Abstract
Assessing the extent of phosphoinositol 3-kinase (PI3K) pathway activity in cancer is vital to predicting sensitivity to PI3K-targeting drugs but the best biomarker of PI3K pathway activity in archival tumor specimens is unclear. Here PI3K pathway activation was assessed, in clinical tissue from 1,021 men with prostate cancers, using multiple pathway nodes that include: PTEN, phosphorylated AKT (pAKT), phosphorylated ribosomal protein S6 (pS6), and stathmin. Based on these markers a 9-point score of PI3K activation was created using the combined intensity of the 4-markers and analyzed its association with proliferation (Ki67), apoptosis (TUNEL), and androgen receptor (AR) status, as well as pathologic features and cancer-specific outcomes. In addition, the PI3K activation score was compared to mRNA expression profiling data for a large subset of men. Interestingly, those tumors with higher PI3K activation scores also had higher Gleason grade (p=0.006), increased AR (r=0.37; p<0.001) and Ki67 (r=0.24; p<0.001), and decreased TUNEL (r= −0.12; p=0.003). While the PI3K activation score was not associated with an increased risk of lethal outcome, a significant interaction between lethal outcome, Gleason and high PI3K score (p=0.03) was observed. Finally, enrichment of PI3K-specific pathways was found in the mRNA expression patterns differentiating the low and high PI3K activation scores; thus, the 4-marker immunohistochemical score of PI3K pathway activity correlates with features of PI3K activation.
Implications
The relationship of this activation score to sensitivity to anti-PI3K agents remains to be tested but may provide more precision guidance when selecting patients for these therapies.
Keywords: phosphoinositide 3-kinase, prostate neoplasm, immunohistochemistry, PTEN, stathmin
Introduction
The phosphoinositol-3 kinase (PI3K) pathway is thought to be central in the development and progression of prostate cancer (1). PI3K pathway activation is associated with cellular proliferation, decreased apoptosis, decreased androgen receptor (AR) signaling, and disruption of DNA repair (2, 3). Agents targeting various points in the PI3K pathway are currently under development for the treatment of prostate cancer (4). The growing experience with targeted drugs in oncology such as erlotinib, suggest that not all men will respond to pathway-targeted agents highlighting the importance of identifying predictive markers. One possible approach to achieving this goal is to identify tumors with activation of the PI3K pathway.
Characterizing activation of the PI3K pathway in prostate cancer has been typically performed by assessing the status of individual key pathway nodes including the loss of the tumor suppressor phosphatase and tensin homolog (PTEN) (5), phosphorylation of AKT (6), and phosphorylation of the downstream marker ribosomal protein S6 (7). Previously used assays have included immunohistochemistry (7), analysis of the phosphoproteome (8), mRNA profiling (9), characterization of copy number aberrations (CNAs) (10) and identification of exome mutations (11). The sample sizes, assay quality and approaches used in prior studies have varied widely.
We hypothesized that given the heterogeneity in how the pathway might be activated in clinical samples, characterizing multiple nodes in the pathway simultaneously may provide a more global assessment of a tumor's PI3K activity than focusing on individual markers. Further, we hypothesized that protein expression would be a robust way of assessing the cellular state of PI3K activity. Using archival tissue samples from men with prostate cancer, we assessed PI3K activation using an IHC score containing four markers. Lacking a gold standard for pathway activity, we compared this score to tumor features, clinical outcomes and mRNA expression profiling.
Materials and Methods
Patients
We nested this study within three cohorts of men with localized prostate cancer: the Swedish Watchful Waiting Study (12), the US Physicians’ Health Study (PHS) (13), and Health Professionals Follow-up Study (HPFS) (14). Details of the cohorts have been described elsewhere. Briefly, the Swedish group (1977-1998) is comprised of men diagnosed with prostate cancer incidentally on transurethral resection of the prostate (TURP) or benign prostatic hypertrophy enucleation who were followed initially with watchful waiting. The men have been followed for mortality through linkage with the Swedish Death Register through March 2008 and the included group represents a case-control set. We also included two U.S. cohorts: the PHS randomized trials of aspirin and supplements in the primary prevention of cardiovascular disease and cancer and the HPFS cohort study of 51,000 US male health professionals. From these two U.S. cohorts, tumor samples from participants diagnosed with prostate cancer between 1983 and 2004 were collected from the treating institution and medical records were abstracted for clinical information and outcomes. From HPFS and PHS, 5% and 9% of the samples respectively were from TURPs, with the remainder from prostatectomy. All men from PHS and HPFS were followed for the development of lethal disease, defined by distant metastases or prostate cancer-specific death through May 2011. A physician committee confirmed causes of death through medical record and death certificate review. This study was approved by the Institutional Review Boards of Harvard School of Public Health and Partners Healthcare.
Marker Selection
We reviewed previously published immunohistochemical markers of PI3K pathway activity to select targets for inclusion with an emphasis on validated antibodies and significant prior literature supporting the marker's inclusion. PTEN function, altered through mutation (11), copy number aberration (10) or post translational regulation, leads to accumulation of the second messenger PIP3 and subsequent recruitment of AKT for activation. In mice with conditional pten loss in the prostate, premalignant lesions can develop, a process heightened to invasive and even metastatic disease with the addition of other molecular changes (1). Loss of tumor PTEN staining has been observed with increasing frequency in higher-grade and stage prostate cancers (5, 15). At the cell membrane, AKT is phosphorylated by either PDK1 (Thr308) or mTOR kinase complex 2 (Ser473) and phospho-antibodies directed at phospho-AKT (pAKT) show reversal of activation with inhibition of the PI3K pathway (16). Further, prostate tumor expression of pAKT has been associated with higher grade disease (6) and worsened prognosis (17). Downstream of AKT, mTOR activates S6 kinase leading to phosphorlylation of ribosomal protein S6 (pS6). Both in cell lines (16) and patient specimens (4), pS6 is lost with PI3K inhibition. In addition, loss of mTOR or akt1 in mice with conditional pten loss reduces the initiation of prostate cancer (18). Stathmin, a microtubule regulating phosphoprotein, has been associated with PTEN loss in vitro (19) as well as in human prostate (20, 21) and breast cancer (9). Stathmin expression is regulated by PI3K inhibitors (8, 9) and in breast cancer, was identified as a robust marker of PTEN loss (9).
Immunohistochemistry
Hematoxylin and eosin slides were reviewed by study pathologists (MF, SF, GF, ML) to identify tumor tissue and systematically assess Gleason grade. Using tumor tissue microarrays (TMAs) constructed from triplicate 0.6 mm cores for each case, we performed immunohistochemical staining to assess tumor expression of cytoplasmic PTEN (Zymed cat. #18-0256; 1:200), cytoplasmic pAKT (Ser473; Cell Signaling cat. #4060; 1:50), cytoplasmic pS6 (Ser240/Ser244; Cell Signaling cat. #2215; 1:50), cytoplasmic stathmin (Cell Signaling cat. #3352; 1:50), and Ki67 (polyclonal anti Ki67 antibody; Vector Labs; 1:2,000) and AR (Upstate [Millipore] cat. #06-680; 1:100). For each marker, a 4-μm section of the TMA was mounted on a glass slide, deparaffinized and microwaved for antigen retrieval in citrate-based buffer. Diaminobezandine (DAB) was used to visualize the immunohistochemistry, with hematoxylin as a counterstain. To estimate the percentage of cells undergoing apoptosis, we used the terminal deoxynucleotide transferase dUTP nick end label (TUNEL) assay with the Apoptag Peroxidase In situ kit (Chemicon International). Antibodies were routinely validated by Western blot and have been published previously.
