Abstract
A major remaining question in glycobiology is how a glycosyltransferase (GT) that retains the anomeric linkage of a sugar catalyzes the reaction. Xylosideα1–3 Xylosyltransferase (XXYLT1) is a retaining GT that regulates Notch receptor activation by adding xylose to the Notch extracellular domain. Here, using natural acceptor and donor substrates and active Mus musculus XXYLT1, we report a series of crystallographic snapshots along the reaction, including an unprecedented natural and competent Michaelis reaction complex for retaining enzymes. These structures strongly support the SNi-like reaction as the retaining mechanism for XXYLT1. Unexpectedly the Epidermal Growth Factor-like repeat acceptor substrate undergoes a large conformational change upon binding to the active site, providing a structural basis for substrate specificity. Our improved understanding of this retaining enzyme will accelerate the design of retaining GT inhibitors that can modulate Notch activity in pathological situations where dysregulation of Notch is known to cause cancer or developmental disorders.
Introduction
Notch signaling plays essential roles in development of all metazoans, and defects in the Notch pathway lead to a variety of human diseases including several cancers and developmental disorders1,2. Notch activation can be modulated by differential O-linked glycosylation of the Notch extracellular domain (NECD)3. O-Linked glucose is added to a subset of Epidermal Growth Factor-like (EGF) repeats in the NECD by Protein O-glucosyltransferase 1 (POGLUT1)4–6 and can be extended to a xyloseα1–3xyloseα1–3glucose trisaccharide by sequential action of Glucosideα1–3 Xylosyltransferase (GXYLT1/2)7 and Xylosideα1–3 Xylosyltransferase (XXYLT1)8. Xylosylation by the retaining glycosyltransferase (GT) XXYLT1 negatively regulates Notch activation9. Detailed knowledge of the mechanism of these enzymes is needed to help design inhibitors/drugs that could be used to modulate Notch activity and to probe Notch’s role in many diseases.
Mammalian XXYLT1 is a type II membrane protein located in the endoplasmic reticulum (ER) with its catalytic domain protruding into the lumen8. XXYLT1 belongs to glycosyltransferase family 8 (GT8) in the CAZY database10, and is a retaining GT, i.e. the α-linked xylose in the donor UDP-xylose retains its stereochemistry after being transferred to the acceptor xylose. The mechanism of retaining GTs remains one of the unresolved core issues in glycobiology due to the paucity of structures of intact ternary complexes11–13. Given the significant structural variances among different retaining GT families, it remains elusive whether there is a universal mechanistic behavior. Two mechanisms have been proposed: double displacement and SNi-like (i for internal return) (Supplementary Fig. 1a, b). Double displacement retains configuration by proceeding through a covalent enzyme-substrate intermediate, whereas SNi-like retains configuration by having the acceptor nucleophile directly attack the donor anomeric carbon from the same side as the leaving phosphate group. The SNi-like mechanism has been gaining traction recently11–14, supported by a recent kinetic isotope study of OtsA15 and a recent study showing the structures of ternary complexes of GalNAc-T2 (including a Michaelis complex trapped using incompetent acceptor peptide)16. Structural studies using ligand analogues or incompetent ligands usually need to make assumptions and need careful interpretation16–18. Thus, though difficult to trap, knowing detailed geometry of the active site with natural substrates would greatly contribute to illuminating the retention of anomeric stereochemistry that is still under much debate. Moreover, mechanistic insights into XXYLT1, a new retaining GT, will help define the generality of possible retaining mechanisms.
Here a comprehensive set of reaction states of Notch-regulating XXYLT1 were visualized using natural donor substrates, competent acceptor ligand and active enzyme. To our knowledge, we trapped the first natural and competent Michealis ternary complex for a retaining GT, which provides a detailed view of the reaction geometry for retention of anomeric configuration and strongly supports the SNi-like mechanism for XXYLT1’s xylose transfer. Furthermore, the use of a folded disaccharide-modified EGF acceptor in our structural study revealed an unusual acceptor recognition mode involving both the disaccharide moiety and EGF, and also demonstrated an unexpected flexibility of EGF module structure. Together with structure-based mutagenesis and in vitro glycosylation assays, these data revealed detailed insights into XXYLT1’s retaining xylose transfer to Notch. Based on our structural study, we analyzed the XXYLT1 alterations in several cancer types, which indicate XXYLT1 may play an important role in Notch-related tumorigenesis in specific cancer types, for example, lung squamous cell carcinoma.
RESULTS
XXYLT1 structure and its orientation to ER membrane
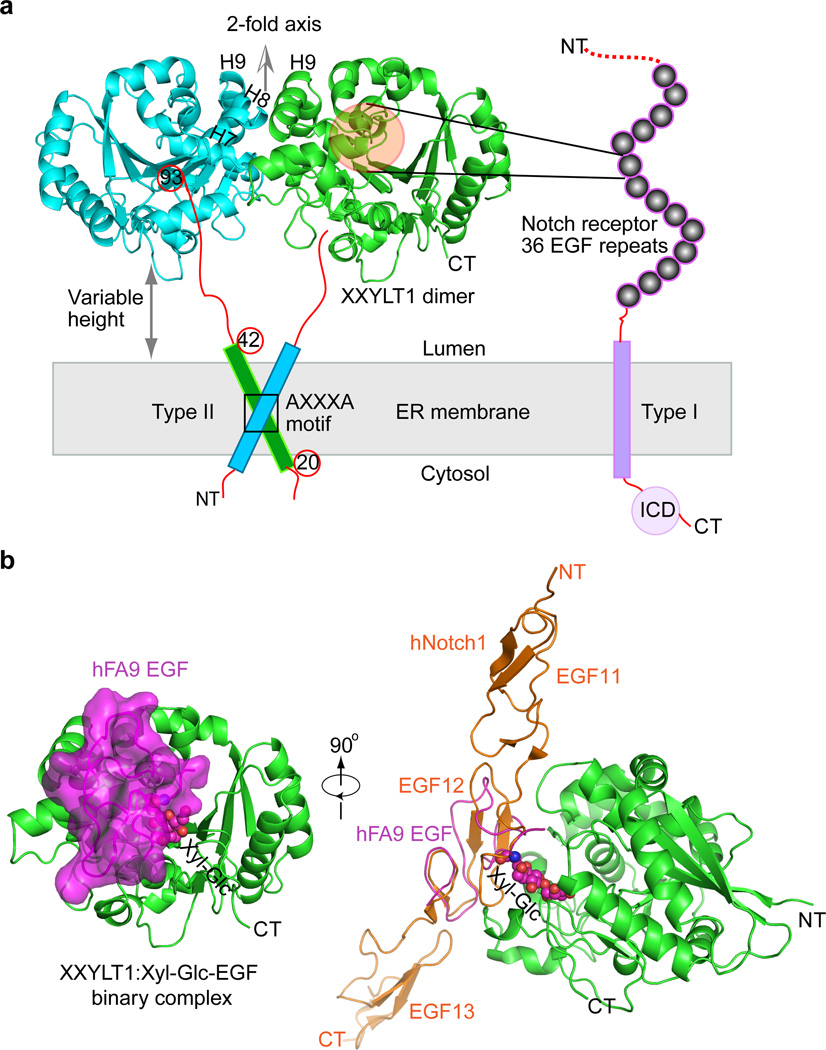
XXYLT1 is a type II membrane protein well conserved among Mus musculus, Homo sapiens and Drosophila melanogaster (Supplementary Fig. 2), the three organisms commonly used to study Notch. We truncated the N-terminal transmembrane anchor helix and expressed the soluble extracellular domain (S43–D392) of mouse XXYLT1 in human embryonic kidney (HEK) 293T cells. Purified protein transferred xylose from the donor UDP-xylose to Xyl-Glc disaccharide-modified EGF repeats from Notch1, Notch2 or human factor IX (hFA9)8,19,20. We first crystallized and solved the structure of XXYLT1 bound with Mn2+ (Fig. 1a, Supplementary Table 1) since XXYLT1 requires a divalent cation for its activity21. The overall structure consists of ~300 residues (V93–E391), lacking the N-terminal unstructured loop (~S43–V92) that was removed by limited proteolysis (See Online Methods). As expected, XXYLT1 had a GT-A fold with the glycosyltransferase signature DXD motif (residues 225–227) coordinating a Mn2+ ion in the active site pocket (Supplementary Fig. 2c)22. Unexpectedly, XXYLT1 formed a dimer in the crystal lattice via the kinked tandem helixes 7–9 (Fig. 1a). Because the full-length XXYLT1 was previously shown to form an SDS-resistant dimer via the AXXXA dimerization motif in the transmembrane helix8, the interface observed in our catalytic domain structure likely provides additional dimerization contact. If the dimer structure is oriented with its two-fold axis perpendicular to the ER membrane as expected in the native environment, the enzyme active pocket faces sideways, ideal for making lateral contact with the luminally oriented EGF repeats of the NECD (Fig. 1a). Interestingly, the disordered stem region (S43–V92) appeared to give the enzyme some freedom to move close to or away from ER membrane in the lumen, enabling XXYLT1 to modify the many Notch EGFs as they are translated, emerge into the ER lumen and fold.
