indicator groups were defined based on hierarchical clustering of microbial communities from the different coalbed samples using the average linkage method and Bray-Curtis dissimilarity implemented in R. The statistical significance of the indicator value (i.e., P value) was calculated using a randomization procedure based on Monte Carlo simulations.
Metagenomic sequencing, assembly, and binning.
Total genomic DNA was extracted from a coal cutting sample as described above and sequenced on a Roche 454 GS FLX Titanium platform (454 Life Sciences, Branford, CT, USA) according to the manufacturer's instructions. Resulting reads were quality filtered using the Metagenomics Analysis Server (MG-RAST) pipeline (25). Reads were dereplicated and filtered using a minimum quality score of 20, a minimum length of 100 bp, and allowing for no ambiguous bases. Filtered reads were subsequently assembled into contigs using Newbler with overlap parameters of 95% minimum identity and a minimum length of 40 bp (26). Resulting contigs were binned into population genomes based on differential coverage, tetranucleotide frequency, and single-copy gene analysis with the software MaxBin, version 2.0, using default parameters (27). Population genome bins were then annotated using MetaPathways, version 2.0, as described below, and taxonomic summaries for each bin were generated using PhyloSift, version 1.0.1, based on phylogenetic analysis of 37 core gene families (28).
Metagenome annotation and pathway analysis.
Annotation of ORFs and metabolic pathway inference were performed using MetaPathways, version 2.0 (29, 30). Briefly, open reading frames (ORFs) were predicted from both filtered sequences and contigs using Prodigal (Prokaryotic Dynamic Programming Genefinding Algorithm) version 2.0, using a minimum nucleotide length of 60 and queried against the Kyoto Encyclopedia of Genes and Genomes (KEGG, accessed 2013), SEED subsystems (accessed January 2014), Clusters of Orthologous Groups (COG), RefSeq (version 62), and MetaCyc (version 18.0) protein databases using the optimized LAST algorithm (E value, E−6) for functional annotation (29). Taxonomic annotation of predicted ORFs was accomplished using MEGAN, version 5 (31). Nucleotide sequences were also queried against the SILVA 111 database to identify SSU rRNA genes. Environmental Pathway/Genome Databases (ePGDBs) were constructed from annotated ORFs using Pathway Tools (32) and MetaCyc, a highly curated database of 2,151 pathways and 14,084 reactions representing all domains of life (33). Pathway inference was based on a set of rules used by the Pathway Tools prediction algorithm “Pathologic” (32), including the presence of all “key reactions,” and on a reaction coverage of at least 50% for a particular pathway. All pathways were then manually curated to verify predictions made by Pathologic.
Genome comparisons.
Comparison of the binned population genome with isolate genomes was accomplished using the protein BLAST (BLASTP) search. Isolate genome sequences were downloaded from the National Center for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov), and ORFs for all genomes were predicted using Prodigal, version 2.0 (34). Predicted ORFs between the population and isolate genomes were compared using BLASTP, and the percent amino acid similarity between best reciprocal BLASTP hits was plotted to identify regions of low similarity. Low-amino-acid similarity regions that were flanked by mobile genetic element (MGE) signatures (e.g., transposases or integrases) and had variable taxonomic annotations were considered putative genomic islands. ORFs were queried against NCBI's RefSeq or nonredundant (nr) database using BLASTP for annotation. Genome-wide average nucleotide identity (gANI) and the alignment fraction (AF) between genomes were calculated using the Microbial Species Identifier (MiSI) method (35).
Sequence data accession numbers.
Amplicon sequences from this study can be accessed through the NCBI's Sequence Read Achieve (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR573852 to SRR573862 and SRR573863 to SRR573891. Metagenome sequences for sample CT-W2-VB-1047 can be accessed through the SRA under accession number SRR573886.
RESULTS
Site description.
Samples were recovered from 10 wells penetrating distinct geological formations in the Alberta Basin (Fig. 1). Wells were characterized by different coal ranks, including subbituminous, high-volatile bituminous, and volatile bituminous coals (Fig. 1; Table 1). Moreover, samples from individual wells were collected at multiple depths associated with different physicochemical conditions, including temperature, pressure, and salinity gradients. A summary of the geological features for each well is presented in Table S1 in the supplemental material. As industrial partners were ultimately responsible for sample collection, either coal cores or cuttings were provided from each well for downstream analysis (Fig. 1; see also Table S1). Drilling fluids and production waters associated with cores and cuttings, respectfully, were also collected for a subset of samples and used for downstream analysis, as indicated in Table S1.
Microbial community structure.
Pyrotag sequencing yielded a total of 763,488 SSU rRNA gene sequences recovered from 8 core, 12 cutting, 4 production water, and 12 core-associated drilling fluid samples. These sequences were clustered with a 97% similarity cutoff into 25,941 operational taxonomic units (OTUs) after singleton removal. Resulting OTUs encompassed 16 different phyla affiliated with 292 families spanning all three domains of life. Table S1 in the supplemental material summarizes the total number of SSU rRNA gene sequences recovered from each sample. Abundant OTUs were defined as having a frequency of >1% in at least one sample, intermediate OTUs as having a frequency between 1% and 0.1% in at least one sample, and rare OTUs as having a frequency of <0.1% in all samples (36). With the exception of OTUs collected from subbituminous coal samples and volatile bituminous coal production waters, rarefaction curves generated from individual samples approached an asymptote, indicating that the total OTU richness was nearly sampled (see Fig. S1 in the supplemental material). Consequently, less-abundant OTUs recovered from subbituminous coals may be underrepresented in the data set. Taxonomic diversity of microbial communities was also investigated by calculating richness (Chao1) and diversity (Simpson and Shannon) indices across each coal sample (see Table S2 in the supplemental material). Chao1 richness estimates indicated that drilling fluid samples from high-volatile bituminous coals contained more unique OTUs than cores recovered from the same coal rank, a result potentially influenced by microorganisms inhabiting the drilling fluid waters used. Simpson and Shannon indices revealed that the taxonomic diversity of microbial communities from across the various coalbed ecosystems was lower on average than that from other natural and engineered ecosystems (e.g., soils, aquatic, and activated sludge) (37, 38). Moreover, microbial communities recovered from deep coalbeds (>1,000 m) were less diverse than those from more shallow ones (see Table S2 in the supplemental material). To determine similarities in community structure between samples at the OTU level, nonmetric multidimensional scaling (NMDS) with Bray-Curtis dissimilarity was used (see Fig. S2 in the supplemental material). While several environmental conditions may have influenced community structure, sampling method, coal rank, and depth appeared to have the strongest impact, based on observed clustering patterns (Fig. 2; see also Fig. S2).
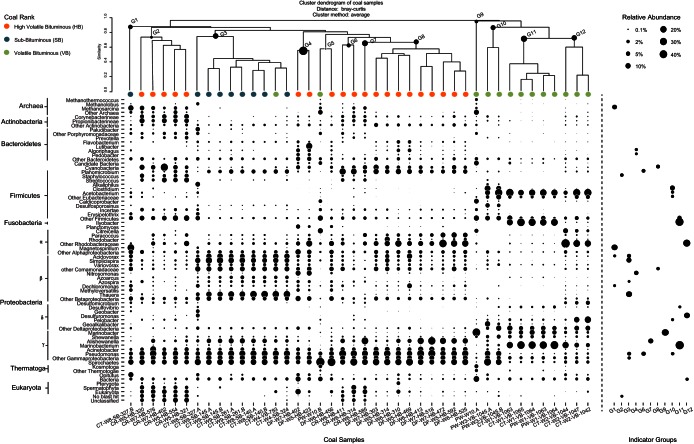
FIG 2.
