Abstract
Type IV P-type ATPases (P4-ATPases) translocate phospholipids from the exoplasmic to the cytoplasmic leaflets of cellular membranes. We and others previously showed that ATP11C, a member of the P4-ATPases, translocates phosphatidylserine (PS) at the plasma membrane. Twenty years ago, the UPS-1 (uptake of fluorescent PS analogs) cell line was isolated from mutagenized Chinese hamster ovary (CHO)-K1 cells with a defect in nonendocytic uptake of nitrobenzoxadiazole PS. Due to its defect in PS uptake, the UPS-1 cell line has been used in an assay for PS-flipping activity; however, the gene(s) responsible for the defect have not been identified to date. Here, we found that the mRNA level of ATP11C was dramatically reduced in UPS-1 cells relative to parental CHO-K1 cells. By contrast, the level of ATP11A, another PS-flipping P4-ATPase at the plasma membrane, or CDC50A, which is essential for delivery of most P4-ATPases to the plasma membrane, was not affected in UPS-1 cells. Importantly, we identified a nonsense mutation in the ATP11C gene in UPS-1 cells, indicating that the intact ATP11C protein is not expressed. Moreover, exogenous expression of ATP11C can restore PS uptake in UPS-1 cells. These results indicate that lack of the functional ATP11C protein is responsible for the defect in PS uptake in UPS-1 cells and ATP11C is crucial for PS flipping in CHO-K1 cells.
Keywords: uptake of fluorescent phosphatidylserine analogs, adenosine triphosphatases, membrane bilayer, phospholipid, plasma membrane, flippase
The lipid bilayers of cellular membranes exhibit asymmetric lipid distributions. In the plasma membrane, the aminophospholipids phosphatidylserine (PS) and phosphatidylethanolamine (PE) are abundant in the cytoplasmic leaflet, whereas phosphatidylcholine (PC) and SM are enriched in the exoplasmic leaflet. Type IV P-type ATPases (P4-ATPases) are essential for generation and maintenance of phospholipid asymmetry in lipid bilayers (1, 2). Regulated exposure of PS in the exoplasmic leaflet is critical for certain biological processes, including apoptotic cell death, platelet coagulation, and fusion of muscle cells (3–5), illustrating the importance of lipid asymmetry at steady state. Most mammalian and yeast P4-ATPases must associate with cell division cycle protein 50 (CDC50) family proteins in order to exit the endoplasmic reticulum and reach their appropriate subcellular destinations (6–12). We recently showed that the human P4-ATPases ATP11A and ATP11C flip nitrobenzoxadiazole (NBD)-labeled PS (NBD-PS) and NBD-PE, whereas ATP8B1, ATP8B2, and ATP10A flip NBD-PC specifically at the plasma membrane (13, 14). Phospholipid asymmetry regulated by P4-ATPases is indispensable for homeostasis of multicellular organisms. Mutations in the human FIC1/ATP8B1 gene cause progressive familial intrahepatic cholestasis (PFIC) (15, 16). Some ATP8B1 mutants found in type 1 PFIC fail to flip PC, indicating that PC-flipping activity at the bile canaliculi is critical for proper bile excretion in liver (13). ATP11C deficiency causes a defect in B-cell maturation, altered erythrocyte shape, and anemia (17, 18). Moreover, ATP11C undergoes caspase-mediated cleavage and is consequently inactivated, resulting in PS exposure on the cell surface during apoptosis (19).
UPS-1 (uptake of fluorescent PS analogs) cells were isolated by screening mutants of CHO-K1 cells defective in nonendocytic uptake of NBD-PS (20) and have been widely used in an assay for PS-flipping activity due to their defect in PS uptake (8, 9, 16, 21, 22). However, the gene(s) responsible for the defect have not been previously identified. In this study, we found that the expression level of ATP11C mRNA was substantially decreased in UPS-1 cells, whereas the level of ATP11A or CDC50A mRNA was not affected. Importantly, we found a nonsense mutation in the ATP11C gene in UPS-1 cells. We further demonstrated that uptake of PS was restored upon exogenous expression of ATP11C in UPS-1 cells. These results indicate that the defect in PS uptake in UPS-1 cells is ascribed to the lack of the functional ATP11C protein.
MATERIALS AND METHODS
Plasmids
P4-ATPase cDNAs were cloned separately into the pENTR3C vector (Invitrogen), as described previously (12).
