Abstract
Objectives:
We aimed to study the potential clinical relevance of 9p allelic loss, with or without copy number variation, in 1p/19q codeleted anaplastic oligodendroglial tumors (AOTs).
Methods:
This study enrolled 216 patients with 1p/19q codeleted AOT. The prognostic value of 9p allelic loss was investigated using a French nation-wide prospective registry, POLA (prise en charge des tumeurs oligodendrogliales anaplasiques) and high-density single nucleotide polymorphism arrays. We validated our results using the Repository of Molecular Brain Neoplasia Data (REMBRANDT) dataset.
Results:
The minimal common region of allelic loss in chromosome arm 9p was 9p21.3. Allelic loss of 9p21.3, detected in 41.7% of tumors, was associated with shorter progression-free and overall survival rates in univariate (p = 0.008 and p < 0.001, respectively) and multivariate analyses (p = 0.009 and p = 0.009, respectively). This finding was validated in the REMBRANDT dataset in univariate and multivariate analysis (p = 0.01 and p = 0.01, respectively).
Conclusion:
Our study highlights a novel potential prognostic biomarker in 1p/19q codeleted AOT. Further prospective studies are warranted to investigate our finding.
Anaplastic or World Health Organization (WHO) grade III oligodendroglial tumors (AOTs), including anaplastic oligodendrogliomas and anaplastic oligoastrocytomas, form a heterogeneous subgroup of diffuse gliomas in adults. So far, 3 major histomolecular subgroups with major clinical relevance have been individualized based on 2 molecular biomarkers: (1) 1p/19q codeleted AOT in approximately 80% of the cases, (2) non-1p/19q codeleted and IDH-mutated AOT in approximately 10% of the cases, and (3) non-1p/19q codeleted and IDH wild-type AOT in approximately 10% of the cases.1,2 Of note, almost all 1p/19q codeleted AOTs are also IDH1/IDH2 mutated and TERT promoter mutated.3
Patients with 1p/19q codeleted AOTs have better prognosis and better response to radiotherapy (RT) plus procarbazine-CCNU-vincristine (PCV) chemotherapy compared with their non-1p/19q codeleted counterparts.4,5 However, in both studies, the median overall survival (OS) in the 1p/19q codeleted subgroup had a broad range of 95% confidence interval (from 6.4 to more than 12 years in one study and a 5-year OS rate from 60.3% to 86.4% in the other study),4,5 suggesting that some patients would have a less favorable prognosis. Identification of novel biomarkers in this tumor subgroup would help to better stratify patients with 1p/19q codeleted AOT.
Although promising novel somatic mutations in CIC (capicua transcriptional repressor) and FUBP1 (far upstream element binding protein 1) have been identified in 1p/19q codeleted AOT, their clinical significance remains unclear.6 Additional prognostic markers are still needed to better stratify the largest group of patients with AOT (i.e., 1p/19q codeleted AOT).
We have recently shown that 9p allelic loss, including monoallelic loss (i.e., hemizygous deletion), monoallelic loss without copy number change (i.e., copy neutral loss of heterozygosity or acquired uniparental disomy [CN-LOH]), and biallelic loss (i.e., homozygous deletion), is frequent in AOT.7 Since 1p/19q codeletion confers a homogeneous glioma phenotype, we have only analyzed AOT harboring this genomic aberration.1 In the present study enrolling a larger number of patients, we addressed the prognostic value of 9p allelic loss in 1p/19q codeleted AOT from the POLA (prise en charge des tumeurs oligodendrogliales anaplasiques) Network, a French nationwide cohort prospectively enrolling all newly diagnosed patients with AOT. The clinical impact of 9p allelic loss has been validated using the publicly available Repository of Molecular Brain Neoplasia Data (REMBRANDT) dataset.8
METHODS
After local diagnosis of newly (with no history of brain tumor) diagnosed AOT, patients were enrolled in the POLA registry. The tumor was then centrally reviewed and characterized for 1p/19q and IDH statuses.7 Participating centers were invited to follow medical guidelines consisting of RT alone that was subsequently changed to RT plus PCV when the results of the European Organization for Research and Treatment of Cancer and the Radiation Therapy Oncology Group phase III clinical trials were updated.4,5 However, participating centers were allowed to select other options (chemotherapy alone or RT plus chemotherapy).