Microbial community structure and indicator taxa for each coalbed sample and indicator group. Bubble size indicates the relative abundance of abundant taxa. The cluster dendrogram (top) was generated using the average-linkage method and Bray-Curtis dissimilarity. Colored bubbles denote sample coal rank. G, group.
Community composition.
To further examine clustering patterns, the taxonomic affiliations of OTUs recovered from across the Alberta Basin were compared. Although many phyla were detected in all coalbed samples, most notably, the Gammaproteobacteria and Spirochaetes, the relative abundance of individual phyla varied considerably with sampling method and coal rank (Fig. 2). This was consistent with observed NMDS clustering patterns (see Fig. S2 in the supplemental material). Cores and associated drilling fluids collected from the high-volatile bituminous coals displayed more similar abundance patterns than cuttings and production waters collected from the subbituminous and volatile bituminous coals (Fig. 2). Abundant taxa (SSU rRNA gene frequency of >1%) recovered from the high-volatile bituminous coals were affiliated with the phyla Proteobacteria (37.4%), Firmicutes (16.3%), Spirochaetes (11.0%), Actinobacteria (8.0%), cyanobacteria (7.5%), Fungi (3.2%), Euryarchaeota (2.6%), and Metazoa (1.3%). Dominant proteobacterial classes were affiliated with Gammaproteobacteria (28.0%), Alphaproteobacteria (5.0%), and Betaproteobacteria (3.6%), whereas dominant Euryarchaeota were affiliated with methanogenic Methanomicrobia (2.2%) (Fig. 2). Abundant taxa (SSU rRNA gene frequency of >1%) recovered from associated drilling fluid samples were affiliated with similar phyla, including Proteobacteria (66.3%), Spirochaetes (15.6%), Bacteroidetes (5.2%), Firmicutes (4.7%), cyanobacteria (2.8%), and Actinobacteria (1.5%) (Fig. 2). Indeed, similar bacterial and archaeal phyla have previously been detected in coalbeds from the Illinois Basin (8) and the Waikato coalfields in New Zealand (9), with the exception of cyanobacteria. The presence of some plant and metazoan reads found in coal cores may represent sample contamination rather than intrinsic populations (Fig. 2). It is also possible that reagent or laboratory contamination may have biased the sequencing results (39), albeit our results remain consistent with those of other studies conducted in separate laboratories and with different DNA extraction procedures (40).
Microbial communities associated with cuttings collected from the subbituminous and volatile bituminous coals displayed further quantitative differences. Abundant taxa recovered from the subbituminous coals were affiliated with the phyla Proteobacteria (82.1%), Spirochaetes (11.19%), and Firmicutes (5.33%). Dominant proteobacterial classes included Betaproteobacteria (48.1%), Gammaproteobacteria (26.6%), Alphaproteobacteria (3.0%), and Deltaproteobacteria (2.1%). Abundant taxa recovered from the volatile bituminous coals were affiliated with the phyla Firmicutes (27.0%) and Spirochaetes (1.2%), as well as with the proteobacterial classes Gammaproteobacteria (24.6%) and Deltaproteobacteria (12.3%). Observed differences between subbituminous and volatile bituminous coal microbial communities also appeared to stratify by depth. For example, deep volatile bituminous coal communities (>1,000 m) harbored considerably more Firmicutes and Fusobacteria (wells W1, W2, and W3), whereas shallower subbituminous coal communities harbored more Betaproteobacteria (wells W4, W8, and W9) (Fig. 2). Interestingly, deep coal samples also harbored significant amounts of the families Oceanospirillaceae and Alteromonadaceae (Fig. 2; see also the supplemental material), which encompass several halophilic representatives. As changes in depth directly correlated with variations in physicochemical conditions, including pressure, temperature, and salinity (see Table S1 in the supplemental material), these factors, in addition to sampling method and coal rank, likely shaped microbial community structure as well.
Indicator analysis.
To identify OTUs diagnostic of the different microbial community structures observed across the Alberta Basin, indicator analysis was performed. Here, indicator groups were defined based on samples with similar taxonomic compositions using hierarchical cluster analysis (Fig. 2). Consistent with NMDS, coal rank and sampling method appeared to be the dominant features stratifying indicator groups (Fig. 2). OTUs were considered indicators of a predefined group if their relative frequency in that group was >50% compared to that of other groups. The distributions of indicator values and P values for all 25,941 OTUs are shown in Fig. S3A in the supplemental material. Based on an indicator value of >0.5 and a P value of <0.05, 1,810 significant indictor OTUs were identified. The distribution of indicator OTUs among abundant, intermediate, and rare classes was 63, 441, and 1,306, respectively (see Fig. S3B).
The taxonomic composition of each indicator group is shown in Fig. 2. Abundant indicator OTUs from the high-volatile bituminous coals (group 2) were affiliated with the genera Staphylococcus and Propionibacterium (Fig. 2). High-volatile bituminous coals also harbored populations of methanogenic archaea affiliated with the genus Methanosarcina. Abundant indicator OTUs from associated drilling fluids (group 8) were affiliated with the genus Synechococcus and uncultured members of the families Comamonadaceae and Burkholderiaceae (Fig. 2). As drilling fluid waters were not sterilized prior to use, these indicators may have originated from ex situ surface water sources. Abundant indicator OTUs from the subbituminous coals (group 3) were affiliated with the genera Thauera, Simplicispira, and Acidovorax. This was consistent with the large presence of Betaproteobacteria recovered from the shallower subbituminous coals (Fig. 2). Abundant populations of the genera Acinetobacter, Streptococcus, Planomicrobium, and Methanosarcina were also detected (Fig. 2). In contrast, abundant indicators from the volatile bituminous coals were affiliated with the genera Marinobacterium and Propionigenium (group 11), as well as with acetogenic Pelobacter and an uncultured member of the Rhodobacteraceae family (group 12). These volatile bituminous coals represented the deepest wells sampled. Group 12 also contained abundant OTUs affiliated with acetogenic Acetobacterium, while associated production waters harbored methanogenic archaea affiliated with acetoclastic Methanosarcina. Indeed, the high abundance of potential acetogenic and methanogenic microbes agrees with previous reports implicating these taxa in syntrophic coal-to-methane transformations (41), where uncultured Rhodobacteraceae members identified here may play important roles in initial aromatic hydrocarbon degradation, as previously reported for representative isolates (42, 43).
Metagenome sequencing and binning.
Information on microbial communities inhabiting deep coal seams (>1,000 m) across the Alberta Basin is scant. In particular, very little information exists on specific metabolic pathways and adaptive strategies required for survival in deep coal seams, often characterized by high pressures, salinities, and temperatures (see Table S1 in the supplemental material). This is especially true for initial aromatic hydrocarbon degradation, which is often predicted to be the rate-limiting step in biogenic CBM production (3). As many identified well sites for enhanced CBM recovery across the Alberta Basin penetrate into deep coal seams and were observed to contain taxa previously implicated in coal-to-methane transformations based on SSU rRNA amplicon sequencing results, metagenomic sequencing was employed to obtain further insight into microbial community function. Here, environmental DNA was extracted from a volatile bituminous coal sample from the Mannville Group (CT-W2-VB-1047) (group 12) and sequenced on the 454 pyrosequencing platform. A total of 439,418 quality-filtered reads were assembled into contigs using Newbler, resulting in 6,495 contigs with an average length of 412 bp and a maximum length of 276,644 bp.