RT-PCR and quantitative RT-PCR
Total RNA was isolated from CHO or UPS-1 cells using Isogen (Nippon Gene) or RNeasy Mini Kit (Qiagen) and then subjected to RT-PCR analysis using the SuperScript III One-Step RT-PCR system (Invitrogen). For quantitative RT-PCR (qRT-PCR), total RNA was subjected to reverse transcription using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). The resultant cDNA was used as a template for PCR using LightCycler FastStart DNA MasterPLUS SYBR Green I (Roche Applied Science); fold changes in gene expression were normalized to the β-actin mRNA level. Chinese hamster ATP11C cDNA was amplified using the following primer pairs: set 1 (sense, 5′-ATACTGAGCTCTTAGAACTGACC-3′; antisense, 5′-ATCACTATTCCTGGCTGCTTGG-3′), set 2 (sense, 5′-GAACAGCACATCAACGTTGATAC-3′; antisense, 5′-CTGATAAATATGAGGAGAATTATGG-3′), and set 3 (3′ untranslated region (UTR); sense, 5′-GTATAGGGTTCAGAATAAATGTCC-3′; antisense, 5′-GATATTAGACCAAGACAATTAGTC-3′). The ATP11A, CDC50A, and β-actin cDNAs were amplified using the following primer pairs: sense, 5′-CATGGAAGTGCTCAAGAGAGAC-3′/antisense, 5′-GAGCAGGCTGACAGTGACAAG-3′; sense, 5′-GCCAGTTAAATGGAGACCCTAG-3′/antisense, 5′-GTCCAGCTGGTAATGTTGGATG-3′; and sense, 5′-CTGTATGCCTCTGGTCGTAC-3′/antisense, 5′-GCCATCTCCTGCTCGAAGTC-3′, respectively.
Sequencing of ATP11C cDNA and its chromosomal gene in CHO-K1 and UPS-1 cells
Total RNA was isolated from UPS-1 cells using RNeasy Mini Kit (Qiagen). Nine fragments of ATP11C cDNA covering full length of ATP11C were amplified by RT-PCR, cloned into TA cloning vector (BioDynamics Laboratory), and sequenced at least four independent clones of each fragment. Nine fragments of ATP11C cDNA was amplified using the following primer pairs: F1 (5′-ATGTCGTGTGCTGGAGAAGAG-3′; 5′-CTCGGAGGTTATCAATGGATTC-3′), F2 (5′-CAGCCAGTCTTGATGGTGAATC-3′; 5′-CCATCAACTTCCTGAGTAGTGC-3′), F3 (5′-CAGATAAGACTGGAACACTCAC-3′; 5′-GTAAGCACCCAGACTTTCAAGC-3′), F4 (5′-GGAGCCACTGCTGTGGAAGAC-3′; 5′-GTAAGCAGCATCATAAAGTGGC-3′), F5 (5′-CAATATTTCTTCTATAAGAACCTTTG-3′; 5′-CTTTAGCTGCAATCAGAGTCAATATG-3′), F6 (5′-CAGATAAGACTGGAACACTCAC-3′; 5′-GATTTGGCTGTTTCCATCTTGTC-3′), F7 (5′-CAGCCAGTCTTGATGGTGAATC-3′; 5′-GTAAGCACCCAGACTTTCAAGC-3′), F8 (5′-CAGATAAGACTGGAACACTCAC-3′; 5′-GTAAGCAGCATCATAAAGTGGC-3′), and F9 (5′-CAATATTTCTTCTATAAGAACCTTTG-3′; 5′-CTGTTACATTATATTAGATTCGTCTG-3′). Genomic DNA was prepared from CHO-K1 and UPS-1 cells using Isogen (Nippon Gene) and 362 bp DNA fragment carrying the exon 19 and its 5′ intron region was amplified by PCR with following primers: 5′-CAGAGACAGCACAACTCTGATAAG-3′; 5′-CACTCAGCAACTTCTTACGATATTC-3′. The PCR products were purified and sequenced with a primer of 5′-GAATTGCTAGTGAGCTATAAGGATG-3′ and analyzed with an ABI PRIZM 3100 Genetic Analyzer.
Antibodies and reagents
The sources of antibodies used in the present study were as follows: monoclonal rabbit anti-sodium potassium ATPase (Na/K-ATPase), Abcam; monoclonal rat anti-HA (3F10), Roche Applied Science; monoclonal mouse anti-β-tubulin, Millipore; Cy3- and horseradish peroxidase-conjugated secondary antibodies, Jackson ImmunoResearch Laboratories; Alexa Fluor 488-conjugated secondary antibody, Invitrogen. The NBD-labeled phospholipids (Avanti Polar Lipids) used were NBD-PS (1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]sn-glycero-3-phosphoserine), NBD-PE (1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]sn-glycero-3-phosphoethanolamine), NBD-PC (1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]sn-glycero-3-phosphocholine), and NBD-SM (N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]sphingosine-1-phosphocholine).