We downloaded the clinical data from the REMBRANDT dataset at https://caintegrator.nci.nih.gov/rembrandt/legal.jsp on February 1, 2015.
Patients.
From the POLA registry, 216 patients with a centrally reviewed diagnosis of newly diagnosed AOT were included in the present study. For all individuals, fresh-frozen tumor tissues were available for genomic and pathologic investigations.
From the REMBRANDT dataset, we used, as a validation cohort, 241 gliomas (WHO grades II and III) including 51 samples exhibiting 1p/19q codeletion.
Standard protocol approvals, registrations, and patient consents.
All patients provided their written informed consent for molecular analysis. The study was approved by the ethics committee of the Hôpital Universitaire La Pitié-Salpêtrière.
Single nucleotide polymorphism array.
DNA extraction was performed as previously reported.8 Single nucleotide polymorphism (SNP) array hybridizations were outsourced to Integragen, as previously reported.7 Two types of platforms were used: HumanCNV370-Quad and Human610-Quad from Illumina (San Diego, CA). Using these normalized log2(ratio) and B-allele frequency values, copy number abnormalities were identified with the SNP-FASST2 (fast adaptive states segmentation technique 2) segmentation algorithm implemented in the Nexus software. Single copy gains and losses were defined with log2(ratio) values of 0.25 and −0.25, respectively, while 2 or more than 2 copies of gains and losses were defined by log2(ratio) values of 0.7 and −1.1, respectively. A chromosome region was identified as allelic loss if ≥95% of the SNP probes in a DNA segment of at least 500 kb exhibited B-allele frequencies of ≥0.8 or ≤0.2. Allelic loss, with a copy number of 2, was considered to be CN-LOH. The 1p/19q codeletion was defined as loss of at least 90% of the probes mapped on 1p and 19q with concurrent chromosomes 1 and 19 centromeric breakpoints. Minimal common regions (MCRs) of gain and loss were identified using the cghMCR package (v1.18) by aligning samples based on chromosome segments defined by genes using the CNTools package (v1.22). Briefly, MCRs are defined as contiguous spans having at least a recurrence rate of 0.75 across samples. If 2 or more altered segments are separated by less than 500 kb, the entire region spanned by the segments is considered to be one altered span. In addition, to further confirm these results, we used an independent approach to reduce copy number dimensions of the SNP array set without losing information, using the CGHregions package (v1.24),8 and we performed 10,000 permutations to obtain a false discovery rate p value from the log-rank p value related to every genomic aberration using the CGHtest package (v1.1).9
We further explored our findings using a validation cohort from the REMBRANDT dataset.10 The CEL files from the Affymetrix GeneChip Human 100K SNP array set were downloaded from http://array.nci.nih.gov and clinical data were downloaded from https://caintegrator.nci.nih.gov/rembrandt/legal.jsp on February 1, 2015. We obtained total copy number from CEL files using the aroma.affymetrix package (v2.13.1) with the CRMAv2 algorithm.11 When paired (tumor and germline) DNA was available, we estimated allele-specific copy number using parent-specific copy number segmentation (PSCBS package v0.43).12 We performed allele-specific copy number segmentation in nonpaired samples (only tumor DNA) using the CalMaTe algorithm (v0.11).13
Statistical analyses.
All analyses were performed with R-project (v3.0.2).14 Comparisons of the clinical features of 1p19q codeleted AOT, with vs without 9p allelic loss, were performed with Fisher exact test for discrete variables, t test for continuous variables, and otherwise, the Mann–Whitney U test was used.