Immunohistochemistry Interpretation
All interpretation of immunohistochemistry was performed blinded to outcomes. Tissue microarray slides were analyzed using the Ariol instrument SL-50 (Applied Imaging, San Jose, CA), a semi-automated image analysis software system. Each core was reviewed by a study pathologist to ensure matching to the TMA map and to manually circle tumor and exclude normal prostatic glands. After appropriate thresholding for each TMA, image analysis was performed using the Ariol MultiStain Assay to generate the following variables: percentage of nuclei positive (Ki67) and average percentage of cytoplasm staining per cell, a measure which approximates the percentage of cells positive for cytoplasmic staining. For pAKT, pS6 and stathmin, a continuous value was obtained after averaging across the replicate cores. For AR, the Ariol and ChromaVision (ChromaVision Medical Systems, Inc., San Juan Capistrano, CA) systems were used. For PTEN, the Nuance system (CRi, Woburn, MA) was used. Histological images were analyzed using a cytoplasmic algorithm, wherein multi- spectral imaging allows the software to segment the nuclei using the unmixed spectra of the nuclear counterstain (hematoxylin) and then assessing the DAB immunohistochemical stain in the cytoplasm (outside of the nucleus). A final score, based on the average percentage of the cytoplasmic tumor area that was positively stained, was determined from each set of three subject cores. This method has been shown to correlate highly with semiquantitative pathologist assessments and there is good correlation between the Ariol and Nuance platforms (22). For the TUNEL assay, every tumor core was evaluated manually to determine the number of positive cells among the total number of tumor cells with two study pathologists independently assessing these samples.
mRNA expression profiling
mRNA expression profiling data was available for 233 of the men from the Swedish cohort and 76 men from the PHS cohort for whom we also had protein expression of the 4 markers. The RNA was extracted from archival specimens from the same tumor-enriched nodule as the tissue microarray cores or a nodule of the same Gleason grade. The techniques for RNA extraction and expression analysis using a custom 6,100 cancer-related gene panel have been previously reported (12).
Statistical Methods
Immunohistochemical assessment
We used quantile normalization to efficiently adjust for potential batch effects of protein expression staining across each of the tissue microarrays using the limma package in R (23). Spearman correlations between markers were calculated. The continuous score for PTEN, pAKT, pS6 and stathmin were divided into tertiles and assigned a value of −1, 0, or 1 for low, intermediate and high staining respectively. As loss of PTEN is associated with pathway activation, the high staining and low staining values were assigned −1 and 1, respectively. Next, we generated a composite score of PI3K activation by summing these tertile values for each case resulting in a 9-point scale ranging from −4 to 4. We performed univariate and multivariate analyses correlating the immunohistochemical PI3K score to baseline characteristics. We examined the association between the PI3K score with lethal prostate cancer outcome during follow up using logistic regression. In light of the previously identified association between PI3K activity and Gleason score (5), we investigated whether the association between PI3K and lethal outcomes differed as a function of Gleason grade. We further correlated the score with apoptosis measured by TUNEL and proliferation measured by Ki67. Given identified associations between AR and PTEN signaling (3), we compared AR staining to the score.
mRNA Expression Profiling
To examine the relationship between the immunohistochemical score and mRNA signatures of pathway activation, we combined the lowest three and highest three categories of scores to denote low and high PI3K activity respectively. Gene sets significantly differentially expressed between these extreme cases were assessed using mean-rank gene set enrichment (24). In this analysis, we utilized all KEGG sets (n=186) along with 13 extracted sets of genes from the literature associated with PI3K pathway activation (8, 9, 19, 25-34) and cell cycle progression (35). To identify a set of genes differentially expressed between the cases in the extreme, we used the significance analysis of microarrays (SAM) approach (36). We applied the published signature of PTEN loss in breast cancer (9) to the extremes samples from our immunohistochemical score to determine an association of our score to existing mRNA signatures from the literature. To test the relationship of flux through the AR pathway to PI3K pathway activity (3), we investigated expression of a 17-gene AR signature (37) relative to our score. Our a priori hypothesis was that the biology of PI3K activation would be independent of sample source and we therefore combined the three cohorts in our primary analysis.
In an exploratory analysis, we used a published mRNA signature of PTEN loss in breast cancer (9) as a gold standard to determine which marker or combination of markers was most closely correlated with PI3K activity defined in this way. We compared all first order combinations of markers to find the best fitting model based on Akaike information criterion.
Results
Cohort characteristics
The clinical and pathologic characteristics of the 1,021 prostate cancer patients for whom immunohistochemical staining data were available are shown in Table 1. The cohort had a median age of 68.2 years (interquartile range [IQR] 63.9-73.4) and more than half of cases were Gleason 7. Over a median follow up of 144 months (IQR 94 – 181), 204 (20%) of men had developed distant metastatic disease or had died of prostate cancer. We did not observe major differences between the U.S. and Swedish cohorts in terms of their baseline clinical and pathologic characteristics.
Table 1.
