Summary
Expression of Pitx2 on the left side of the embryo patterns left-right (LR) organs including the dorsal mesentery (DM), whose asymmetric cell behavior directs gut looping. Despite the importance of organ laterality, chromatin-level regulation of Pitx2 remains undefined. Here we show that genes immediately neighboring Pitx2 in chicken and mouse, including a long noncoding RNA, Playrr (Pitx2 locus asymmetric regulated RNA), are expressed on the right side and repressed by Pitx2. CRISPR/Cas9 genome editing of Playrr, 3D fluorescent in situ hybridization (FISH) and variations of chromatin conformation capture (3C), demonstrate that mutual antagonism between Pitx2 and Playrr is coordinated by asymmetric chromatin interactions dependent on Pitx2 and CTCF. We demonstrate that transcriptional and morphological asymmetries driving gut looping are mirrored by chromatin architectural asymmetries at the Pitx2 locus. We propose a model where Pitx2 auto-regulation directs chromatin topology to coordinate LR transcription of this locus essential for LR organogenesis.
INTRODUCTION
The external symmetry of vertebrates conceals left-right (LR) asymmetries of the internal organs essential for function and placement within the body cavity. LR patterning initiates early during gastrulation via transient signaling by Nodal, which yields persistent expression of the transcription factor Pitx2 throughout the left lateral mesoderm (Logan et al. 1998; Shiratori et al. 2001). Pitx2 then specifies left identity within individual organ primordia. The control of laterality by Pitx2 is very conserved, functioning even in basal deuterostomes such as sea urchin and non-bilaterians such as hydra (Duboc et al. 2005; Watanabe et al. 2014).
Regulation of Pitx2 in vertebrates involves multiple isoforms. The Pitx2a and 2b splice variants are expressed bilaterally, while Pitx2c is transcribed from a separate promoter and is exclusively left-sided (Shiratori et al. 2006; Liu et al. 2001). Pitx2 null embryos die mid-gestation with global laterality defects (Lu et al. 1999). Moreover, PITX2 is mutated in humans with Axenfeld-Rieger Syndrome (ARS), characterized by mental retardation, craniofacial and body wall defects, and umbilical hernias (Semina et al. 1996). Importantly, some ARS patients bear no mutations in Pitx2 coding sequences, but harbor lesions within a conserved gene desert adjacent to PITX2, suggesting cis-regulatory elements within the desert are essential for Pitx2 expression (Flomen et al. 1998; Volkmann et al. 2011; Reis et al. 2012). Furthermore, single-nucleotide polymorphisms (SNPs) associated with PITX2-linked atrial fibrillation (AF), map to the same gene desert (Lubitz et al. 2010; Kent et al. 2002). Despite much available knowledge of early laterality control, specific genomic mechanisms governing the expression of its master effector gene remain unexplored.
Here we employ the unique binary (L vs. R) organization of the gut dorsal mesentery (DM), a suspensory mesodermal bridge with asymmetries directed by left-sided Pitx2 (Fig. 1A, top). Condensation of the left and expansion of the right side causes the DM to deform, tilting the attached gut tube leftward (Davis et al. 2008; Kurpios et al. 2008). This tilt biases asymmetric gut rotation, the disruption of which randomizes gut looping (Davis et al. 2008) (Fig. 1A, top).
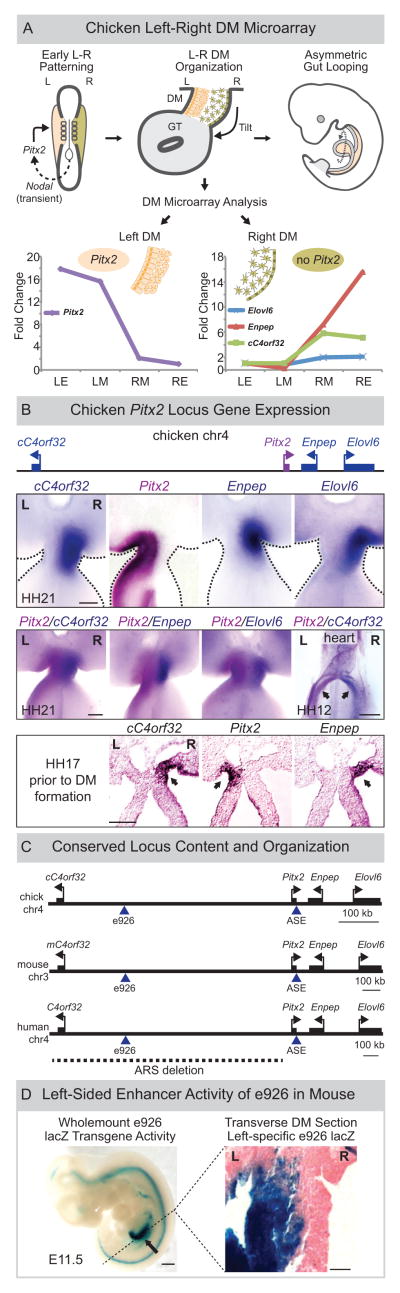
Figure 1. LR asymmetric gene expression at the Pitx2 locus.
(A) Pitx2 initiates gut looping directed by LR changes in the DM cellular architecture (tan, left; green, right). DM microarray (LE, left epithelium; LM, left mesenchyme; RM, right mesenchyme; RE, right epithelium) identifies genes linked to Pitx2 with right side-restricted expression, validated in (B) via whole mount ISH (double labeling Pitx2, magenta; right-specific genes, blue) at HH21 (DM), at HH17 (lateral mesoderm, DM precursor, arrows) and at HH12 (sinus venosus of the primitive heart, arrows). (C) Pitx2 locus conservation in chicken, mouse, and human (dashed line, human ARS deletion). Note, human locus is inverted relative to its orientation on chr4. (D) e926 directs left-specific reporter gene expression (LacZ) in the left DM of transgenic mouse embryos. Scale bars: (B) 100μm, HH21 and HH17, 500μm, HH12; (D) 500μm, whole mount, 25μm, section. See also Figure S1.
To define the molecular composition of the DM, we laser dissected and microarray profiled the left (Pitx2-positive) and right (Pitx2-negative) sides of the DM when these asymmetries are apparent (chicken HH21, akin to mouse embryonic [E] day 10.5) (Hamburger and Hamilton 1951; Welsh et al. 2013). Here we reveal that genes neighboring Pitx2 and the adjacent conserved gene desert are expressed in a right-specific pattern opposite to left-specific Pitx2. Within the gene desert, we identified a conserved sequence element e926 that functions endogenously as a promoter for a conserved long noncoding RNA (lncRNA) we named Playrr, that is exclusively transcribed in the right DM. Importantly, genetic analyses found Pitx2 and Playrr expression are mutually antagonistic. We employed fluorescent in situ hybridization (FISH) and variations of chromatin conformation capture (3C) to correlate this binary LR expression with nuclear architecture in the DM and identified conserved LR differences in nuclear proximity of e926/Playrr and Pitx2 that are dependent on Pitx2 and CTCF. Our data demonstrate chromatin-level regulation that mirrors LR organogenesis and that tissue-specific cis-regulatory topology establishes LR transcription among higher vertebrates.
RESULTS
Right-sided expression of genes at the Pitx2 locus is opposite to Pitx2 on the left
Our previous microarray analysis of the left and right chicken DM (Welsh et al., 2013) aimed to identify Pitx2 target effector genes responsible for cellular asymmetries in the DM. We first confirmed that Pitx2 is the most differentially expressed gene on the left side of the DM, with ~19-fold higher expression in the left vs. right DM (Fig. 1A, graph). Our analyses also revealed that glutamyl aminopeptidase A (Enpep) is the most differentially expressed gene in the right DM, with ~17-fold higher expression (Fig. 1A, graph). Remarkably, these two genes of highly contrasting transcription are located immediately adjacent to each other in the genome. Chicken Pitx2 is flanked proximally by a large (~600kb) gene desert and 27kb distally by the convergently transcribed Enpep (Fig. 1B). We confirmed these findings in situ and demonstrated that DM tissue asymmetry is mirrored by exclusive LR expression patterns of these genes (Fig. 1B HH21).