Cell culture, establishment of stable cell lines, and immunofluorescence analysis
UPS-1 (20) cells and parental CHO-K1 cells were grown in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F-12 medium supplemented with 10% fetal calf serum, 2.5 mM l-glutamine. UPS-1 cell line, originally established by Kentaro Hanada (National Institute of Infectious Diseases, Tokyo, Japan), was provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT (Tsukuba, Japan). For retroviral production, pMXs-neo-derived vectors for expression of HA-tagged P4-ATPases were cotransfected with pEF-gag-pol and pCMV-VSVG-RSV-Rev into HEK293T cells. The pMXs-neo vectors and the pEF-gag-pol plasmid were kind gifts from Toshio Kitamura (The University of Tokyo, Tokyo, Japan). The pCMV-VSVG-RSV-Rev plasmid was a kind gift from Hiroyuki Miyoshi (RIKEN BRC, Tsukuba, Japan). The resultant retroviruses were concentrated as described previously (23) and then used to infect UPS-1 cells. For stable cell lines, the infected cells were selected in medium containing G418 (400 μg/ml) and isolated as clones. A mixed population of retrovirus-infected cells or cloned stable cell lines were used for flippase assay, RT-PCR, and immunofluorescence analysis. Immunofluorescence staining was performed as described previously (24, 25) and visualized using an Axiovert 200MAT microscope (Carl Zeiss, Thornwood, NY). Briefly, cells were fixed, permeabilized and immunostained with anti-sodium potassium ATPase (Na/K-ATPase) and anti-HA (3F10) antibodies followed by Alexa Fluor 488-conjugated anti-rabbit and Cy3-conjugated anti-rat secondary antibodies.
Flippase assay
Incorporation of NBD-phospholipids was analyzed by flow cytometry as described (13). In brief, CHO-K1 or UPS-1 cells were detached from dishes in PBS containing 5 mM EDTA and then harvested by centrifugation. Cells (1 × 106 cells per sample) were washed and equilibrated at 15°C for 15 min in 500 μl of HBSS (pH 7.4) containing 1 g/l glucose (HBSS-glucose). An equal volume of 2 μM NBD-phospholipid in HBSS-glucose was added to the cell suspension and incubated at 15°C. At each time point, 200 μl of cell suspension was collected and mixed with 200 μl of ice-cold HBSS-glucose containing 5% fatty acid-free BSA (Wako Pure Chemical) in order to extract NBD-lipids incorporated into the exoplasmic leaflet of the plasma membrane, as well as unincorporated NBD-lipids. Next, 10,000 cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences) to measure fluorescence of NBD-lipids translocated into the cytoplasmic leaflet of the plasma membrane. Mean fluorescence intensities per cell were calculated. Propidium iodide-positive cells (i.e., dead cells) were excluded from the analysis.
RESULTS AND DISCUSSION
A nonsense mutation is found in ATP11C gene in UPS-1 cells
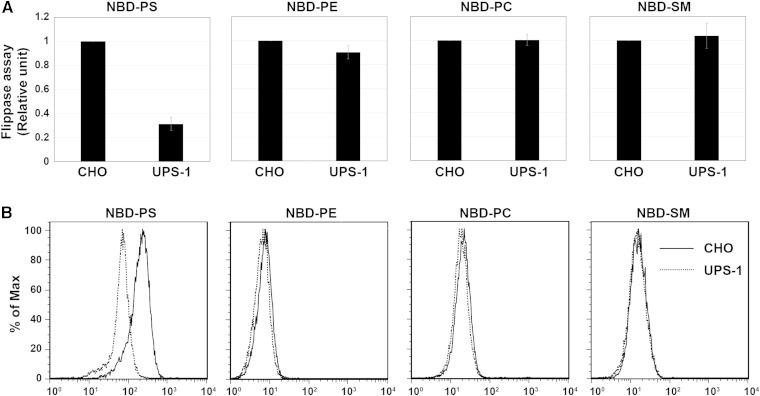
UPS-1, a mutant cell line of CHO-K1 (hereafter CHO), is defective in nonendocytic uptake of NBD-PS (20). Prior to the experiments described below, we confirmed that the uptake of NBD-PS is defective in UPS-1 cells. To this end, we assayed the flippase activities by incubating CHO and UPS-1 cells in the presence of NBD-PS, -PE, -PC, or -SM at 15°C, followed by extraction with fatty acid-free BSA of fluorescent phospholipids that were unincorporated or retained in the exoplasmic leaflet of the plasma membrane. As shown in Fig. 1A, B, uptake of NBD-PS was lower in UPS-1 cells than in parental CHO cells, whereas uptake of NBD-PE, -PC, or -SM did not change. These results are consistent with those of an original report (20).