Baseline clinical characteristics and outcomes for the three prostate cancer cohorts
| Entire Cohort | mRNA Cohort | ||||||
|---|---|---|---|---|---|---|---|
| HPFS (n=375) | PHS (n=288) | Swedish (n=358) | Combined (n=1021) | PHS (n=76) | Swedish (n=233) | Combined (n=309) | |
| Year of Diagnosis - median (IQR) | 1995 (1992 - 1997) | 1997 (1993 - 2000) | 1992 (1989 - 1994) | 1994 (1991 - 1997) | 1993 (1991 - 1995) | 1991 (1989 - 1994) | 1992 (1989 - 1994) |
| Age at diagnosis - median (IQR), yrs | 66.0 (62.0 - 70.0) | 66.0 (62.2 - 69.8) | 73.8 (69.1 - 79.1) | 68.2 (63.9 - 73.4) | 65.5 (61.6 - 69.1) | 73.9 (69.6 - 79.4) | 72.6 (66.6 - 77.7) |
| Gleason - n (%)* | |||||||
| ≤6 | 58 (15) | 76 (26) | 105 (29) | 239 (23) | 8 (11) | 64 (27) | 72 (23) |
| 7 | 239 (64) | 165 (57) | 165 (46) | 569 (56) | 49 (64) | 106 (45) | 155 (50) |
| 8-10 | 77 (21) | 47 (16) | 83 (23) | 207 (20) | 19 (25) | 63 (27) | 82 (27) |
| PSA at diagnosis# - median (IQR), ng/mL | 7.6 (5.2 - 12.1) | 6.5 (4.7 - 9.3) | NA | 7.0 (5.0 - 11.0) | 8.2 (5.4 – 13.1) | NA | 8.2 (5.4 - 13.1) |
| Stage - n (%)¶ | |||||||
| pT2 | 244 (65) | 179 (62) | -- | 423 (41) | 38 (50) | -- | 38 (12) |
| pT3 | 97 (26) | 59 (20) | -- | 156 (15) | 21 (28) | -- | 21 (7) |
| pT4 or N1 or M1 | 11 (3) | 10 (3) | -- | 21 (2) | 5 (7) | -- | 5 (2) |
| T1a | -- | -- | 92 (26) | 92 (9) | -- | 63 (27) | 63 (20) |
| T1b | -- | -- | 264 (74) | 264 (26) | -- | 170 (73) | 170 (56) |
| Follow up - median (IQR), mo | 172 (143 - 200) | 137 (108 - 182) | 101 (53 - 150) | 144 (94 - 181) | 190 (127 - 220) | 94 (45 - 135) | 114 (64 - 165) |
| Lethal§ - n (%) | 41 (12) | 23 (9) | 142 (47) | 206 (22) | 17 (22) | 126 (54) | 143 (46) |
| IHC Scoreɜ - n (%) | |||||||
| Low (−4, −3, −2) | 83 (22) | 65 (23) | 78 (21) | 226 (22) | 15 (20) | 48 (21) | 63 (20) |
| Intermediate (−1, 0, 1) | 214 (57) | 155 (54) | 210 (59) | 579 (57) | 48 (63) | 141 (61) | 189 (61) |
| High (2, 3, 4) | 78 (21) | 68 (24) | 70 (20) | 216 (21) | 13 (17) | 44 (19) | 57 (18) |
Gleason missing on 1 from HPFS and 5 from the Swedish cohort
PSA missing from 64 in HPFS and 40 in PHS
Stage missing in 23 from HPFS, 40 from PHS, and 2 from the Swedish cohort
IHC Score is the combined 4 marker IHC signature of PI3K activation
Lethal outcome missing in 118 missing due to insufficient follow-up (minimum of 8 years without a lethal or metastatic event)
Immunohistochemistry Score
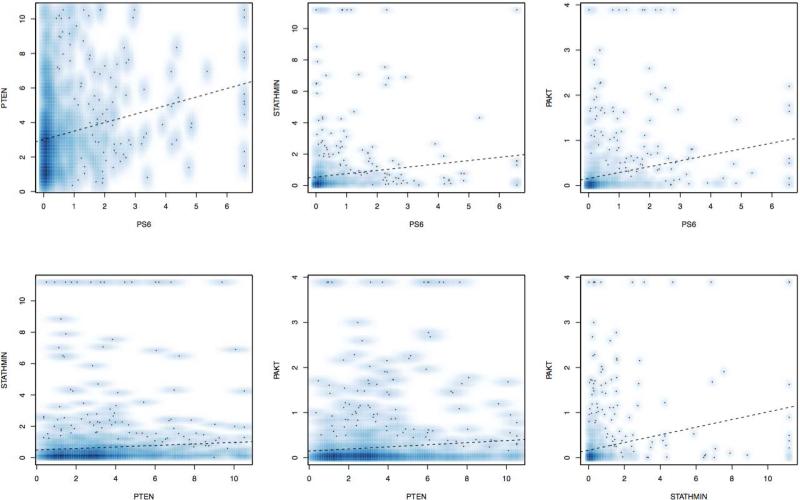
Positive correlations in the staining between individual markers in the pathway (Figure 1) were statistically significant but generally weak. The strongest correlations were between pAKT and stathmin (r=0.24; 95% CI: 0.18 – 0.30; p<0.001) and pAKT and pS6 (r=0.23; 95% CI: 0.17 – 0.29; p<0.001). A negative correlation between PTEN staining and other markers in the pathway was not observed.
Figure 1.
Correlation plots of the normalized immunohistochemical staining plots for each marker along with Spearman correlation coefficient.
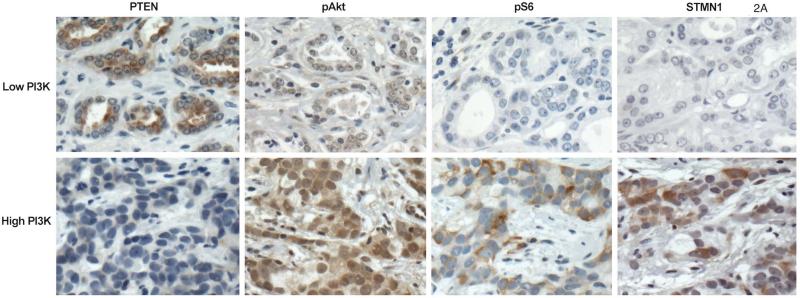
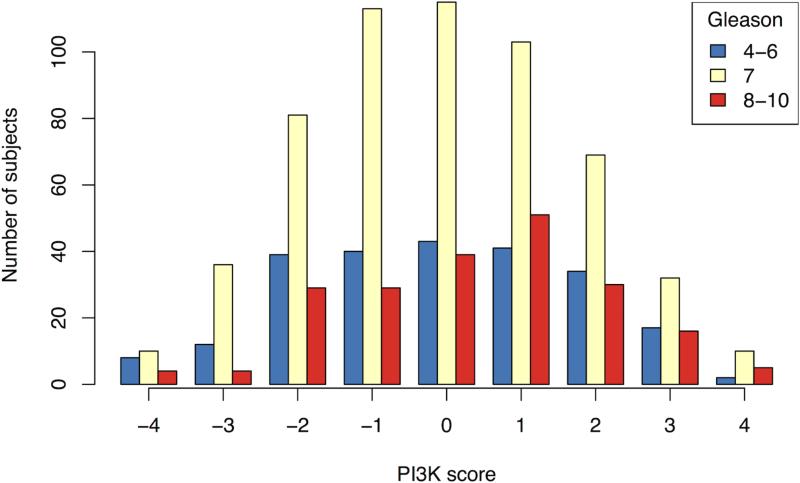
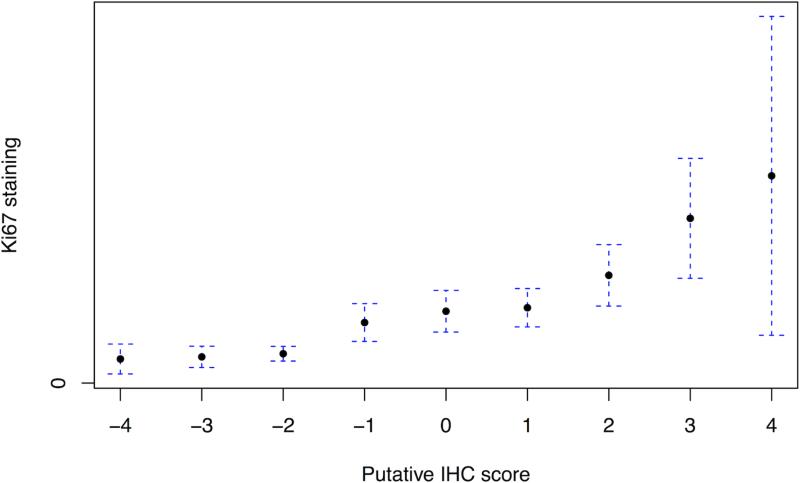
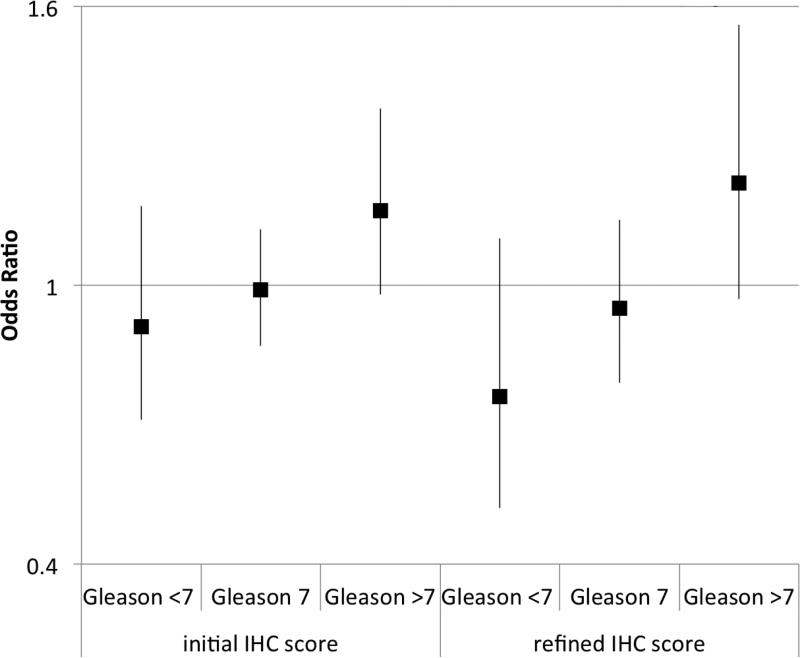