Figure 1. The mouse XXYLT1 is a dimer and has a GT-A fold with its active site facing sideways to facilitate lateral modification of Notch.
(a) The overall structure of type II membrane protein XXYLT1, with its truncated amino terminal transmembrane domain shown as a cylinder. The non-crystallographic two-fold axis is oriented upward, perpendicular to the sketched ER membrane in gray. The type I membrane protein Notch receptor is sketched with its 36 EGF repeats as spheres. NT, N-terminus; CT, C-terminus; ICD, intracellular domain; H7, Helix 7; H8, Helix 8; H9, Helix 9. (b) Left, overall structure of XXYLT1 in complex with hFA9 Xyl-Glc-EGF; right, superposition of the crystal structure of human Notch1 EGF11–13 (PDB ID 2VJ3) with the acceptor EGF, showing that the enzyme interacts laterally with the protein substrates. XXYLT1 is in green cartoon and in the same orientation as the green apo structure in (a); Xyl-Glc-EGF is in magenta cartoon with a semi-transparent surface view, and the covalently linked disaccharide is in spheres. The human Notch1 EGF11–13 is shown in orange cartoon.
Binary structure of XXYLT1 with acceptor Xyl-Glc-EGF
To better understand how the enzyme recognizes this acceptor substrate, we incubated XXYLT1 with the folded Xyl-Glc disaccharide-modified acceptor substrate hFA9 Xyl-Glc-EGF, purified the binary complex by size exclusion chromatography (Supplementary Fig. 3), and then crystallized and solved the binary complex structure. When the crystal structure of the binary complex XXYLT1:Xyl-Glc-EGF was oriented like apo-XXYLT1 (as in Fig. 1a), we found that the acceptor substrate Xyl-Glc-EGF indeed bound to the side of the enzyme (Fig. 1b). Superimposition of the human Notch1 EGF11–13 crystal structure on the XXYLT1-bound hFA9 EGF oriented the EGF11–13 N-terminus away from and the C-terminus toward the membrane (Fig. 1b). This orientation is consistent with the membrane topology of the Notch receptor. Therefore, the binary structure provided a glimpse into how XXYLT1 modifies a newly synthesized Notch receptor with multiple tandem EGF repeats.
Unprecedented conformational change of recognized EGF
The EGF domain is an evolutionarily conserved protein domain of ~30–40 residues featuring three disulfide bonds and a two-stranded β-sheet23,24,25. Surprisingly, we found the EGF repeat in the binary complex underwent a large conformational change compared to the isolated EGF motif: the two β-strands were separated and converted to loops (Fig. 2a). Consistent with the significant changes, a least square fitting of our enzyme-bound hFA9 EGF structure with the structure of the same but unbound protein (PDB ID 1EDM) revealed a large root-mean-square-deviation (RMSD) of 6.27 Å. Our finding suggested that individual EGF modules are more flexible than previously thought. This flexibility may be amplified in the NECD with 36 such repeats, and may contribute to ligand binding or subsequent endocytosis-mediated NECD pulling event1,3.
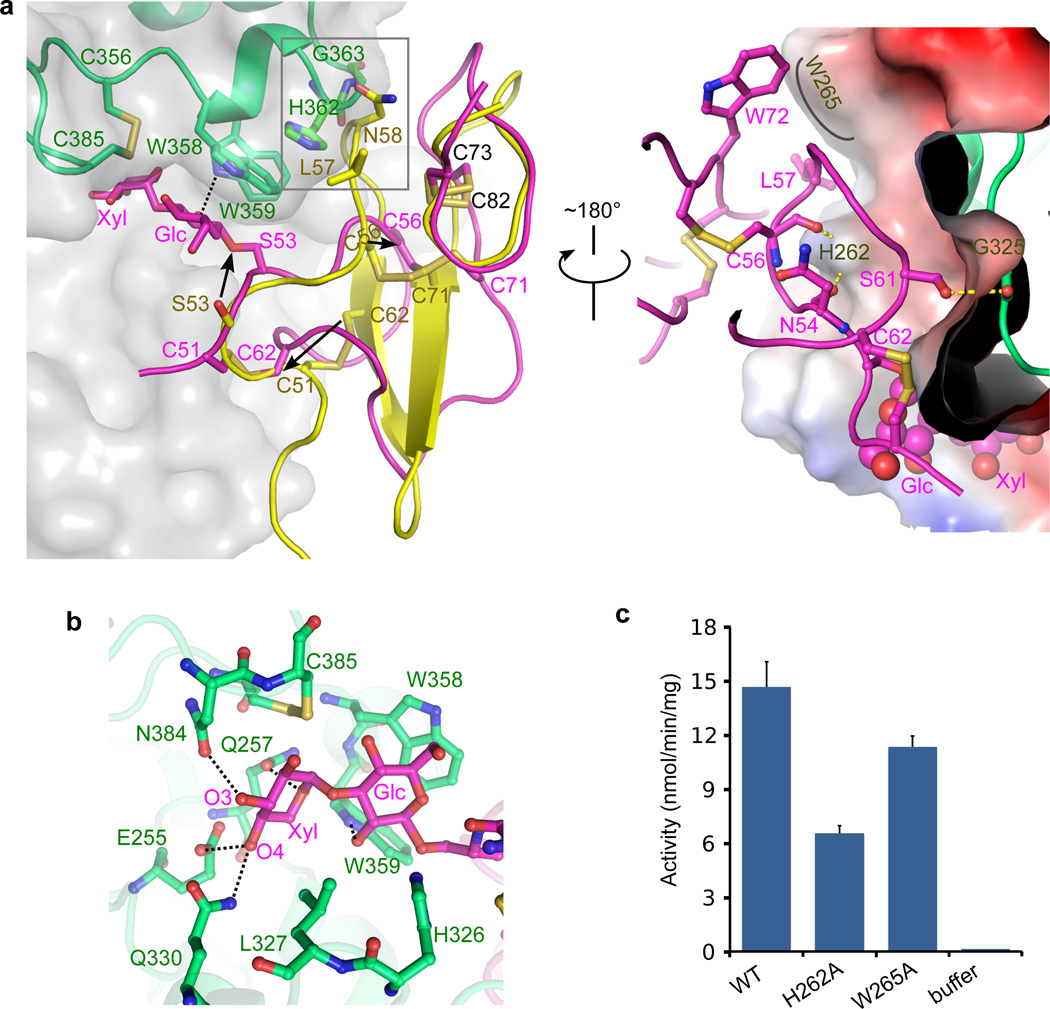
Figure 2. The Xyl-Glc-EGF acceptor substrate undergoes a large conformational change when bound to XXYLT1.
(a) Front (left) and back (right) view of the interface between XXYLT1 and the bound Xyl-Glc-EGF. In the front view, the apo-hFA9 EGF (PDB ID 1EDM, yellow) is superimposed with the bound Xyl-Glc-EGF in magenta cartoon view to show large conformational changes and dissolution of the two β-strands upon binding to XXYLT1. The enzyme is in semitransparent gray surface and partly in green cartoon. The black arrows highlight the movement of EGF C51, S53, and C56 during the conformational change. The gray rectangle marks the potential steric clash with the enzyme if EGF did not change its conformation. In the back view, the superimposed apo-hFA9 EGF in the front view is no longer shown and the electrostatic surface potential of the enzyme is shown. (b) Recognition of the disaccharide portion of the acceptor by XXYLT1 active site residues. Dashed black lines indicate H-bonds between acceptor and enzyme. (c) In vitro transferase activity of the wild type and interface residue mutated enzymes. The data were from three independent assays. The bars indicate mean ± S.E.M.
