Abstract
Eukaryotic genomes are dynamically regulated through a host of epigenetic stimuli. The substrate for these epigenetic transactions, chromatin, is a polymer of nucleosome building blocks. In native (i.e. cellular) chromatin, each nucleosome can differ from its neighbors through the localized installation of covalent modifications to both the genomic DNA and the histone packaging proteins. The heterotypic nature of chromatin presents a formidable obstacle to biochemical studies seeking to understand the role of context on epigenetic regulation and that, as a consequence, must employ compositionally defined chromatin substrates. Here, we introduce a chemical approach to the production of heterotypic ‘designer’ chromatin that can be used in such studies. Our method involves attachment of a user-defined modified histone peptide to a designated nucleosome within the polymer by using a Peptide Nucleic Acid (PNA) targeting compound. We apply this strategy to dissect the role of chromatin context on both the activation and inhibition of the histone methyltransferase, PRC2, which methylates Lys 27 of histone H3 (H3K27). Our studies show that PRC2 can be stimulated to produce de novo H3K27 methylation from a defined nucleation site. More generally, this technology promises to facilitate biochemical studies that require the use of heterotypic chromatin substrates.
Keywords: chromatin, epigenetics, Peptide nucleic acids
Dynamic organization of the eukaryotic genome is critical for proper cellular differentiation and is achieved through a complex system of compaction of DNA around histone proteins. Histones undergo extensive post-translational modifications (PTMs), which facilitate regulation of chromatin transactions.[1] A quintessential example of this regulation is the methylation of H3K27 by the enzyme Polycomb Repressive Complex 2 (PRC2), a modification that is associated with gene silencing.[2] PRC2 is a large multi-subunit protein complex that is itself regulated by pre-existing PTMs on the chromatin substrate.[3, 4] Critically, this includes the product of its own activity, H3K27me3, which allosterically activates the methyltransferase activity of PRC2.[5] This positive feedback loop is thought to allow propagation of the silencing mark into nearby nucleosomes, although it remains unclear whether any additional physical or biochemical factors control the extent of this PTM propagation.[3] Given that both hyper- and hypo-PRC2 activity have been linked to human cancers,[6] there is considerable need to fully understand how this enzyme is regulated in both normal and disease cells.
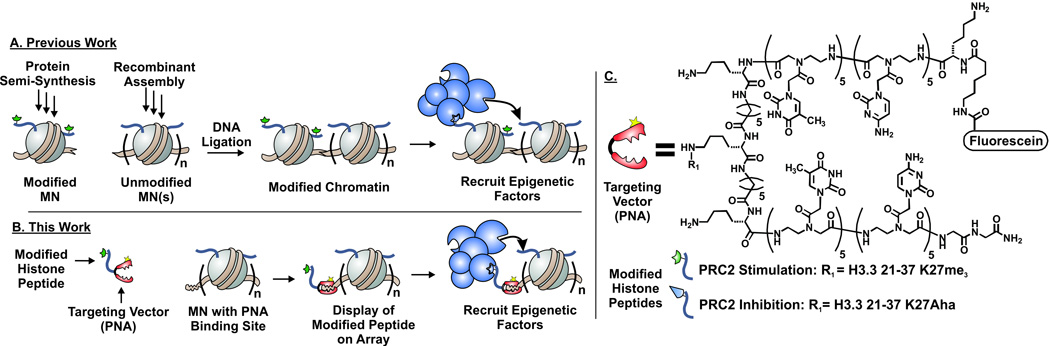
Understanding epigenetic phenomena such as PTM ‘spreading’ at a molecular level requires access to heterotypic chromatin substrates containing a nucleation site (i.e. a nucleosome with a pre-installed PTM) and a substrate region (i.e. lacking the PTM in question). Only when the position of the nucleation site is defined is it possible to gauge the extent of PTM propagation. Isolation of homogeneous (i.e. compositionally defined) chromatin substrates from natural sources is currently impossible, and so substrates suitable for mechanistic studies have to be assembled by in vitro reconstitution methods.[7] Access to site-specifically modified histones using methods such as semi-synthesis and amber suppression is now well established.[8] These PTM-containing histones can be deposited on to DNA sequences with strong nucleosome positioning elements (e.g. Widom 601[9]) such that mononucleosomes (MNs) and homotypic oligonucleosome arrays can be accessed in a straightforward fashion. However, extending these procedures to the generation of heterotypic oligonucleosome arrays is non-trivial and currently involves inefficient, and technically demanding, nucleosome-level ligation strategies[8] (Figure 1A). As a consequence, the molecular mechanisms underlying PTM spreading processes remain poorly understood.[2] Herein, we report the use of a convergent chemical biology strategy that allows modified histone-derived peptides to be targeted to a specific region within an oligonucleosome array, thereby creating nucleation sites for exploring PTM propagation mechanisms. To highlight this technology, we investigated the spatial constraints associated with the regulation of PRC2 activity. Our studies show that the methyltransferase can install the H3K27me modification on nucleosomes adjacent to a targeted nucleation site on a chromatin array.