We created a 9-point signature of PI3K pathway activation using the combined staining of PTEN, pAKT, pS6 and stathmin (Figure 2A). Of the entire cohort, 2.3% and 1.7% fell into the lowest and highest categories respectively, highlighting the rare concordance of all four markers defining pathway inactivation or activation. On univariate analysis, the score was not significantly associated with age, body mass index or PSA at diagnosis, but was positively associated with increasing Gleason score (p=0.006; Figure 2B). The median IHC score for Gleason <7 was −0.10 (95%: .34 to .13), for Gleason 7 it was −0.08 (95% −.23 to .06), and for >7 it was 0.31 (95% .07 to .55). Increasing immunohistochemical score was associated with decreased apoptosis, Pearson correlation −0.12 (n=578; 95% CI: −0.20 – −0.04; p=0.003) and increased cellular proliferation as measured by Ki67 staining, Pearson correlation +0.24 (n=964; 95% CI: 0.18 – 0.30; p<0.001; Figure 2C). On univariate analysis, a one-point increase in the score had an odds ratio (OR) of 1.07 for lethality (95% CI: 0.98 – 1.17; p=0.14). We found a significant interaction between Gleason, the score and lethality (p=0.03). Compared to men with lower Gleason scores, the immunohistochemical score was more associated with lethal outcome for cases with Gleason scores of ≥8 (Figure 2D).
Figure 2.
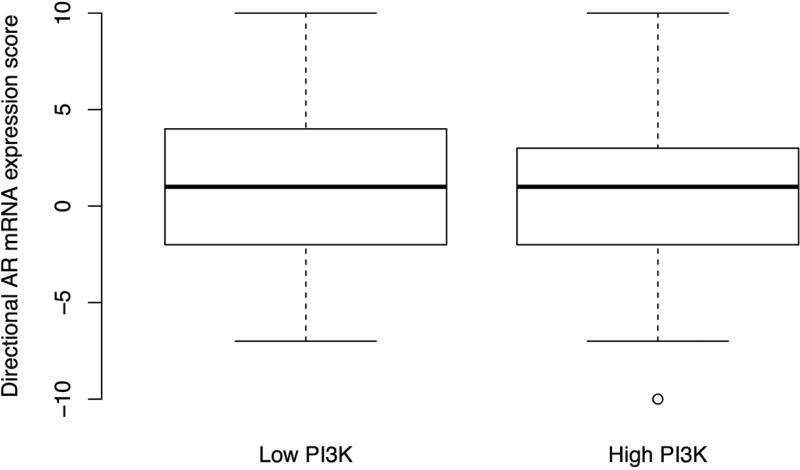
The immunohistochemical score of PI3K pathway activation is associated tumor and clinical features suggestive of more aggressive disease; (A) immunohistochemical staining for PTEN, pAKT, pS6, and stathmin in patterns reflecting high and low pathway activity; (B) distribution of immunohistochemical scores by Gleason score; (C) correlation between the immunohistochemical score and Ki67 staining; (D) ORs for lethal outcome for a one-point increase in the initial 4-marker and refined 2-marker score by Gleason showing a significant interaction; and (E) box plots for an mRNA signature of PI3K activation from breast cancer showing a statistically lower score in the low (−4 to −2) immunohistochemical score cases relative those with a high (2 to 4) score.
In the 309 cases for which we had both GEP and the 4-marker score, we assessed genes, and subsequently gene sets, differentially expressed between cases with low scores (−4 to −2; 20%) and high scores (2 to 4; 18%). Using SAM analysis, the genes most differentially expressed between high and low immunohistochemistry scores were PMAIP1, FANCC, RLF, RREB1, NUSAP1, TK1, DPT and WFS1 (Table 2). Only the cell cycle progression gene set (35) significantly differentiated the two groups (p<0.001) but to illustrate the remaining top 10, we have included them in Table 2. The second and third most differential sets were the KEGG mismatch repair and the set identified in PTEN deficient breast samples (9) (Table 2). Among the top 25 differential genes sets, there were three PI3K specific (8, 9, 25) and three KEGG DNA-repair sets: mismatch repair, base-excision repair and homologous recombination. Using the directional expression of 98 available genes from a signature of PTEN loss in breast cancer (9) we found the signature to be significantly associated with the low and high immunohistochemistry scores (p=0.006; Figure 2E). Looking for correlation between individual mRNA and immunohistochemical expression, we found significant correlation for STMN1 (r=0.24; 95% CI: 0.13 – 0.34; p<0.01) but not for PTEN (p=0.5), AKT1 (p=0.2), or RPS6 (p=0.3).
Table 2.