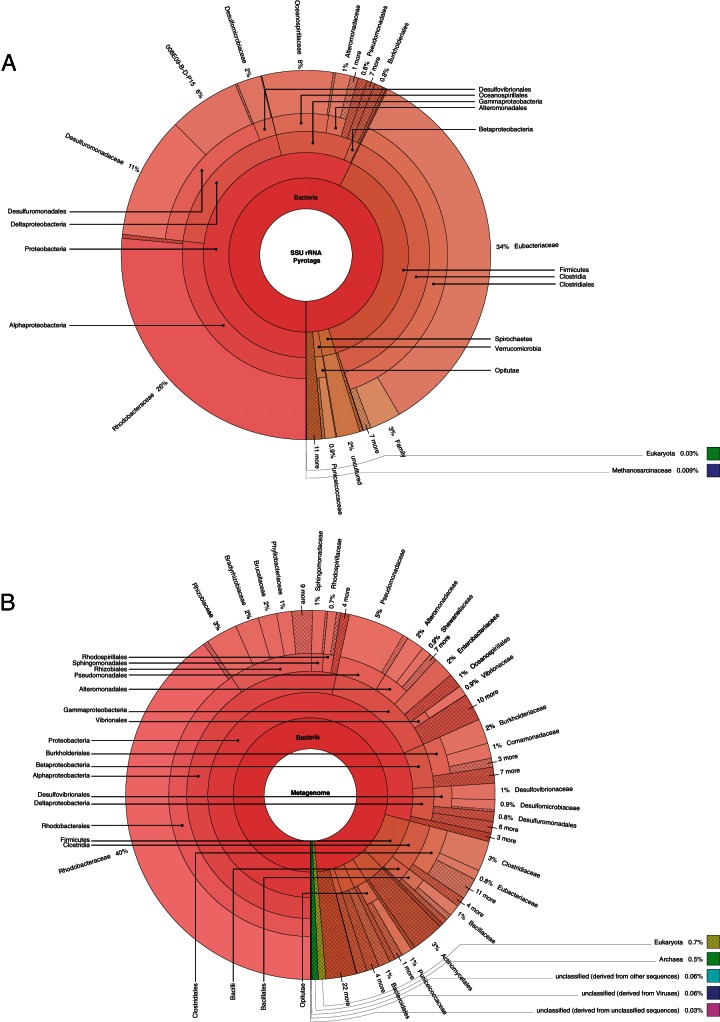
The taxonomic distribution of predicted open reading frames (ORFs) identified on contigs displayed quantitative differences compared to the results obtained by SSU rRNA gene amplicon sequencing (Fig. 3). Specifically, considerable differences were observed among bacteria affiliated with the families Eubacteriaceae, Rhodobacteraceae, Burkholderiaceae, Desulfuromonadaceae, Oceanospirillaceae, and Pseudomonadaceae. It is possible that the lack of agreement in community structure between sequencing methods reflects the limited number of reference genomes available in sequence databases (44). Such biases appear in our SSU rRNA gene data set, as many SSU rRNA gene sequences were affiliated with uncultured bacteria at the family level (16% on average). Additional caveats associated with pyrotag analysis may have influenced our results; these include differences in rrn operon copy number between taxa and PCR priming biases (45, 46). Nonetheless, both approaches identified a high abundance of uncultured bacteria affiliated with the Rhodobacteraceae family: 41.8% based on metagenomic methods and 26.5% based on pyrotags (Fig. 3).
FIG 3.
Comparison of taxonomies using pyrotag and metagenome analyses for the CT-W2-VB-1047 coal sample. Pyrotag abundance is based on SSU rRNA gene abundance (A); metagenome abundance is based on ORF counts (B).
To further examine the functional content of the CT-W2-VB-1047 metagenome, assembled reads were binned into population genomes based on tetranucleotide frequency, differential coverage, and single-copy gene analysis using MaxBin (27). This resulted in one near-complete population genome (bin 1) and one partial population genome (bin 2) with greatest similarity to Celeribacter baekdonensis (Rhodobacteraceae) and Acetobacterium dehalogenans (Eubacteriaceae) (see Table S3 in the supplemental material), respectively, reinforcing ORF taxonomy results (Fig. 3). Single-copy gene analysis revealed that the Celeribacter sp. bin contained 106 of 107 marker genes (99%), whereas the Acetobacterium sp. bin contained only 67 of 107 (63%) (see Table S3). Genome contamination (i.e., the presence of misplaced contigs) of the Celeribacter sp. and Acetobacterium sp. bins was estimated to be 8% and 33%, respectively, based on the presence of multiple single-copy genes (see Table S3). Only the Celeribacter sp. bin was used for further analysis because of its high estimated completeness and low contamination. This population genome encompassed 513 contigs (N50 = 41,884 bp) and had the highest representation in the CT-W2-VB-1047 metagenome (Fig. 3; see also Table S3). Given the high coverage of bin 1 sequences in the CT-W2-VB-1047 metagenome and interest in functions relevant to CBM in the deep coal seems, subsequent efforts focused on analyzing metabolic potential and genomic variation within Celeribacter sp. populations inhabiting the deep coalbed milieu.
Coalbed Celeribacter sp. metabolic potential.
To identify core functions encoded by the C. baekdonensis population genome, predicted ORFs were first annotated using the MetaPathways pipeline. A large proportion of annotated ORFs in the Celeribacter sp. population genome were affiliated with transport systems, in particular, cation/multidrug efflux pumps and ABC transporters for sugars, amino acids, and spermidine/putrescine. C4-dicarboxylate tripartite ATP-independent periplasmic (TRAP) transport systems and other chemotaxis proteins were also abundant (see File S1 in the supplemental material). Annotated ORFs were subsequently used by Pathway Tools to generate a Celeribacter sp. pathway/genome database (PGDB). This PGDB contained 467 pathways involved in biosynthesis, degradation, energy generation, and other cellular functions (see File S2 in the supplemental material). Central carbon metabolic pathways encoded both autotrophic and heterotrophic growth modes. Pathways for inorganic carbon fixation included the Calvin-Benson-Bassham (CBB) cycle (via ribulose 1,5-bisphosphate carboxylase/oxygenase [RubisCO]); pathways for growth on organic carbon included the Embden-Meyerhof-Parnas (glycolysis) pathway, pentose phosphate pathway, gluconeogenesis, and tricarboxylic acid (TCA) cycle. Other noteworthy organic carbon metabolic pathways were fatty acid beta-oxidation, the glyoxylate cycle, and acidogenic fermentation (see File S2). These pathways would enable coalbed Celeribacter sp. populations to utilize both long-chain and short-chain fatty acids (e.g., acetate) to satisfy cellular carbon requirements (47), as well as to generate energy anaerobically through substrate-level phosphorylation.
Dominant catabolic pathways encoded in the Celeribacter sp. population genome were associated with carbohydrate, carboxylate, and aromatic compound degradation (see File S2B in the supplemental material). The high abundance of ABC-type sugar and C4-dicarboxylate TRAP transporters together with encoded central carbon pathways (see Files S1 and S2) would enable coalbed Celeribacter sp. to utilize both organic sugars (glucose, xylose, and sucrose) and organic acids (acetate, succinate, and fumarate) as carbon and energy sources. These substrates could be respired under aerobic conditions, whereas sugar compounds (e.g., glucose) could be fermented to methanogenic substrates (e.g., acetate) under anaerobic conditions. Pathways for glycogen biosynthesis and degradation were also identified in the population genome (see File S2), which would allow Celeribacter sp. to accumulate carbon and energy reserves necessary for coping with periods of nutrient starvation.