Figure 1.
A. Schematic of how chromatin with site-specific PTMs (green symbols) is conventionally assembled from protein semi-synthesis and recombinant expression, followed by DNA ligation. B. Illustration of the display of a histone peptide by a PNA targeting vector (red) which recognizes a specific DNA sequence, resulting in the surrogate of native chromatin used in this study. C. Structure of the PNA vector and the modified histone peptides. Aha = (S)- aminoheptanoic acid.
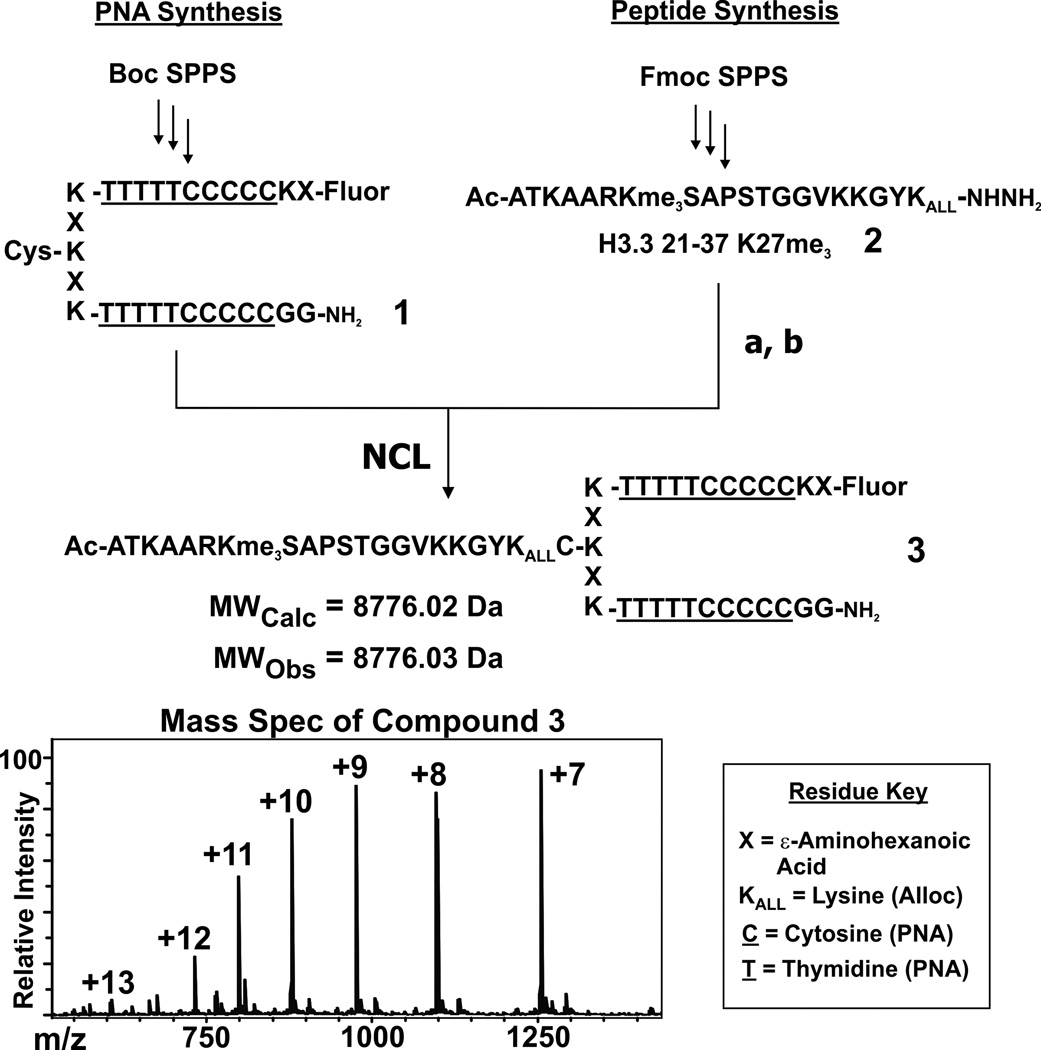
We imagined using the DNA within chromatin as a convenient recognition platform for targeting vectors linked to histone-derived peptides (Figure 1B). Tremendous progress has been made in the design of molecules that bind DNA in a sequence-specific manner.[10] We chose to use Peptide Nucleic Acids (PNAs) in the current study, although we note that other DNA recognition agents could be employed (e.g. hairpin polyamides[11]). PNAs, synthetic analogs of DNA with a peptide-like backbone, have been used extensively to invade double-stranded DNA sequences (see Supporting Information for additional references).[12] We appended the PNA target sequence to one end of the nucleosomal DNA sequence (designated hereafter as DNAtarget), a location similar to the site of protrusion of the H3 tail from the nucleosome core.[13] PNA constructs were synthesized via Boc-SPPS using established procedures[14] and were functionalized with both a cysteine residue (for native chemical ligation, NCL) and a fluorescein moiety (to facilitate imaging of the complexes) as shown in Scheme 1.