Genes significantly differentially expressed between the low and high PI3K tumors as defined by the 4-marker immunohistochemical score and the top 10 gene sets differentially expressed between the two groups.
| Genes | |||
|---|---|---|---|
| gene | full name | Pathways | |
| up | PMAIP1 | phorbol-12-myristate-13-acetate-induced protein 1 | P53, DNA damage response, apoptosis |
| FANCC | Fanconi anemia, complementation group C | DNA repair, | |
| RLF | rearranged L-myc fusion | transcription regulation | |
| RREB1 | ras responsive element binding protein 1 | Ras signaling; transcription regulation | |
| NUSAP1 | nucleolar and spindle associated protein 1 | cell cycle regulation | |
| TK1 | thymidine kinase 1, soluble | pyrmidine metabolsm | |
| down | DPT | dermatopontin | cell proliferation, cell adhesion |
| WFS1 | Wolfram syndrome 1 | protein processing | |
| Gene sets | |
|---|---|
| Set | adjusted p |
| Cell cycle progression (35) | <0.001 |
| KEGG mismatch repair | 0.15 |
| Saal (9) | 0.15 |
| KEGG oocyte meiosis | 0.34 |
| KEGG progesterone mediate oocyte maturation | 0.34 |
| KEGG regulation of actin cytoskeleton | 0.34 |
| KEGG prostate cancer | 0.34 |
| KEGG dilated cardiomyopathy | 0.48 |
| KEGG hypertrophic cardiomyopathy | 0.49 |
| KEGG sphingolipid metabolism | 0.56 |
Androgen Receptor
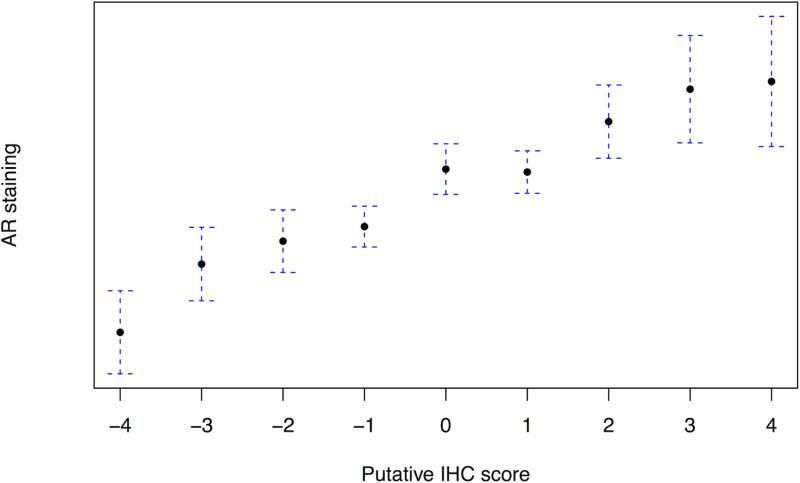
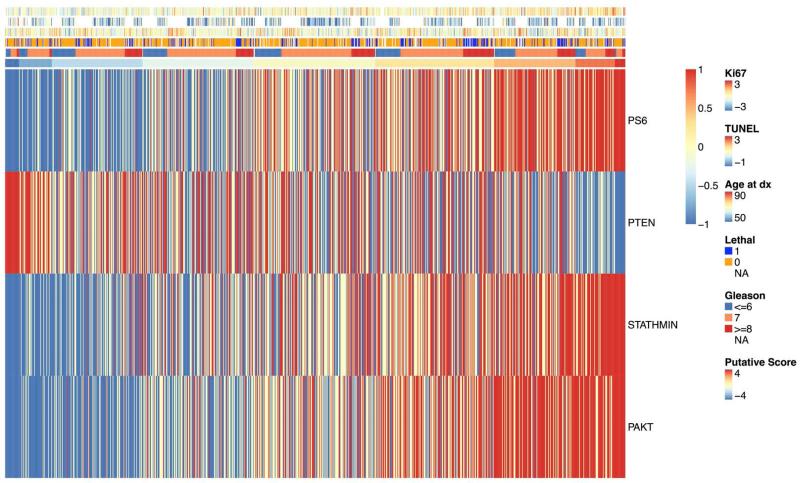
In light of the previously reported reciprocal relationship between PI3K pathway activation and flux through the AR pathway (3), we assessed how the 4-marker immunohistochemical score correlated to AR staining (n=657) and expression of 17 AR-targeted genes (37). As shown in Figure 3A, the 4-marker immunohistochemical score was positively correlated with tumor AR staining, r=0.37 (95% CI: 0.30 – 0.43; p<0.001). Among the 309 cases for which we had mRNA expression, we observed no association between expression of the AR responsive gene signature and the immunohistochemical score (Figure 3B). AR expression was not associated with expression of the 17-gene signature of AR signaling. AR expression relative to the tertile of immunohistochemical scores for all four markers along with clinical and pathologic features is shown in Figure 3C. The heat map, ordered by 4-marker score, illustrates the overall pattern of immunohistochemistry staining for the four markers relative to proliferation (Ki67), apoptosis (TUNEL), age at diagnosis, lethality, and Gleason score.
Figure 3.
The 4-marker immunohistochemical score for PI3K activation is positively correlated with AR staining; (A) 4-marker immunohistochemical score of PI3K activation relative to AR staining; (B) 17-gene mRNA signature of AR flux score in low (−4 to −2) and high (2 to 4) immunohistochemical score cases showing no significant differences; and (C) heatmap of tertile of immunohistochemical staining four all four marker with corresponding annotation of (top to bottom) Ki67, TUNEL, age at diagnosis, lethal outcome, Gleason score, and the 4-marker score.
Immunohistochemical Score Refinement
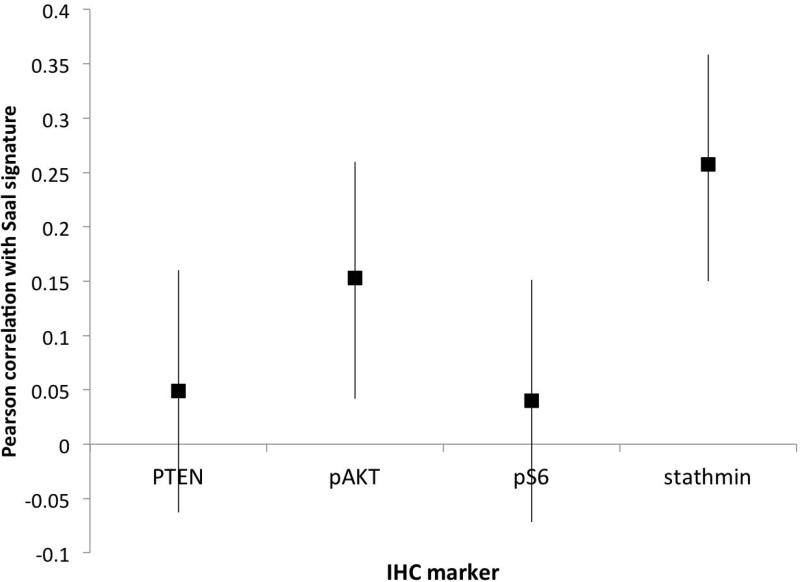
We next sought to determine the relative importance of PTEN, pAKT, pS6 and stathmin in capturing activation of the PI3K pathway. Given its correlation with our 4-marker score, we assessed which marker, or set of markers, were most correlated with the signature from Saal et al shown to be associated with PTEN loss in breast cancer (9). Individually, both stathmin (r=0.26; 95% CI: 0.15 – 0.36; p<0.001) and pAKT (r=0.15; 96% CI: 0.04 – 0.26; p=0.007) were significantly correlated with the signature (Figure 4A). Comparing all possible first-order models of fit, the model containing pS6 and stathmin was identified as most strongly correlating with the mRNA signature of PI3K activation. Using the same tertiles of staining of stathmin and pS6 used for the 4-marker score, we created a “refined” 5-point scale of PI3K activation with only these two markers.