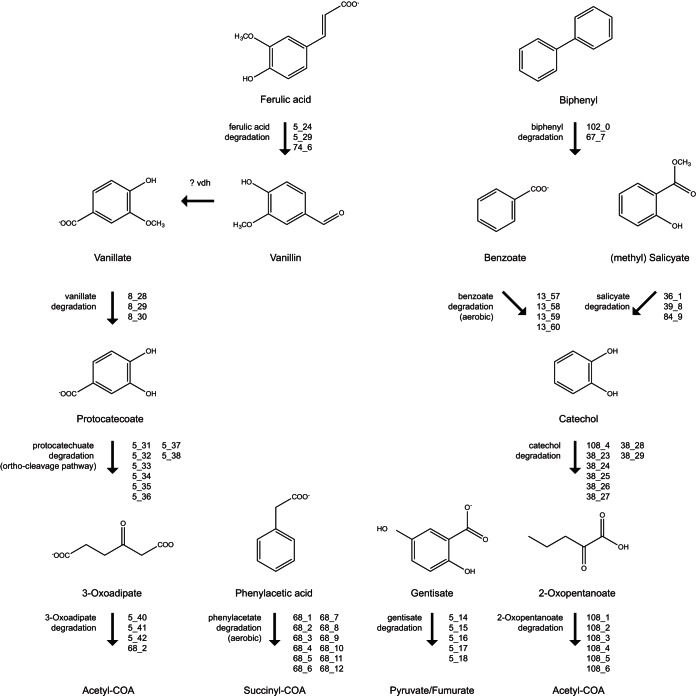
The population genome also encoded enzymes for degrading a wide range of aromatic compounds, including ferulate, vanillate, biphenyl, benzoate, salicylate, and phenylacetate (Fig. 4). These substrates would serve as carbon and energy sources for Celeribacter sp., which are transformed to TCA cycle intermediates through the intermediary aromatic compounds protocatechuate, catechol, and gentisate and through the carboxylic acid intermediates 3-oxoadipate and 2-oxopentenoate (Fig. 4). Such aromatic compounds are expected to be available in coalbed environments as they are common by-products of coal and lignin degradation (48). Unexpectedly, all aromatic degradation pathways were aerobic, requiring mono- and dioxygenases that incorporate oxygen atoms into their substrates (49). This is inconsistent with conventional ideas on microbial decomposition of coal and aromatic hydrocarbons, where anaerobic conversion pathways are believed to be prevalent (3, 50). Nonetheless, recent metagenomic investigations also observed high proportions of genes involved in aerobic metabolism of aromatic compounds across multiple coalbed environments (14).
FIG 4.
Summary of known aromatic compound degradation pathways annotated in the Celeribacter sp. population genome (bin 1). Specific ORFs assigned to aromatic degradation pathways are provided in Table S4 in the supplemental material. The breakdown products, including acetyl-coenzyme A (CoA), succinyl-CoA, pyruvate, and fumarate, can subsequently enter the TCA cycle.
Reactions that utilize inorganic substrates for energy metabolism were also encoded in the Celeribacter sp. population genome. Annotation of the Celeribacter sp. population genome revealed a putative pathway for carbon monoxide (CO) oxidation to CO2 via CO dehydrogenase (CODH) cox genes. Sequence analysis of the recovered coxL gene encoding the large catalytic subunit indicated that Celeribacter sp. contains the putative form II CODH (51, 52). Consistent with aromatic degradation pathways, these enzymes were also aerobic and may allow Celeribacter sp. to use CO as a supplementary energy and carbon source (together with the CBB cycle) (52).
Comparative genomics with closely related bacteria.
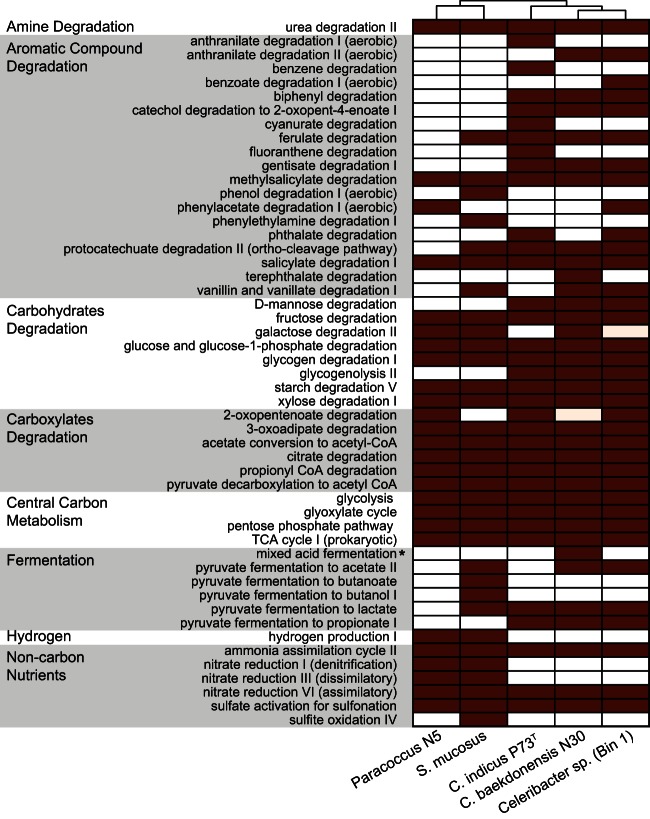
To identify pathways potentially unique to coalbed Celeribacter sp. populations, PGDBs were subsequently generated for four closely related Rhodobacteraceae genomes isolated from activated sludge, saline soil, marine, and deep-sea sediment environments: Paracoccus sp. N5 (53), Salipiger mucosus (54), Celeribacter baekdonensis B30 (55), and Celeribacter indicus P73T (56, 57). Genomes were selected based on their high homology with and taxonomic relatedness to bin 1 ORFs. Although most central carbon and energy generation pathways described above were conserved across all genomes, some variation in aromatic-compound-degrading pathways was observed (Fig. 5). For example, the coalbed Celeribacter sp. population genome (bin 1) was the only genome that encoded oxygenase enzymes for phenylacetate and benzoate degradation. Other aromatic degradation pathways encoded by coalbed Celeribacter sp. that had variable presence across other isolates included anthranilate, ferulate, phthalate, salicylate, and vanillate degradation (Fig. 5). Significant variation was also observed in genome-wide average nucleotide identity (gANI) between the three Celeribacter genomes, which ranged from approximately 76 to 80% over an alignment fraction (AF) of 0.5 to 0.6 (see File S3 in the supplemental material).
FIG 5.
Overview of central carbon and energy metabolic pathways present in Paracoccus sp. N5, S. mucosus, C. indicus P73T, C. baekdonensis N30, and bin 1 (Celeribacter sp.) pathway/genome databases (PGDBs). A filled box indicates that the pathway is present; a white box indicates that the pathway is absent. Genomes were ordered using Hclust in R (dendrograms were generated using ward clustering). *, the pyruvate formate-lyase sequence was identified at the end of a bin 1 contig and was truncated to 166 amino acids.