In contrast to the large-scale conformational changes found in the EGF repeat, the XXYLT1 structure remained largely unchanged upon acceptor binding (Supplementary Fig. 4), with an overlay RMSD of 0.61 Å between the apo and the acceptor bound enzyme structures. This observation suggested that XXYLT1 has a relatively rigid EGF binding surface.
It is interesting to ask what caused the conformational changes in EGF. We know that addition of the Xyl-Glc disaccharide onto hFA9 EGF repeat does not by itself cause a structural change26, and we did not think crystal contact was the reason since there were only two weak contacts around the EGF with adjacent enzyme molecules in the crystal lattice (Supplementary Fig. 5). More importantly, the XXYLT1:Xyl-Glc-EGF binary complex was formed as a stable complex in solution before crystallization (Supplementary Fig. 3). Therefore, the observed conformational change in the disaccharide modified EGF was likely caused by its specific recognition by and binding to the XXYLT1. In the binary complex structure, both the disaccharide and the EGF were recognized by and in extensive interaction with the enzyme. In the acceptor disaccharide region, the distal xylose formed four strong H-bonds with the enzyme, and the proximal glucose ring stacked against W358 and H326 and H-bonds with W359 of XXYLT1 (Fig. 2b). Thus, the enzyme appeared to pull the disaccharide into its active site, driving the conformational changes in EGF (Supplementary Movie 1). Because the EGF binding interface of XXYLT1 was rigid, as described above, the protruding helix-turn-helix motif (H362–Y364) forced the EGF to change its conformation in order to avoid the otherwise extensive steric clashes (Fig. 2a). The three disulfide bonds in the EGF remained intact in the enzyme-bound conformation. These disulfide bonds likely facilitate rapid refolding of the EGF into its native structure after being released from XXYLT1 following completion of the xylose transfer reaction (Supplementary Movie 1). The importance of the disulfide bonds may explain our previous observation that XXYLT1 modifies the Xyl-Glc disaccharide on an EGF repeat with intact disulfides more efficiently than a disulfide-reduced EGF repeat19.
The EGF conformation in the binary complex structure was unstable, because the hydrophobic residues P55 and Y69, a part of the core of the natively folded EGF, were exposed (Supplementary Movie 1). This EGF conformation was stabilized by XXYLT1 via three H-bonds (EGF N54 with XXYLT1 H262, EGF C56 with XXYLT1 H262, and EGF S61 with XXYLT1 G325), as well as hydrophobic stacking between EGF L57 and W72 and XXYLT1 W265 (Fig. 2a). Therefore, the three XXYLT1 residues (H262, W265, and G325) appeared to be important for the enzyme activity. Substituting residues H262 and W265 with alanines in the acceptor-binding platform significantly reduced the XXYLT1 activity in vitro (Fig. 2c). XXYLT1 G325 faced EGF S61 in the crystal structure, which is known to be O-fucosylated on native hFA927. We found that the presence of O-fucose mono- or disaccharide on EGF S61 did not affect XXYLT1 activity in vitro (Supplementary Fig. 6b). Therefore, XXYLT1 modified the EGF repeat regardless of the O-fucosylation status of S61. Thus, we suggest that absence of a side chain in XXYLT1 G325 allows the enzyme to modify O-fucosylated EGF repeats.
Glucoside xylosyltransferases 1 and 2 (GXYLT1/2), which act upstream of XXYLT1 in the Notch O-glucosylation pathway, are homologous to XXYLT18 but with a low sequence identity of ~ 9% or 14% between XXYLT1 and GXYLT1 or GXYLT2, respectively. Sequence alignment with ClustalW showed that their EGF recognition surfaces were not conserved: H262, W265, and G325 in XXYLT1 are changed to G, N/S, and I, respectively, in GXYLT1/2 (Supplementary Fig. 2b). Therefore, GXYLT1/2 may recognize folded EGF repeats in a manner that is different from XXYLT1. This is not surprising considering the GXYLT1/2 acceptor substrate is a mono-glucosylated EGF.
Different reaction states trapped with natural ligands
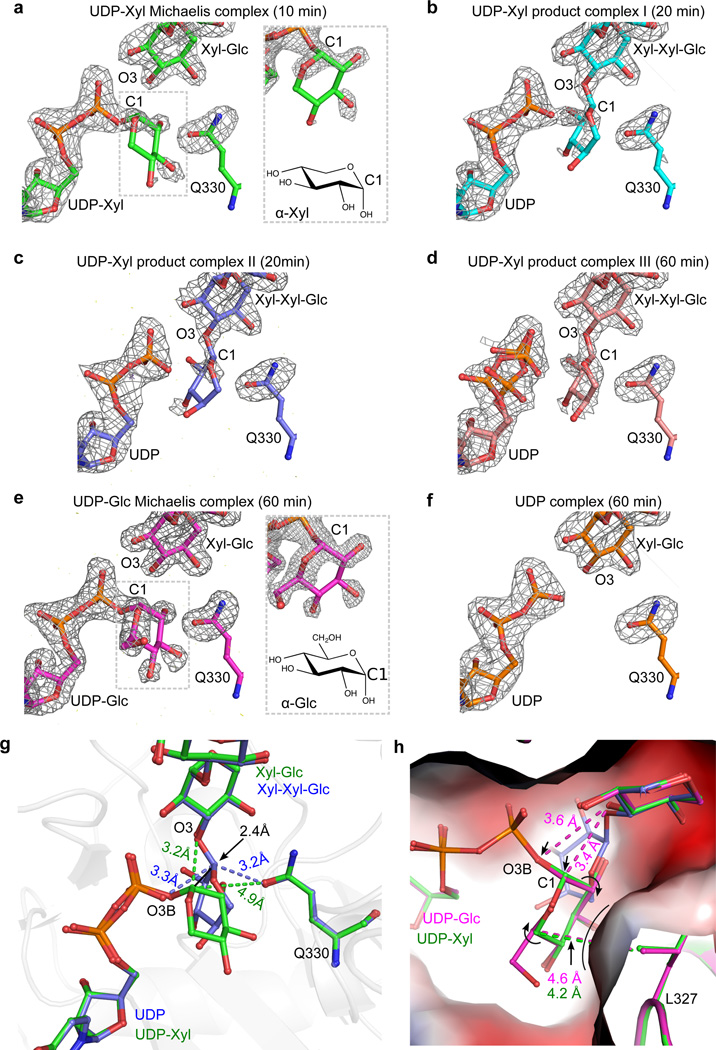
Though difficult to capture, the structure of a competent Michaelis ternary complex would be valuable for distinguishing the two possible GT retaining mechanisms, as the structure would be devoid of factors that may complicate mechanistic determination when non-natural substrates are used. We trapped functional ternary complexes containing natural donor ligand, natural acceptor ligand and active enzyme by soaking the donor substrate UDP-xylose into the XXYLT1:Xyl-Glc-EGF binary complex crystals for 10, 20, or 60 min, respectively. We solved four structures of these soaked crystals. Based on their respective soaking time and substrate density features, these structures could be sorted into three stages of the transfer reaction (Fig. 3a–d). In the 10-min complex at 1.95-Å resolution, the unbiased Fo-Fc difference density map calculated before ligand modeling showed an almost intact UDP-xylose near the distal acceptor xylose and enzyme residue Q330 (Fig. 3a). Part of the donor xylose ring had no density probably due to the lack of stabilizing interactions. This structure had both natural donor substrate and competent acceptor substrate in place prior to catalysis, representing the first natural Michaelis complex of retaining XXYLT1. Furthermore, with increasing soaking time, we observed the transfer reaction proceeding. In one 20-min complex crystal (Product complex I), the donor xylose density was connected to both UDP and acceptor xylose, suggesting a mixed state (Fig. 3b). In another 20-min complex (Product complex II), the donor xylose had weak densities and was apparently connected only to the acceptor xylose, indicating that the transfer reaction was near completion (Fig. 3c). The UDP in Product complex II adopted a similar conformation to the UDP moiety in the UDP-Xyl Michaelis complex, but its pyrophosphate group shifted slightly away from the active site (Supplementary Fig. 7). In the 1-hr complex (Product complex III), the well-defined difference density of separated UDP and the trisaccharide product indicated that the transfer reaction was complete (Fig. 3d). Several hydrogen bonds stabilized the transferred terminal xylose (Supplementary Fig. 7a), but the space between the UDP and Q330 was tight, leading to further shift of pyrophosphate away from the active site. Thus, two UDP conformations were identified (Supplementary Fig. 7b). In fact, the progression of the transfer reaction apparently coincided with a gradual shift of the pyrophosphate (Supplementary Fig. 7c–e). The bulky nature of the UDP leaving group and the product trisaccharide in the narrow enzyme active site may promote the departure of the leaving group and release of the product. Indeed, binary complex crystals soaked with UDP-Xyl for over 5 hours were all cracked. This catalytic competence in our crystallography condition can be clearly illustrated by morphing the density evolution between the four different structures (Supplementary Movie 2).