Further examination identified two more genes at this locus with right-sided expression: fatty acid elongase, (Elovl6), and Loc422694 (hereafter cC4orf32, ortholog of human C4ORF32) (Fig. 1A–C). This asymmetric pattern of expression was not exclusive to the DM: asymmetries were present well prior to DM formation in the lateral mesoderm (precursor to the DM, Fig. 1B HH17) and in the heart, where Pitx2 also plays an essential role (Fig. 1B, HH12) (Franco and Campione 2003). The importance of the Pitx2 locus during embryogenesis is underscored by strict conservation of its gene content, order, and orientation from humans to frogs, suggesting that functional and regulatory constraints maintain such synteny (Kikuta et al. 2007; Nobrega et al. 2003) (Fig. 1C).
Identification of asymmetric regulatory element e926 at the Pitx2 locus
Asymmetric Pitx2c expression requires the ASE enhancer element located in the last intron of Pitx2 (Shiratori et al., 2006). ASE functional activity is highly conserved, and homologous ASE sequences from fish, frog, chicken, mouse, and human drive left-specific reporter gene activity in mice (Shiratori et al. 2001, 2006). We hypothesized that additional elements exist in the gene desert that govern robust LR expression of neighboring genes. Such deserts are devoid of protein coding genes, and commonly contain regulatory elements (REs) that determine spatiotemporal expression of nearby genes.
We searched the Vista Enhancer Database (Visel et al. 2007) in the area of the gene desert and identified hs926, a human-derived sequence highly conserved among human, mouse, and chicken (Fig. 1C). Element 926 (e926, 1614 base pairs) showed reproducible enhancer-like activity in the midgut of wholemount E11.5 transgenic reporter embryos (Fig. 1D, n=4/7), and drove robust lacZ reporter activity specifically in the left DM of the midgut and duodenum (Fig. 1D and S1 n=3/3 (transgenic embryos kindly provided by the Vista Enhancer group). Interestingly, right-specific e926 enhancer activity was noted in the heart (Fig. S1, n=2/3). Thus, e926 functions as an operational enhancer in transgenesis assays, and responds both to the left and right regulatory environments in the DM and other asymmetric organs.
Right-specific regulatory activity of e926 in vivo
Since enhancers may act differently in trans as single elements or in the cis-context of endogenous loci (Marinić et al. 2013; Ruf et al. 2011; West et al. 2002), we sought to characterize e926 function in the DM. We used genome-wide global run-on sequencing, GRO-seq, of the left vs. right DM (Fig. 2A) (Core et al. 2008). GRO-seq maps nascent transcripts engaged by RNA polymerase II (RNA Pol II) by collecting strand-specific reads to identify regions of divergent transcription, a signature of active REs (Core et al. 2008; Melgar et al. 2011; Core et al. 2014). We used discriminative Regulatory Element detection from GRO-seq (dREG), which recognizes the pattern of divergent transcription at active REs, to characterize the regulatory state of both nascent transcription and RE activation (Fig. 2A, green) (Danko et al. 2014).
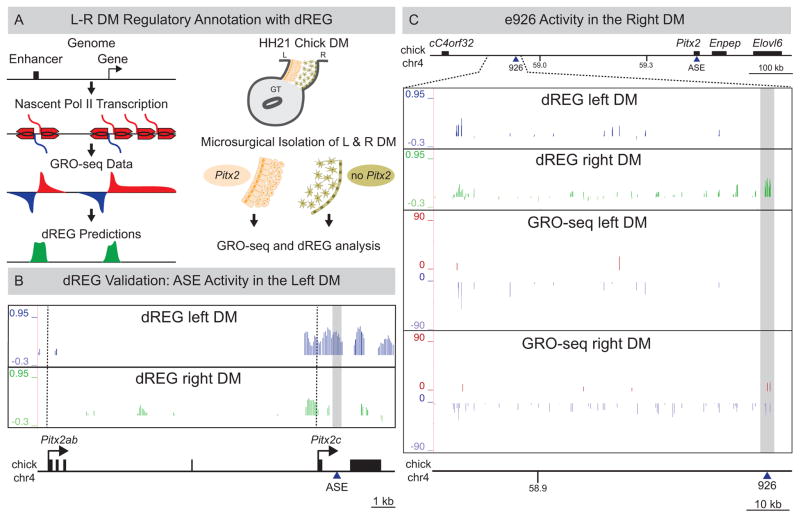
Figure 2. LR DM GRO-seq/dREG analyses.
(A) Left: GRO-seq and dREG. Right: Libraries were prepared from HH21 LR DM samples (B) dREG peaks corresponding to ASE are left-specific (grey box). Dashed vertical lines mark Pitx2ab and Pitx2c TSS. (C) dREG peaks overlapping e926 are right-specific (grey box) in contrast to left-specific e926 transgenic reporter activity. GRO-seq detects asymmetric transcription from the minus strand at the proximal gene desert in the right DM. See also Figure S2 and S3.
Consistent with our microarray data, both GRO-seq and dREG analysis confirmed the binary LR gene expression from the Pitx2 locus (Fig. S2). For example, GRO-seq reads mapped to the asymmetric Pitx2c at the distal end of the desert only in the left DM samples, and dREG peaks at the ASE enhancer were also exclusively observed in the left samples (Fig. 2B, grey box; Fig. S3). In contrast, GRO-seq and dREG detected extensive transcription of the proximal desert preferentially in the right DM (Fig. 2C, Fig. S2), with dREG peaks at the e926 enhancer (Fig. 2C, grey box). This confirms e926 as an asymmetrically responsive cis-RE, but contrasts starkly with the transgenesis assay, which suggested a left-specific activity (Fig. 1D).
Identification of a conserved lncRNA, Playrr, transcribed from e926 on the right side
An alternative explanation for the discrepancy in asymmetric activity of e926 is that this RE functions not as an enhancer, but as a gene promoter in the right DM. However, no such genes are annotated within this region of the chicken gene desert. Interestingly, GRO-seq reads extended a considerable distance from e926, suggesting a potential gene transcribed from the minus strand towards cC4orf32 (Fig. 2C, GRO-seq right DM). Supporting this hypothesis, the syntenic region of the mouse genome is annotated with D030025E07Rik, a long noncoding RNA (lncRNA) with a transcriptional start site (TSS) contained within e926 that is also transcribed from the minus strand of mouse chromosome 3 (Fig. 3A). We generated clones and RNA probes from D030025E07Rik and chicken ESTs from this transcribed region. In both species, expression of this lncRNA was restricted to the right side of the DM consistent with the GRO-seq data (Fig. 3B), suggesting a conserved lncRNA is expressed on the right side, opposite to Pitx2. We refer to this lncRNA as Playrr (Pitx2 locus asymmetric regulated RNA).
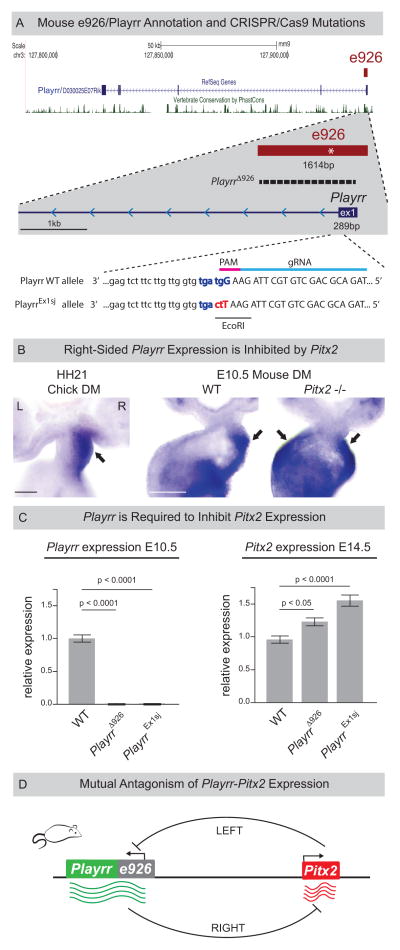
Figure 3. Identification of a conserved lncRNA, Playrr, asymmetrically transcribed from e926.