Survival analysis was performed using the survival package (v2.37-7) in R.15 Progression-free survival (PFS) was calculated from initial diagnosis to tumor progression, death, or last follow-up. OS was calculated from initial diagnosis to death or last follow-up. Radiologic tumor progression was defined as any increase of contrast-enhancing tumor area more than 25% or an additional contrast-enhancing area. Clinical tumor progression was defined as significant deterioration of neurologic status. Survivals were plotted using the Kaplan-Meier method and compared using the log-rank test and Cox proportional hazard ratio. Variables with p value <0.2 in univariate analysis were included as covariates in the multivariate analysis: (1) age, as categorical variable, dichotomized at the median of the whole cohort, i.e., 50 years; (2) Karnofsky performance status, as categorical variable ≥70% vs <70%; and (3) treatments received, i.e., RT plus PCV, RT alone, RT plus temozolomide, and other chemotherapies alone. Interactions were not considered. Patients with no missing data were retained for multivariate analysis. The POLA cohort is prospective starting in 2009 with median follow-up of 2.5 years; thus, to allow more robust comparisons of medians, we have selected PFS and OS rate at 3 years.
In the REMBRANDT dataset analysis, the variables 1p/19q codeletion and phenotype (unknown, mixed, oligodendroglioma, and astrocytoma) were included as covariates in the Cox proportional hazard ratio. Interactions were not considered.
Statistical significance was defined as a p value <0.05; all tests were 2-sided.
RESULTS
Population.
Two hundred sixteen patients with 1p/19q codeleted AOT enrolled in the POLA registry between 2009 and 2013 were included in the present study. The distribution of the main clinical features of the patients is summarized in table 1. The frequency of chromosome aberrations is represented in figure e-1 on the Neurology® Web site at Neurology.org. We used the REMBRANDT dataset as a validation series (241 gliomas, 51 harboring 1p/19q codeletion).
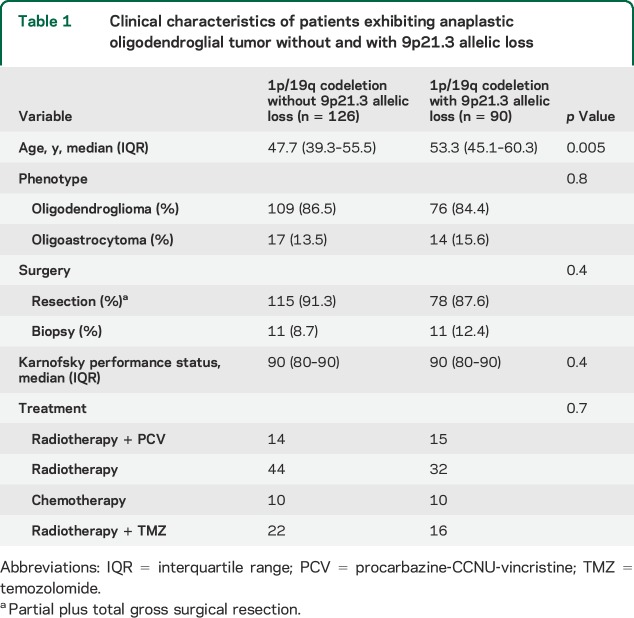
Table 1.
Clinical characteristics of patients exhibiting anaplastic oligodendroglial tumor without and with 9p21.3 allelic loss
MCR of allelic loss in chromosome arm 9p MCR(9p).
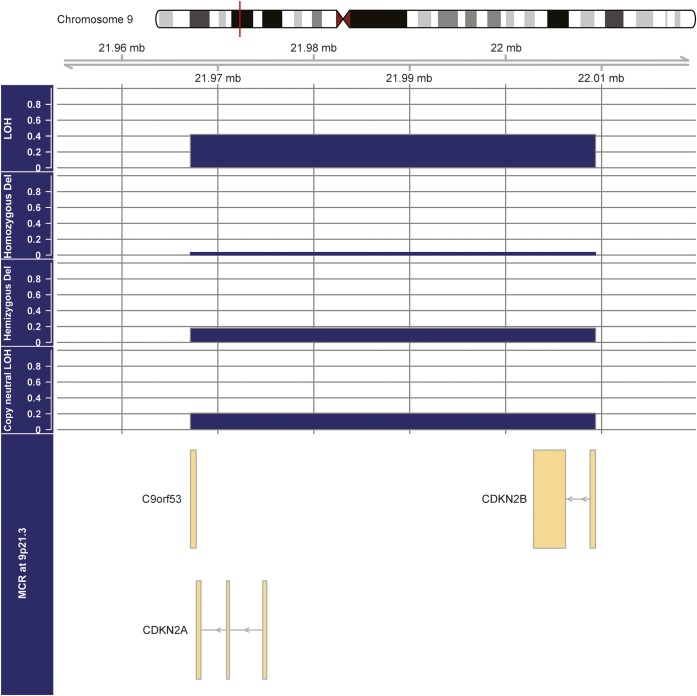
The MCR(9p) is the genomic region 9p21.3 (from 21957144 to 21999312) including C9orf53, CDKN2A, and CDKN2B (figure 1).