Fig. 1.
UPS-1 cells have a defect in NBD-PS uptake. A, B: Parental CHO-K1 (CHO) and UPS-1 cells were incubated with the indicated NBD-lipids at 15°C for 10 min (NBD-PS) or 15 min (NBD-PE, NBD-PC, or NBD-SM). After extraction with fatty acid-free BSA, the residual fluorescence intensity associated with the cells was determined by flow cytometry. (A) Graphs are averages of three independent experiments ± SD. B: Histogram of a representative experiment displaying the differences in fluorescence intensity between CHO (solid lines) and UPS-1 (dashed lines) cells.
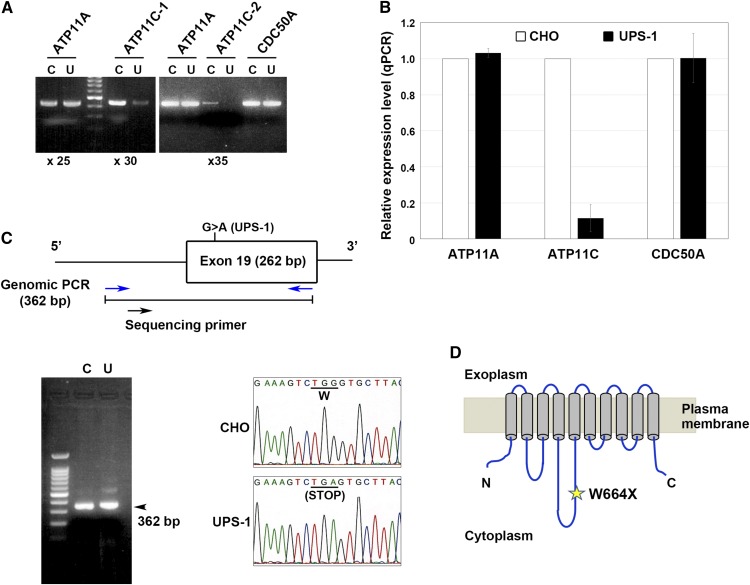
We recently elucidated flippase activities and substrate specificities of the plasma membrane-localized human P4-ATPases ATP8B1, ATP8B2, ATP10A, ATP11A, and ATP11C using NBD-labeled phospholipids. ATP11A and ATP11C translocate NBD-aminophospholipids (PS and PE), whereas ATP8B1, ATP8B2, and ATP10A preferentially flip NBD-PC (13, 14). We also previously showed that CDC50A is required for the plasma membrane localization of ATP11A and ATP11C (12). Knockdown by RNA interference or clustered regularly interspaced short palindromic repeat/CRISPR-associated protein-9 nuclease (CRISPR/Cas9) knockout of CDC50A dramatically decreases PS-flipping activity in several cell types (our unpublished observations) (19). Moreover, ATP11C deficiency also decreases PS-flipping activity (17–19). Therefore, we hypothesized that the defect in the uptake of PS in UPS-1 cells might result from a defect in plasma membrane-localizing PS-flipping P4-ATPases (ATP11A and ATP11C) or a defect in CDC50A, which is required for delivery of P4-ATPases to the plasma membrane. To examine this hypothesis, we first performed RT-PCR to measure the mRNA levels of ATP11A, ATP11C, and CDC50A in CHO and UPS-1 cells (Fig. 2A). All primer sets were designed to include at least one predicted intron to avoid amplification from genomic DNA. As shown in Fig. 2A, the mRNA level of ATP11C was substantially lower in UPS-1 cells (U) than in CHO cells (C). We confirmed the low level of ATP11C mRNA in UPS-1 cells using three independent primer sets (Figs. 2A and 4E). By contrast, the mRNA levels of ATP11A and CDC50A were comparable between UPS-1 and CHO cells (Fig. 2A). We also examined the mRNA levels of ATP11A, ATP11C, and CDC50A by qRT-PCR. As shown in Fig. 2B, the mRNA level of ATP11C was much lower in UPS-1 cells than in the parental CHO cells. By contrast, the ATP11A and CDC50A mRNA levels in UPS-1 cells were comparable to those in CHO cells.
Fig. 2.
Reduced expression and mutation of ATP11C in UPS-1 cells. A: RT-PCR was performed using total RNA isolated from CHO (C) and UPS-1 (U) cells. Two different primer sets for ATP11C were designed (×25, 25 cycles; ×30, 30 cycles; ×35, 35 cycles). B: qRT-PCR results from two independent experiments (averages are shown). C: Genomic PCR analysis of ATP11C. A region encompassing exon 19 and a preceding intron (362 bp) was amplified using a primer set (blue arrows) for genomic DNA from CHO (C) and UPS-1 (U) cells. The arrowhead indicates the position of the amplified genomic PCR product (362 bp). Direct sequencing data indicate the presence of a homozygous point mutation (G to A) in the ATP11C gene in UPS-1 cells. D: Schematic representation of the ATP11C protein and the position of the nonsense mutation found in UPS-1 cells.