Scheme 1.
Synthesis of PNA 1 (left) and histone peptide 2 (right), followed by the ligation to create PNA-peptide chimera 3. Reaction conditions: (a). 20 mM NaNO2, 6 M GnHCl, 0.2 M phosphate buffer pH = 3, 0° C, 20 min. (b). 50 mM MESNa, 100 mM thiophenol, 6 M GnHCl, 0.2 M phosphate buffer pH = 7.5. Below is the structure of the full construct 3, and the mass spectrum of the purified product. Note that histone peptide 2 has the tripeptide GYK appended to it; see Supporting Information for synthetic details and compound characterization.
Histone H3.3 peptides were synthesized as C-terminal acyl hydrazides[15] and ligated to the PNA molecules using NCL (see Supporting Information for synthetic details). Modifications associated with both PRC2 activation (constructs 2–4) and inhibition (constructs 5–8) were synthesized so as to explore orthogonal aspects of enzyme regulation.
An electrophoretic mobility shift assay was employed to characterize the recognition of nucleosomal DNA by our PNA-based targeting vectors. Incubation of MNs (50 nM) containing the DNAtarget with five equivalents of PNA constructs (250 nM) gave a quantitative gel-shift of the nucleosomal band (see Supporting Information Figure S1). A fluorescent signal was also seen at the same mobility as the nucleosomal band, providing evidence that the PNA construct had formed a complex with the target-DNA. Control experiments with MNs lacking the PNA binding motif showed no gel shift or fluorescence signal, indicating that the PNA specifically recognized the appended DNAtarget sequence.
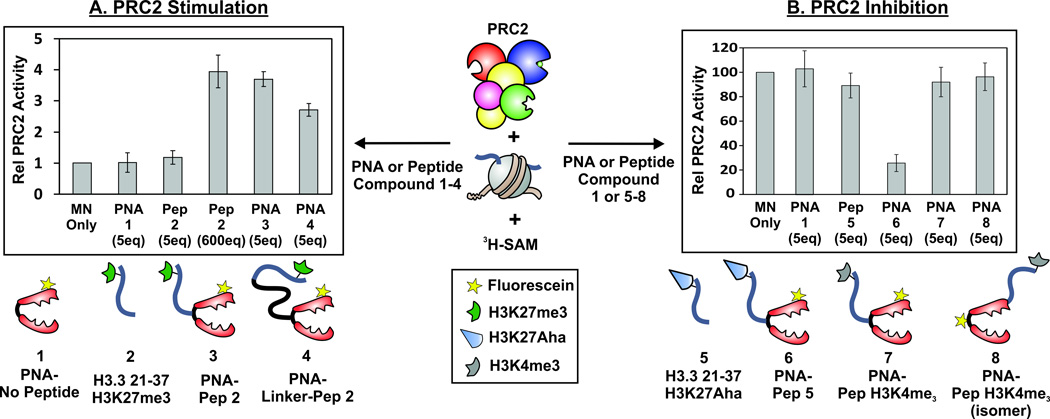
Next, we assessed whether our PNA-peptide constructs could modulate the methyltransferase activity of PRC2. We employed a radiolabeling assay using MN substrates, purified PRC2 enzyme and3H-SAM. Inclusion of construct 3 (which contains both the targeting vector and the activating H3K27me3 peptide) at a concentration of 250 nM (corresponding to 5 eq. compared to MN substrate) in the assay mix led to a 3.5-fold stimulation of PRC2 activity compared to the MN-only control reaction (Figure 2A). This activation was not observed when construct 1 (which contains the targeting vector but no activating peptide) was used, nor was it seen when construct 3 was added to MN substrate lacking the DNAtarget sequence (Figure S2). Importantly, addition of construct 2 (which contains the activating peptide but not the targeting vector) had no effect on PRC2 activity when added at a concentration of 250 nM. Indeed, a concentration of 30 µM (600 eq.) had to be employed before peptide 2 stimulated PRC2 activity to a similar degree as construct 3. To further assess the effects of spatial constraints on the stimulation, we synthesized an additional construct, 4, that included a PEG chain spacer between the PNA moiety and the histone peptide. Construct 4 provided a slightly lower level of PRC2 activation as compared to 3 (Figure 2A), suggesting the enzyme is sensitive to the distance between the activating peptide and the substrate. Collectively, these results show that our targeting system can be used to display a histone-derived peptide on a MN substrate such that PRC2 activity is stimulated.