Figure 3. Snapshots of XXYLT1 along its retaining reaction pathway.
(a–f) Distinct states trapped and visualized by crystallography using active binary enzyme-acceptor complex with different donors. The donor substrate is either UDP-Xyl, UDP-Glc, or UDP. The acceptor substrate is hFA9 Xyl-Glc-EGF. Unbiased Fo-Fc difference electron density map of the active site was calculated before modeling donor, acceptor, and the carboxamide group of the XXYLT1 Q330 (gray mesh, contoured at 3.0σ except for Product complex I at 2.6σ). (a–d) Four stages of the transfer reaction in which the XXYLT1:Xyl-Glc-EGF binary complex crystals have been soaked with UDP-Xyl for 10, 20, 20, and 60 min, respectively. (e) UDP-Glc Michaelis complex after the binary complex crystal has been soaked with UDP-Glc for 60 min. (f) The XXYLT1:Xyl-Glc-EGF:UDP ternary complex after soaking UDP into binary complex crystal for 60 min. The zoomed window in (a) and (e) provides a slightly different view of the donor Xyl or Glc density, along with their respective chemical structures below. (g) Comparison of the active site configurations of UDP-Xyl Michaelis complex (Green) and the trisaccharide product complex (Product complex II, slate blue). (h) Comparison of the UDP-Xyl Michaelis ternary complex (Green) with the UDP-Glc Michaelis ternary complex (Magenta) and the Product complex II (Slate blue). The enzyme active site pocket is shown in electrostatic surface view.
UDP-Glc is a less favored donor substrate than UDP-Xyl of XXYLT18. The slower reaction with UDP-Glc provided a better opportunity than UDP-xylose for capturing an intact donor substrate in the crystal. We soaked UDP-Glc into the XXYLT1:Xyl-Glc-EGF binary complex crystals for 1 hr, and indeed observed essentially intact density for UDP-Glc (Fig. 3e). Interestingly, there was a well-resolved density for the hydroxymethyl group of donor Glc, compared to the density map where UDP-Xyl was used (Fig. 3a). This was consistent with the small chemical difference between UDP-Xyl and UDP-Glc, and allowed us to further validate the natural Michaelis complexes we captured. As a control, we determined the structure of XXYLT1:Xyl-Glc-EGF binary complex crystal soaked with UDP. As expected, the active site pocket was clearly devoid of a donor sugar, and the UDP was in the relaxed conformation (Fig. 3f).
XXYLT1 does not contribute the nucleophile
The UDP-Xyl Michaelis ternary complex and Product complex II (Fig. 3a,c) represented the beginning and the end of the transfer process. Superimposition of these two structures revealed that upon transfer, the donor xylose rotated by ~90°, and the anomeric carbon moved upward by 2.4 Å to form a glycosidic bond with the acceptor xylose (Fig. 3g). In the beginning complex, the donor sugar xylose was stabilized by interaction of its O2 hydroxyl with enzyme Q330 and its O3 hydroxyl with enzyme L327 backbone and S289 (Supplementary Fig. 8). The donor xylose O4 hydroxyl was not stabilized, accounting for the weak density near this region (Fig. 3a).
In all these structures, the residue closest to the anomeric carbon was Q330, the only candidate enzymatic residue that could function as the catalytic nucleophile. However, Q330 was 4.9 Å away from the anomeric carbon of the donor xylose, too far away to be a nucleophile (Supplementary Fig. 8). Also, in all four XXYLT1:Xyl-Glc-EGF:UDP-Xyl ternary complex structures (Fig. 3a–d), no density was observed connecting the donor sugar and the Q330 in the unbiased Fo-Fc difference maps even at a lower sigma level (2.25σ). Therefore, these observations were inconsistent with a double displacement mechanism, which requires an enzymatic nucleophile (e.g. Q330) for the first inversion reaction (Supplementary Fig. 1a). Furthermore, no residues around the active site underwent any significant changes during the transfer reaction (Fig. 3a–d). This observation argued against a possible conformational change to allow active site residues (including Q330 or other possible residues) to function as nucleophile, which was previously proposed as another possibility for double displacement28.
Competent Michaelis complex supports SNi-like mechanism
In contrast to double displacement, an SNi-like reaction has been proposed as an alternative mechanism for retaining GTs11–13, which is further supported by the UDP-Xyl Michaelis complex structure of XXYLT1. In this competent Michaelis complex, the O3 hydroxyl (OH) of the acceptor xylose was 3.2 Å above the anomeric carbon and on the same side as the leaving UDP β-phosphate, in an ideal position to function as the nucleophile (Supplementary Fig. 8). The O3 OH was nearly perpendicular (92.3°) to the anomeric carbon and the leaving group of the donor ligand, ideally positioned to displace the anomeric carbon orthogonally and consistent with the recently proposed orthogonal displacement configuration29. Importantly, the identified nucleophile Xyl O3 OH was within 3.5 Å of several groups, including the O2 OH on the donor xylose ring, and the β-phosphate oxygen O1B and O3B, and XXYLT1 N384. These interactions constrained the nucleophile on the same side as the leaving group, a hallmark of the SNi mechanism. We suggest that the β-phosphate oxygen O3B is the most likely base to deprotonate the acceptor xylose O3 OH, as the O2 OH of the donor xylose and O1B of β-phosphate oxygen are both involved in two hydrogen bonds, compromising their basicity. These observations together strongly support a substrate-assisted catalytic mode without the involvement of enzymatic general bases.
Our observation of nearly intact UDP-Glc in the XXYLT1:Xyl-Glc-EGF:UDP-Glc ternary complex structure (Fig. 3e) provided further support for the above-described mechanism. The binding mode of UDP-Glc was similar to that of UDP-xylose, except for the extra hydroxymethyl group that stacks against L327, shifting the anomeric carbon by 0.2 Å away from the nucleophile and concomitantly weakening the H-bond between β-phosphate oxygen O3B of donor and the acceptor nucleophile (Fig. 3h). These subtle shifts appear to be sufficient to distort the reaction geometry to prevent the transfer of glucose, potentially explaining why the enzyme greatly prefers UDP-Xyl over UDP-Glc8. In fact, no product density was observed even when UDP-Glc was soaked into the binary complex crystals for two weeks.
Water molecules did not appear to play an important role in the XXYLT1 transfer reaction. Both UDP-Xyl and UDP-Glc Michaelis complexes had essentially the same water pattern in their respective active sites (Supplementary Fig. 9): the nearest water to the anomeric carbon was on the ‘back face’ and was still ~3.8 Å away from the anomeric carbon and, therefore, too distant to hydrolyze the donor substrate. The fact that donor UDP-Glc remained intact in the UDP-Glc Micahaelis complex after two weeks of soaking further supported our proposal that water molecule was not involved in the transfer reaction.
