Abstract
There has been increasing interest in utilizing bottom-up approaches to develop synthetic cells. A popular methodology is the integration of functionalized synthetic membranes with biological systems, producing “hybrid” artificial cells. This Concept article covers recent advances and the current state-of-the-art of such hybrid systems. Specifically, we describe minimal supramolecular constructs that faithfully mimic the structure and/or function of living cells, often by controlling the assembly of highly ordered membrane architectures with defined functionality. These studies give us a deeper understanding of the nature of living systems, bring new insights into the origin of cellular life, and provide novel synthetic chassis for advancing synthetic biology.
Keywords: artificial cell, vesicle, polymersome, drug delivery, synthetic biology
Introduction
The fabrication of artificial cells from purely synthetic components could provide a revolutionary methodology to reconstruct life's functions within unnatural materials. Advances in synthetic biology and systems chemistry have enabled the construction of structures that are inspired by and made in the likeness of living materials. Artificial cells have the potential to shed light on biological processes such as gene expression, energy transduction, and self-assembly. They also offer an opportunity to organize chemical reactions in unique, nanoscale compartments. From an application standpoint, artificial cells hold promise as next-generation liposomal drug delivery systems and as a possible alternative technology for replacing biological cells. While approaches to create synthetic cells vary, and contain a further hierarchal division into top-down or bottom-up techniques, in this Concept, we will specifically focus on bottom-up approaches that assemble “hybrid” artificial cells from synthetic membranes. These methods are well suited for a range of applications, such as integrating highly ordered and functionalized vesicles with biological machinery and creating “hybrid” minimal cells using nonbiological chassis. We cover recent fundamental advances that have improved the state-ofthe-art of novel synthetic cell membranes, and have opened up exciting potential applications in biosensing, catalysis and pharmacology.
Recent Advances in Synthetic Cell Membranes
Fatty Acid, Phospholipid and Synthetic Lipid Vesicles
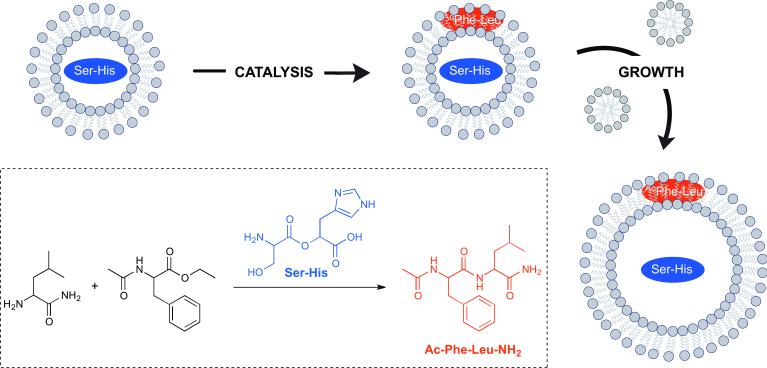
Fatty acid-based vesicles have several physical, chemical and functional properties that make them attractive for the design and development of synthetic cells with minimally evolved biochemical machinery.[1] Free fatty acids spontaneously form membrane bilayers in a highly cooperative way, which is characterized by a rapid exchange between vesicle membranes, micelles, and dissolved monomers. This dynamic exchange makes fatty acids well suited as components of protocell membranes because such vesicles can grow and divide in several ways without the loss of contents, such as nucleic acids. Many elegant methods have been developed to induce and repeat cycles of growth and division with fatty acid vesicles, for instance, by the concentration of a lipid solution by solvent evaporation.[2] The Szostak group has conducted several sophisticated studies with fatty acid vesicles as protocells. Recently, they showed that an encapsulated dipeptide catalyst can drive vesicle growth through the synthesis of additional peptides, which bind to the membrane and promote the recruitment of additional amphiphiles from the environment (Figure 1).[3] The group has also coupled prebiotically plausible fatty acid protocells with RNA replication by utilizing magnesium binding citrate chelating agents.[4] Past efforts have been hindered by the ability to free divalent cations to disrupt fatty acid vesicles and degrade RNA. While a prebiotically plausible citrate synthesis pathway is not currently known, it seems likely that alternative small molecule metal chelators, even short peptides, could also protect RNA replication and fatty acid membranes from decomposition.
Figure 1.
Fatty acid-based vesicles as protocells. Fatty acid vesicles containing a dipeptide catalyst seryl-histidine (Ser-His), which catalyzes the formation of a second dipeptide, N-Acetyl-L-phenylalanine-leucinamide (AcPheLeuNH2). The newly-formed hydrophopic dipeptide localizes to the vesicle membranes, imparting enhanced affinity for fatty acids, thus promoting vesicle growth.