Figure 2.
PRC2 methyltransferase activity using MNs and PNA-histone peptide constructs. A. PRC2 stimulation with peptides/PNAs containing the H3K27me3 motif. B. PRC2 inhibition with peptides/PNAs that include either the H3 21–37 K27Aha (aminoheptanoic acid residue) or the H3K4me3 motif. Scintillation counts shown are the average of at least three independent replicates, with error bars representing one standard deviation. PRC2 is depicted with the following color scheme: EZH2 (blue), EED (green), AEBP2 (magenta), RbAp46/8 (red), Suz12 (yellow).
Next we turned our attention to histone modifications that inhibit the activity of PRC2. It has recently been discovered that lysine 27 of histone H3 is frequently mutated to methionine in pediatric brain cancers. The H3K27M mutation creates a potent orthosteric inhibitor of PRC2 and even short H3-derived peptides can inhibit the enzyme, albeit more weakly compared to when the mutation is presented in a nucleosomal context.[16, 17] Given this, we asked whether targeting H3-derived inhibitory peptides to a MN would lead to an improvement in potency against PRC2. A PNA construct, 6, was synthesized containing an H3-derived peptide (5) in which lysine 27 was replaced with (S)-aminoheptanoic acid (Aha), a residue that we have previously shown improves the inhibitory activity compared to methionine.[16] Incubation of MNs (50 nM) with construct 6 (250 nM) led to a ~75% reduction in PRC2 activity as measured by the3H-SAM radioassay (Figure 2B). As expected, no inhibition of PRC2 activity was seen when MNs were incubated with the same concentrations of PNA 1 or peptide 5. Indeed, the latter had to be added at a concentration of 20 µM in order to achieve an equivalent level of inhibition (Supporting Information Figure S2). Thus, our nucleosomal targeting system significantly increases the potency of this class of inhibitory peptides against PRC2. Presumably this boost results from additional binding interactions between the enzyme and the MN that are furnished by the nucleosomal presentation of the inhibitory peptide. As an additional probe of PRC2 inhibition, we generated constructs 7 and 8, which contain short peptides bearing the H3K4me3 modification in different topologies. Previous studies indicate that H3K4me3 inhibits PRC2 activity, but only when present in the same histone as the substrate lysine 27 (i.e. cis inhibition).[3] Thus, we were curious whether targeting the H3K4me3 inhibitory modification to a nucleosome using the PNA vector would mimic this mode of inhibition and hence lead to a reduction in H3K27 methylation by PRC2. However, addition of construct 7 or 8 to MNs had no effect on H3K27 methylation by PRC2, despite forming the expected complex with the MN (Figure 2B and Figure S1). These results highlight the stringent spatial requirements associated with H3K4me3-mediated inhibition of PRC2, a feature that presumably prevents spurious inhibition through trans-modes of regulation.