Figure 1. Minimal common region of allelic loss in a 1p/19q codeleted anaplastic oligodendroglial tumor harboring 9p21.3 allelic loss.
The different panels show the frequencies of the different mechanisms of loss of heterozygosity (LOH). MCR = minimal common region.
Overall, 9p21.3 allelic loss was detected in 90 of 216 patients (41.7%). The mechanisms of 9p21.3 allelic loss were CN-LOH, hemizygous deletion, and homozygous deletion in 44 cases (48.9%), 39 cases (43.3%), and 7 cases (7.8%), respectively (figure 1). Accordingly, in the REMBRANDT dataset, 9p21.3 allelic loss was detected in 21 of 51 1p/19q codeleted samples (41.1%). The mechanisms of 9p21.3 allelic loss in 1p/19q codeleted samples from the REMBRANDT dataset were CN-LOH, hemizygous deletion, and homozygous deletion in 12 cases (57.1%), 8 cases (38.1%), and 1 case (4.8%), respectively (data not shown).
Clinico-molecular correlations.
Patients with 1p/19q codeleted AOT with 9p21.3 allelic loss were older than their 9p21.3 intact counterparts (p = 0.005). The distribution of histologic phenotype, surgical procedure (biopsy vs surgery including partial and total gross resection), and medical treatments according to the 9p21.3 status was similar (table 1).
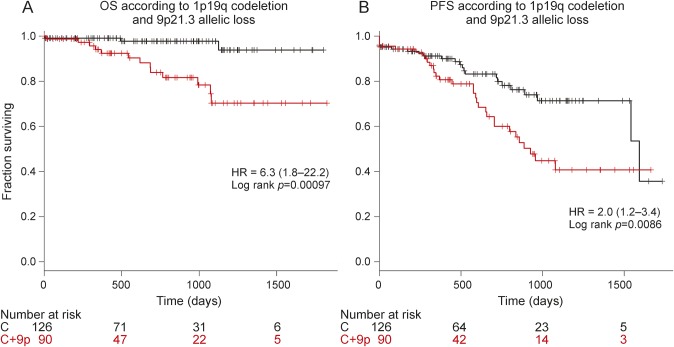
Allelic loss of 9p21.3 was associated with shorter PFS and OS in univariate analysis (p = 0.008 and p < 0.001, respectively) (table 2, figure 2, A and B). We have corrected log-rank p value in order to identify additional potential genomic aberrations associated with prognosis in 1p/19q codeleted AOT performing 10,000 permutations. Of note, the sole genomic alteration with a false discovery rate log-rank p value <0.05 was CDKN2A locus (figure e-1).