(A) Annotation of mouse lncRNA Playrr (D030025E07Rik). e926 contains the TSS and exon 1 of Playrr and a conserved Pitx2 binding motif (asterisk). CRISPR/Cas9 mutation of e926/Playrr (PlayrrΔ926, 1420bp dashed line; PlayrrEx1sj, red and blue letters). Exon 1 (upper case), intron 1 (lower case), relative guide RNA (gRNA, light blue) and PAM motif (pink) were used to produce PlayrrEx1sj. (B) Whole mount ISH showing right-specific Playrr in WT HH21 chicken and E10.5 mouse embryos vs. bilateral Playrr expression in Pitx2-null mouse DM. (C) qRT-PCR expression in PlayrrΔ926 and PlayrrEx1sj mutants (n=3) relative to WT (n=3) of Playrr (left) and Pitx2 (right) at E10.5 or E14.5, respectively, error bars ± S.E.M. (D) Mutual antagonism of Playrr and Pitx2 expression. Scale bars in (B) 100μm. See also Figure S1 and S4.
Mutual antagonism of Pitx2 and Playrr expression in vivo
We found conserved Pitx2 binding sites within e926 suggesting that Pitx2 regulates Playrr expression (Fig. S1B). To test this in vivo, we examined Playrr expression in the DM of E10.5 Pitx2−/− mouse embryos. In the absence of Pitx2, we found that Playrr is expressed bilaterally in the DM (Fig. 3B). Using qRT-PCR we confirmed upregulation of Playrr expression in whole Pitx2−/− embryos, while other locus genes were unaffected. This indicates that in the left DM, Pitx2 specifically represses Playrr expression (Fig. S4C).
To uncouple e926 and Playrr function during Pitx2 locus expression (Fig. 3A), we used CRISPR/Cas9 genome editing. We first targeted the proximal and distal ends of e926, simultaneously deleting e926 and disrupting Playrr transcription (PlayrrΔ926). Second, to disrupt Playrr while leaving e926 intact, we mutated 3 base pairs of the U1 snRNP recognition sequence (ggtagt) at the 3′ end of Playrr exon 1 (PlayrrEx1sj, Fig. 3A), leading to intron retention and exosomal degradation (Almada et al. 2013). Loss of detectable Playrr RNA was confirmed by qRT-PCR in both PlayrrΔ926 and PlayrrEx1sj E10.5 embryos (Fig. 3C). Importantly, significant upregulation of Pitx2 was found in E14.5 organ primordia of both PlayrrΔ926 and PlayrrEx1sj embryos compared to WT littermates (Fig. 3C, E14.5). These data demonstrate mutual antagonism between Pitx2 and Playrr expression and establish that function of the lncRNA Playrr is required to modulate Pitx2 expression levels (Fig. 3D).
Asymmetric chromatin looping of the Pitx2 locus in the chicken DM
The contralateral expression of Pitx2 and Playrr, their mutual antagonism, and evolutionary conservation of their arrangement on the chromosome, suggest they may be regulated by chromosome structure. To address this, we employed multi-color 3D DNA FISH on chicken DM sections (3D tissue-FISH). We used DNA probes (15–20 kb) to label cC4orf32 (Cy5, blue) at the proximal gene desert, Playrr (DIG, green) within the desert, and Pitx2 (Cy3, red) at the distal desert (Fig. 4A). Quantified interprobe distances (i.e. cC4orf32-Playrr, cC4orf32-Pitx2, and Playrr-Pitx2) were used to determine the position of these loci in 3D space relative to each other and to the intervening desert.
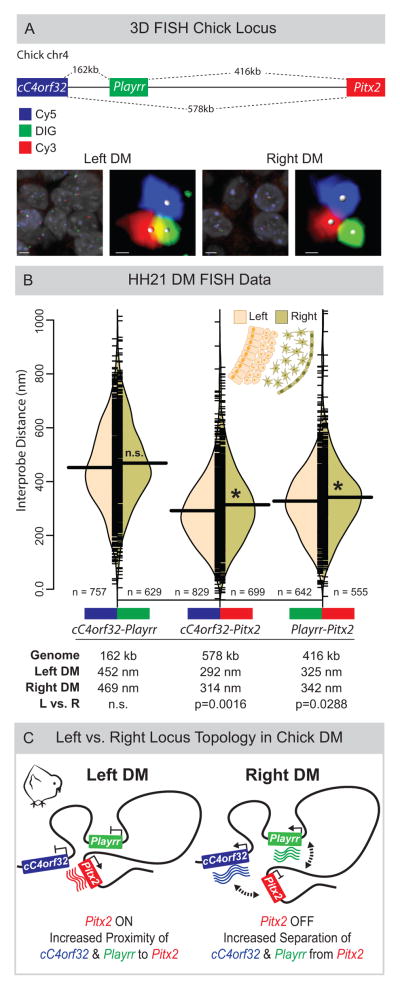
Figure 4. Asymmetric chromatin interactions of the Pitx2 locus in the chicken LR DM.
(A) Linear genomic distances in the chicken separating cC4orf32, Playrr, and Pitx2, marked with Cy5, DIG, and Cy3 labeled DNA probes, respectively. FISH labeled nuclei and individual loci in the left and right DM. (B) Split bean plots of interprobe distances for each probe pair (WT, HH21). Individual measurements are plotted as short horizontal ticks, the density trace and mean are plotted as the filled curve and large horizontal bar, respectively. Table summary of distances separating each probe pair in the linear genome or nucleus. (C) Model of Pitx2 locus organization and gene expression in the chicken DM. Scale bars: (A) 2μm and 300nm for nuclei and individual loci, respectively. See also Figure S4.
Within the genome, cC4orf32 and Playrr are nearest each other, separated by 162kb (compare to 578kb between cC4orf32 and Pitx2, or 416kb between Playrr and Pitx2 (Fig. 4A). However, our FISH revealed that within DM nuclei, both cC4orf32 and Playrr were significantly closer to Pitx2 than to each other (Fig. 4B). This striking interaction brings far-separated genes (416 kb and 578 kb) into closer spatial proximity than genes separated by just 160 kbs. Our data therefore establish that long-range looping of the Pitx2 locus positions the proximal and distal ends of the gene desert in close 3D proximity.
The global architecture of the Pitx2 locus is similarly organized in the left and right DM (Fig. 4BC). Importantly however, we identified subtle but statistically significant LR differences in interprobe distances for both cC4orf32-Pitx2 and Playrr-Pitx2, demonstrating closer proximity of both cC4orf32 and Playrr to Pitx2 in the left DM compared to the right (Fig. 4B, Fig. S4A). LR differences in these distances were reproducible across five replicate experiments (Fig. S4). Hence, mirroring the asymmetric gene expression and cellular architecture in the DM, proximity of cC4orf32, Playrr, and Pitx2 DNA in nuclei of the left DM is associated with preferential transcription of Pitx2. Conversely, in the right DM, where cC4orf32 and Playrr are expressed, they are further separated from Pitx2 (Fig. 4C). Thus, LR differences in 3D looping of the Pitx2 locus are characterized by subtle local shifts, rather than global differences, in the positioning of genes within a constant overall locus topology.