It has been shown that C. indicus P73T is capable of degrading a wide range of polycyclic aromatic hydrocarbons (PAHs), such as fluoranthene (56, 57). However, no homologs for the dioxygenases predicted to be involved in C. indicus P73T PAH metabolism were identified in the coalbed Celeribacter sp. genome, similar to findings for C. baekdonensis B30 (57). While this suggests that coalbed Celeribacter sp. may not be capable of PAH degradation, experimental evidence is still needed to confirm this prediction. Moreover, this indicates that other community members are likely responsible or specialized in the degradation and activation of PAH.
Habitat-specific gene pools.
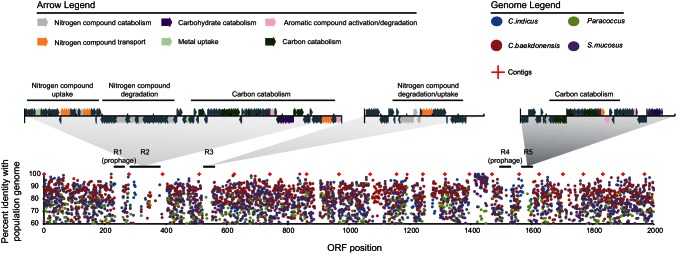
In an attempt to understand local selection pressures driving genomic differentiation events in the deep coalbed milieu, fine-scale differences between the Celeribacter sp. population genome and the closely related isolate genomes stated above were examined. Large variations in genome content and architecture were hypothesized to originate from adaptive habitat-specific gene pools, based on previous observations in marine ecosystems (58, 59). Bin 1 contigs were compared to isolate genomes using the protein Basic Local Alignment Search Tool (BLASTP) (21), and the homology between best reciprocal hits was plotted to identify regions of low similarity (Fig. 6; see also File S3 in the supplemental material). This resulted in the identification of 224 ORFs unique to the Celeribacter sp. population genome. Indeed, many of these ORFs formed discrete clusters, often located on putative genomic islands, suggesting that they encode functions acquired via lateral gene transfer (59). Annotation of the ORFs in these clusters revealed putative genes for nitrogen and carbohydrate catabolism, as well as genes encoding aromatic compound degradation proteins (Fig. 6; see also File S3). Moreover, the presence of several spermidine/putrescine ABC transporters and TonB-dependent siderophore receptors was identified within variable genomic regions, in agreement with the high abundance of ORFs encoding these proteins in the Celeribacter sp. population genome. Two prophages were also found in regions of low genomic similarity (see File S3). Interestingly, one prophage carried ORFs encoding genes involved in benzoate degradation, consistent with the unique potential of the Celeribacter sp. population genome to utilize these substrates (Fig. 5).
FIG 6.
Fine-scale comparisons of Paracoccus sp. N5, S. mucosus, C. baekdonensis N30, and bin 1 (Celeribacter sp.) genomes. The x axis indicates the ORF position in the bin 1 population genome (by ORF number); the y axis indicates the percent amino acid similarity of isolate genomes with the bin 1 population genome as determined by best reciprocal BLASTP hits. Arrows indicate predict ORFs identified within variable genomic regions. ORF annotations for each variable region (R) can be found in File S3 in the supplemental material.
DISCUSSION
Patterns of endemism and syntrophy.
Microbial communities recovered from across the Alberta Basin displayed patterns of endemism, wherein different microbial consortia were unique to defined geographical regions that exhibited variation in coal rank and physicochemical condition correlating with depth. This result was consistent with reports from other coalbed ecosystems that also observed significant diversity between microbial communities spanning different coal formations (8, 9, 11, 60, 61). Indeed, differences in coal rank are expected to influence CBM production as low-maturity coals (e.g., subbituminous coals) are less recalcitrant and more biodegradable than higher-maturity coals (e.g., volatile bituminous coals) due to changes in carbon and oxygen content (8). It should be noted that considerable differences in microbial CBM production have been observed from incubations with identical low-rank coals (62, 63), suggesting that local well conditions also contribute to biogenic CBM production potential. Such conditions could include the availability and type of methanogenic substrates (62), the hydrogeological regime of basin waters (4, 64), and coal basin permeability (65).
Despite the large variation in microbial community structure observed across the Alberta Basin, all indicator groups contained unique consortia with a potential to transform coal into methane. For example, specific OTUs previously implicated in aromatic compound degradation, fermentation, and methanogenesis, respectively, affiliated with Thauera (66), Streptococcus, and Methanosarcina were found in the subbituminous coals indicator group, whereas OTUs with similar functional potentials affiliated with uncultured Rhodobacteraceae, Pelobacter, and Methanosarcina were detected in the volatile bituminous coals indicator group. While this suggests that local selection pressures may determine niche partitioning across various taxonomic affiliations, thermodynamic constraints promote recurring patterns of syntrophic interactions that drive CBM production through metabolic cooperation (67). The extent to which cooperative interactions change over time or succeed one another during the coal conversion process remains to be determined. Recent anaerobic-digester studies suggest that syntrophic populations of fermenting bacteria and methanogenic archaea remain stable and resilient for a particular function, in contrast to nonsyntrophic community members, both over time and under perturbation (68). However, other studies monitoring similar syntrophic populations involved in anaerobic conversion of municipal solid waste to methane have reported successive interactions, where hydrolysis of larger polymeric substances occurs initially, followed by acidification and finally methanogenesis (69, 70). We posit that similar successional patterns transpire in the coalbed milieu, albeit over longer times scales, governed by a given coal formation's decomposition state.
Aromatic compound degradation in a deep coalbed.
Published metagenomic sequence information from coalbed ecosystems is conspicuously underrepresented in public databases. Sequence information from the San Juan Basin in New Mexico focused on different stages of CBM production can be gleaned from a patent application filed in 2011 (G. V. Toledo, T. H. Richardson, U. Singl, E. J. Mathur, and J. C. Venter, U.S. patent application US20100047793). More recently, Strachan and colleagues used coalbed-derived fosmid libraries to study lignin-transforming activities from Rockyford Standard (CO182) and Basal (CO183) coal zones in the Alberta Basin (71). In the current study, we produced a metagenome from a volatile bituminous coal sample dominated by Rhodobacteraceae sequences that could be assembled and binned into a nearly complete population genome related to Celeribacter sp. (bin 1). Metabolic pathways inferred from the Celeribacter sp. population genome identified routes for both autotrophic and heterotrophic growth, as well as chemolithotrophy via CO oxidation, reflective of a mixotrophic lifestyle. The genome also encoded pathways for acidogenic fermentation, which would allow coalbed Celeribacter sp. populations to survive under both aerobic and anaerobic conditions. Importantly, the products of such fermentation reactions (i.e., acetate, H2, and CO2) could serve as primary substrates for methanogens, thereby supporting biogenic CBM production. This versatile energy metabolism would be advantageous for life close to fluctuating oxic-anoxic interfaces that experience variable nutrient availabilities (72). Such dynamic conditions may be occurring in the Mannville Group coal seam due to mixing of fluids from lower, more saline, aquifers that may carry dissolved oxygen (64).