Encouraged by the results with MNs, we next asked whether the targeting system could be used with oligonucleosome arrays, in principle providing an expedient and modular route to heterotypic ‘designer’ chromatin. We designed a DNA sequence that included four copies of the 601 nucleosome positioning sequence as well as the PNA targeting site, DNAtarget, on one terminus as well as unique restriction sites (see Supporting Information for details). An oligonucleosome array was generated using this template DNA via standard protocols[18] and the ability of this array to form complexes with the PNA-peptide constructs was confirmed using the electrophoretic mobility shift assay (Figure S3). With this surrogate of heterotypic chromatin in place, we first asked whether the PRC2 activity could be stimulated on a nucleosomal array using our PNA targeting system. Accordingly, we incubated the nucleosome array with five equivalents of stimulatory PNA construct 3 and then measured PRC2 activity using3H-SAM. As in the case of MN substrates, we observed a stimulation of methyltransferase activity in the presence of this PNA construct (Figure S4). No such stimulation was detected when we used similar amounts of control constructs 1 or 2 (500 eq. of the latter was required to give a comparable level of stimulation to 5 eq. of 3).
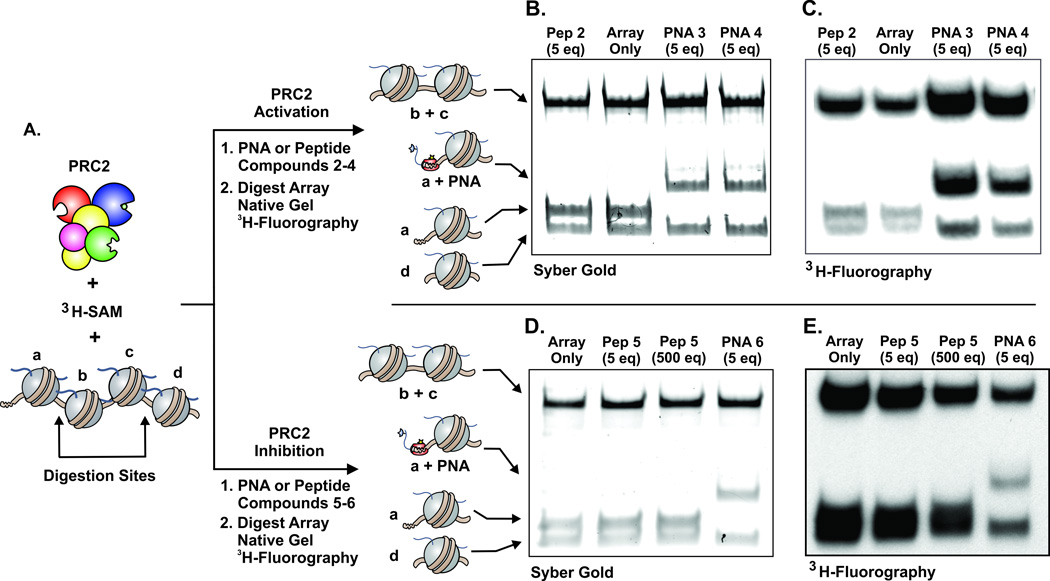
The presence of unique restriction sites within the array afforded us the opportunity to ask where de novo methylation was taking place, i.e. in which nucleosomes (designated a–d, Figure 3A). PRC2 methylated arrays were digested with endonucleases, resolved by native gel electrophoresis and then analyzed by fluorography. In the absence of any added construct, as well as the presence of low concentrations of peptide 2, we observed de novo methylation across the array (Figure 3B–C, Figure S5). The addition of PNA construct 3 resulted in an increase in methylation on all four nucleosomes in the array, with the largest boost being observed on nucleosome a, the PNA target (Figure 3B–C, Figure S5). Increased methylation on the nucleation site is expected based on our results with MNs above. However, the detection of higher signal in nucleosomes b–d is consistent with an increased PRC2 activity that radiates from the nucleation site. Similar results were obtained with PNA conjugate 4, which contains the PEG spacer between the targeting vector and the stimulatory peptide. The observed PTM propagation can be accounted for either by a tethering mechanism, in which activated PRC2 anchored at the first nucleosome can reach adjacent nucleosomes, and/or by a processive type process.
Figure 3.
A. PRC2 methyltransferase assays using oligonucleosome arrays with the designated PNA or peptide, followed by restriction digestion and native gel electrophoresis (B,D), 3H-fluorography (C,E). The upper portion shows PRC2 activation using compounds 2–4 and the distribution of de novo methylation, with the PNA-bound MN a showing a significant increase in methylation relative to MN d. The lower panel is the measurement of PRC2 inhibition with compounds 5 and 6, showing dramatic reduction of methylation to the PNA-bound MN a. Note that all assays used an excess of unlabeled SAM prior to endonuclease digestion to stop the radiolabeling, and all PRC2 inhibition assays had peptide 2 at a final concentration of 25 µM to increase the activity or PRC2. See Supporting Information for full assay details and additional gels and fluorographs (Figure S5).