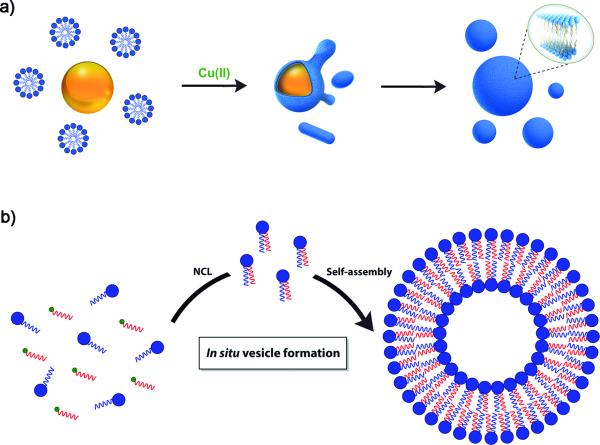
Despite their successful use in protocell research, fatty acids still inhibit the function of key enzymes such as polymerases, and fatty acid vesicles are not stable in the presence of excess free divalent cations, so other membrane compositions are likely required in order to construct highly efficient and functional synthetic cells using modern and complex biochemical machinery.[5] As natural membranes are composed primarily of phospholipids, it is perhaps not surprising that phospholipid vesicles have been shown to be appropriate compartments for creating artificial cells from biological components. In addition to their biocompatibility, phospholipid membranes are attractive due to their stability and ease of formation with a wide range of techniques.[6] However, phospholipids are essentially insoluble in aqueous solution and are highly resistant to exchange when embedded within membranes. These features hinder studies aiming to explore the effect of phospholipid addition on vesicle morphology, and makes de novo phospholipid vesicle generation difficult. Given the importance of mimicking biological systems, several groups have recently explored methods for the de novo generation of synthetic lipid membranes from reactive amphiphilic precursors.[7] The Devaraj group has described a biomimetic coupling reaction capable of driving de novo self-assembly of phospholipid membranes (Figure 2a).[8] This system utilizes a copper catalyzed azide-alkyne cycloaddition (CuAAC) to join an oleyl azide and an alkyne modified lysolipid, which affords a nonbiological triazole-containing phospholipid. In situ membrane formation occurs spontaneously and does not require preexisting membranes to house catalysts or precursors. In particular, bioorthogonal click chemistry reactions occur at the interface of insoluble oleyl azide emulsion droplets and the coating monolayers of alkyne lysolipid. The CuAAC approach has been recently employed to design a self-reproducing system that can drive the repeated synthesis and growth of phospholipid membranes.[9] The regeneration of membrane-bound autocatalysts continually induces the formation of triazole phospholipids, mimicking natural membrane generation. Remarkably, the native chemical ligation (NCL) has been reported to spontaneously generate phospholipid membranes from water-soluble starting materials (Figure 2b).[10] This non-enzymatic approach was used to form phospholipids in a highly specific and chemoselective way from long-chain thioesters. The lipids are capable of in situ self-assembly into vesicles that can grow to several microns in diameter. Moreover, the high reaction rate, the orthogonality and the biocompatibility of this methodology are key features that make it a powerful option for the efficient encapsulation of relevant biological materials, such as proteins. Proposed applications of this novel membrane assembly approach include drug delivery, bioreactors and reconstitution of functional membrane proteins. Additionally, a novel and straightforward strategy for the preparation of functional phospholipid membranes is through the use of thiolyne click chemistry reactions.[11] This methodology provides a powerful tool to rapidly access biologically relevant phospholipid analogs.
Figure 2.
De novo self-assembly of phospholipid membranes. a) Membrane assembly driven by a copper-catalyzed biomimetic reaction. Azide oil droplets (orange) interact with alkyne lysolipid micelles (blue) in aqueous solution to form an emulsion. Copper catalyst (green) addition triggers azide-alkyne coupling via bioorthogonal triazole formation, which occurs at the interface between insoluble azide oil droplets and the alkyne-containing aqueous solution. This azide-alkyne cycloaddition spontaneously drives de novo phospholipid vesicle formation. b) Spontaneous vesicle assembly induced by NCL-based phospholipid synthesis.
While some investigators have focused on the use of either synthetic amphiphiles or phospholipids, Sugawara et al. have recently achieved membrane vesicle growth and division by cleverly using a combination of phospholipids and unnatural lipids which can be synthesized in situ using small-molecule catalysts.[12] The inclusion of phospholipids enabled the function of embedded polymerases. Eventually, new lipids dilute out the phospholipid, making the system incapable of indefinite propagation. The combination of catalyzed lipid synthesis and subsequent vesicle growth and division in the presence of nucleic acids is an exciting advance toward the self-replication of hybrid synthetic cells.