The Celeribacter sp. population genomic potential to degrade and utilize a diverse range of aromatic compounds, including benzoate, salicylate, and vanillate, is relevant to CBM production. Indeed, benzoate, salicylate, and vanillate are all by-products of coal and lignin degradation (73, 74), and their utilization likely results in the production of key organic intermediates that can serve as substrates for fermenting bacteria and methanogens (75). Interestingly, encoded pathways for aromatic degradation employed enzymes exhibiting high sequence similarity to aerobic homologs, such as different mono- and dioxygenases. Similar observations have been made across diverse hydrocarbon resource environments (14), where it has been suggested that aerobic enzyme activity is supported via slow diffusion of oxygen from the solid coal matrix or recharge from oxygen-bearing meteoric waters flowing from fractured coal seams (64). Strictly speaking, aerobic degradation of aromatic compounds is at odds with biogenic CBM production because primary fermentation of the resulting organic compounds to methanogenic substrates and smaller organics is not thought to occur under aerobic conditions. However, based on inferred pathways that would allow coalbed Celeribacter sp. to accumulate glycogen and ferment, we hypothesize that exposure of Celeribacter sp. to oxic-anoxic cycling may promote the biodegradation of coal to methane. Here, during recharge of coalbeds with oxygen, Celeribacter sp. would degrade aromatic compounds for biomass synthesis and glycogen storage. During subsequent periods of anoxia, accumulated glycogen would be fermented by Celeribacter sp. for carbon and energy requirements via acidogenic fermentation pathways, resulting in excreted by-products that support methanogenesis. Similar physiological adaptations have been observed in other bacteria that experience periodic oxygen cycling, such as organisms responsible for biological phosphorus removal in activated sludge ecosystems (76). Indeed, the high abundance of anaerobic bacteria, such as Acetobacterium- and Pelobacter-related organisms, in both the amplicon and metagenomic sequencing data sets is consistent with the production of Celeribacter sp. fermentation by-products, where Acetobacterium and Pelobacter may facilitate acetogenic and secondary fermentation processes that also drive methanogenesis. Nonetheless, future experiments are needed to decipher specific interactions between these organisms and Celeribacter sp. that result in the degradation of coal to methane.
Genomic differentiation between Celeribacter sp.
Comparative analysis of the Celeribacter sp. population genome with cultivated reference genomes from activated sludge, saline soil, and marine environments revealed potentially adaptive habitat-specific traits. Our results showed that variable regions within the Celeribacter sp. population genome harbored genes involved in aromatic compound degradation, consistent with the high availability of these substrates in the coalbed milieu. A recent study identified numerous gene clusters sourced from coalbeds that acted on lignin-derived aromatic polymers that were associated with MGEs, raising the possibility that coal and lignin transformation is a highly specialized process (71). Interestingly, one MGE encoded a prophage containing enzymes for benzoate degradation. It is possible that this virus-encoded benzoic acid dioxygenase would allow infecting phage to reprogram the host carbon metabolism (77, 78); however, such events require further characterization in coalbed environments.
The presence of nitrogen and carbohydrate catabolism genes within Celeribacter sp. variable genomic regions may also reflect adaptive metabolic strategies. The ability to rapidly assimilate and degrade carbohydrates or organic acids that periodically become available in the coalbed environment may be preferentially used by Celeribacter sp., as has been shown for other Proteobacteria (79–81). While such compounds are not expected to be abundant in deep coalbeds, they may become available via biomass decay (82). Moreover, scavenged carbohydrate or organic acid compounds could also provide fermentation substrates for Celeribacter sp. under anaerobic conditions, thereby stimulating biogenic methane production. Genes involved in nitrogen acquisition identified within Celeribacter sp. variable regions included ABC-type transporters for polyamines and nitrate. These systems are known to play important roles in nitrogen scavenging (83) and may be key adaptive functions for survival in deep coalbeds manifesting limited nitrogen resources.
Concluding remarks.
In summary, our results show that microbial community structure displayed patterns of endemism and habitat selection across coalbed formations within the Alberta Basin, consistent with observations for other geographical locations (8, 9, 11, 60, 61). While some phylum-level taxonomic patterns were observed, the relative abundance of specific taxonomic groups was localized to discrete wells likely shaped by habitat-specific environmental conditions, such as coal rank and depth-dependent physicochemical conditions. This was reinforced by the Celeribacter sp. population genome recovered using metagenomic sequencing, where encoded metabolic pathways for the utilization of numerous aromatic compounds and the production of fermentation by-products may support specialization within the biodegradation of coal to methane. Genomic comparisons between coalbed Celeribacter sp. and closely related species displayed a continuum of differentiation, likely reflective of habitat-specific selection pressures (58, 84). These findings implicate nitrogen availability and the ability to utilize diverse organic substrates as potential drivers of selection in deep coalbeds, which should be considered in the development of biostimulation strategies to enhance microbial CBM production in nonproductive or declining wells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Melanie Scofield for assistance with laboratory work.
This work was carried out under the auspices of Genome Canada, Genome British Columbia, Genome Alberta, the Natural Sciences and Engineering Research Council of Canada, Canada Foundation for Innovation, the Tula Foundation-funded Centre for Microbial Diversity and Evolution, the Canadian Institute for Advanced Research (CIFAR), and Innovates Centre of Research Excellence, Alberta, through grants awarded to S.J.H. and K.B. The Western Canadian Research Grid provided access to high-performance computing resources.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01737-15.
REFERENCES
- 1.Tiwary RK. 2001. Environmental impact of coal mining on water regime and its management. Water Air Soil Pollut 132:185–199. doi: 10.1023/A:1012083519667. [DOI] [Google Scholar]
- 2.Bian Z, Inyang HI, Daniels JL, Otto F, Struthers S. 2010. Environmental issues from coal mining and their solutions. Miner Sci Technol 20:215–223. [Google Scholar]
- 3.Strąpoć D, Mastalerz M, Dawson K, Macalady J, Callaghan AV, Wawrik B, Turich C, Ashby M. 2011. Biogeochemistry of microbial coal-bed methane. Annu Rev Earth Planet Sci 39:617–656. doi: 10.1146/annurev-earth-040610-133343. [DOI] [Google Scholar]
- 4.Scott AR. 2002. Hydrogeologic factors affecting gas content distribution in coal beds. Int J Coal Geol 50:363–387. doi: 10.1016/S0166-5162(02)00135-0. [DOI] [Google Scholar]
- 5.Jones EJ, Voytek MA, Corum MD, Orem WH. 2010. Stimulation of methane generation from nonproductive coal by addition of nutrients or a microbial consortium. Appl Environ Microbiol 76:7013–7022. doi: 10.1128/AEM.00728-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penner TJ, Foght JM, Budwill K. 2010. Microbial diversity of western Canadian subsurface coal beds and methanogenic coal enrichment cultures. Int J Coal Geol 82:81–93. doi: 10.1016/j.coal.2010.02.002. [DOI] [Google Scholar]