Roles of enzyme active site residues in transfer reaction
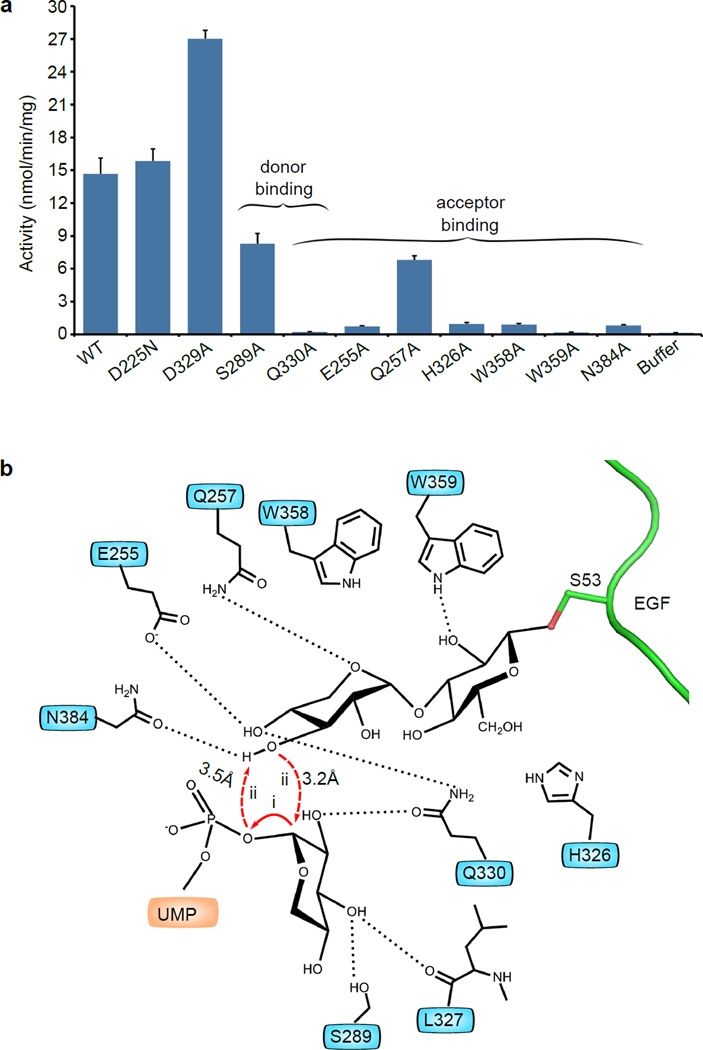
The requirement of a strict geometry of the donor and acceptor substrates in the SNi-like mechanism predicted that a retaining transferase is highly sensitive to both the donor chemistry and the enzyme active site composition. Transferase sensitivity to the donor was borne out by the crystal structure of the UDP-Glc ternary complex. To investigate if XXYLT1 activity is sensitive to the active site composition, we substituted eight residues that make contact with the donor and/or acceptor with alanine and confirmed their protein expression and folding (Supplementary Fig. 6a and 10, Supplementary Table 2). We found by in vitro activity assay that xylose transfer activity was either abolished by Q330A and W359A substitutions, or profoundly reduced by E255A, H326A, W358A, and N384A substitutions, or reduced by Q257A, and S289A substitutions (Fig. 4a). The crucial role of Q330 was expected from the crystal structure, because it formed a strong H-bond (2.6 Å) with the O2 OH of donor xylose, and a second H-bond with the O4 OH of acceptor xylose, bringing the two xylose rings together and orienting them for the transfer reaction (Supplementary Fig. 8). W359 was particularly important for the enzyme activity, perhaps because this residue formed a H-bond with the glucose of the acceptor disaccharide. We further hypothesized that substituting residues that were close to but do not make contact with the donor xylose (for example, XXYLT1 D225 or D329) may not adversely affect the XXYLT1 activity. We produced XXYLT1 with either D225N or D329A substitution, and found that these proteins had either similar (D225N) or even increased (D329A) in vitro activity compared to the wild type enzyme (Fig. 4a). These results supported our proposal that the enzyme active site functions to accurately position the donor and acceptor substrates in an ideal geometry for catalysis, consistent with the substrate-assisted SNi-like mechanism (Fig. 4b). Precise orientation of the donor and acceptor is important for a number of possible reasons: it may properly orient the acceptor nucleophile, stabilize the positive charge on the anomeric carbon15 and the negatively charged phosphate in the transition state30, and may promote the leaving-group departure.
Figure 4. Proposed retaining mechanism of XXYLT1.
(a) In vitro transferase activity of wild type XXYLT1 and donor and acceptor-binding site mutants. The data were from three independent assays. The bars indicate mean ± S.E.M. (b) Schematics of the proposed retaining reaction mechanism of XXYLT1, depicting a front-side SNi-like catalysis mechanism. The reaction geometry revealed by the natural UDP-Xyl Michaelis complex indicated some dissociative features where the cleavage of UDP leaving group (step i) may precede the nucleophilic attack by the deprotonated O3 hydroxyl of acceptor (step ii). Depicted residues orient and position the donor and acceptor substrates.
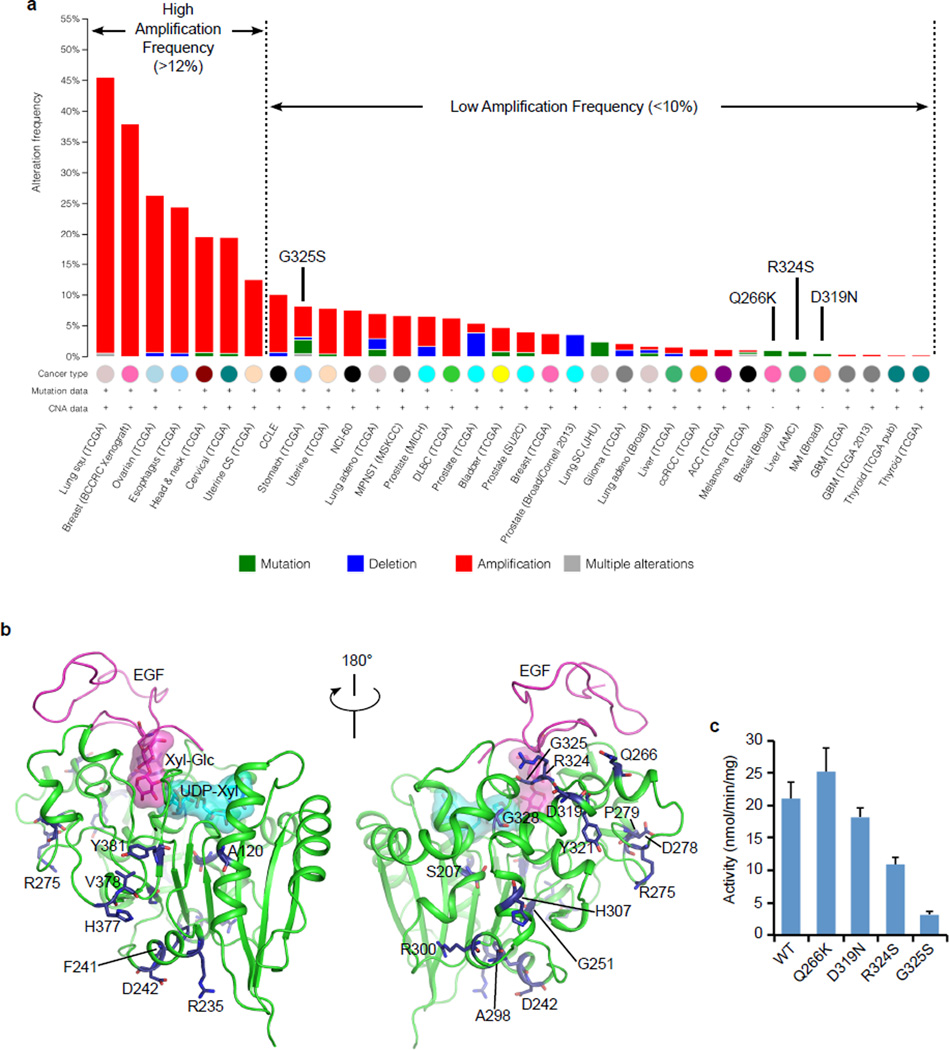
XXYLT1 alterations in cancers
Because XXYLT1 negatively regulates Notch9, and Notch aberrations have been linked to myeloid cancers31 and squamous cell carcinoma of lung32, head and neck squamous cell carcinoma33, we wondered if the enzyme XXYLT1 is implicated in cancer. We analyzed the publicly available cancer genomic data in cBioportal34 and found that XXYLT1 is amplified in several cancer types, especially in lung squamous cell carcinoma (Fig. 5a). We also identified 22 cancer-associated missense mutations (Supplementary Table 3), although mutation occurred at a much lower frequency than amplification. Mapping these missense mutations onto the crystal structure showed that many were solvent exposed and likely benign (Fig. 5b). Four mutations (Q266K, D319N, R324S and G325S) were close to the bound EGF acceptor. We examined their effects on enzyme using in vitro assays (Supplementary Fig. 10, and 11, Supplementary Table 2). We found that Q266K and D319N retained the enzymatic activity while R324S and G325S reduced the activity by ~ 50–80 % (Fig. 5c). Notably, the activity-reducing mutations occurred in cancers with much lower incidence of XXYLT1 amplification (Fig. 5a).