Peptide- and Protein-based Artificial Membranes
In recent years, the library of materials that can reproducibly form membrane assemblies has expanded tremendously. There has been increasing interest in designing peptides and proteins that can spontaneously self-assemble into complex cell mimetic structures, which can have several attractive properties. For example, peptide vesicular structures (peptosomes) carry the advantage of being biocompatible, and highly stable toward physical degradation.[13] Typically, hydrophilic peptides are linked to a synthetic hydrophobic segment, which drives self-assembly. The Tomich group has recently established a new peptide motif using branched molecules composed entirely of amino acids (Figure 3).[14] These molecules contain a cationic head region attached to hydrophobic segments thus mimicking bilayer-forming amphiphiles such as phosphoglycerides. These relatively short sequences undergo self-assembly to adopt solvent-filled vesicles, which are bilayer-delimited spheres with 50-200 nm diameters, stabilized by a β-like structure. The peptosomes are able to efficiently encapsulate fluorescent molecules and are readily internalized by epithelial cells, strongly suggesting that peptide-based vesicles could act as biodegradable drug delivery devices. Proteinosomes are also possible using modified proteins. The Mann group has designed intricate proteinosome microcompartments utilizing a membrane of cross-linked amphiphilic bovine serum albumin.[15] These protocells have the ability to rapidly degrade in the presence of proteases, which causes release of entrapped genetic polymers.
Figure 3.
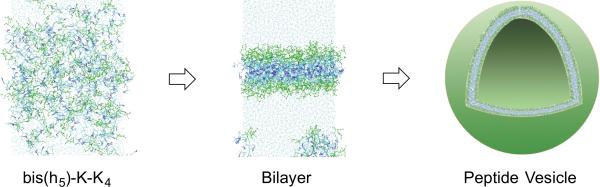
Snapshots from a coarse-grained-simulation of the self-assembly process for the amphiphilic peptide bis(h5)-K-K4, which leads to stable β-like assemblies to adopt a vesicular morphology [Adapted from reference 14].
Drawing inspiration from nature, biotechnology labs have recognized that several protein-based building blocks can self-assemble to form compartmentalized structures with potential applications in medicine, optics, or electronics. Among the wide collection of available self-assembling protein architectures, bacterial encapsulins are of particular interest since they form highly stable icosahedral structures and can be readily modified both on the outer or inner surface.[16] Interestingly, these nonviral protein microcompartments can act as potential artificial organelles by confining enzymes and substrates within the protein cage. Recently, Koay et al. have reported the molecular self-sorting and selective packaging of teal fluorescent protein (TFP) cargo. The TFP was engineered with a native C-terminal docking sequence, leading to in vivo assembly with bacterial encapsulins.[17] Such self-assembling systems may provide exciting opportunities as scaffolds for in situ catalytic reactions.
Membrane-forming Polymeric Systems
Vesicles composed of polymeric materials often possess greater stability than liposomes, and, with appropriate modification, have low immunogenicity.[18] Polymer-modified vesicles can mimic specific cellular architectures by spanning a wide range of properties including size, membrane permeability, and target specificity. Over the last few years, several biomedical applications of polymer-based vesicles, which can form artificial membranes through coating, blending with other polymers, and grafting to polymer substrates, have been described. Remarkably, polymeric vesicles known as polymersomes, generated by the self-assembly of amphiphilic copolymers, offer both the membrane and inner cavity as sites for inserting or encapsulating functional biomolecules. This design flexibility has enabled the construction of artificial bioreactors and blood-compatible materials with cell membrane-like surfaces. Therefore, polymersomes can act as an excellent interface between artificial and biological systems. For instance, the Leermakers group has described a versatile methodology for constructing highly stable multifunctional protein polymersomes by triggered templated self-assembly.[19] These compartments have potential drug- and gene-delivery applications. The Nallani and Mori groups have jointly shown that blending cationic phospholipids and block copolymers during vesicle self-assembly enhances cellular interaction and antigen delivery of the corresponding hybrid polymersomes.[20] Furthermore, the Liedberg group has combined porous polymersomes with fluorescent conjugated polymers to monitor an ATP-dependent enzymatic reaction in vesicular compartments.[21]
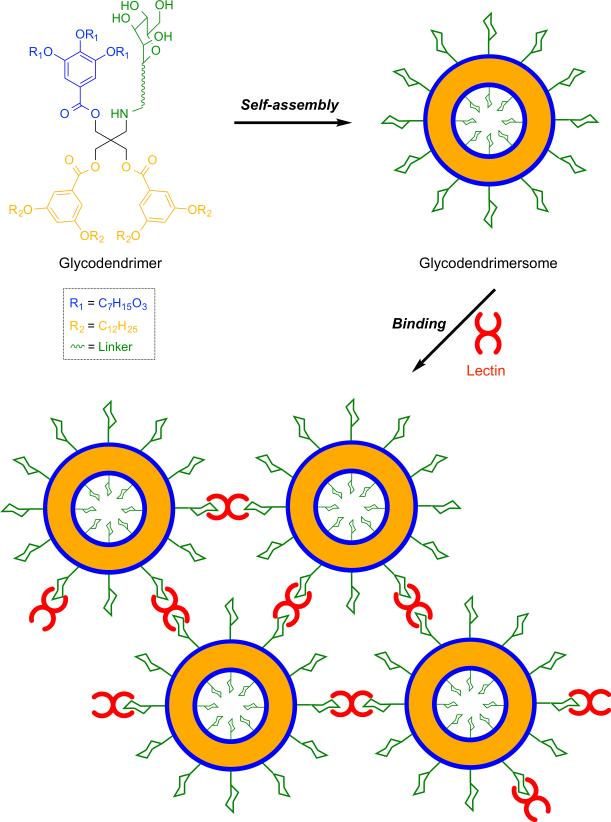
Glycans are critical components of cells membranes involved in cellular functions such as immune recognition and cellular adhesion.[22] Therefore, there is significant interest in using carbohydrate-containing vesicles for mimicking the supramolecular multivalency of glycan modified biological membranes. Recently, the Percec group has reported a straightforward method for the assembly of amphiphilic Janus glycodendrimers to produce monodisperse, stable, and impermeable soft unilamellar multivalent glycodendrimersomes (Figure 4).[23] These membrane mimics are promising tools for various fundamental and technological areas of nanomedicine, such as targeted drug delivery and vaccination.