Figure 4.
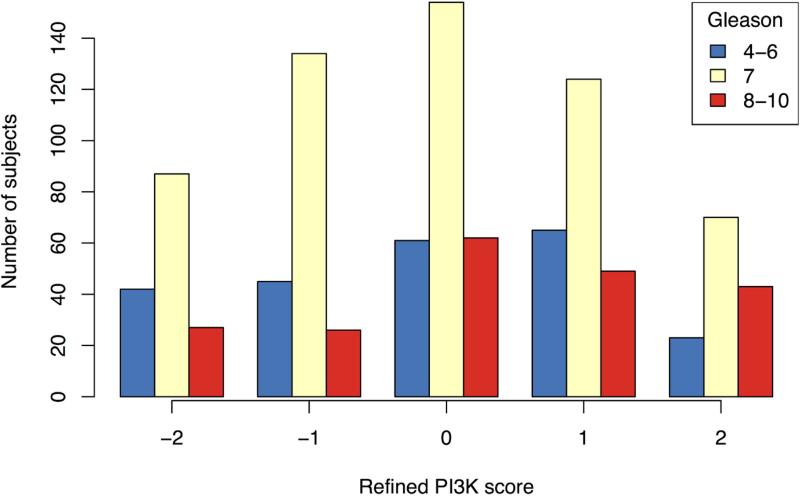
Identification of a refined immunohistochemical score built against an mRNA signature of PI3K activation (Saal et al, 2007); (A) correlation between PTEN, pAKT, pS6 and stathmin and the mRNA signature of PI3K individually; and (B) distribution of 2-marker refined score using pS6 and stathmin by Gleason score.
As with the 4-marker score, the refined 2-marker score was positively correlated with Ki67 staining (r=0.23; 95% CI: 0.17 – 0.29; p<0.001) but was not significantly associated with apoptosis (r=0.02; 95% CI: −0.06 – 0.10; p=0.6). Also similar to the 4-markers score, there was no significant association with age, PSA level, BMI, or the development of lethal disease. The 2-marker score was positively correlated with Gleason score (p=0.002; Figure 4B) and on univariate analysis, a one-point increase in the refined score had an OR of 1.08 for lethality (95% CI: 0.96 - 1.23; p=0.20). Similar to the relationship observed with the 4-marker score, the refined score and Gleason had a significant interaction with lethality (p=0.008; Figure 2D).
Discussion
Using immunohistochemical assessment of PTEN, pAKT, pS6 and stathmin, we developed a signature of PI3K pathway activation in prostate cancer. Lacking a gold standard marker in human tissue to compare to, we used mRNA expression, cellular proliferation, apoptosis, and correlation with clinical factors to support the interpretation that the combination of these four markers is capturing PI3K activity.
Several lines of evidence suggest that our immunohistochemistry score is associated with PI3K activity. First, an increase in the immunohistochemistry score was associated with tumor features previously suggested to be associated with PI3K pathway activation. We found that increased PI3K activation was associated with increasing Gleason score, a finding similar to prior studies in prostate cancer (5, 15, 16, 38, 39). Further, we found increased Ki67 staining among tumors with higher PI3K immunohistochemistry scores, an anticipated biologic finding and one similar to prior studies. Increased PI3K activation was also significantly associated with decreased apoptosis, a finding in line with the known biology of the pathway (8).
Second, we found that genes differentially expressed between cases with high and low immunohistochemical scores were associated with cellular proliferation and DNA repair (Table 2). The PI3K pathway has been associated with DNA repair (40) including through mechanisms related to cell cycle regulation (2). Additionally, pathway activation is correlated with cellular proliferation in vitro and in model systems as well as in clinical samples (41). Similarly to the individual genes, the gene sets identified as differentiating those tumors with high and low immunohistochemistry scores were enriched for processes associated with PI3K pathway activation. The most differentially expressed was the gene set associated previously with cell cycle progression (35) though sets associated with PI3K activation, prostate cancer and DNA repair were also highly represented. These findings are similar to other studies investigating genes differentially expressed between cells with and without PI3K pathway activation (19, 28). Finally, the immunohistochemical score was directly correlated with an established mRNA signature of PTEN deficient breast cancers (9).
The ability of PI3K pathway activation to predict clinical outcome after prostatectomy has depended on which marker was investigated and what outcome was studied; generally higher activation has been associated with a higher likelihood of disease recurrence following surgery (9, 15, 17, 38, 39, 42, 43). In our data, the immunohistochemical signature of PI3K activation was not significantly associated with the development of clinically relevant lethal disease on univariate analysis though there was evidence of a significant interaction with Gleason with the trend towards the high PI3K activity being more associated with lethality in high Gleason grades. The implications of this are unclear though this finding may support the rationale for the use of PI3K inhibitors in higher-grade tumors. Further research will be required to determine whether high relative to low Gleason grade tumors rely on different pathways to drive prostate cancer mortality. Given the heterogeneity of the three populations and non-random manner in which cases were included in the cohorts, caution is warranted in interpreting the outcomes related to lethality in this study. Ultimately, our goal in this study was not to develop a new prognostic signature in prostate cancer but rather to move towards signatures which may be predictive of response to PI3K-targeted agents.
It remains unclear which immunohistochemical markers and which combination are most important for capturing PI3K activity, and therefore presumed inhibitor sensitivity, in archival prostate cancer samples. To explore this question, we used the mRNA signature of PI3K activity derived from breast samples (9) as a gold standard and investigated which immunohistochemical markers were most correlated to the extremes identified using that approach. We found that the combination of pS6 and stathmin staining best correlated to this signature. The inclusion of stathmin is unsurprising insofar as this marker was found as the immunohistochemical target best correlated with the signature in breast cancer (9). The absence of PTEN and the inclusion of pS6 in this correlation is notable given that the signature was initially derived from clinical samples which lacked PTEN immunohistochemical staining. One explanation for this finding is the robustness of the antibodies for stathmin and pS6 in these archival samples relative to the other two markers. It is known for example that there are significant challenges with phosphoantibodies in the setting of differential fixation approaches (44), and pS6 appears to be more resistant to this effect compared to other markers (45). Additionally, there is reported variability in the quality of PTEN antibodies (46). A more recent PTEN antibody has been shown to have excellent performance in cell lines and clinical samples (15, 46). We have reviewed this newer antibody on a limited subset of these cases and found a staining to be highly concordant with our current PTEN antibody so it is not clear that its use would change our results. In our data, there was a no significant correlation between PTEN mRNA and immunohistochemical expression. In both the 4-marker and refined 2-marker scores, each constituent marker was given equal weighting. In our analysis this was necessary though arguably a true signature of pathway activation, or inhibitor sensitivity, would likely weigh markers quite differently based on their individual contribution relative to a clinical outcome. With only a subset of the tumors having mRNA available, this aspect must be interpreted with appropriate caution.