Spatial proximity of Playrr and Pitx2 is a conserved feature of locus topology
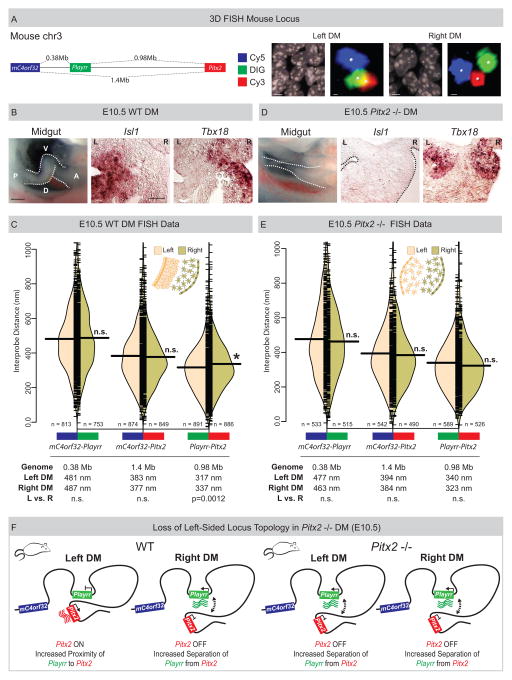
To address the degree to which asymmetric nuclear architecture of the chicken Pitx2 locus is conserved, we used the same FISH labeling scheme on mouse DM sections and analyzed pairwise interprobe distances for 5730508B09Rik (mC4orf32, the mouse ortholog of C4ORF32, Cy5, blue) in the proximal desert, Playrr (DIG, green) within the desert, and Pitx2 (Cy3, red) at the distal desert (Fig. 5A). In the mouse genome, the linear genomic distance separating the FISH probe pairs mC4orf32-Playrr, mC4orf32-Pitx2, and Playrr-Pitx2 is 380kb, 1.36Mb, and 980kb respectively (Fig. 5C).
Figure 5. Spatial proximity of Playrr-Pitx2 chromatin interactions are dependent on Pitx2.
(A) Linear genomic distances in the mouse separating mC4orf32, Playrr, and Pitx2, marked with Cy5, DIG, and Cy3 labeled DNA probes, respectively. FISH labeled nuclei and individual loci in the left and right DM. (B) Chiral looping of the WT midgut is accompanied by left-specific Isl1 and right-specific Tbx18. A, anterior; P, posterior; D, dorsal; V, ventral. (C) Split bean plots (WT E10.5). Table summary of distances separating each probe pair in the linear genome or nucleus. (D) Altered midgut looping and right-isomerized DM in Pitx2-null embryos is evidenced by loss of left-specific Isl1 and bilateral Tbx18 expression. (E) Split bean plots (Pitx2-null E10.5). (F) Model of Pitx2 locus organization and gene expression in the mouse DM. Scale bars: (A) 2μm and 300nm for nuclei and individual loci, respectively; (B) 250μm, whole mount, 25μm, section. See also Figure S4.
Consistent with the chicken DM, long-range looping across the mouse gene desert positions mC4orf32 and Playrr in significantly closer proximity to Pitx2 than to each other (Fig. 5C). Remarkably, though mouse Playrr is located nearly twice the genomic distance proximal to Pitx2 compared to chicken, the interprobe distance measured for Playrr-Pitx2 was nearly identical to that measured in chicken (Fig 4B and Fig. 5C). Moreover, Playrr-Pitx2 spatial proximity in the left vs. the right DM was maintained in the mouse, further highlighting our observation that proximity of Playrr and Pitx2 on the left is associated with preferential Pitx2 transcription (Fig. 5C). In contrast, the position of mC4orf32 relative to both Playrr and Pitx2 is farther than observed in chicken and not different in nuclei on the left vs. right (Fig. 5CF), a finding accompanied by the bilateral expression of mC4orf32 in mouse DM (Fig. S6A). Thus, small differences in the position of genes relative to each other have direct consequences on their asymmetric expression. Collectively, LR asymmetry in nuclear proximity of Playrr to Pitx2 is a conserved feature of Pitx2 locus topology.
Altered LR patterning disrupts asymmetric Pitx2 locus topology
In mice lacking Pitx2, the left DM fails to condense, all LR DM asymmetry is lost and stereotypical gut looping is randomized (a ‘double-right’ phenotype) (Fig. 5D) (Davis et al. 2008; Kurpios et al. 2008). Pitx2-null embryos lose left-specific expression of Pitx2 target genes such as Islet1 (Fig. 5D), and show bilateral expression of right-sided genes, such as Tbx18 (Fig. 5D) and Playrr (Fig. 3C). To investigate whether the subtle LR differences of locus topology are similarly affected, we performed 3D tissue-FISH on Pitx2-null mouse DM. Importantly, we found that the global locus topology was maintained while the proximity of Playrr and Pitx2 in the left WT DM was lost in Pitx2-null embryos (Fig. 5E). Hence, in the absence of Pitx2, Playrr and Pitx2 are significantly farther apart in the left DM compared to the WT left DM, and indistinguishable from WT interprobe distances in the right DM (Fig. 5CE, Fig. S4). Thus, a double-right phenotype of the Pitx2-null DM is accompanied by isomerized nuclear architecture at the Pitx2 locus (Fig. 5F). These data indicate that Pitx2 is required for asymmetric positioning of individual loci within the overall locus topology (Fig. 5F).
Analysis of Playrr-Pitx2 chromatin interactions in mouse ES cells
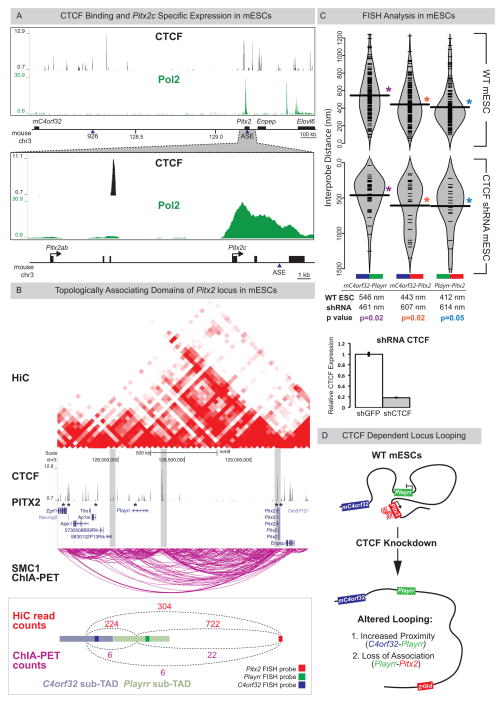
The chromatin regulatory landscape within mouse embryonic stem cells (mESCs) has been extensively characterized. Re-analyses of existing RNA Pol II ChIP-seq data (Kagey et al. 2010) demonstrate that mESCs express the LR asymmetric Pitx2c isoform (Fig. 6A), while mC4orf32 is expressed at low levels and Enpep and Playrr are undetectable. Pitx2c expression in these cells is accompanied by activation of the asymmetric ASE enhancer (Fig. S7) (Creyghton et al. 2010). We therefore leveraged available chromatin interaction data sets to analyze locus topology associated with Pitx2c expression in mESCs (Shen et al. 2012; Dowen et al. 2014) to model the left-specific transcriptional and regulatory profile of the locus.
Figure 6. Playrr-Pitx2 chromatin interactions in mouse ES cells are dependent on CTCF.
(A) Top: CTCF binding at the Pitx2 locus (CTCF ChIP-seq); Bottom: mESCs preferentially express the asymmetric Pitx2c isoform (RNA Pol II ChIP-seq). (B) A topologically associating domain (TAD) spans the Pitx2 locus (HiC, red; Smc1 ChIA-PET, magenta). Vertical grey bars highlight boundaries between sub-TADs. CTCF binding sites (ChIP-seq) at both Pitx2 and Playrr are associated with Pitx2 binding (Pitx2-FLAG ChIP-seq, asterisks). Quantification of HiC (red) and Smc1 ChIA-PET (magenta) interactions between the genomic interval containing the Pitx2 FISH probe and the Playrr sub-TAD (light green) compared to the mC4orf32 sub-TAD (light blue). (C) FISH analysis of Pitx2 locus topology in WT (top) or shRNA mediated CTCF knockdown mESCs (bottom, y-axis has been inverted); table shows summary of distances separating each probe pair in the nucleus. CTCF expression via qRT-PCR in control vs CTCF k.d. mESCs, error bars ± S.E.M. (D) Summary of Pitx2 locus topology in mESCs. See also Figure S7.