Figure 4.
Self-assembly of amphiphilic glycodendrimers into uniform unilamellar vesicles (glycodendrimersomes), which exhibit specific and potent bioactivity in binding biomedically relevant lectins.
Approaches to Compartmentalization
It is thought that membrane compartmentalization was a key step in life's origins, and sophisticated compartmentalization strategies are a hallmark of eukaryotic cells.[24] In recent years, there has been tremendous effort to entrap cellular functions within artificial biomimetic compartments. Much attention has focused on a wide collection of single-compartment, lipid-based systems capable of DNA replication and protein production. A recognition that multicompartmentalization provides spatiotemporal control over reactions, high local reactant concentrations, concentration gradients, and the separation of incompatible reaction components has motivated developments in novel microreactor architectures.[25] These structures are often based on a liposome-in-liposome design or built using liposomes contained within layer-by-layer (LbL) capsules. For example, the Möhwald and Sukhorukov groups have jointly reported a concentric, two-compartment, LbL capsule-in-capsule architecture based on polyelectrolyte systems (Figure 5).[26] Near-infrared laser light illumination causes the disruption of the inner shell, affording an efficient tool to achieve the inter-compartmentalized mixing of the contents. This approach offers an elegant method for triggering bioreactions in confined spaces.
Figure 5.
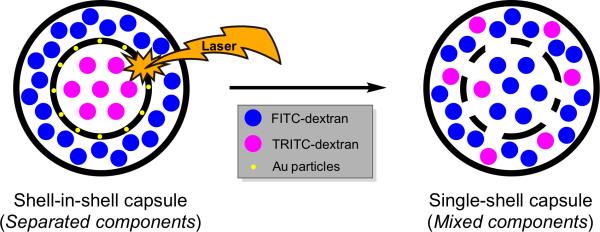
Schematic representation of a shell-in-shell polyelectrolyte multilayer capsule obtained by LbL assembly. Laser induced inter-compartmentalized mixing results due to light absorption by gold nanoparticles embedded into the inner shell.
Polymersomes have also been utilized for the construction of multicompartmentalized architectures.[27] Smaller polymersomes can be specifically used to create a polymersomes-in-polymersome (PiP) system, which mimics the structure of advanced cells.[28] For instance, the Lecommandoux group has developed a fast and simple approach for generating multicompartmentalized polymeric materials using emulsioncentrifugation.[29] The technique allows the encapsulation of multiple compartments containing unique contents, which could provide great advantages in multi-step catalysis as well as biomedical applications.