Figure 5. Notch signaling in certain cancers may be inhibited by alterations in the XXYLT1.
(a) The human XXYLT1 is frequently amplified in specific types of cancers. 22 individual point mutations have also been reported in certain cancer cells, which are listed in Supplementary Table 3. Gene alteration data were obtained from cancer genomics site: cBioPortal. (b) The cancer-associated mutations in human XXYLT1 were mapped onto the crystal structure of the mouse UDP-Xyl Michaelis ternary complex and shown as blue sticks. Most mutations were solvent exposed and away from the donor and acceptor binding sites, indicating they are likely benign. (c) In vitro xylosyltransferase activities of the wild type and four cancer-related mutants with hFA9 Xyl-Glc-EGF as the acceptor. The data were from three independent assays. The bars indicate mean ± S.E.M.
DISCUSSION
There are over 90 GT families with over 20,000 members (www.CAZY.org)10, about one third of which are retaining enzymes. However, the catalytic mechanism for the retention of anomeric stereochemistry is still debated11–13. We trapped two Michaelis ternary complexes with natural ligands and wild type enzyme (Fig. 3a, e) at high resolution (1.95 Å and 1.62 Å, respectively), and solved the structures of several in situ reaction states (Fig. 3b–d, Supplementary Movie 2). These structures provided strong structural support for the SNi-like retention mechanism. In hindsight, our success in trapping the UDP-Xyl Michaelis complex was serendipitous. The lifetime of a natural Michaelis complex is usually too short to be captured via the crystallographic approach. Luckily, XXYLT1:Xyl-Glc-EGF binary complex was crystallized in a Bis-Tris buffer at pH 6.5. We later found that XXYLT1 had low activity in this buffer (Supplementary Fig. 6c), providing a potential explanation for why this transient yet competent Michaelis complex was trapped in the crystals.
Notably, the observed donor and acceptor geometry in our UDP-Xyl Michaelis complex structure appeared to support an asynchronous reaction mode with a dissociative character, whereby UDP might dissociate prior to the nucleophilic attack (Supplementary Fig. 8, Fig. 4b). On one hand, both distances between the nucleophile and the anomeric carbon (3.2 Å) and between nucleophile and the O3B catalytic base (3.5 Å) were a little long, suggesting that a nucleophilic attack may not be imminent in this configuration. On the other hand, the UDP moiety of UDP-Xyl was under severe stress: in the absence of xylose, the UDP moiety alone would relax towards the acceptor disaccharide by as much as 0.7 Å, shortening the distance between O3B and nucleophile from 3.5 Å to 2.8 Å (Supplementary Fig. 7e). Furthermore, as we mentioned earlier, the space between UDP and XXYLT1 Q330 appeared to be too narrow to comfortably accommodate the xylose ring (Supplementary Fig. 7a, b). For these reasons, we suggest that dissociation of the bulky UDP moiety from xylose may occur before the nucleophilic attack on the anomeric carbon by deprotonated O3 hydroxyl of acceptor (Fig. 4b). During this process, an oxocarbenium ion character would likely develop, in which case the transient positive charge could be stabilized by the nearby Q330. Similar modes of asynchronous reaction were previously suggested based on a kinetic isotope effect study of trehalose-6-phosphate synthase OtsA15 and quantum mechanics calculations of the transferases OtsA and LgtC30–35, and now are further supported by our structural work on XXYLT1.
While this manuscript was under preparation, a Michaelis complex structure of the retaining transferase GalNAc-T2 trapped using an incompetent acceptor peptide and an analogue donor UDP-5SGalNAc was reported16. Although this complex structure also supports the SNi-like retaining mechanism, significant differences exist between it and our UDP-Xyl Michaelis complex. First, the displacement angle between the nucleophile and the C1-O3B bond is 79.8° in the GalNAc-T2 ternary complex, compared to 92.3° in our structure that ideally fits orthogonal displacement configuration29 (Supplementary Fig. 8). Second, the structure of the Michaelis complex of GalNAc-T2 shows less dissociative property, as the potential nucleophile is very close to both the anomeric carbon of the analogue donor (2.5 Å) and one phosphate oxygen (2.7 Å). This was dramatically different from the 3.2–3.5 Å distances observed in our natural Michaelis complex structure that suggested that phosphate dissociation may precede nucleophilic attack. Thus, though the catalytically competent natural Michaelis complex shown here revealed features of a retaining mechanism for XXYLT1, and some advances have been achieved for other GTs through kinetic and structural methods, the general application of this mechanism for all retaining enzymes remains to be clarified.
The retaining transferase GalNAc-T2 undergoes a conformational change upon donor binding, suggesting that the donor binds the enzyme before the acceptor16. However, XXYLT1 did not undergo major conformational changes upon substrate binding, and the bound acceptor substrate did not obstruct the entrance of the donor substrate into the active site of XXYLT1 in any of the ternary complex structures. This raised a question about the order of ligand binding. In our UDP-Xyl ternary complex structures obtained by soaking the donor substrates into the preformed crystals of the enzyme-acceptor binary complex, the transfer reaction was able to proceed to completion (Fig. 3a–d, Supplementary Movie 2). Thus, we believe that in principle either donor or acceptor could bind XXYLT1 first. However, when the acceptor substrate binds the XXYLT1 first in solution, followed by binding of the donor substrate, non-productive hydrolysis of donor substrate can be reduced as compared to when the donor binds first.
The Notch signaling pathway has been an attractive target in cancer research, although the effects of Notch signaling on tumor behavior are dependent on the type of cancer2. In most cases, aberrant Notch activation is oncogenic. However, in some cases such as squamous carcinomas derived from the epidermis, lung, and head and neck, and acute myeloid leukemia, inactivation of Notch signaling is oncogenic2. Our analysis revealed that XXYLT1 is highly amplified with few and benign mutations in specific cancer types, notably including lung squamous cell carcinoma, and head and neck squamous cell carcinoma (Fig. 5a). While a few mutations (e.g. R324S and G325S) caused reduced XXYLT1 activity, which may lead to enhanced Notch signaling, many more cancers show amplified XXYLT1, which would lead to reduced Notch signaling. Although much work needs to be done, considering that XXYLT1 can negatively regulate Notch signaling9, our findings suggest that Notch-inhibitory xylosylation by XXYLT1 may play an important role in Notch-related tumorigenesis in specific cancer types, for example, lung squamous cell carcinoma. In this sense, the Notch-modifying XXYLT1 may be a target for anticancer drug development, and our detailed structure and function studies will facilitate such efforts. Few GT inhibitors have been developed due to the extended GT catalytic site as relatively large oligosaccharide substrates and products have to be accommodated36,37. In this regard, the recognition interface between XXYLT1 and the acceptor substrate described here may provide a new venue away from the catalytic site for small inhibitor/drug development.
Online methods
Preparation of XXYLT1 acceptor substrate Xyl-Glc-EGF
The procedure was performed as previously described19. Briefly, human factor IX (hFA9) EGF repeat was expressed in E coli and purified by Ni-NTA affinity chromatography followed by reverse phase HPLC. The purified hFA9 EGF was sequentially incubated with the enzymes (POGLUT1 and GXYLT1) together with donor substrates (UDP-Glc, Sigma-Aldrich, >98% purity) and UDP-xylose (Complex Carbohydrate Research Center, The University of Georgia, >98% purity) in order to add the disaccharide Xyl-Glc to Ser53 of the protein. For addition of the O-fucose monosaccharide (fucose) or disaccharide (N-acetylglucosamine (GlcNAc)-fucose) to Ser61 of the hFA9, recombinant enzymes (protein O-fucosyltransferase-1 and Lunatic fringe GlcNAc-trasnsferase) and appropriate donor substrates (GDP-fucose, AccendaTech, >98% purity) and/or UDP-GlcNAc (Sigma-Aldrich, >98% purity)) were added to the reaction mixture. The final product was purified by reverse phase HPLC and lyophilized. Product mass was analyzed by LC-MS/MS.