We defined looping interactions using HiC, a genome-wide variation of chromatin conformation capture (3C), and by paired-end tag sequencing (ChIA-PET) (de Wit and de Laat 2012). Both HiC and Smc1 ChIA-PET demonstrated enriched long-range interactions at the Pitx2 locus compared to flanking regions, data consistent with a topologically associating domain (TAD). Importantly, these data suggest the division of the TAD into sub-TADs that correlate with the position of our DNA FISH probes (Fig. 6B).
To determine if proximity of Playrr to Pitx2 vs. mC4orf32 accompanies Pitx2c expression in mESCs as observed in vivo, we quantified HiC & ChIA-PET reads linking the mC4orf32 and Playrr sub-TADs with each other and to Pitx2 (Fig. 6B, bottom). Significantly, although Playrr is 2.5 times farther from Pitx2 than from mC4orf32, the Playrr sub-TAD interaction with Pitx2 is nearly 4 times greater than with the adjacent mC4orf32 sub-TAD (Fig. 6B, bottom).
Finally, we performed FISH on WT mESCs. We found that the 3D organization of the Pitx2 locus in mESCs is nearly identical to mouse DM, with Playrr-Pitx2 closest and mC4orf32-Playrr furthest separated (Fig. 6C top). Thus, genome-wide molecular analysis and direct visualization together confirm the preferential association of Playrr with Pitx2 that accompanies asymmetric Pitx2 expression.
CTCF is essential for maintaining the long-range Playrr-Pitx2 interaction
CCCTC-binding factor, CTCF, is a sequence-specific scaffold protein that, by interaction with proteins such as Cohesin and Mediator, directs long-range looping to shape cell-type specific chromatin architecture (Splinter et al. 2006; Phillips-Cremins et al. 2013).
CTCF ChIP-seq data from mESCs identified CTCF binding peaks in the proximal gene desert near mC4orf32 and Playrr (Fig. 6B) (Kagey et al. 2010). Both Smc1 ChIA-PET and CTCF ChIP-seq highlighted that the Playrr-Pitx2 interaction is mediated by CTCF sites within the Pitx2 gene and upstream of e926/Playrr (Fig. 6AB). Significantly, Pitx2-FLAG ChIP-seq in vivo established that the CTCF binding sites at both Pitx2 and Playrr may also be associated with Pitx2 binding (Fig. 6B) (Tao et al. 2014).
To test this hypothesis we knocked down CTCF in mESCs (Fig. 6C), which abrogated long-range looping across the gene desert and disrupted global chromatin organization of the locus (Fig. 6CD), compared to WT mESCs. Significantly, CTCF knockdown also decreased the distance separating mC4orf32-Playrr (mean separation 546nm vs. 461nm in WT vs CTCF knockdown) and abolished the close proximity of Playrr to Pitx2 (mean separation 412nm vs. 614nm in WT vs CTCF knockdown). This demonstrates a requirement for CTCF during long-range chromatin looping at the Pitx2 locus, a prerequisite for Pitx2-dependent regulation of laterality (Fig. 6D).
DISCUSSION
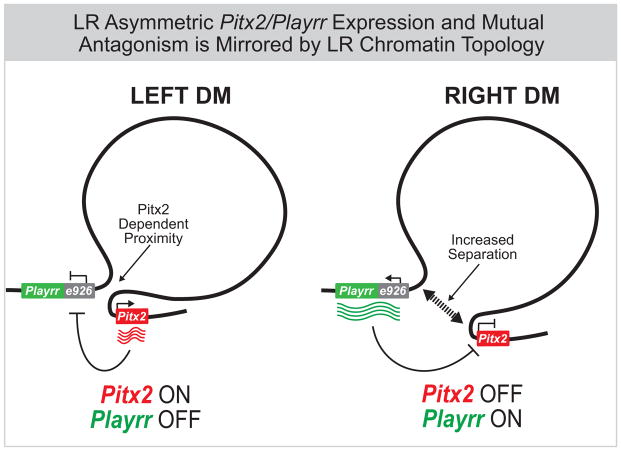
Pitx2 expression governs transcriptional programs that determine asymmetric organ morphology. In the DM, left-sided Pitx2 regulates molecular and cellular asymmetry required to loop and vascularize the gut (Davis et al. 2008; Kurpios et al. 2008; Welsh et al. 2013; Mahadevan et al. 2014). We now show this binary LR tissue organization is mirrored by transcriptional output and chromatin architecture at the Pix2 locus in the left vs. right DM (Fig. 7).
Figure 7. Summary of Pitx2 locus asymmetric expression and chromatin topology.
In chicken and mouse, left-specific Pitx2 expression and right-sided expression of flanking genes at the Pitx2 locus, including the conserved lncRNA Playrr is accompanied by Pitx2 dependent asymmetric chromatin topology in the DM. In vivo, Pitx2 and Playrr expression are mutually antagonistic.
Our 3D tissue FISH analyses characterize how the conserved genomic interval harboring Pitx2 is organized in 3D space to effect asymmetric gene expression and LR patterning. The topology of the Pitx2 locus, as measured optically, demonstrates both global and local organizational principles consistent with HiC and ChIA-PET data from mESCs, an in vitro model for left-specific Pitx2 locus transcription. In chicken and mouse, we establish that long-range chromatin looping across a large gene desert is a conserved and constitutive feature of Pitx2 locus organization, dependent on CTCF. Within this constitutive topology, we show that Playrr-Pitx2 are in closer proximity in the left DM where Pitx2 is preferentially expressed but positioned further apart in the right DM where Playrr is expressed and Pitx2 is not (Fig. 7). While LR differences in interprobe distances are notably small, they are reproducible across biological replicates, models, and organisms. Moreover, the mean Playrr-Pitx2 interprobe distance in the left DM of Pitx2-null mice is similar to that on the right side of both Pitx2−/− and WT mice, showing these LR differences are Pitx2-dependent. Our FISH analysis only marks the relative position of asymmetrically expressed genes in the left or right DM and it is likely that the subtle LR proximity differences are actually secondary to interactions between promoters and sequences that we have not labeled. Indeed, both HiC data and Smc1 ChIA-PET in mESCs demonstrate that the preferential interaction between Playrr and Pitx2 likely involves looping between CTCF sites near e926/Playrr and within the Pitx2 gene (Fig. 6B).
TAD boundaries are enriched in CTCF binding (Dixon et al. 2012; Nora et al. 2013; Phillips-Cremins et al. 2013). TAD structure also correlates with blocks of genomic synteny, and thus changes in their structure are likely evolutionarily constrained (Dixon et al. 2012; Nora et al. 2013). We show that CTCF is required for maintenance of the megabase-scale topology of the Pitx2 locus, characterized via 3D FISH. However, different gene expression patterns in chicken vs. mouse are consistent with significant differences in positioning of the proximal but not distal gene desert. Interestingly, in mESCs, binding of CTCF downstream of exon 3 of the bilaterally expressed Pitx2a overlaps a conserved CTCF recognition sequence in chicken (Fig. 6B) while CTCF binding sites proximally in the vicinity of C4orf32 appear to lack strict conservation. Interestingly, the gene desert is conserved in zebrafish, but they lack a C4ORF32 ortholog, suggesting evolutionary divergence of the proximal gene desert (Fig. S6C) (Volkmann et al. 2011). Furthermore, while Elovl6 is present, the zebrafish Pitx2 locus lacks an Enpep ortholog, and although both chick and mouse loci possess Enpep, this gene is not expressed in the mouse DM compared to its right-sided expression in chicken. Hence despite changes in gene content and regulatory refinement at the Pitx2 locus during evolution, control over Pitx2 spatial expression has been maintained. Consistent with this hypothesis, the HiC interaction map of mESC suggests that Pitx2 and Enpep are partitioned into separate regulatory domains (Fig. 6B). Thus, while the Pitx2 locus in chicken and mouse exhibits conserved architecture, species-specific differences have consequences on spatial gene expression and may result from the acquisition or turnover of CTCF binding.