Recent Applications Combining Synthetic Membranes with Biological Systems
Drug and Gene Delivery
An extremely important and actively pursued application of synthetic membranes is the development of highly efficient drug and gene delivery systems that integrate with appropriate biological systems. Peptide- and polymer-based vesicles have recently become an attractive option as drug delivery vehicles for anti-cancer drugs. Lecommandoux et al. have successfully created doxorubicin (DOX)-loaded self-targeting vesicles from diblock copolymer polymersomes, comprised of a poly(γ-benzyl-L-glutamate) core conjugated to a natural glycosaminoglycan, hyaluronan (PBLG23-b-HYA10).[30] DOX delivered to cancer cells using polymersomes reduces tumor volumes more effectively than free DOX, while also reducing cardiotoxicity, a well-established DOX side effect. The authors attribute the improved performance of DOX-delivered polymersomes to a receptor-mediated endocytic pathway that is not possible with the free drug. More recently, the group has extended this work by implementing drug delivery in animal models.[31] DOX-loaded vesicles are highly stable in serum and circulate in the bloodstream for extended periods of time. Additionally, multifunctional polymersomes have been developed by coupling DOX with iron oxide nanoparticles, leading to materials that can be exploited as magnetic resonance imaging (MRI) contrast agents or physically manipulated using a magnetic field gradient.[32]
Assembling synthetic cells to execute DNA programs is also an active area of research. Siegal-Gaskins et al. have established a cell-free “breadboard” platform that allows synthetic biocircuits to operate in an environment similar to that found within live cells.[33] The simplicity of this system may shed light on fundamental aspects of biocircuit operation normally masked by cellular complexity. While membranes are typically thought of as necessary compartments for synthetic cells, Karzbrun et al. have recently developed a novel 2D DNA silicon-based biochip compartment capable of metabolism, programmable protein synthesis and gene expression, offering a versatile dynamic system for studying bioprocesses in artificial compartments.[34]
Bioreactors
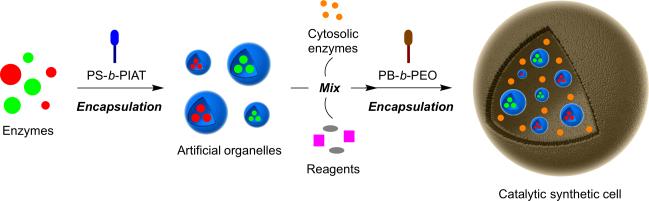
Synthetic membranes fabricated by self-assembly have attracted considerable attention for possible applications in biocatalysis. The Nallani group has constructed a multicompartment cascade bioreactor containing passive, size-selective protein channels.[35] The synthetic membranes house a porin, outer-membrane protein F (ompF), in an interior nanocompartment, which also encapsulates soluble glucose oxidase. The nanocompartment is then enclosed in an outer, semi-permeable membrane containing horseradish peroxidase. When glucose and a hydrogen peroxide-sensitive fluorogenic dye are added to the bioreactor, the fluorescent signal is significantly higher when the porin is present versus control bioreactors without the membrane channel. Recently, the van Hest and Lecommandoux groups have jointly described the combination of both membrane-containing PiP's and enzymatic polymersome organelle mimics in a single system to study multistep confined bioreactions (Figure 6).[36] The corresponding multicompartmentalized system mimics cellular reactions that take place in multiple membrane-bound compartments. These synthetic systems could provide a simplified model platform for studying the effect of compartmentalization on cellular processes.
Figure 6.
Multicompartmentalized catalytic synthetic cells. Initially, encapsulation of enzymes occurs in polystyrene-b-poly(3-(isocyano-lalanyl-amino-ethyl)-thiophene) (PS-b-PIAT) nanoreactors, followed by mixing of the artificial organelles, cytosolic enzymes, and reagents, and subsequent encapsulation of the mixture in polybutadiene-b-poly(ethylene oxide) (PB-b-PEO) vesicles to create the functional synthetic cell, inside which enzymatic multicompartment catalysis occurs.
Bayley et al. has also successfully developed a versatile technique for assembling whole networks of contiguous bioreactors, called multisomes.[37] Aqueous nanocompartments are inserted in an oil droplet immersed in water and stabilized with a monolayer of lipid. Nanocompartments quickly interface with each other and the surface of the oil droplet, forming bilayers that can house membrane channel proteins. The result is a set of connected reactors that are also capable of interfacing with the larger bulk environment and can be precisely tuned to rupture and mix upon temperature or pH changes. Recently, this technology has been utilized to engineer sophisticated 3D-patterned networks, providing groundwork towards the goal of preparing synthetic tissues.[38]
Transmembrane Channels
Functional channel-forming proteins equip artificial cells with a powerful tool to transport cargo. Substrate-specific transport can be tailored by judicious choice of pore proteins. However, incorporating integral membrane proteins into minimal cells can often be challenging. Recently, the Ueda group has taken an important step toward reliable insertion of membrane proteins by successfully synthesizing and incorporating the E. coli Sec translocon into synthetic cells.[39] The functional translocon, which is the natural machinery utilized to insert membrane proteins into lipid bilayers, was then successfully utilized to reconstitute functional peptidase into the vesicle membranes. This work should lay a solid foundation for future development of highly sophisticated artificial cells capable of synthesizing and incorporating complex transmembrane proteins into their membranes. Oftentimes, in vesicle translation and transcription studies, the bilayer is treated as an inert compartment, serving only to sequester and concentrate the machinery responsible for creating additional proteins or nucleic acids. Soga et al. have recently investigated the link between vesicle size and membrane-localized translation products.[40] After ruling out potential effects of compartment size on protein synthesis, the team demonstrated that a translated membrane transporter is less apt to localize to the bilayer of a giant unilamellar vesicle (GUV) as the size of the vesicle increases. The result likely stems from the increased ratio of lipid membrane to vesicle volume in smaller vesicles. While this achievement is intriguing in its own right, it should immediately lend itself to optimizing localization of functional, translated products to the surface of vesicles for model membrane-protein studies and for the development of more sophisticated and efficient artificial cells.