- 7.Klein DA, Flores RM, Venot C, Gabbert K, Schmidt R, Stricker GD, Pruden A, Mandernack K. 2008. Molecular sequences derived from Paleocene Fort Union Formation coals vs. associated produced waters: implications for CBM regeneration. Int J Coal Geol 76:3–13. doi: 10.1016/j.coal.2008.05.023. [DOI] [Google Scholar]
- 8.Strapoc D, Picardal FW, Turich C, Schaperdoth I, Macalady JL, Lipp JS, Lin Y-S, Ertefai TF, Schubotz F, Hinrichs K-U, Mastalerz M, Schimmelmann A. 2008. Methane-producing microbial community in a coal bed of the Illinois basin. Appl Environ Microbiol 74:2424–2432. doi: 10.1128/AEM.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fry JC, Horsfield B, Sykes R, Cragg BA, Heywood C, Kim GT, Mangelsdorf K, Mildenhall DC, Rinna J, Vieth A, Zink K-G, Sass H, Weightman AJ, Parkes RJ. 2009. Prokaryotic populations and activities in an interbedded coal deposit, including a previously deeply buried section (1.6–2.3 km) above ∼150 Ma basement rock. Geomicrobiol J 26:163–178. doi: 10.1080/01490450902724832. [DOI] [Google Scholar]
- 10.Midgley DJ, Hendry P, Pinetown KL, Fuentes D, Gong S, Mitchell DL, Faiz M. 2010. Characterisation of a microbial community associated with a deep, coal seam methane reservoir in the Gippsland Basin, Australia. Int J Coal Geol 82:232–239. doi: 10.1016/j.coal.2010.01.009. [DOI] [Google Scholar]
- 11.Tang Y-Q, Ji P, Lai G-L, Chi C-Q, Liu Z-S, Wu X-L. 2012. Diverse microbial community from the coalbeds of the Ordos Basin, China. Int J Coal Geol 90–91:21–33. [Google Scholar]
- 12.Ünal B, Perry VR, Sheth M, Gomez-Alvarez V, Chin K-J, Nüsslein K. 2012. Trace elements affect methanogenic activity and diversity in enrichments from subsurface coal bed produced water. Front Microbiol 3:175. doi: 10.3389/fmicb.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaton A, Langenberg W, Pană C. 2006. Coalbed methane resources and reservoir characteristics from the Alberta Plains, Canada. Int J Coal Geol 65:93–113. doi: 10.1016/j.coal.2005.04.013. [DOI] [Google Scholar]
- 14.An D, Caffrey SM, Soh J, Agrawal A, Brown D, Budwill K, Dong X, Dunfield PF, Foght J, Gieg LM, Hallam SJ, Hanson NW, He Z, Jack TR, Klassen J, Konwar KM, Kuatsjah E, Li C, Larter S, Leopatra V, Nesbø CL, Oldenburg T, Pagé AP, Ramos-Padron E, Rochman FF, Saidi-Mehrabad A, Sensen CW, Sipahimalani P, Song YC, Wilson S, Wolbring G, Wong M-L, Voordouw G. 2013. Metagenomics of hydrocarbon resource environments indicates aerobic taxa and genes to be unexpectedly common. Environ Sci Technol 47:10708–10717. doi: 10.1021/es4020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luef B, Frischkorn KR, Wrighton KC, Holman H-YN, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH, Comolli LR, Banfield JF. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 16.Foght J, Aislabie J, Turner S, Brown CE, Ryburn J, Saul DJ, Lawson W. 2004. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb Ecol 47:329–340. [DOI] [PubMed] [Google Scholar]
- 17.Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- 18.Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ, Sullivan MB. 2013. Diversity and population structure of marine group A bacteria in the northeast subarctic Pacific Ocean. ISME J 7:256–268. doi: 10.1038/ismej.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 23.Dufrene M, Legendre P. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345–366. doi: 10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2. [DOI] [Google Scholar]
- 24.De Cáceres M, Legendre P, Moretti M. 2010. Improving indicator species analysis by combining groups of sites. Oikos 119:1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x. [DOI] [Google Scholar]
- 25.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J-B, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt K, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y-W, Tang Y-H, Tringe SG, Simmons BA, Singer SW. 2014. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. 2014. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konwar KM, Hanson NW, Pagé AP, Hallam SJ. 2013. MetaPathways: a modular pipeline for constructing pathway/genome databases from environmental sequence information. BMC Bioinformatics 14:202. doi: 10.1186/1471-2105-14-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson NW, Konwar KM, Hawley AK, Altman T, Karp PD, Hallam SJ. 2014. Metabolic pathways for the whole community. BMC Genomics 15:619. doi: 10.1186/1471-2164-15-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res 17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karp PD, Latendresse M, Caspi R. 2011. The pathway tools pathway prediction algorithm. Stand Genomic Sci 5:424–429. doi: 10.4056/sigs.1794338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A. 2015. Microbial species delineation using whole-genome sequences. Nucleic Acids Res 43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. 2009. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- 37.Gihring TM, Green SJ, Schadt CW. 2012. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14:285–290. doi: 10.1111/j.1462-2920.2011.02550.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim B-C, Kim S, Shin T, Kim H, Sang B-I. 2013. Comparison of the bacterial communities in anaerobic, anoxic, and oxic chambers of a pilot A2O process using pyrosequencing analysis. Curr Microbiol 66:555–565. doi: 10.1007/s00284-013-0311-z. [DOI] [PubMed] [Google Scholar]
- 39.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H, Yu Z, Liu R, Zhang H, Zhong Q, Xiong Z. 2012. Methylotrophic methanogenesis governs the biogenic coal bed methane formation in Eastern Ordos Basin, China. Appl Microbiol Biotechnol 96:1587–1597. doi: 10.1007/s00253-012-3889-3. [DOI] [PubMed] [Google Scholar]
- 41.Beckmann S, Lueders T, Krüger M, von Netzer F, Engelen B, Cypionka H. 2011. Acetogens and acetoclastic methanosarcinales govern methane formation in abandoned coal mines. Appl Environ Microbiol 77:3749–3756. doi: 10.1128/AEM.02818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MJ, Allen CCR, Kulakov LA, Lipscomb DA. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J Bacteriol 181:6200–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar M, Khanna S. 2010. Diversity of 16S rRNA and dioxygenase genes detected in coal-tar-contaminated site undergoing active bioremediation. J Appl Microbiol 108:1252–1262. doi: 10.1111/j.1365-2672.2009.04523.x. [DOI] [PubMed] [Google Scholar]
- 44.Albertsen M, Saunders AM, Nielsen KL, Nielsen PH. 2013. Metagenomes obtained by “deep sequencing”—what do they tell about the enhanced biological phosphorus removal communities? Water Sci Technol 68:1959–1968. doi: 10.2166/wst.2013.441. [DOI] [PubMed] [Google Scholar]
- 45.Crosby LD, Criddle CS. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques 34:790–794, 796, 798. [DOI] [PubMed] [Google Scholar]
- 46.Pinto AJ, Raskin L. 2012. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7:e43093. doi: 10.1371/journal.pone.0043093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alber BE, Spanheimer R, Ebenau-Jehle C, Fuchs G. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol Microbiol 61:297–309. doi: 10.1111/j.1365-2958.2006.05238.x. [DOI] [PubMed] [Google Scholar]
- 48.Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. 2011. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- 49.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 50.Head IM, Jones DM, Larter SR. 2003. Biological activity in the deep subsurface and the origin of heavy oil. Nature 426:344–352. doi: 10.1038/nature02134. [DOI] [PubMed] [Google Scholar]