Our study demonstrates the utility of combining genome-wide analyses in vivo to characterize nascent transcription and RE activity within a single sample. An unanticipated finding from this approach was the identification of Playrr as an asymmetrically expressed lncRNA derived from the conserved e926 RE. Rather than matching the left-specific enhancer activity of e926 observed in transgenic embryos, we show that Playrr is exclusively expressed in the right DM. Thus, dREG mapping of active REs in situ highlights one caveat of testing putative cis-regulatory sequences in trans, isolated from their endogenous context (Marinić et al. 2013).
A coherent understanding of the biological roles of lncRNAs is still lacking, although one of transcriptional regulation is an emerging theme (Rinn and Chang 2012; Vance and Ponting 2014). Recent analysis of the trans-acting lncRNA Fendrr, which interacts with the polycomb repressive complex 2 (PRC2), establishes a role for lncRNAs in regulation of Pitx2 expression (Grote et al. 2013). Although the impact on LR patterning was not addressed, mutation of Fendrr causes loss of Pitx2 silencing and altered differentiation of the lateral mesoderm (Grote et al. 2013; Grote and Herrmann 2013). We now show that Playrr, a lncRNA asymmetrically expressed from the Pitx2 locus itself, also acts to modulate Pitx2 expression. Interestingly, expression of Fendrr is restricted to gastrulation stages of development while Playrr is expressed later during organogenesis. Significantly, evidence for auto-regulation of asymmetric gene expression by Pitx2 has been documented during heart and limb development (Shiratori et al. 2006, 2014). The contralateral expression and mutual antagonism of Playrr and Pitx2 expressed from the same locus suggest e926/Playrr is an integrated regulatory module that provides such auto-regulation. Since the function of many lncRNAs likely depends on structural conformations permissive of DNA, RNA, or protein interactions (Rinn and Chang 2012; Nitsche et al. 2015; Johnsson et al. 2014), further characterization of our PlayrrEx1sj mutant provides a unique means to address the specific mechanisms through which Playrr influences Pitx2 locus output. Finally, Pitx2 expression is required in dynamic spatial patterns during development, where individual isoforms play dose-dependent roles in diverse tissues (Liu et al. 2001; Waite et al. 2013). Notably, midgut looping requires high levels of Pitx2c and is sensitive to bilateral misexpression of Pitx2c (Liu et al. 2001). Thus reciprocal interactions between Pitx2 and e926/Playrr likely provide an organ-intrinsic mechanism for tight Pitx2 dosing.
Mutations of human PITX2 result in a spectrum of ARS-associated birth defects and predispose to cardiac fibrillation and arrhythmia (Semina et al. 1996; Tao et al. 2014; Wang et al. 2014). Conservation of the gene desert at the locus and the number of noncoding mutations identified in human ARS patients likely reflects the regulatory complexity required to fine tune Pitx2 expression. Knowledge of the mechanisms that direct Pitx2 spatiotemporal expression is as critical as defining its downstream targets. Our work begins to shed light on the cis-regulatory mechanisms and etiology of ARS and Pitx2 associated AF, offering unprecedented potential to develop mouse models of these important human diseases.
EXPERIMENTAL PROCEDURES
Animals
WT and Pitx2 mutant embryos, Pitx2hd allele (Lu et al., 1999), were collected from timed matings with the morning of the plug defined as E0.5. Fertile eggs (White Leghorn) obtained from the Cornell Poultry Farm were incubated at 38°C and staged (Hamburger and Hamilton, 1992). Experiments adhered to guidelines of the Institutional Animal Care and Use Committee of Cornell University, under the Animal Welfare Assurance on file with the Office of Laboratory Animal Welfare.
Laser capture microdissection (LCM) and microarray
Detailed methods and full microarray results are in preparation to be published elsewhere. Briefly, cryosections of unfixed/flash frozen WT HH21 chicken DM were arrayed on membrane slides (Leica, 11505189). The asymmetric morphology of the left and right DM was used to discriminate and capture separately each compartment. Three separate biological experiments of four cell compartments were performed, giving 12 microarray samples (biological triplicate). RNA was isolated using the PicoPure RNA Isolation kit (Arcturus, KIT0202), and cDNA was prepared using the WT-Ovation Pico kit (NuGen). Affymetrix cRNA target labeling reactions were carried out per manufacturer instructions, and GCOS output files were loaded into GeneSpringGX 7.3 or GeneSpring 13 software packages (Agilent) for expression analyses.
RNA in situ hybridization
250μm thick embryo slices for whole mount RNA in situ hybridization (ISH) were collected with a McIlwain tissue chopper (Campden Instruments), fixed in 4% PFA/PBS ON, dehydrated, and stored in 100% methanol prior to processing. Whole mount ISH followed standard protocols as previously described (Welsh et al. 2013).
Statistical analysis
Measurement data were analyzed with R and Mann–Whitney–Wilcoxon test was used to compare interprobe distances in FISH experiments, and data was plotted with the beanplot package in R (Kampstra 2008). Box plots of FISH data in supplemental figure 4 were generated using JMP Pro 11. Student’s t-test was used for comparison of QRT-PCR data, error bars show ± S.E.M.
LR GRO-seq and dREG analysis
Whole embryos (n=250) were chopped into 250μm transverse slices using a McIlwain tissue chopper, followed by manual microdissection of the left and right DM. Collected tissues were pooled, snap frozen in liquid nitrogen and stored at −80°C until processing (Core et al. 2008). We also performed GRO-seq on whole embryos (HH21, n=2), on embryonic heads (HH12, n=250) and on the left and right hemisected embryos (HH12, n=250). dREG analysis of GRO-seq data sets are available via GitHub (https://github.com/Danko-Lab/dREG).
Cloning, plasmids, and oligonucleotides
Full-length cDNAs and RNA probes were cloned using TA cloning (Invitrogen) and oligo-dT primed cDNA reverse transcription (Superscript III, Invitrogen) from RNA pooled from HH19 and HH21 whole chicken, or from E8.5–18.5 whole mouse embryos. Cloned DNA was sequence-verified.
CRISPR/Cas9 targeting of e926/Playrr
The following Guide RNAs were designed (http://crispr.mit.edu/) (Hsu et al. 2013) and cloned into BbsI cut pX330: Playrr, TAGACGCAGCTGTGCTTAGAAGG; e926 proximal GTGGCGGACTCATGTTAAAAAGG; e926 distal GTGATTCCCACCACGCTTTGAGG. For Playrr targeting, the 156bp ssODN used as a repair template carries a 3bp change to disrupt the 5′ splice site of Playrr exon 1 and introduce an EcoRI restriction site for genotyping (see Fig. 3). sgRNA was in vitro transcribed (Ambion MEGAshortscriptT7 kit) from PCR generated template and purified (Ambion MEGAclear kit). Each sgRNA (50ng/μl), ssODN (10ng/μl) and Cas9 mRNA (100ng/μl) were injected into F1 hybrid (C57BL/6J x FvB/N) 1 cell embryos. Injected embryos were cultured to the 2 cell stage prior to transfer to recipient females. Two independent lines were established and analyzed for each mutation.
qRT-PCR
Embryonic tissue was isolated in cold PBS, stored in RNAlater until extraction (Qiagen RNeasy miniprep kit). For cultured cells, RNA was extracted using 4–6 × 106 cells. 2μg of RNA was reverse transcribed using the ABI high-capacity cDNA archive kit and diluted to 20ng/μl. The following TaqMan gene expression assays were used for relative quantification (ABI7500): Actb, Mm00607939_s1; 5730508B09Rik (mC4orf32), Mm02375228_s1; D030025E07Rik (Playrr), Mm03937997_m1; CTCF, Mm00484027_m1; Enpep, Mm00468278_m1; GAPDH, Mm99999915_g1; Pitx2ab, Mm00660192_g1; Pitx2c, Mm00440826_m1; pan-Pitx2, Mm01316994_m1.