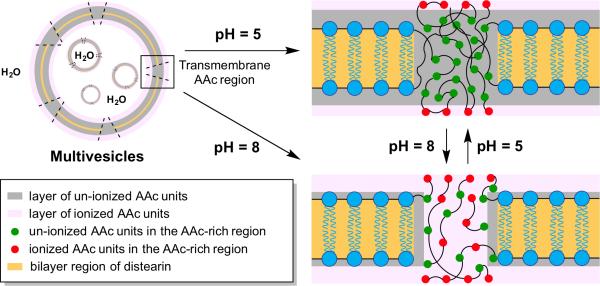
Additionally, it is also possible to utilize completely synthetic materials that can insert themselves into membranes and act as channels. The Chiu group has created multivesicle assemblies in which the membranes are outfitted with abiotic polymeric channels based on acrylic acid (AAc) units (Figure 7).[41] The pH-responsive polymeric channels are permeable to hydrophilic solutes. Equipping artificial vesicles with transmembrane proteins capable of controlling absorption and release processes holds promise for more advanced nanoreactor technology.
Figure 7.
Multicompartmentalized vesicles feature pH-responsive transmembrane channels. Multivesicle assemblies comprised of acrylic acid (AAc) and distearin acrylate (DSA) were prepared using a two-stage double emulsion. Increasing pH allows AAc residues to ionize, opening hydrophilic transmembrane channels.
Models for cell biology studies
Integrating synthetic constructs with biological systems has opened up novel opportunities for exploring fundamental biological properties and processes, such as the effect of macromolecular crowding or the trafficking of proteins across endosomal and mitochondrial membranes. LeDuc and colleagues recently reported the effect of molecular crowding on gene expression within lipid vesicles.[42] While crowding has been extensively studied for simple biochemical reactions, this work showed that crowding could increase the robustness of gene expression within synthetic cells. In a remarkable study, Huck and coworkers have used picoliter-sized droplets to create crowded coacervate environments.[43] The coacervate creates an artificial cell-like environment with remarkably improved transcription rates. Their results demonstrate the importance of confinement and compartmentalization for the organization of biochemical components. With the aim to control membrane localization of expressed proteins, a robust modular system was developed for targeting proteins to phospholipid membranes using SNAP-tag reactive lipid anchors.[44] This approach can be conveniently used to trigger membrane curvature events induced by natural biomolecules. Furthermore, photocaged lipid anchors enabled spatiotemporal control over protein modification.
The Bayley group has recently directed attention towards investigating the unfolding energetics of a model protein, thioredoxin, as it translocates across a membrane bilayer via the model protein nanopore, α-hemolysin.[45] Energetics were obtained by measuring current changes across rabbit, red blood cell membranes while electrophoretically threading the N-terminus of thioredoxin, conjugated to an oligonucleotide, through the hemolysin nanopore. The established model is especially relevant for the investigation of translocation systems.
Summary and Outlook
The scientific community has been exploring numerous powerful methodologies for the self-assembly of molecular components into minimal cells. The tools of synthetic biology are making possible the reconstitution of the necessary machinery to create synthetic membranes, as well as the intracellular constituents to generate specific functions, enabling the efficient construction of artificial cells. Novel approaches have been developed for the incorporation and/or integration of biological components in synthetic capsules to facilitate signaling responses, drug and gene delivery, and encapsulation and extended expression of biological species. We have surveyed recent advances and the current state-of-the-art of these compartmentalized systems. In particular, we have described relevant aspects of the design and preparation of minimal supramolecular architectures that can faithfully mimic or reconstruct the structure and/or function of living cells. These “hybrid” artificial cells are being actively investigated due their potential applications in different fields such as chemistry, medicine, pharmacology and materials science.
Acknowledgements
This work was partially supported by the University of California, San Diego, the National Science Foundation [CHE-1254611] and the Department of Defense (Army Research Office) [MURIW911NF-13-1-0383]. Roberto J. Brea thanks the Human Frontier Science Program (HFSP) for his Cross-Disciplinary Fellowship. Michael D. Hardy acknowledges support from an NIH T32 training grant (GM007240).