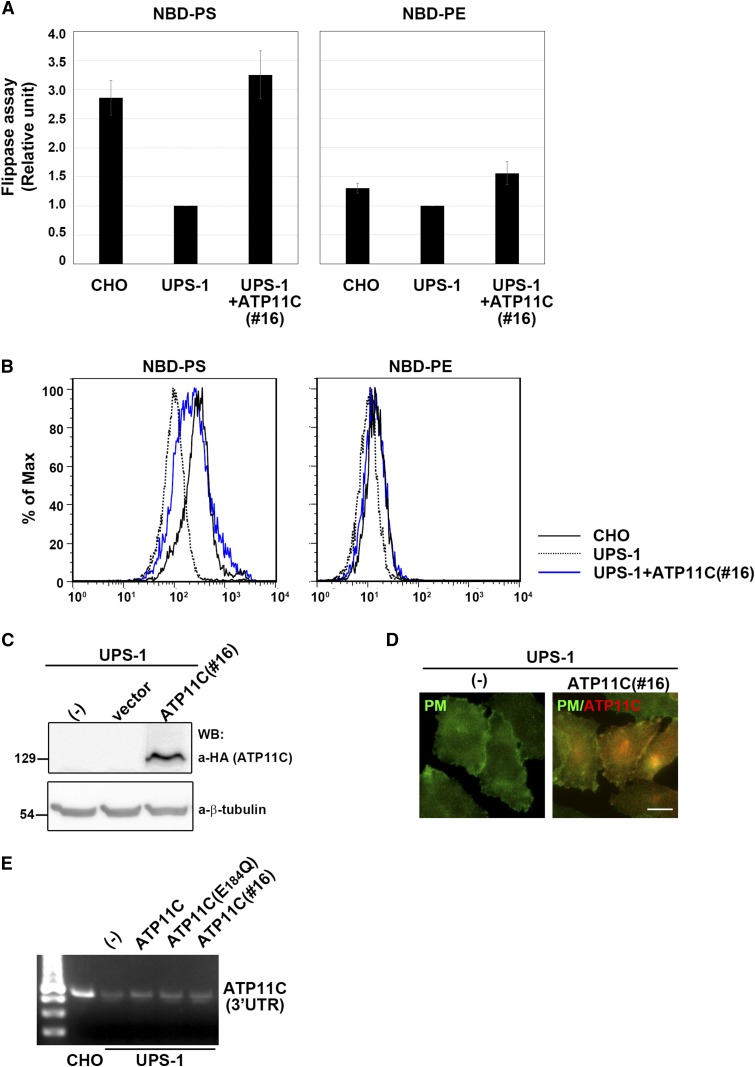
Fig. 4.
Stable expression of ATP11C in UPS-1 cells restored PS-flipping activity comparable to parental CHO cells. A: An ATP11C-expressing UPS-1 cell line (clone 16) was isolated. The flippase activities of CHO, UPS-1, and UPS-1(clone16) cells were measured as described in Fig. 3A. Graphs are averages of four independent experiments ± SD. B: Histogram of a representative experiment showing the differences in the fluorescence intensity among CHO (solid lines), UPS-1 (dashed lines), and an ATP11C-expressing UPS-1 cell line (clone 16) (blue lines) in the presence of the indicated NBD-lipids. C: Expression level of ATP11C in UPS-1 cells (clone 16) was analyzed by immunoblotting with antibodies against HA and β-tubulin (as an internal control). D: Cells were fixed, permeabilized, and immunostained with antibody against Na/K-ATPase (a marker for the plasma membrane, PM) and anti-HA antibody followed by Alexa488-conjugated anti-rabbit and Cy3-conjugated anti-rat antibodies, respectively. Bar indicates 10 μm. E: RT-PCR was performed using total RNA isolated from indicated cells. A primer set targeting the 3′UTR of the Chinese hamster ATP11C was designed.