FISH
The following BACs spanning either the chicken or mouse Pitx2 locus were used: chicken – CH261-95I8, CH261-66N5, CH261-187K8, CH261-34B16, CH261-110J5, CH261-91C24, CH261-134M23; mouse – RP23-306C6, RP23-328J13, RP23-225C17, RP24-98F15, RP23-266N9, RP23-106J9, RP23-150F9, RP24-156B21, RP24-100G2, RP23-307L21. The genomic distances analyzed in this study are resolvable via interphase FISH (50–100 kb); to further maximize resolution we designed probes to be less than 25kb (Ronot and Usson 2001; Trask et al. 1991). Genomic intervals spanned by FISH probes and probe sizes: chicken (galGal4): cC4orf32 (23.8kb), chr4:56,825,232–56,849,123; e926 (16kb), chr4:56,989,104–57,005,206; Pitx2 (17.5kb), chr4:57,398,355–57,415,925; mouse (mm10): mC4orf32 (15.5kb), chr3:127,846,052–127,861,644; e926 (16kb), chr3:128,221,875–128,237,854; Pitx2 (21kb), chr3:129,199,824–129,221,272. Probes were labeled with dUTP-Cy3, -Cy5, or -DIG, via nick translation of 1μg of DNA (Roche). Embryos were fixed overnight in 4% paraformaldehyde and embedded in paraffin following standard protocol. 6μm sections were collected on Suprerfrost plus slides, dried overnight at 37°C, baked at 60°C for 20 minutes, cooled, and then dewaxed in xylene with 2 washes for 10 minutes each, followed by 2 washes in 100% ethanol, and 1 wash in 70% ethanol, 5 minutes each. Sections were treated for 5 minutes with 0.2N NaOH in 70% ethanol to remove RNAs, washed twice in 70% ethanol, rehydrated, and washed 10 minutes in 0.1M citrate buffer at 80°C. Sections were washed in water, equilibrated in 2X SSC for 5 minutes, denatured for 2.5 minutes at 79°C in 70% formamide/2X SSC and hybridized overnight at 37°C with 50ng of probe, 10μg species specific Cot-1 DNA, 10μg salmon sperm DNA, and 10μg tRNA. Sections were washed with 2X SSC/50% formamide at 37°C, 2X SSC 37°C, 1X SSC at room temperature, for 15 minutes each. DIG labeled probes were detected with AF488 anti-DIG conjugated antibodies diluted 1:500 in 4X SSC/1% BSA. Mouse ESCs cells were seeded onto coverslips, fixed in 4% paraformaldehyde, then permeabilized following published protocols (Kurz et al., 1996). Hybridization followed the same protocol with the exception that the citrate unmasking step was omitted.
Quantification of HiC and Smc1 ChIA-PET interactions
Previously published HiC (Dixon et al. 2012, GEO accession GSE35156) or ChIA-PET (Dowen et al. 2014, GEO accession GSE57911) data sets were used to analyze chromatin interactions in mESCs linking the following genomic intervals (mm9): mC4orf32 sub-TAD, chr3:127279136–127674264; Playrr sub-TAD, chr3:127674265–128090964, Pitx2 FISH probe, chr3:128902742–128924190. The boundaries of sub-TADs were defined based on CTCF peaks (Roadmap data). To quantify HiC interactions, the virtual 4C tool (http://promoter.bx.psu.edu/hi-c/) was used with rs47546564, rs31670515, and Pitx2 as anchor points for the mC4orf32 sub-TAD, Playrr sub-TAD, and Pitx2 FISH probe, respectively. Read counts across each genomic interval connected to an individual anchor point were summed. To quantify ChIA-PET interactions, we counted the number of PETs that had one end overlapping with Pitx2, mC4orf32 or mPlayrr FISH probes and the other end overlapping with individual sub-TADs by at least 1 bp. The count results were similar from two Smc1 ChIA-PET replicates; we therefore took the sum of the two and visualized the data in WashU genome browser.
Image acquisition and analysis for 3D FISH
Slides were imaged on a Zeiss 710 scanning laser microscope using a 63X/1.4 NA Plan-APOCHROMAT oil immersion objective and Z-series data were acquired using the optimal step size. Imaris image analysis software (Bitplane) was used to quantify interprobe distances. Following image filtering using baseline subtraction, FISH signals for each channel were defined using the built in spot function which segments the signal using a Gaussian filter based background subtraction method and calculates the center of image mass of each segmented spot. The position of each spot object in (x, y, z) is equal to the value of the center of homogeneous mass of the object. Distance measurements between the center of mass for each signal was determined pairwise for each probe pair using the measurement point tool and verified by 2 independent investigators.
Lentiviral production and transduction
293FT cells were transfected with pLKO.1 lentiviral plasmids containing either shRNAs targeting CTCF or GFP, along with the packaging plasmids pLP1, pLP2, and pLP/VSV-g, using Lipofectamine 2000. Viral media was collected and centrifuged at 3000 rpm to pellet debris and then filtered through a 0.45μm PVFD filter and frozen until use. Virus containing media was supplemented with additional FBS, LIF, and 6μg/ml polybrene, prior to use. WT ES cells, expanded on gamma irradiated feeder cells, were trypsinized, plated for 30 minutes onto gelatin-free tissue culture plates to remove feeders prior to transduction. Following seeding ES cells at 2 × 106 cells/100mm plate and incubation for 24 hours, media was exchanged with ES media supplemented with 6μg/ml polybrene and plates were incubated for an additional15 minutes at 37°C prior to exchanging media with virus containing ES media. After transduction, media was exchanged for normal ES cell media and cells were incubated for an additional 24 hours prior to selection with 2μg/ml puromycin. After 3 days of selection, ES cells were trypsinized and plated for FISH or collected for RNA.
Supplementary Material
Acknowledgments
We thank Dr. Len Pennacchio and Jennifer Akiyama (http://enhancer.lbl.gov) for providing archived embryos, Drs. Victor Corces, Stephen Dalton, and Yuhua Sun for shRNA constructs, David Gludish for technical expertise on shRNA and for microarray analyses, and Dr. John Schimenti for discussions and stocks of mES cells. Jason Crossley, Michelle Miller, and Sriharsha Ponna provided technical assistance. We are grateful to Aravind Sivakumar, Aparna Mahadevan, and other members of the Kurpios lab for feedback on the manuscript. We thank Cornell Biotechnology Resource Center Imaging Facility and Carol Bayles for assistance with microscopy (NIH 1S10RR025502-01) and Rob Munroe and Chris Abratte of the Cornell Stem Cell and Transgenic Core Facility.
Footnotes
Accession Numbers
The microarray data have been deposited in the NIH Gene Expression Omnibus (GEO accession number GSE71117).
AUTHOR CONTRIBUTIONS
ICW and NAK designed the study. ICW performed all the experiments with additional contributions from LSS for FISH guidance and data analysis; HK for performing GRO-seq with guidance from JTL; CGD for dREG analysis with guidance from HK; FLC for double label RNA ISH (Fig. 1B) and CRISPR/Cas9 deletion of e926 (Fig. 3AC); MZ, and JFM for HiC and ChIA-PET computation analysis and quantifications (Fig. 6B). ICW and NAK wrote the manuscript with input from all coauthors.