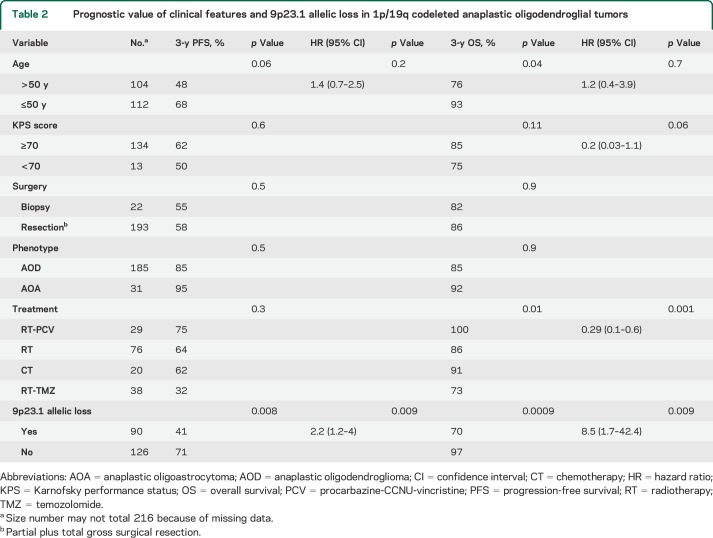
Table 2.
Prognostic value of clinical features and 9p23.1 allelic loss in 1p/19q codeleted anaplastic oligodendroglial tumors
Figure 2. Kaplan-Meier curves in 1p/19q codeleted anaplastic oligodendroglial tumor according to 9p21.3 loss.
Progression-free survival (PFS) (A) and overall survival (OS) (B) according to 9p23.1 allelic status. C = 1p/19q codeletion without 9p21.3 allelic loss; C+9p = 1p/19q codeletion with 9p21.3 allelic loss; HR = hazard ratio.
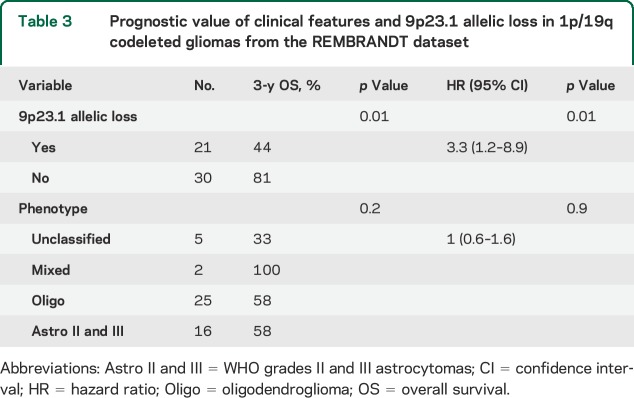
In multivariate analysis for PFS, 9p21.3 allelic loss was the sole factor associated with a shorter 3-year PFS rate (41% vs 71%, p = 0.009, hazard ratio [HR] = 2.2 [1.2–4]). For OS, 9p21.3 allelic loss was associated with shorter 3-year OS rate (70% vs 97%, p = 0.009, HR = 8.5 [1.7–42.4]) (table 2). Accordingly, in the REMBRANDT dataset, 9p21.3 allelic loss was also an independent prognostic marker associated with shorter 3-year OS rate in 1p/19q codeleted gliomas (44% vs 81%, p = 0.01, HR = 3.3 [1.2–8.9]) (table 3). Although it is in the setting of a large heterogeneous pathologic group that does not integrate molecular features, 9p21.3 allelic loss was also an independent prognostic biomarker in WHO grades II and III gliomas from the REMBRANDT dataset (table e-1).
Table 3.
Prognostic value of clinical features and 9p23.1 allelic loss in 1p/19q codeleted gliomas from the REMBRANDT dataset
DISCUSSION
Up to now, no additional genomic biomarker has been identified to stratify 1p/19q codeletion AOTs. The present study highlights 9p21.3 allelic loss (including homozygous deletion, hemizygous deletion, and CN-LOH) as an independent prognostic factor for PFS and OS in patients with 1p/19q codeleted AOT. The independent prognostic value of 9p21.3 allelic loss was validated in an independent cohort using the REMBRANDT dataset. In addition, patients exhibiting 1p/19q codeleted with 9p21.3 allelic loss tend to be older. The most frequent mechanism of 9p21.3 allelic loss was CN-LOH (48.9%) while the 9p21.3 hemizygous deletion represented 43.3% of the cases. These mechanisms of 9p21.3 allelic loss were also validated in the REMBRANDT dataset. This result may explain why this biomarker was not identified in previous studies analyzing exclusively copy number changes (i.e., comparative genomic hybridization, quantitative PCR) or allelic status (i.e., microsatellites analysis).