Next, we examined a possibility that the coding sequence of ATP11C is changed in UPS-1 cells. To this end, we cloned nine cDNA fragments for ATP11C from UPS-1, which altogether cover the entire coding sequence of ATP11C and its C-terminal splicing variant, and sequenced at least four independent clones for each fragment. Interestingly, we found a nonsense mutation in the region covered by exon 19 (Fig. 2C). Direct sequencing of a chromosomal DNA region (362 bp) encompassing exon 19 and the preceding intron indicated that the ATP11C gene in UPS-1 cells carried a G to A homozygous mutation in exon 19, resulting in a change in the Trp664 codon (TGG) to a nonsense codon (TGA) (Fig. 2C, D). Although chromosomal heterogeneity has been observed in CHO cell lines (26, 27), the raw data of direct sequencing of the chromosomal DNA showed a single peak at the mutation site (Fig. 2C), indicating a homozygous mutation in the ATP11C gene. We cannot exclude a possibility that the locus of ATP11C gene in UPS-1 cells is hemizygous. These results indicate that the defect of PS uptake in UPS-1 cells is caused by the lack of the functional ATP11C protein, although we could not examine the ATP11C protein level because an antibody, which can detect endogenous ATP11C in CHO cells, is not available.
The mutation site was located at 205 nucleotides upstream from the junction between exon 19 and exon 20 (Fig. 2C). Nonsense-mediated mRNA decay (NMD) is a well-characterized posttranscriptional quality control mechanism to ensure transcription fidelity (28). NMD can be activated when a nonsense codon appears more than 50–55 nucleotides upstream from an exon-exon junction. Thus, in UPS-1 cells, the ATP11C mRNA might be degraded by the NMD mechanism, and thus the level of ATP11C was dramatically reduced (Fig. 2A, B).
We previously showed that expression of ATP11A and ATP11C in HeLa cells increases the PS- and PE-flipping activities, although the PE-flipping activity of ATP11C is lower than that of ATP11A (13). Because PE-flipping activity was not significantly affected in UPS-1 cells (Fig. 1A, B), ATP11C is primarily responsible for PS-flipping in CHO cells. The residual PS-flipping activity observed in UPS-1 cells might be due to the presence of ATP11A or other flipping activities (Fig. 1A). ATP11C appears to be a critical protein for PS flipping at the plasma membrane in multiple cell types: 1) some phenotypes of the ATP11C mutant mice cannot be suppressed by the presence of endogenous ATP11A (17, 18), and 2) degradation of ATP11C by caspase is responsible for PS exposure to the outer leaflet of the plasma membrane in apoptotic cells (19).
Exogenous expression of ATP11C complements the defect in PS-flipping activity in UPS-1 cells
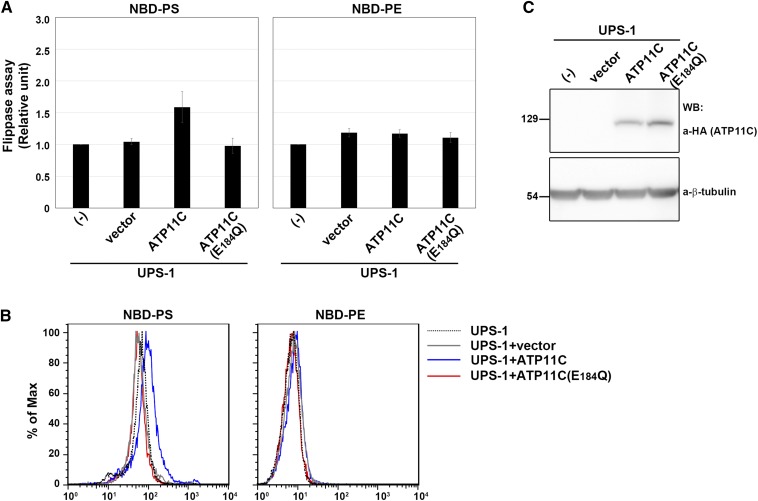
Next, we asked whether PS-flipping activity can be recovered by exogenous expression of ATP11C in UPS-1 cells. To this end, we expressed C-terminally hemagglutinin (HA)-tagged ATP11C or its ATPase-deficient Glu-to-Gln mutant (E184Q) (13) by infection of recombinant retrovirus and subjected the infected cells to the flippase assay. ATP11C and the ATP11C(E184Q) mutant were expressed at comparable levels, as confirmed by immunoblot analysis (Fig. 3C). By expressing ATP11C but not ATP11C(E184Q), the PS-flipping activity was significantly increased, relative to vector-infected UPS-1 cells (Fig. 3A, B), whereas the PE-flipping activity was not changed. When a UPS-1 cell line stably expressing ATP11C was examined (clone 16), PS-flipping activity was approximating to that in parental CHO cells (Fig. 4A, B). We confirmed ATP11C expression in clone 16 by immunoblot analysis (Fig. 4C) and its localization to the plasma membrane by immunofluorescence (Fig. 4D). Thus, in CHO cells, ATP11C is crucial for PS flipping, but not PE flipping. Because there was a subtle, but significant, increase in the PE-flipping activity in stably ATP11C-expressing UPS-1 cells (Fig. 4A), exogenous ATP11C may also flip NBD-PE as we described previously (13).