BIBLIOGRAPHY
- Anderson E, Devenney PS, Hill RE, Lettice LA. Mapping the Shh long-range regulatory domain. Development. 2014 Oct;141(20):3934–3943. doi: 10.1242/dev.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342(6157):1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KSJ, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci. 2011;108:15751–15756. doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA. Bystander gene activation by a locus control region. EMBO J. 2004;23:3854–3863. doi: 10.1038/sj.emboj.7600365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Hyland SL, Core LJ, Martins AL, Waters CT, Lee HW, Cheung VG, Kraus WL, Lis JT, Siepel A. Accurate Identification of Active Transcriptional Regulatory Elements from Global Run-On and Sequencing Data. bioRxiv. 2014 doi: 10.1038/nmeth.3329. http://www.biorxiv.org/content/early/2014/11/12/011353.abstract. [DOI] [PMC free article] [PubMed]
- Davis NM, Kurpios NA, Sun X, Gros J, Martin JF, Tabin CJ. The Chirality of Gut Rotation Derives from Left-Right Asymmetric Changes in the Architecture of the Dorsal Mesentery. Dev Cell. 2008;15:134–145. doi: 10.1016/j.devcel.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra R-J, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Röttinger E, Lapraz F, Besnardeau L, Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell. 2005;9:147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Flajollet S, Rachez C, Ploton M, Schulz C, Gallais R, Métivier R, Pawlak M, Leray A, Issulahi AA, Héliot L, et al. The Elongation Complex Components BRD4 and MLLT3/AF9 Are Transcriptional Coactivators of Nuclear Retinoid Receptors. PLoS One. 2013;8(6):e64880. doi: 10.1371/journal.pone.0064880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flomen RH, Vatcheva R, Gorman PA, Baptista PR, Groet J, Barišić I, Ligutic I, Nižetić D. Construction and Analysis of a Sequence-Ready Map in 4q25: Rieger Syndrome Can Be Caused by Haploinsufficiency ofRIEG, but Also by Chromosome Breaks ≈ 90 kb Upstream of This Gene. Genomics. 1998;47:409–413. doi: 10.1006/geno.1997.5127. [DOI] [PubMed] [Google Scholar]
- Franco D, Campione M. The role of Pitx2 during cardiac development: Linking left-right signaling and congenital heart diseases. Trends Cardiovasc Med. 2003;13:157–163. doi: 10.1016/s1050-1738(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EEM. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512(7512):96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- Grote P, Herrmann BG. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10:1579–1585. doi: 10.4161/rna.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The Tissue-Specific lncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen C-A, Schmitt AD, Espinoza Ca, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4(9):e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampstra P. Beanplot: A Boxplot Alternative for Visual Comparison of Distributions. J Stat Softw. 2008;28:1–9. [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engström PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Jung I, Lee H, Kang K, Kim M, Jeong K, Kwon CS, Han YM, Kim YS, Kim D, et al. Human histone H3K79 methyltransferase DOT1L methyltransferase binds actively transcribing RNA polymerase II to regulate gene expression. J Biol Chem. 2012;287:39698–39709. doi: 10.1074/jbc.M112.384057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpios NA, Ibañes M, Davis NM, Lui W, Katz T, Martin JF, Belmonte JCI, Tabin CJ. The direction of gut looping is established by changes in the extracellular matrix and in cell: cell adhesion. Proc Natl Acad Sci. 2008;105:8499–8506. doi: 10.1073/pnas.0803578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJH, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci U S A. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu W, Lu MF, Brown NA, Martin JF. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development. 2001;128:2039–2048. doi: 10.1242/dev.128.11.2039. [DOI] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Pagán-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Mahadevan A, Welsh IC, Sivakumar A, Gludish DW, Shilvock AR, Noden DM, Huss D, Lansford R, Kurpios NA. The Left-Right Pitx2 Pathway Drives Organ-Specific Arterial and Lymphatic Development in the Intestine. Dev Cell. 2014;31:690–706. doi: 10.1016/j.devcel.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinić M, Aktas T, Ruf S, Spitz F. An Integrated Holo-Enhancer Unit Defines Tissue and Gene Specificity of the Fgf8 Regulatory Landscape. Dev Cell. 2013;24:530–542. doi: 10.1016/j.devcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2011;12(11):R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, De Laat W, Spitz F, Duboule D. A regulatory archipelago controls hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning Human Gene Deserts for Long-Range Enhancers. Sci. 2003;302(5644):413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: A basis for structural and regulatory chromosomal neighborhoods? BioEssays. 2013;35:818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, May G, Soneji S, Hong D, Enver T. MLLT3 Regulates Early Human Erythroid and Megakaryocytic Cell Fate. Cell Stem Cell. 2008;2:264–273. doi: 10.1016/j.stem.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Rainger JK, Bhatia S, Bengani H, Gautier P, Rainger J, Pearson M, Ansari M, Crow J, Mehendale F, Palinkasova B, et al. Disruption of SATB2 or its long-range cis-regulation by SOX9 causes a syndromic form of Pierre Robin sequence. Hum Mol Genet. 2014;23(10):2569–2579. doi: 10.1093/hmg/ddt647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin SV, Gavrilov AA, Pichugin A, Lipinski M, Iarovaia OV, Vassetzky YS. Transcription factories in the context of the nuclear and genome organization. Nucleic Acids Res. 2011;39:9085–9092. doi: 10.1093/nar/gkr683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis LM, Tyler RC, Volkmann Kloss BA, Schilter KF, Levin AV, Lowry RB, Zwijnenburg PJG, Stroh E, Broeckel U, Murray JC, et al. PITX2 and FOXC1 spectrum of mutations in ocular syndromes. Eur J Hum Genet. 2012;20(12):1224–1233. doi: 10.1038/ejhg.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Peña J, Sabbagh W, Greenwald J, Choe S, et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- Schaukowitch K, Joo J-Y, Liu X, Watts JK, Martinez C, Kim T-K. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. Two-step regulation of left-right asymmetric expression of Pitx2: Initiation by nodal signaling and maintenance by Nkx2. Mol Cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Yashiro K, Shen MM, Hamada H. Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development. 2006;133:3015–3025. doi: 10.1242/dev.02470. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Von Hase J, Stein S, Vincent S, Molloy KR, Kreth G, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, De Laat W. CTCF mediates long-range chromatin looping and local histone modification in the β??-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim J-E, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Tao Y, Zhang M, Li L, Bai Y, Zhou Y, Moon AM, Kaminski HJ, Martin JF. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet. 2014;7:23–32. doi: 10.1161/CIRCGENETICS.113.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Werken HJG, Landan G, Holwerda SJB, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Öz Y, Bouwman BAM, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser - A database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35 (Database issue):D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann BA, Zinkevich NS, Mustonen A, Schilter KF, Bosenko DV, Reis LM, Broeckel U, Link BA, Semina EV. Potential novel mechanism for Axenfeld-Rieger syndrome: Deletion of a distant region containing regulatory elements of PITX2. Investig Ophthalmol Vis Sci. 2011;52:1450–1459. doi: 10.1167/iovs.10-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite MR, Skidmore JM, Micucci JA, Shiratori H, Hamada H, Martin JF, Martin DM. Pleiotropic and isoform-specific functions for Pitx2 in superior colliculus and hypothalamic neuronal development. Mol Cell Neurosci. 2013;52:128–139. doi: 10.1016/j.mcn.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB, Tao Y, Chen Y, Wehrens XHT, Martin JF. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci U S A. 2014;111:9181–9186. doi: 10.1073/pnas.1405411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Schmidt HA, Kuhn A, Hoger SK, Kocagoz Y, Laumann-Lipp N, Ozbek S, Holstein TW. Nodal signalling determines biradial asymmetry in Hydra. Nature. 2014;515 (7525):112–115. doi: 10.1038/nature13666. [DOI] [PubMed] [Google Scholar]
- Welsh IC, Thomsen M, Gludish DW, Alfonso-Parra C, Bai Y, Martin JF, Kurpios Na. Integration of left-right Pitx2 transcription and Wnt signaling drives asymmetric gut morphogenesis via Daam2. Dev Cell. 2013;26:629–644. doi: 10.1016/j.devcel.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, Paquette D, Dostie J, Bickmore WA. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28:2778–2791. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A, Michos O, Spitz F, Haramis A-PG, Panman L, Galli A, Vintersten K, Klasen C, Mansfield W, Kuc S, et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 2004;18:1553–1564. doi: 10.1101/gad.299904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.