Cloning, protein expression and purification of XXYLT1
Cloning, protein expression, and purification of N-terminally truncated mouse XXYLT1 (S43–D392) were performed as previously described8. Briefly, pSecTag2c vector (Invitrogen) encoding Myc/His6-tagged mouse XXYLT1 was transiently transfected in HEK293T cells. The protein was purified from the culture media of the transfected cells by using Ni-NTA affinity chromatography, dialyzed against TBS containing 20% glycerol, and stored at –80°C until use. Protein expression was confirmed by Western blot analysis with anti-Myc antibody (Clone: 9E10, Stony Brook University, Cell Culture/Hybridoma Facility, 1:1000) and anti-β-actin antibody as control (Clone: AC-15, Abcam, 1:5000). Protein purity and concentration were estimated by Coomassie stain with BSA as standard. Computational secondary structure analysis using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) predicted a largely unstructured loop at the N-terminus spanning ~S43–P95. We therefore subjected the purified protein to limited proteolysis by a panel of ten proteases (Proti-Ace &Proti-Ace 2 kit, Hampton Research). Based on the favorable cleavage pattern, we chose trypsin for preparative treatment with protein to trypsin ratio of 500:1 (w/w) and 4°C overnight incubation. The trypsinized sample was further purified by size-exclusion chromatography (Superdex 200, GE Healthcare) in 20 mM HEPES, pH 7.5, 150 mM NaCl. Mass spectrometry detected peptides from S87 to R373 in the purified sample, indicating a stable domain, which was subsequently shown to retain the enzyme activity. The sample was concentrated to 11 mg/ml for apo-XXYLT1 crystallization.
For preparation of the binary complex of XXYLT1 and the disaccharide modified hFA9 acceptor ligand (Xyl-Glc-EGF), the purified XXYLT1 was mixed with 3 fold molar excess of Xyl-Glc-EGF, incubated at 4°C for 2 hr, then the mixture was applied to a gel filtration column (Superdex 200, GE Healthcare) to separate the binary complex from the unbound component proteins. Complex formation was confirmed by Tricine-SDS-PAGE analysis followed by silver staining. The purified binary complex was concentrated to 6 mg/ml for crystallization.
Crystallization, ligand soaking, and heavy atom soaking
For XXYLT1 crystallization, the hanging-drop diffusion method was used to produce initial micro-crystals in mother liquor containing 20 mM HEPES, pH 7.5, 1.5 M Li2SO4 after approximately 4 weeks at 20°C. Seeding improved the crystals with maximum dimensions reaching ~30 µm × 30 µm × 150–300 µm. Mn2+ at the final concentration of 20 mM was added to crystal-containing drops and incubated for 1 hr. Well solution containing 1.5 M Li2SO4 turned out to be a good cryoprotectant, so the Mn2+-soaked crystals were directly flash frozen in liquid nitrogen.
Crystals of the XXYLT1:Xyl-Glc-EGF binary complex were obtained several days after setting up the hanging-drop vapor diffusion plates at 20°C using a reservoir solution containing 0.2 M Li2SO4, 0.1 M Bis-Tris, pH 6.5, and 21% PEG3350. For ligand soaking in the XXYLT1:Xyl-Glc-EGF crystals, UDP (Sigma-Aldrich, >96%), UDP-xylose (Complex Carbohydrate Research Center, The University of Georgia, >98% purity), or UDP-Glc (Sigma-Aldrich, >98% purity) at the final concentration of 20 mM together with 20 mM Mn2+ were added to crystal-containing drops for indicated time period (10 min–1 hr) before crystals were picked up and flash-frozen in liquid nitrogen. The XXYLT1:Xyl-Glc-EGF binary complex or ligand soaked crystals were frozen in the cryo-protectant consisting of the well solution with increased PEG3350 concentration (30%).
For heavy atom derivatives of XXYLT1/ Mn2+ crystals, we screened several heavy atom compounds and found that the following conditions in the original well solution as soaking solution gave derivatized crystals that diffracted to >4 Å resolution with useful anomalous signals: (1) 1 min soaking in 0.75 M KI; (2) 10 min soaking in 10 mM K2PtCl4; (3) 4 hr soaking in 20 mM SmCl3.
X-ray diffraction data collection, structural determination and refinement
All datasets were collected at the NSLS beamline X25 or X29 or X6A in Brookhaven National laboratory at 1.1 Å wavelength, except for the heavy atom derivative datasets that were collected at the wavelength of 1.7 Å, 1.0715 Å or 1.8446 Å for KI, K2PtCl4, or SmCl3 derivatized crystals, respectively. Diffraction images were processed and scaled in HKL200038. The XXYLT1/Mn2+ crystals belonged to the space group P3121 with two molecules in the asymmetrical unit (ASU). The XXYLT1:Xyl-Glc-EGF binary complex crystals had a space group of P3 with one complex in the ASU.
Substructure determination with KI and K2PtCl4 derivative datasets both failed, likely due to a low occupancy. For the 3.6 Å SmCl3-derivative dataset, two samarium atoms in the ASU were found and the initial SAD phases with figure of merit of 0.346 were obtained using the PHENIX AutoSol program39. With the initial phase, a crude electron density map was calculated and a poly-alanine model with limited α-helixes and β-strands was manually built in COOT40. The starting model was then used to generate phase for the 3 Å XXYLT1/Mn2+ native dataset by molecular replacement with the program MOLREP41. With improved electron density map, the XXYLT1/Mn2+ model was partially built by PHENIX AutoBuild, and further corrected and completed in several iterations by manual building in COOT followed by refinement by REFMAC in CCP442,43.
The XXYLT1:Xyl-Glc-EGF binary complex structure was solved by molecular replacement with program MOLREP using the XXYLT1/Mn2+ structure as the search model. For all ligand soaked datasets, structures were determined following a similar strategy: we used XXYLT1:Xyl-Glc-EGF binary complex structure with removed Xyl-Glc disaccharide and Q330 carboxamide as search model for molecular replacement in MOLREP, followed by one round of automatic refinement in REFMAC without building the soaked ligand. Then the Fo-Fc difference maps and the 2Fo-Fc electron density maps were carefully analyzed before building the ligand(s) into the density map. Mn2+ was fit into the map first, followed by the building of donor ligand, Xyl-Glc disaccharide (or Xyl-Xyl-Glc trisaccharide) and carboxamide group of XXYLT1 Q330. After ligand building, several additional rounds of refinement were carried out in REFMAC. Water molecules were added at last. The crystallographic statistics for data collection and refinement are presented in Supplemental Table 1. The structure-related figures or movies were made using PyMOL44 or Chimera45.
Mutagenesis and enzyme activity measurement and mass spectrometry
Site-directed mutagenesis was performed by a conventional PCR-based method with the pSecTag vector encoding wild type XXYLT1 as template. The primers used in this study are presented in Supplemental Table 4. Introduced mutations were confirmed by direct DNA sequencing. The enzymatic assay with radio-labelled UDP-[14C]xylose (PerkinElmer, >97%) was performed as previously described19. Briefly, the standard 10-µl reaction mixtures contained 50 mM HEPES pH 6.8, 10 mM MnCl2, 10 µM Xyl-Glc-EGF repeat, 10 µM UDP-[14C(U)]xylose (7.14 GBq/mmol), 20 ng XXYLT1 enzymes, and 0.1% Nonidet P-40. The reaction was performed at 37°C for 20 min and stopped by adding 900 µl of 100 mM EDTA pH 8.0. The sample was loaded onto a C18 cartridge (100 mg, Agilent Technologies). After the cartridge was washed with 5 ml H2O, the EGF repeat was eluted with 1 ml of 80% methanol. Incorporation of [14C(U)]xylose into the Xyl-Glc-EGF repeats was determined by scintillation counting of the eluate. Reactions without enzymes were used as background control. Data were from three independent assays. The values indicate mean ± S.E.M.