Fig. 3.
Exogenous expression of human ATP11C (WT) but not ATP11C(E184Q) rescued PS-flipping activity in UPS-1 cells. A: UPS-1 cells (-) infected with empty retrovirus vector (vector) or recombinant virus vector encoding HA-tagged ATP11C(WT) or ATP11C(E184Q) were incubated with the indicated NBD-lipids at 15°C for 15 min (10 min for NBD-PS). After extraction with fatty acid-free BSA, the residual fluorescence intensity associated with the cells was determined by flow cytometry. Graphs show the results from three independent experiments (averages ± SD). B: Histogram of a representative experiment showing the differences in fluorescence intensity among UPS-1 (dashed lines), UPS-1 infected with empty retrovirus vector (gray lines), or UPS-1 infected with retrovirus vector encoding ATP11C (blue lines) or ATP11C(E184Q) (red lines) in the presence of the indicated NBD-lipids. C: Expression levels of ATP11C and ATP11C(E184Q) in UPS-1 cells were analyzed by immunoblotting with antibodies against HA and β-tubulin (as an internal control).
In order to exclude possible contamination of parental CHO cells in the ATP11C-expressing UPS-1 cells, we performed RT-PCR analysis using a set of primers targeting the 3′-UTR of the Chinese hamster ATP11C. The endogenous mRNA level of ATP11C did not change among the exogenous ATP11C- or ATP11C(E184Q)-expressing UPS-1 cells (Fig. 4E). Thus, the recovery of PS-flipping activity in UPS-1 cells is due to the exogenous expression of ATP11C rather than contamination of parental CHO cells.
Based on these results, we conclude that the defect in PS uptake observed in UPS-1 cells is ascribed to the lack of functional ATP11C. It is likely that ATP11C is a major P4-ATPase involved in PS flipping in CHO cells. ATP11C-deficient mice exhibit anemia, hyperbilirubinemia, abnormal differentiation of B cells, and hepatocellular carcinoma (17, 18, 29). Hopefully, UPS-1 cells will be useful for understanding the physiological and pathophysiological roles of ATP11C.
Footnotes
Abbreviations:
- CDC50
- cell division cycle protein 50
- CHO
- Chinese hamster ovary
- CRISPR/Cas9
- clustered regularly interspaced short palindromic repeat/CRISPR-associated protein-9 nuclease
- HA
- hemagglutinin
- NBD
- nitrobenzoxadiazole
- NMD
- nonsense-mediated mRNA decay
- P4-ATPase
- type IV P-type ATPase
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PFIC
- progressive familial intrahepatic cholestasis
- PS
- phosphatidylserine
- qRT-PCR
- quantitative RT-PCR
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan; the Takeda Science Foundation; the Inamori Foundation; and the Research Foundation for Pharmaceutical Sciences.
REFERENCES
- 1.Graham T. R. 2004. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 14: 670–677. [DOI] [PubMed] [Google Scholar]
- 2.Pomorski T., Holthuis J. C. M., Herrmann A., van Meer G. 2004. Tracking down lipid flippases and their biological functions. J. Cell Sci. 117: 805–813. [DOI] [PubMed] [Google Scholar]
- 3.Zwaal R. F., Schroit A. J. 1997. Pathophysiologic implications of membrane phospholipid asymmetry in blood cells. Blood. 89: 1121–1132. [PubMed] [Google Scholar]
- 4.van den Eijnde S. M., van den Hoff M. J., Reutelingsperger C. P., van Heerde W. L., Henfling M. E., Vermeij-Keers C., Schutte B., Borgers M., Ramaekers F. C. 2001. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 114: 3631–3642. [DOI] [PubMed] [Google Scholar]
- 5.Williamson P., Schlegel R. A. 2002. Transbilayer phospholipid movement and the clearance of apoptotic cells. Biochim. Biophys. Acta. 1585: 53–63. [DOI] [PubMed] [Google Scholar]
- 6.Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. 2004. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 15: 3418–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. 2007. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell. 18: 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulusma C. C., Folmer D. E., Ho-Mok K. S., de Waart D. R., Hilarius P. M., Verhoeven A. J., Oude Elferink R. P. 2008. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology. 47: 268–278. [DOI] [PubMed] [Google Scholar]