The MCR(9p) encompasses CDKN2A and CDKN2B. We have recently shown in a smaller series of AOTs that 9p CN-LOH is associated with CDKN2A underexpression.7 The CDKN2A locus encodes, by alternate splicing, for 2 major cell cycle–regulating proteins, p16INK4A and p14ARF. Protein p16INK4A inactivates cyclin-dependent kinases that phosphorylate the retinoblastoma protein, resulting in a G1 phase arrest, while p14ARF stabilizes p53 by inducing MDM2 degradation.16 CDKN2A acts therefore as a “double” tumor suppressor gene and its silencing promotes oncogenesis and tumor growth in various cancers including diffuse gliomas. For instance, CDKN2A homozygous deletion is observed in 52% of glioblastomas, a more aggressive subtype of diffuse glioma.17 The present study also supports a clinical value of 9p21.3 allelic loss in AOT.
A practical consequence is that patients with 1p/19q codeleted AOTs with 9p21.3 allelic loss should be monitored closely. While the very long survival of AOTs with 1p/19q codeletion alone raises the question of delaying RT to preserve long-term cognitive functions, this strategy could be questionable in patients exhibiting 1p/19q codeleted AOTs with 9p21.3 allelic loss.
Additional studies investigating the significance of the 9p21.3 allelic loss in patients with 1p/19q codeleted AOT are warranted to investigate and validate our findings. Indeed, although our study is based on a multicentric prospective cohort, the follow-up of patients remains short.
Supplementary Material
GLOSSARY
- AOT
anaplastic oligodendroglial tumor
- CN-LOH
copy neutral loss of heterozygosity
- HR
hazard ratio
- MCR
minimal common region
- OS
overall survival
- PCV
procarbazine-CCNU-vincristine
- PFS
progression-free survival
- POLA
prise en charge des tumeurs oligodendrogliales anaplasiques
- REMBRANDT
Repository of Molecular Brain Neoplasia Data
- RT
radiotherapy
- WHO
World Health Organization
Footnotes
Supplemental data at Neurology.org
Contributor Information
Collaborators: POLA Network, Clovis Adam, Marie Andraud, Marie-Hélène Aubriot-Lorton, Claire Blechet, Mario Campone, Dominique Cazals-Hatem, Marie-Pierre Chenard, Dan Chiforeanu, Marie-Danièle Diebold, Sandrine Eimer, Guillome Gauchotte, Stéphane Gaillard, Anne Jouvet, Jean-Louis Kemeny, Toufic Khalil, Emmanuèle Lechapt-Zalcman, Caroline Le Guérinel, Pierre-Marie Levillain, Delphine Loussouarn, Nadine Martin-Duverneuil, Claude-Alain Maurage, Michel Peoc’h, Marc Polivka, Isabelle Quintin-Roue, Philippe Richard, Valérie Rigau, Audrey Rousseau, Gwenaelle Runavot, Henri Sevestre, Raoulin Soulard, Emmanuelle Uro-Coste, Marie-Claire Tortel, Fanny Vandenbos, Gabriel Viennet, and Chiara Villa
AUTHOR AFFILIATIONS