Intrinsic fluorescence spectra of XXYLT1 and its mutants
To determine if any of the mutations disturbed the folding of the protein, the spectra of the native tryptophan residues were recorded for both the wild type and each of the mutants. Purified protein samples were prepared the day of their use at a concentration of 3 µg per 800 µl in either PBS (Invitrogen) or PBS with 2 M or 4 M guanidinium chloride (99% pure, Sigma). Freshly prepared samples were kept on ice until at least 20 min before reading, at which point they were placed in a 37°C water bath. After acclimation to 37°C, samples were transferred to quartz cuvettes in a holder that was maintained at 37°C with a circulating water bath (Neslab RTE-110). The fluorescence spectra of tryptophan were obtained using a SPEX Fluorlog τ2 with a detector connected to a liquid heat-exchanged thermoelectric cooler (Products for Research, Inc.). Tryptophan residues were excited at 280 nm, and their fluorescence was recorded for 3 sec/nm over the range 320–380 nm. Resulting spectra had their blanks, PBS with or without 2 M or 4 M guanidinium chloride, subtracted and the result was smoothed using the Savitzky-Golay algorithm. Values for fluorescence at 340 nm and 350 nm were extracted from the data. In PBS buffer, all proteins had a 340/350 nm ratio of ~1.00 and an emission maxima (λ max) of ~344 nm (Supplementary Figure 10 and Supplementary Table 2), values typical for properly folded proteins46. This suggests that these mutations had no significant effect on the solvent accessibility of tryptophan residues. As a protein unfolds, the tryptophan is more exposed and the λ max shifts to longer wavelengths47. In PBS buffer with guanidinium chloride, all proteins experienced a similar red shift as guanidinium chloride concentration increased from 0 to 4 M. In 2 M guanidinium chloride, average λ max was 348 nm with the 340/350 nm ratio of 0.96. In PBS with 4M guanidinium chloride, average λ max was 355 nm with the 340/350 nm ratio of 0.87, revealing a guanidinium chloride-concentration dependent gradual unfolding. This unfolding pattern is similar for all proteins tested. This experiment shows that the mutant proteins are as well folded as the WT.
Supplementary Material
Acknowledgments
We thank members of the Li and Haltiwanger labs for critical comments on this work. The work is supported by NIH grants GM061126 (to R.S.H) and AG029979 (to H.L.), SBU-BNL Seed Grant (to R.S.H. and H.L.), DFG grant BA4091/5-1 (to H.B.) and NSF grant DMR 1404985 (to E.L.). We acknowledge access to beamlines X25, X29, X6A at NSLS, Brookhaven National Laboratory and thank the staff at these beamlines. NSLS were supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. UDP-xylose isolation by the Carbosource Services at the Complex Carbohydrate Research Center, University of Georgia was supported in part by the U.S. Department of Energy grant DE-FG02-93ER20097. The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Footnotes
Accession codes
The coordinates have been deposited at the PDB with accession codes 4WLM, 4WM0, 4WMA, 4WMB, 4WMI, 4WMK, 4WN2 and 4WNH.
Author contributions
H.Y., H.B., R.S.H., H.L. and H.T. designed research. H.Y., M.T., J.L., J.K. and H.T. performed experiments. H.Y., J.L., E.L., R.S.H., H.L., and H.T. analyzed data and wrote the paper. J.L., J.K., and E.L. joined this project after the original submission of the manuscript, and contributed to the biophysical experiments regarding the folding status of wild type and mutated XXYLT1 proteins, which was essential for the revision of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25:318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi H, Haltiwanger RS. Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 2014;453:235–242. doi: 10.1016/j.bbrc.2014.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acar M, et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez-Valdivia R, et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138:1925–1934. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi H, et al. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16600–16605. doi: 10.1073/pnas.1109696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethi MK, et al. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J. Biol. Chem. 2010;285:1582–1586. doi: 10.1074/jbc.C109.065409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethi MK, et al. Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch. J. Biol. Chem. 2012;287:2739–2748. doi: 10.1074/jbc.M111.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TV, et al. Negative regulation of notch signaling by xylose. PLoS Genet. 2013;9:e1003547. doi: 10.1371/journal.pgen.1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 12.Breton C, Fournel-Gigleux S, Palcic MM. Recent structures, evolution and mechanisms of glycosyltransferases. Curr. Opin. Struct. Biol. 2012;22:540–549. doi: 10.1016/j.sbi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Hurtado-Guerrero R, Davies GJ. Recent structural and mechanistic insights into post-translational enzymatic glycosylation. Curr. Opin. Chem. Biol. 2012;16:479–487. doi: 10.1016/j.cbpa.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Chaikuad A, et al. Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21028–21033. doi: 10.1073/pnas.1113921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SS, et al. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat. Chem. Biol. 2011;7:631–638. doi: 10.1038/nchembio.628. [DOI] [PubMed] [Google Scholar]
- 16.Lira-Navarrete E, et al. Substrate-guided front-face reaction revealed by combined structural snapshots and metadynamics for the polypeptide N-acetylgalactosaminyltransferase 2. Angew. Chem. Int. Ed. Engl. 2014;53:8206–8210. doi: 10.1002/anie.201402781. [DOI] [PubMed] [Google Scholar]
- 17.Persson K, et al. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat. Struct. Biol. 2001;8:166–175. doi: 10.1038/84168. [DOI] [PubMed] [Google Scholar]
- 18.Errey JC, et al. Mechanistic insight into enzymatic glycosyl transfer with retention of configuration through analysis of glycomimetic inhibitors. Angew. Chem. Int. Ed. Engl. 2010;49:1234–1237. doi: 10.1002/anie.200905096. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi H, Kantharia J, Sethi MK, Bakker H, Haltiwanger RS. Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats. J. Biol. Chem. 2012;287:33934–33944. doi: 10.1074/jbc.M112.401315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor P, et al. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7290–7295. doi: 10.1073/pnas.1319683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamida S, et al. Detection of UDP-D-xylose: alpha-D-xyloside alpha 1-->3xylosyltransferase activity in human hepatoma cell line HepG2. J. Biochem. 1996;120:1002–1006. doi: 10.1093/oxfordjournals.jbchem.a021492. [DOI] [PubMed] [Google Scholar]
- 22.Wiggins CA, Munro S. Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wouters MA, et al. Evolution of distinct EGF domains with specific functions. Protein Sci. 2005;14:1091–1103. doi: 10.1110/ps.041207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao Z, et al. The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell. 1995;82:131–141. doi: 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 25.Cordle J, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitamura M, Hojo H, Nakahara Y, Ishimizu T, Hase S. Synthesis and conformational characterization of the epidermal growth factor-like domain of blood coagulation factor IX carrying xylosyl-glucose. Glycoconj. J. 2004;21:197–203. doi: 10.1023/B:GLYC.0000045092.37417.0f. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura H, Takao T, Hase S, Shimonishi Y, Iwanaga S. Human factor IX has a tetrasaccharide O-glycosidically linked to serine 61 through the fucose residue. J. Biol. Chem. 1992;267:17520–17525. [PubMed] [Google Scholar]
- 28.Lairson LL, et al. Intermediate trapping on a mutant retaining alpha-galactosyltransferase identifies an unexpected aspartate residue. J. Biol. Chem. 2004;279:28339–28344. doi: 10.1074/jbc.M400451200. [DOI] [PubMed] [Google Scholar]
- 29.Schuman B, Evans SV, Fyles TM. Geometric attributes of retaining glycosyltransferase enzymes favor an orthogonal mechanism. PLoS One. 2013;8:e71077. doi: 10.1371/journal.pone.0071077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez H, Polyak I, Thiel W, Lluch JM, Masgrau L. Retaining glycosyltransferase mechanism studied by QM/MM methods: lipopolysaccharyl-alpha-1,4-galactosyltransferase C transfers alpha-galactose via an oxocarbenium ion-like transition state. J. Am. Chem. Soc. 2012;134:4743–4752. doi: 10.1021/ja210490f. [DOI] [PubMed] [Google Scholar]
- 31.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ardevol A, Rovira C. The molecular mechanism of enzymatic glycosyl transfer with retention of configuration: evidence for a short-lived oxocarbenium-like species. Angew. Chem. Int. Ed. Engl. 2011;50:10897–10901. doi: 10.1002/anie.201104623. [DOI] [PubMed] [Google Scholar]
- 36.Jung KH, Schmidt RR. Glycosyltransferase Inhibitors. In: Wong CH, editor. Carbohydrate-Based Drug Discovery. Wiley-VCH; 2003. pp. 609–659. [Google Scholar]
- 37.Gloster TM, Vocadlo DJ. Developing inhibitors of glycan processing enzymes as tools for enabling glycobiology. Nat. Chem. Biol. 2012;8:683–694. doi: 10.1038/nchembio.1029. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 38.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 42.Pannu NS, Murshudov GN, Dodson EJ, Read RJ. Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- 43.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 45.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bram O, et al. Relaxation dynamics of tryptophan in water: A UV fluorescence up-conversion and molecular dynamics study. J. Phys. Chem. A. 2010;114:9034–9042. doi: 10.1021/jp101778u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.