In mice conditionally null for pten, observed low prostate AR protein expression was reversed following PI3K directed therapy (3). Additionally, this study found that the mRNA signature of AR activity (37) was elevated following PI3K pathway inhibition in the model system. Among 106 prostatectomy samples, the AR signature was repressed in cases with PTEN loss defined by either copy number change or transcript loss. Based on these data, our hypothesis was that high PI3K pathway activation would have been associated with both low AR staining and low AR pathway activity measured by mRNA expression. Instead, we found a strong positive correlation between our 4-marker PI3K signature and AR protein expression. This observation is in line with results from a small cohort of men in which pAKT staining was associated with increased AR staining in prostatectomy samples (47). Additionally, in LNCaP cells, inhibition of the PI3K pathway leads to decreased AR expression (48). Finally, in a cohort of more than 600 prostatectomy samples, AR was correlated with Ki67 staining (49) similar to our results. Our data from 309 prostate cancer samples demonstrated no relationship between the AR mRNA signature and protein staining or the immunohistochemical signature of PI3K activation. The explanation for this discordance is unclear, though may be related to our definition of PI3K activity or tumor heterogeneity.
Across 1,021 samples, we found only 1.7% showed a completely concordant picture of low staining for PTEN and high staining for the remaining markers and 2.3% showing the reversed picture. Depending on the marker used, there have been wide ranging estimates of PI3K dysfunction in prostate cancer. It seems likely that the very extremes of our 4-marker score are truly capturing some aspect or aspects of PI3K activation but it is unclear what threshold should be used to call that pathway “active” or “inactive” when using a combination of immunohistochemical markers which interrogate different portions of the pathway activated in potentially very different ways. Highlighting the possibility that our signature is identifying various pathways active in heterogeneous clinical samples (e.g. PI3K and mTOR) is the lack of expected strong correlation between staining of individual markers. Previous studies have also failed to find correlation between anticipated markers including pS6, pAKT and PTEN (38, 39, 50) suggesting the potential for complex interplay between these important pathways in clinical samples. Further, we were using a combination of samples from TURP and prostatectomy specimens potentially introducing additional heterogeneity.
Lacking data from clinical trials using PI3K-targeted agents, it is unknown whether we selected the ideal panel of immunohistochemical markers to predict treatment response to targeted agents. We showed that pS6 and stathmin were best correlated with an mRNA signature of PTEN loss in breast cancer but comparisons to other published signatures could have also been made (19, 30). While this was a large study using clinically meaningful outcomes, ours is not the first study to use multiple immunohistochemical markers to identify PI3K pathway activation and others have shown that combining markers in the pathway is more strongly associated with outcome than a single marker (43). Previous work has highlighted the heterogeneity of PTEN staining for example within a given prostate cancer (5) and one challenge is that we do not have a way to readily analyze this variability. What this heterogeneity means biologically or clinically and how to measure and record it across many samples is not known.
We hypothesized that we could simply and reliably determine PI3K pathway activation in prostate tumors using the combined staining of PTEN, pAKT, pS6 and stathmin. In over 1,000 archival samples, we found that the extremes of this 4-marker score were associated with both clinical and genetic features suggestive of PI3K activation. For those men with higher Gleason scores, an increase in the PI3K pathway activity defined in this way was associated with an increased risk of developing lethal disease. We sought to identify which the most important markers out of the four were and found that a combination of stathmin and pS6 was most correlated with a prior mRNA signature of pathway activity in breast cancer. This work raises several questions including what threshold should be used to characterize a tumor as having the PI3K pathway active if assessed in this way; what if any clinical implications there are for the identified interaction between Gleason, the immunohistochemical score and lethal outcome; whether other makers would be better at identifying pathway activation; and ultimately whether pathway activity and this signature in particular has any predictive value in men being treated with PI3K inhibitors. To date, clinically used predictive markers have included specific mutations, translocations, mRNA signatures, and immunohistochemistry and further work in combination with clinical trial results will be needed to determine the utility of the markers we propose here.
In conclusion, a combination of PTEN, pAKT, pS6, and stathmin appears to capture PI3K pathway activity in prostate tumors as assessed by molecular and clinical features. Of the four, our data suggest that the combination of pS6 and stathmin may be equally good at determining activation.
Acknowledgements
We are grateful to the participants in the Physicians’ health Study, Health Professionals Follow-up Study, and Swedish Watchful Waiting Cohorts for their ongoing participation in the cohort. We would like to thank Greg Judson, Elizabeth Frost-Hawes, and Lauren McLaughlin for their research roles in follow-up of the cohort. We would like to thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. The authors assume full responsibility for analyses and interpretation of these data.
Financial Support: Support was provided by grants P50CA090381-08 from the Dana-Farber/Harvard Cancer Center Specialized Programs of Research Excellence (SPORE) in Prostate Cancer, UM1CA167552, R01CA141298, R01CA136578, R01CA131945, and P01CA89021 from the National Cancer Institute of the National Institutes of Health, and the Prostate Cancer Foundation Young Investigator awards (NEM, SF, and LAM).
Footnotes
Conflicts of interest: None of the authors have any conflicts of interest.