References
- 1.a Zhu TF, Szostak JW. J. Am. Chem. Soc. 2009;131:5705–5713. doi: 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mansy SS, Schrum JP, Krishnamurthy M, Tobe S, Treco DA, Szostak JW. Nature. 2008;454:122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Zhu TF, Adamala K, Zhang N, Szostak JW. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9828–9832. doi: 10.1073/pnas.1203212109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Budin I, Debnath A, Szostak JW. J. Am. Chem. Soc. 2012;134:20812–20819. doi: 10.1021/ja310382d. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Takakura K, Sugawara T. Langmuir. 2004;20:3832–3824. doi: 10.1021/la049738a. [DOI] [PubMed] [Google Scholar]; d Takakura K, Toyota T, Sugawara T. J. Am. Chem. Soc. 2003;125:8134–8140. doi: 10.1021/ja029379a. [DOI] [PubMed] [Google Scholar]
- 3.Adamala K, Szostak JW. Nat. Chem. 2013;5:495–501. doi: 10.1038/nchem.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adamala K, Szostak JW. Science. 2013;342:1098–1100. doi: 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blain JC, Szostak JW. Annu. Rev. Biochem. 2014;83:615–640. doi: 10.1146/annurev-biochem-080411-124036. [DOI] [PubMed] [Google Scholar]
- 6.Jesorka A, Orwar O. Annu. Rev. Anal. Chem. 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- 7.a Takakura K, Yamamoto T, Kurihara K, Toyota T, Ohnuma K, Sugawara T. Chem. Commun. 2014;50:2190–2192. doi: 10.1039/c3cc47786j. [DOI] [PubMed] [Google Scholar]; b Minkenberg CB, Li F, van Rijn P, Florusse L, Boekhoven J, Stuart MC, Koper GJ, Eelkema R, van Esch JH. Angew. Chem. Int. Ed. 2011;50:3421–3424. doi: 10.1002/anie.201007401. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011;123:3483–3486. [Google Scholar]
- 8.Budin I, Devaraj NK. J. Am. Chem. Soc. 2012;134:751–753. doi: 10.1021/ja2076873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy MD, Yang J, Selimkhanov J, Cole CM, Tsimring LS, Devaraj NK. Proc. Natl. Acad. Sci. U. S. A. 2015 doi: 10.1073/pnas.1506704112. in press. DOI: 10.1073/pnas.1506704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brea RJ, Cole CM, Devaraj NK. Angew. Chem. Int. Ed. 2014;53:14102–14105. doi: 10.1002/anie.201408538. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:14326–14329. [Google Scholar]
- 11.Zhou CY, Wu H, Devaraj N. Chem. Sci. 2015 doi: 10.1039/c5sc00653h. in press. DOI: 10.1039/c1035sc00653h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurihara K, Tamura M, Shohda K, Toyota T, Suzuki K, Sugawara T. Nat. Chem. 2011;3:775–781. doi: 10.1038/nchem.1127. [DOI] [PubMed] [Google Scholar]
- 13.Tian B, Tao X, Ren T, Weng Y, Lin X, Zhang Y, Tang X. J. Mater. Chem. 2012;22:17404–17414. [Google Scholar]
- 14.Gudlur S, Sukthankar P, Gao J, Avila LA, Hiromasa Y, Chen J, Iwamoto T, Tomich JM. PLoS One. 2012;7:e45374. doi: 10.1371/journal.pone.0045374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Patil AJ, Li M, Mann S. J. Am. Chem. Soc. 2014;136:9225–9234. doi: 10.1021/ja504213m. [DOI] [PubMed] [Google Scholar]
- 16.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Nat. Struct. Mol. Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 17.Rurup WF, Snijder J, Koay MS, Heck AJ, Cornelissen JJ. J. Am. Chem. Soc. 2014;136:3828–3832. doi: 10.1021/ja410891c. [DOI] [PubMed] [Google Scholar]
- 18.Tanner P, Baumann P, Enea R, Onaca O, Palivan C, Meier W. Acc. Chem. Res. 2011;44:1039–1049. doi: 10.1021/ar200036k. [DOI] [PubMed] [Google Scholar]
- 19.Li F, de Wolf FA, Marcelis AT, Sudholter EJ, Cohen Stuart MA, Leermakers FA. Angew. Chem. Int. Ed. 2010;49:9947–9950. doi: 10.1002/anie.201004003. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010;122:10143–10146. [Google Scholar]
- 20.Su X, Mohamed Moinuddeen SK, Mori L, Nallani M. J. Mater. Chem. B. 2013;1:5751–5755. doi: 10.1039/c3tb21111h. [DOI] [PubMed] [Google Scholar]