Finally, we probed the effects of PRC2 inhibition using our designer 4-mer array. As in the case of MNs, use of the PNA targeting vector dramatically increased the potency of the inhibitory peptide (Figure S4). To determine where inhibition was occurring, we again turned to digestion of the arrays. As expected, the presence of high concentrations of non-targeted inhibitory peptide 5 resulted in uniform reduction in de novo methylation across the array (Figure 3D–E). The addition of construct 6, on the other hand, led to PRC2 inhibition preferentially on the nucleosome to which it was targeted, i.e. nucleosome a (Figure 3D–E). This type of spatially biased inhibition was expected. However, it is interesting that inhibition at positions b–d occurred under the conditions of the experiment. We note that peptide 5 produces little PRC2 inhibition when applied at this concentration (5 eq, Figure S4), implying that the reduction in methylation in nucleosomes b–d must originate from construct 6 anchored at site a. Thus, while PRC2 inhibition is primarily localized to the PNA targeting site, the enzyme does sense the presence of the inhibitor peptide at nearby nucleosomes. This spatial influence of the inhibitor may be a factor in vivo, where, although only a small fraction of the histone H3 pool bears the K27M mutation, a substantial reduction in global methylation level was found.[17]
In this study, we have outlined a convergent strategy to generate functional surrogates of heterotypic, “designer” chromatin. This technology may provide a modular and expeditious platform to interrogate the molecular mechanisms of histone-modifying enzymes. As an illustration, we investigated both the allosteric activation and orthosteric inhibition of the methyltransferase PRC2 in a MN and oligonucleosome context. In addition to providing a significant increase in the effective concentration of the displayed histone peptide, the targeting method allowed us to explore PTM propagation in an array context. Our results are consistent with the idea that PCR2 can methylate nucleosomes adjacent to a nucleation site through an allosterically activated positive feedback mechanism. Future studies will address the physical basis of this phenomenon. More generally, we envision further dissection of epigenetic phenomena using chromatin surrogates such as these.
Supplementary Material
Acknowledgements
We would like to acknowedge Dr. Miquel Vila-Perello, Dr. Yael David, and Prof. C. David Allis for helpful discussion.
Footnotes
This research was supported by the U.S. National Institutes of Health (grants R37-GM086868 and R01 GM107047) and the STARR foundation (grant I6-A614).
Contributor Information
Zachary Z. Brown, Department of Chemistry, Princeton University, Princeton, NJ 08544 (USA)
Manuel M. Müller, Department of Chemistry, Princeton University, Princeton, NJ 08544 (USA)
Ha Eun Kong, Department of Chemistry, Princeton University, Princeton, NJ 08544 (USA).
Peter W. Lewis, Email: plewis@discovery.wisc.edu, Wisconsin Institute for Discovery, University of Wisconsin, Madison, WI 53715 (USA).
Tom W. Muir, Email: muir@princeton.edu, Department of Chemistry, Princeton University, Princeton, NJ 08544 (USA).
References
- 1.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitges F, Prusty A, Faty M, et al. Molecular Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz YB, Pirrotta V. Nat Rev Genet. 2013;14:853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 5.Margueron R, Justin N, Ohno K, et al. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi P, Allis CD, Wang GG. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang CJ, Hung MC. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allis CD, Muir TW. Chembiochem. 2011;12:264–279. doi: 10.1002/cbic.201000761. [DOI] [PubMed] [Google Scholar]
- 8.Müller MM, Muir TW. Chemical reviews. 2014 [Google Scholar]
- 9.Lowary PT, Widom J. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 10.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Research. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dervan PB, Edelson BS. Current opinion in structural biology. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 12.Kaihatsu K, Janowski BA, Corey DR. Chem Biol. 2004;11:749–758. doi: 10.1016/j.chembiol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 14.Kates SA, Albericio F. Solid-phase synthesis : a practical guide. New York: Marcel Dekker; 2000. [Google Scholar]
- 15.Zheng JS, Tang S, Qi YK, Wang ZP, Liu L. Nat Protoc. 2013;8:2483–2495. doi: 10.1038/nprot.2013.152. [DOI] [PubMed] [Google Scholar]
- 16.Brown ZZ, Müller MM, Jain SU, Allis CD, Lewis PW, Muir TW. J Am Chem Soc. 2014;136:13498–13501. doi: 10.1021/ja5060934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis PW, Müller MM, Koletsky MS, et al. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luger K, Rechsteiner TJ, Richmond TJ. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.