- 51.King GM. 2003. Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl Environ Microbiol 69:7257–7265. doi: 10.1128/AEM.69.12.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King GM, Weber CF. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5:107–118. doi: 10.1038/nrmicro1595. [DOI] [PubMed] [Google Scholar]
- 53.Li K, Wang S, Shi Y, Qu J, Zhai Y, Xu L, Xu Y, Song J, Liu L, Rahman MA, Yan Y. 2011. Genome sequence of Paracoccus sp. strain TRP, a chlorpyrifos biodegrader. J Bacteriol 193:1786–1787. doi: 10.1128/JB.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riedel T, Spring S, Fiebig A, Petersen J, Kyrpides NC, Göker M, Klenk H-P. 2014. Genome sequence of the exopolysaccharide-producing Salipiger mucosus type strain (DSM 16094T), a moderately halophilic member of the Roseobacter clade. Stand Genomic Sci 9:1331–1343. doi: 10.4056/sigs.4909790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee S-Y, Park S, Oh T-K, Yoon J-H. 2012. Celeribacter baekdonensis sp. nov., isolated from seawater, and emended description of the genus Celeribacter Ivanova et al. 2010. Int J Syst Evol Microbiol 62:1359–1364. doi: 10.1099/ijs.0.032227-0. [DOI] [PubMed] [Google Scholar]
- 56.Lai Q, Cao J, Yuan J, Li F, Shao Z. 2014. Celeribacter indicus sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium from deep-sea sediment and reclassification of Huaishuia halophila as Celeribacter halophilus comb. nov. Int J Syst Evol Microbiol 64:4160–4167. doi: 10.1099/ijs.0.069039-0. [DOI] [PubMed] [Google Scholar]
- 57.Cao J, Lai Q, Yuan J, Shao Z. 2015. Genomic and metabolic analysis of fluoranthene degradation pathway in Celeribacter indicus P73T. Sci Rep 5:7741. doi: 10.1038/srep07741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coleman ML, Chisholm SW. 2010. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci U S A 107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polz MF, Alm EJ, Hanage WP. 2013. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29:170–175. doi: 10.1016/j.tig.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimizu S, Akiyama M, Naganuma T, Fujioka M, Nako M, Ishijima Y. 2007. Molecular characterization of microbial communities in deep coal seam groundwater of northern Japan. Geobiology 5:423–433. doi: 10.1111/j.1472-4669.2007.00123.x. [DOI] [Google Scholar]
- 61.Li D, Hendry P, Faiz M. 2008. A survey of the microbial populations in some Australian coalbed methane reservoirs. Int J Coal Geol 76:14–24. doi: 10.1016/j.coal.2008.04.007. [DOI] [Google Scholar]
- 62.Harris SH, Smith RL, Barker CE. 2008. Microbial and chemical factors influencing methane production in laboratory incubations of low-rank subsurface coals. Int J Coal Geol 76:46–51. doi: 10.1016/j.coal.2008.05.019. [DOI] [Google Scholar]
- 63.Jones EJP, Voytek MA, Warwick PD, Corum MD, Cohn A, Bunnell JE, Clark AC, Orem WH. 2008. Bioassay for estimating the biogenic methane-generating potential of coal samples. Int J Coal Geol 76:138–150. doi: 10.1016/j.coal.2008.05.011. [DOI] [Google Scholar]
- 64.Bachu S, Michael K. 2003. Possible controls of hydrogeological and stress regimes on the producibility of coalbed methane in Upper Cretaceous—tertiary strata of the Alberta basin, Canada. AAPG Bull 87:1729–1754. doi: 10.1306/06030302015. [DOI] [Google Scholar]
- 65.Tremain C, Tyler R. 1997. Cleat, fracture, and stress patterns in the Piceance Basin, Colorado: controls on coalbed methane producibility, p 103–114. In Hoak TE, Klawitter AL, Blomquist PK (ed) Fractured reservoirs: characterization and modeling guidebook. Rocky Mountain Association of Geologists, Denver, CO. [Google Scholar]
- 66.Breinig S, Schiltz E, Fuchs G. 2000. Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J Bacteriol 182:5849–5863. doi: 10.1128/JB.182.20.5849-5863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 68.Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, Cummings TA, Beers AR, Knight R, Angenent LT. 2011. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci U S A 108:4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barlaz MA, Schaefer DM, Ham RK. 1989. Bacterial population development and chemical characteristics of refuse decomposition in a simulated sanitary landfill. Appl Environ Microbiol 55:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nayak BS, Levine AD, Cardoso A, Harwood VJ. 2009. Microbial population dynamics in laboratory-scale solid waste bioreactors in the presence or absence of biosolids. J Appl Microbiol 107:1330–1339. doi: 10.1111/j.1365-2672.2009.04319.x. [DOI] [PubMed] [Google Scholar]
- 71.Strachan CR, Singh R, VanInsberghe D, Ievdokymenko K, Budwill K, Mohn WW, Eltis LD, Hallam SJ. 2014. Metagenomic scaffolds enable combinatorial lignin transformation. Proc Natl Acad Sci U S A 111:10143–10148. doi: 10.1073/pnas.1401631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castelle CJ, Hug LA, Wrighton KC, Thomas BC, Williams KH, Wu D, Tringe SG, Singer SW, Eisen JA, Banfield JF. 2013. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat Commun 4:2120. doi: 10.1038/ncomms3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annu Rev Plant Biol 54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 74.Sainsbury PD, Hardiman EM, Ahmad M, Otani H, Seghezzi N, Eltis LD, Bugg TDH. 2013. Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. ACS Chem Biol 8:2151–2156. doi: 10.1021/cb400505a. [DOI] [PubMed] [Google Scholar]
- 75.Orem WH, Voytek MA, Jones EJ, Lerch HE, Bates AL, Corum MD, Warwick PD, Clark AC. 2010. Organic intermediates in the anaerobic biodegradation of coal to methane under laboratory conditions. Org Geochem 41:997–1000. doi: 10.1016/j.orggeochem.2010.03.005. [DOI] [Google Scholar]
- 76.Kristiansen R, Nguyen HTT, Saunders AM, Nielsen JL, Wimmer R, Le VQ, McIlroy SJ, Petrovski S, Seviour RJ, Calteau A, Nielsen KL, Nielsen PH. 2013. A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal. ISME J 7:543–554. doi: 10.1038/ismej.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hurwitz BL, Hallam SJ, Sullivan MB. 2013. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol 14:R123. doi: 10.1186/gb-2013-14-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roux S, Hawley AK, Torres Beltran M, Scofield M, Schwientek P, Stepanauskas R, Woyke T, Hallam SJ, Sullivan MB. 2014. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLife 3:e03125. doi: 10.7554/eLife.03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerischer U. 2002. Specific and global regulation of genes associated with the degradation of aromatic compounds in bacteria. J Mol Microbiol Biotechnol 4:111–121. [PubMed] [Google Scholar]
- 80.Dal S, Steiner I, Gerischer U. 2002. Multiple operons connected with catabolism of aromatic compounds in Acinetobacter sp. strain ADP1 are under carbon catabolite repression. J Mol Microbiol Biotechnol 4:389–404. [PubMed] [Google Scholar]
- 81.Fischer R, Bleichrodt FS, Gerischer UC. 2008. Aromatic degradative pathways in Acinetobacter baylyi underlie carbon catabolite repression. Microbiology 154:3095–3103. doi: 10.1099/mic.0.2008/016907-0. [DOI] [PubMed] [Google Scholar]
- 82.Okabe S, Kindaichi T, Ito T. 2005. Fate of 14C-labeled microbial products derived from nitrifying bacteria in autotrophic nitrifying biofilms. Appl Environ Microbiol 71:3987–3994. doi: 10.1128/AEM.71.7.3987-3994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mou X, Vila-Costa M, Sun S, Zhao W, Sharma S, Moran MA. 2011. Metatranscriptomic signature of exogenous polyamine utilization by coastal bacterioplankton. Environ Microbiol Rep 3:798–806. doi: 10.1111/j.1758-2229.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- 84.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, Chisholm SW. 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.