From AP-HP (A.A., C. Dehais, J.G., J.-Y.D., A.I.), Groupe Hospitalier Pitié-Salpêtrière, Service de neurologie 2-Mazarin, Paris; Sorbonne Universités (A.A., J.-Y.D., A.I.); UPMC Univ Paris 06, UM 75; Inserm (A.A., C.C., K.M., J.-Y.D., A.I.), U 1127, ICM, Paris; CNRS (A.A., C.C., K.M., J.-Y.D., A.I.), UMR 7225, ICM, Paris; ICM (A.A., C.C., K.M., J.-Y.D., A.I.), Paris; Hospices Civils de Lyon (F. Ducray), Hôpital Neurologique, Lyon Neuroscience Research Center INSERM U1028/CNRS UMR 5292 and Université de Lyon-Université Claude Bernard Lyon 1, Service de Neuro-Oncologie, Bron; AP-HP (K.M.), Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie-Raymond Escourolle, Sorbonne Universités, Paris; Hôpital de la Timone, Assistance Publique-Hôpitaux de Marseille, Faculté de Médecine la Timone, Aix Marseille Université and Inserm, CRO2 UMR_S 911, Service de Pathologie et Neuropathologie (D.F.-B.) and Service de Neuro-Oncologie (O.C.), Marseille; Institut Claudius Regaud and INSERM U1037 (E.C.-M.), Service de radiothérapie, Toulouse; CHU de Lille (C.R.), Service de Neurochirurgie, Lille; CHU de Bordeaux-GH Pellegrin (H.L., S.E.-H.), Service de Neurochirurgie, Bordeaux; AP-HP, CHU Lariboisière (S.E.-H.), Service de Neurochirurgie, Paris; CHU Nancy (P.B.), Service de Neuro-oncologie, Nancy; CHU Charles Nicolle (O.L.), Service de Neurochirurgie, Rouen; CHU Amiens (C. Desenclos), Hôpital Nord, Service de Neurochirurgie, Amiens; CHU de Caen (J.-S.G.), Service de Neurologie, Caen; Hôpital de la cavale blanche (P.D.-H.), CHU Brest, Service de Neurochirurgie, Brest; Centre Georges-François Leclerc (F.G.), Service d'oncologie médicale, Dijon; Clinique de Courlancy (P.C.), Service de Radiothérapie, Rheims; CHU Besançon (J.G.), Hôpital Jean Minjoz, Service de Neurochirurgie, Besançon; AP-HP (F.P.), CHU Bicêtre, Service de Neurochirurgie, Le Kremlin-Bicêtre; Institut Gustave Roussy (F. Dhermain), Service de Radiothérapie, Villejuif; AP-HP (A.F.C.), CHU Avicenne, Service de Neurologie, Bobigny; Institut de Cancérologie de l'Ouest (ICO)–CLCC René Gauducheau (J.-S.F.), Service d'oncologie médicale, Saint-Herblain; CHU Angers (P.M.), Service de Neurochirurgie, Angers; CHU de Montpellier (L.B.), Service de Neurochirurgie, Montpellier; AP-HP (T.F.), CHU Beaujon, Service de Neurochirurgie, Clichy; CHR Orléans (M.F.), Service de Radiothérapie, Orléans; CHU Nice (D.F.), Service de Neurochirurgie, Nice; CHU Saint-Étienne (M.-J.M.-F.), Hôpital Nord, Saint-Priest en Jarez; Centre Eugène Marquis (E.V.), Service d'Oncologie Médicale, Rennes; CH Colmar (C.G.), Service de Neurologie, Colmar; CHU Henri Mondor (C.L.G.), Service de Neurochirurgie, Créteil; CHU de Limoges–Hôpital Dupuytren (E.-M.G.), Service de Neurochirurgie, Limoges; Centre Paul Strauss (G.N.), Service de Radiothérapie, Strasbourg; HIA Sainte-Anne (N.D.), Service de Neurochirurgie, Toulon; Centre Jean Perrin (X.D.), Service d'Oncologie Médicale, Clermont-Ferrand; Hôpital Foch (E.B.), Service d'Oncologie Médicale, Suresnes; CHU Poitiers (M.W.), Service de Neurochirurgie, Poitiers; HIA du Val de Grâce (D.R.), Service de Neurologie, Paris; and Clinique des Cèdres (I.C.), Service d'Oncologie Médicale, Cornebarrieu, France.