- 9.Bryde S., Hennrich H., Verhulst P. M., Devaux P. F., Lenoir G., Holthuis J. C. 2010. CDC50 proteins are critical components of the human class-1 P4-ATPase transport machinery. J. Biol. Chem. 285: 40562–40572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Velden L. M., Wichers C. G. K., van Breevoort A. E. D., Coleman J. A., Molday R. S., Berger R., Klomp L. W. J., van de Graaf S. F. J. 2010. Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J. Biol. Chem. 285: 40088–40096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman J. A., Molday R. S. 2011. Critical role of the b-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J. Biol. Chem. 286: 17205–17216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takatsu H., Baba K., Shima T., Umino H., Kato U., Umeda M., Nakayama K., Shin H-W. 2011. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 286: 38159–38167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takatsu H., Tanaka G., Segawa K., Suzuki J., Nagata S., Nakayama K., Shin H. W. 2014. Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J. Biol. Chem. 289: 33543–33556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naito T., Takatsu H., Miyano R., Takada N., Nakayama K., Shin H. W. 2015. Phospholipid flippase ATP10A translocates phosphatidylcholine and is involved in plasma membrane dynamics. J. Biol. Chem. 290: 15004–15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulusma C. C., Groen A., Kunne C., Ho-Mok K. S., Spijkerboer A. L., Rudi de Waart D., Hoek F. J., Vreeling H., Hoeben K. A., van Marle J., et al. 2006. Atp8b1 deficiency in mice reduces resistance of the canalicular membrane to hydrophobic bile salts and impairs bile salt transport. Hepatology. 44: 195–204. [DOI] [PubMed] [Google Scholar]
- 16.Folmer D. E., van der Mark V. A., Ho-Mok K. S., Oude Elferink R. P. J., Paulusma C. C. 2009. Differential effects of progressive familial intrahepatic cholestasis type 1 and benign recurrent intrahepatic cholestasis type 1 mutations on canalicular localization of ATP8B1. Hepatology. 50: 1597–1605. [DOI] [PubMed] [Google Scholar]
- 17.Yabas M., Teh C. E., Frankenreiter S., Lal D., Roots C. M., Whittle B., Andrews D. T., Zhang Y., Teoh N. C., Sprent J., et al. 2011. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat. Immunol. 12: 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yabas M., Coupland L. A., Cromer D., Winterberg M., Teoh N. C., D’Rozario J., Kirk K., Broer S., Parish C. R., Enders A. 2014. Mice deficient in the putative phospholipid flippase ATP11C exhibit altered erythrocyte shape, anemia and reduced erythrocyte lifespan. J. Biol. Chem. 289: 19531–19537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segawa K., Kurata S., Yanagihashi Y., Brummelkamp T. R., Matsuda F., Nagata S. 2014. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 344: 1164–1168. [DOI] [PubMed] [Google Scholar]
- 20.Hanada K., Pagano R. E. 1995. A Chinese hamster ovary cell mutant defective in the non-endocytic uptake of fluorescent analogs of phosphatidylserine: isolation using a cytosol acidification protocol. J. Cell Biol. 128: 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyato Y., Ibuki Y., Ohyama H., Yamada T., Goto R. 2001. Phosphatidylserine induces apoptosis in CHO cells without mitochondrial dysfunction in a manner dependent on caspases other than caspases-1, -3, -8 and -9. FEBS Lett. 504: 73–77. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X., Libby R. T., de Vries W. N., Smith R. S., Wright D. L., Bronson R. T., Seburn K. L., John S. W. 2012. Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8: e1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki J., Umeda M., Sims P. J., Nagata S. 2010. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 468: 834–838. [DOI] [PubMed] [Google Scholar]
- 24.Shin H-W., Kobayashi H., Kitamura M., Waguri S., Suganuma T., Uchiyama Y., Nakayama K. 2005. Roles of ARFRP1 (ADP-ribosylation factor-related protein 1) in post-Golgi membrane trafficking. J. Cell Sci. 118: 4039–4048. [DOI] [PubMed] [Google Scholar]
- 25.Shin H. W., Shinotsuka C., Nakayama K. 2005. Expression of BIG2 and analysis of its function in mammalian cells. Methods Enzymol. 404: 206–215. [DOI] [PubMed] [Google Scholar]
- 26.Xu X., Nagarajan H., Lewis N. E., Pan S., Cai Z., Liu X., Chen W., Xie M., Wang W., Hammond S., et al. 2011. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29: 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wurm F. M., Hacker D. 2011. First CHO genome. Nat. Biotechnol. 29: 718–720. [DOI] [PubMed] [Google Scholar]
- 28.Popp M. W., Maquat L. E. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siggs O. M., Arnold C. N., Huber C., Pirie E., Xia Y., Lin P., Nemazee D., Beutler B. 2011. The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat. Immunol. 12: 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]