- 21.Yildiz UH, De Hoog HP, Fu Z, Tomczak N, Parikh AN, Nallani M, Liedberg B. Small. 2014;10:442–447. doi: 10.1002/smll.201300060. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsubo K, Marth JD. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Moussodia RO, Sun HJ, Leowanawat P, Muncan A, Nusbaum CD, Chelling KM, Heiney PA, Klein ML, Andre S, Roy R, Gabius HJ, Percec V. Angew. Chem. Int. Ed. 2014;53:10899–10903. doi: 10.1002/anie.201403186. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:11079–11083. [Google Scholar]
- 24.Paleos CM, Pantos A. Acc. Chem. Res. 2014;47:1475–1482. doi: 10.1021/ar4002679. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Fu L, Wei L, Kiang J, Binks BP. J. Am. Chem. Soc. 2015;137:1362–1371. doi: 10.1021/ja512337z. [DOI] [PubMed] [Google Scholar]
- 26.Kreft O, Skirtach AG, Sukhorukov GB, Möhwald H. Adv. Mater. 2007;19:3142–3145. [Google Scholar]
- 27.Shum HC, Zhao YJ, Kim SH, Weitz DA. Angew. Chem. Int. Ed. 2011;50:1648–1651. doi: 10.1002/anie.201006023. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011;125:2523–2528. [Google Scholar]
- 28.Kim SH, Shum HC, Kim JW, Cho JC, Weitz DA. J. Am. Chem. Soc. 2011;133:15165–15171. doi: 10.1021/ja205687k. [DOI] [PubMed] [Google Scholar]
- 29.Marguet M, Edembe L, Lecommandoux S. Angew. Chem. Int. Ed. 2012;51:1173–1176. doi: 10.1002/anie.201106410. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012;124:1199–1202. [Google Scholar]
- 30.Upadhyay KK, Bhatt AN, Mishra AK, Dwarakanath BS, Jain S, Schatz C, Le Meins J-F, Farooque A, Chandraiah G, Jain AK, Misra A, Lecommandoux S. Biomaterials. 2010;31:2882–2892. doi: 10.1016/j.biomaterials.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 31.Upadhyay KK, Mishra AK, Chuttani K, Kaul A, Schatz C, Le Meins JF, Misra A, Lecommandoux S. Nanomed. Nanotechnol. 2012;8:71–80. doi: 10.1016/j.nano.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Pourtau L, Oliveira H, Thevenot J, Wan Y, Brisson AR, Sandre O, Miraux S, Thiaudiere E, Lecommandoux S. Adv. Healthc. Mater. 2013;2:1420–1424. doi: 10.1002/adhm.201300061. [DOI] [PubMed] [Google Scholar]
- 33.Siegal-Gaskins D, Tuza ZA, Kim J, Noireaux V, Murray RM. ACS Synth. Biol. 2014;3:416–425. doi: 10.1021/sb400203p. [DOI] [PubMed] [Google Scholar]
- 34.Karzbrun E, Tayar AM, Noireaux V, Bar-Ziv RH. Science. 2014;345:829–832. doi: 10.1126/science.1255550. [DOI] [PubMed] [Google Scholar]
- 35.Siti W, de Hoog H-PM, Fischer O, Shan WY, Tomczak N, Nallani M, Liedberg B. J. Mater. Chem. B. 2014;2:2733–2727. doi: 10.1039/c3tb21849j. [DOI] [PubMed] [Google Scholar]
- 36.Peters RJ, Marguet M, Marais S, Fraaije MW, van Hest JC, Lecommandoux S. Angew. Chem. Int. Ed. 2014;53:146–150. doi: 10.1002/anie.201308141. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:150–154. [Google Scholar]
- 37.Villar G, Heron AJ, Bayley H. Nat. Nanotechnol. 2011;6:803–808. doi: 10.1038/nnano.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wauer T, Gerlach H, Mantri S, Hill J, Bayley H, Sapra KT. ACS Nano. 2014;8:771–779. doi: 10.1021/nn405433y. [DOI] [PubMed] [Google Scholar]
- 39.Matsubayashi H, Kuruma Y, Ueda T. Angew. Chem. Int. Ed. 2014;53:7535–7538. doi: 10.1002/anie.201403929. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:7665–7668. [Google Scholar]
- 40.Soga H, Fujii S, Yomo T, Kato Y, Watanabe H, Matsuura T. ACS Synth. Biol. 2014;3:372–379. doi: 10.1021/sb400094c. [DOI] [PubMed] [Google Scholar]
- 41.Chiu HC, Lin YW, Huang YF, Chuang CK, Chern CS. Angew. Chem. Int. Ed. 2008;47:1875–1878. doi: 10.1002/anie.200704078. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2008;47:1901–1904. [Google Scholar]
- 42.Tan C, Saurabh S, Bruchez MP, Schwartz R, Leduc P. Nat. Nanotechnol. 2013;8:602–608. doi: 10.1038/nnano.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolova E, Spruijt E, Hansen MMK, Dubuc E, Groen J, Chokkalingam V, Piruska A, Heus HA, Huck WTS. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11692–11697. doi: 10.1073/pnas.1222321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudd AK, Valls Cuevas JM, Devaraj NK. J. Am. Chem. Soc. 2015;137:4884–4887. doi: 10.1021/jacs.5b00040. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Larrea D, Bayley H. Nat. Nanotechnol. 2013;8:288–295. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]