References
- 1.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal Feedback Regulation of PI3K and Androgen Receptor Signaling in PTEN-Deficient Prostate Cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, et al. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30:282–90. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 5.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–6. [PubMed] [Google Scholar]
- 6.Malik SN, Brattain M, Ghosh PM, Troyer DA, Prihoda T, Bedolla R, et al. Immunohistochemical demonstration of phospho-Akt in high Gleason grade prostate cancer. Clin Cancer Res. 2002;8:1168–71. [PubMed] [Google Scholar]
- 7.Thomas GV, Horvath S, Smith BL, Crosby K, Lebel LA, Schrage M, et al. Antibody-based profiling of the phosphoinositide 3-kinase pathway in clinical prostate cancer. Clin Cancer Res. 2004;10:8351–6. doi: 10.1158/1078-0432.CCR-04-0130. [DOI] [PubMed] [Google Scholar]
- 8.Andersen JN, Sathyanarayanan S, Di Bacco A, Chi A, Zhang T, Chen AH, et al. Pathway-based identification of biomarkers for targeted therapeutics: personalized oncology with PI3K pathway inhibitors. Sci Transl Med. 2010;2:43ra55. doi: 10.1126/scitranslmed.3001065. [DOI] [PubMed] [Google Scholar]
- 9.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She Q-B, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur SR, Hoshida Y, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penney KL, Sinnott JA, Fall K, Pawitan Y, Hoshida Y, Kraft P, et al. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol. 2011;29:2391–6. doi: 10.1200/JCO.2010.32.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B, et al. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–8. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- 15.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, et al. PTEN Protein Loss by Immunostaining: Analytic Validation and Prognostic Indicator for a High Risk Surgical Cohort of Prostate Cancer Patients. Clin Cancer Res. 2011;17:6563–73. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–30. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 17.Ayala G, Thompson T, Yang G, Frolov A, Li R, Scardino P, et al. High levels of phosphorylated form of Akt-1 in prostate cancer and non-neoplastic prostate tissues are strong predictors of biochemical recurrence. Clin Cancer Res. 2004;10:6572–8. doi: 10.1158/1078-0432.CCR-04-0477. [DOI] [PubMed] [Google Scholar]
- 18.Nardella C, Carracedo A, Alimonti A, Hobbs RM, Clohessy JG, Chen Z, et al. Differential requirement of mTOR in postmitotic tissues and tumorigenesis. Sci Signal. 2009;2:ra2. doi: 10.1126/scisignal.2000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stolarov J, Chang K, Reiner A, Rodgers L, Hannon GJ, Wigler MH, et al. Design of a retroviral-mediated ecdysone-inducible system and its application to the expression profiling of the PTEN tumor suppressor. Proc Natl Acad Sci U S A. 2001;98:13043–8. doi: 10.1073/pnas.221450598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Cima I, Schiess R, Wild P, Kaelin M, Schuffler P, Lange V, et al. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc Natl Acad Sci U S A. 2011;108:3342–7. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiore C, Bailey D, Conlon N, Wu X, Martin N, Fiorentino M, et al. Utility of multispectral imaging in automated quantitative scoring of immunohistochemistry. J Clin Pathol. 2012;65:496–502. doi: 10.1136/jclinpath-2012-200734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. [Google Scholar]
- 24.Michaud J, Simpson KM, Escher R, Buchet-Poyau K, Beissbarth T, Carmichael C, et al. Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genomics. 2008;9:363. doi: 10.1186/1471-2164-9-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci USA. 2010;107:10208–13. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12:R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra5. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushima-Nishiu M, Unoki M, Ono K, Tsunoda T, Minaguchi T, Kuramoto H, et al. Growth and gene expression profile analyses of endometrial cancer cells expressing exogenous PTEN. Cancer Res. 2001;61:3741–9. [PubMed] [Google Scholar]
- 29.O'Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res. 2010;16:3670–83. doi: 10.1158/1078-0432.CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- 30.Mehrian-Shai R, Chen CD, Shi T, Horvath S, Nelson SF, Reichardt JK, et al. Insulin growth factor-binding protein 2 is a candidate biomarker for PTEN status and PI3K/Akt pathway activation in glioblastoma and prostate cancer. Proc Natl Acad Sci U S A. 2007;104:5563–8. doi: 10.1073/pnas.0609139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, Hu Y, Huo Y, Liu M, Freeman D, Gao J, et al. PTEN deletion leads to up-regulation of a secreted growth factor pleiotrophin. J Biol Chem. 2006;281:10663–8. doi: 10.1074/jbc.M512509200. [DOI] [PubMed] [Google Scholar]
- 32.Musatov S, Roberts J, Brooks AI, Pena J, Betchen S, Pfaff DW, et al. Inhibition of neuronal phenotype by PTEN in PC12 cells. Proc Natl Acad Sci U S A. 2004;101:3627–31. doi: 10.1073/pnas.0308289101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson L, Li J, Liaw D, Hennessy I, Oliner J, Christians F, et al. PTEN expression causes feedback upregulation of insulin receptor substrate 2. Mol Cell Biol. 2001;21:3947–58. doi: 10.1128/MCB.21.12.3947-3958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong TM, Yang PC, Peck K, Chen JJ, Yang SC, Chen YC, et al. Profiling the downstream genes of tumor suppressor PTEN in lung cancer cells by complementary DNA microarray. Am J Respir Cell Mol Biol. 2000;23:355–63. doi: 10.1165/ajrcmb.23.3.4002. [DOI] [PubMed] [Google Scholar]
- 35.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–30. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Jendrossek V, Henkel M, Hennenlotter J, Vogel U, Ganswindt U, Mueller I, et al. Analysis of complex protein kinase B signalling pathways in human prostate cancer samples. BJU Int. 2008;102:371–82. doi: 10.1111/j.1464-410X.2008.07703.x. [DOI] [PubMed] [Google Scholar]
- 39.Koumakpayi IH, Le Page C, Mes-Masson AM, Saad F. Hierarchical clustering of immunohistochemical analysis of the activated ErbB/PI3K/Akt/NF-kappaB signalling pathway and prognostic significance in prostate cancer. Br J Cancer. 2010;102:1163–73. doi: 10.1038/sj.bjc.6605571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ming M, Feng L, Shea CR, Soltani K, Zhao B, Han W, et al. PTEN positively regulates UVB-induced DNA damage repair. Cancer Res. 2011;71:5287–95. doi: 10.1158/0008-5472.CAN-10-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh PM, Malik SN, Bedolla RG, Wang Y, Mikhailova M, Prihoda TJ, et al. Signal transduction pathways in androgen-dependent and -independent prostate cancer cell proliferation. Endocr Relat Cancer. 2005;12:119–34. doi: 10.1677/erc.1.00835. [DOI] [PubMed] [Google Scholar]
- 42.Halvorsen OJ, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res. 2003;9:1474–9. [PubMed] [Google Scholar]
- 43.Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, et al. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res. 2007;13:3860–7. doi: 10.1158/1078-0432.CCR-07-0091. [DOI] [PubMed] [Google Scholar]
- 44.Pinhel IF, Macneill FA, Hills MJ, Salter J, Detre S, A'Hern R, et al. Extreme loss of immunoreactive p-Akt and p-Erk1/2 during routine fixation of primary breast cancer. Breast Cancer Res. 2010;12:R76. doi: 10.1186/bcr2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holzer TR, Fulford AD, Arkins AM, Grondin JM, Mundy CW, Nasir A, et al. Ischemic time impacts biological integrity of phospho-proteins in PI3K/Akt, Erk/MAPK, and p38 MAPK signaling networks. Anticancer Res. 2011;31:2073–81. [PubMed] [Google Scholar]
- 46.Sangale Z, Prass C, Carlson A, Tikishvili E, Degrado J, Lanchbury J, et al. A robust immunohistochemical assay for detecting PTEN expression in human tumors. Appl Immunohistochem Mol Morphol. 2011;19:173–83. doi: 10.1097/PAI.0b013e3181f1da13. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu Y, Segawa T, Inoue T, Shiraishi T, Yoshida T, Toda Y, et al. Increased Akt and phosphorylated Akt expression are associated with malignant biological features of prostate cancer in Japanese men. BJU Int. 2007;100:685–90. doi: 10.1111/j.1464-410X.2007.07014.x. [DOI] [PubMed] [Google Scholar]
- 48.Manin M, Baron S, Goossens K, Beaudoin C, Jean C, Veyssiere G, et al. Androgen receptor expression is regulated by the phosphoinositide 3-kinase/Akt pathway in normal and tumoral epithelial cells. Biochem J. 2002;366:729–36. doi: 10.1042/BJ20020585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, Wheeler T, Dai H, Frolov A, Thompson T, Ayala G. High level of androgen receptor is associated with aggressive clinicopathologic features and decreased biochemical recurrence-free survival in prostate: cancer patients treated with radical prostatectomy. Am J Surg Pathol. 2004;28:928–34. doi: 10.1097/00000478-200407000-00013. [DOI] [PubMed] [Google Scholar]
- 50.Antonarakis ES, Keizman D, Zhang Z, Gurel B, Lotan TL, Hicks JL, et al. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer. 2012 doi: 10.1002/cncr.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]