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: A. Alentorn, F. Ducray, J.-Y. Delattre, and A. Idbaih. Performed the experiments: C. Carpentier. Analyzed the data: A. Alentorn, A. Idbaih. Contributed reagents/materials/analysis tools: C. Dehais, F. Ducray, C. Carpentier, K. Mokhtari, D. Figarella-Branger, O. Chinot, E. Cohen-Moyal, C. Ramirez, H. Loiseau, S. Elouahdani-Hamdi, P. Beauchesne, O. Langlois, C. Desenclos, J.-S. Guillamo, P. Dam-Hieu, F. Ghiringhelli, P. Colin, J. Godard, F. Parker, F. Dhermain, A.F. Carpentier, J.-S. Frenel, P. Menei, L. Bauchet, T. Faillot, M. Fesneau, D. Fontaine, M.-J. Motuo-Fotso, E. Vauleon, C. Gaultier, C. Le Guerinel, E.-M. Gueye, G. Noel, N. Desse, X. Durando, E. Barrascout, M. Wager, D. Ricard, I. Carpiuc, J.-Y. Delattre, A. Idbaih. Wrote the manuscript: A. Alentorn, A. Idbaih. Included patients: POLA Network. Review of manuscript: A. Alentorn, C. Dehais, F. Ducray, C. Carpentier, K. Mokhtari, D. Figarella-Branger, O. Chinot, E. Cohen-Moyal, C. Ramirez, H. Loiseau, S. Elouahdani-Hamdi, P. Beauchesne, O. Langlois, C. Desenclos, J.-S. Guillamo, P. Dam-Hieu, F. Ghiringhelli, P. Colin, J. Godard, F. Parker, F. Dhermain, A.F. Carpentier, J.-S. Frenel, P. Menei, L. Bauchet, T. Faillot, M. Fesneau, D. Fontaine, M.-J. Motuo-Fotso, E. Vauleon, C. Gaultier, C. Le Guerinel, E.-M. Gueye, G. Noel, N. Desse, X. Durando, E. Barrascout, M. Wager, D. Ricard, I. Carpiuc, J.-Y. Delattre, A. Idbaih.
STUDY FUNDING
This work is funded by the French Institut National du Cancer (INCa) and part of the national program Cartes d'Identité des Tumeurs (CIT) (http://cit.ligue-cancer.net/) funded and developed by the Ligue Nationale contre le cancer. The research leading to these results received funding from the program “Investissements d'avenir” ANR-10-IAIHU-06. Frozen specimens from Lyon were stored in NeuroBioTec, Groupement Hospitalier Est, Bron, France. Also supported by the Institut Universitaire du Cancer (IUC). Frozen specimens from Marseille were stored in the AP-HM tumor bank (AC 2013-1786).
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Van den Bent MJ, Bromberg JEC. Anaplastic oligodendroglial tumors. Handb Clin Neurol 2012;105:467–484. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Burger P, et al. International Society of Neuropathology–Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 2014;24:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA 2013;110:6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van den Bent MJ, Brandes AA, Taphoorn MJB, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 2013;31:344–350. [DOI] [PubMed] [Google Scholar]
- 5.Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 2013;31:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011;333:1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Idbaih A, Ducray F, Dehais C, et al. SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas. PLoS One 2012;7:e45950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Wiel MA, Wieringen WN. CGHregions: dimension reduction for array CGH data with minimal information loss. Cancer Inform 2007;3:55–63. [PMC free article] [PubMed] [Google Scholar]
- 9.van de Wiel MA, Smeets SJ, Brakenhoff RH, et al. CGHMultiArray: exact P-values for multi-array comparative genomic hybridization data. Bioinformatics 2005;21:3193–3194. [DOI] [PubMed] [Google Scholar]
- 10.Madhavan S, Zenklusen JC, Kotliarov Y, et al. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res 2009;7:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bengtsson H, Wirapati P, Speed TP. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics 2009;25:2149–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olshen AB, Bengtsson H, Neuvial P, et al. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation. Bioinformatics 2011;27:2038–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortiz-Estevez M, Aramburu A, Bengtsson H, et al. CalMaTe: a method and software to improve allele-specific copy number of SNP arrays for downstream segmentation. Bioinformatics 2012;28:1793–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: Available at: http://www.R-project.org/. Accessed April 6, 2014. [Google Scholar]
- 15.Therneau TM. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 16.Haber DA. Splicing into senescence: the curious case of p16 and p19ARF. Cell 1997;91:555–558. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.