Abstract
We report the synthesis and characterization of a bimetallic complex derived from a new family of potent and selective metalloinsertors containing an unusual Rh—O axial coordination. This complex incorporates a monofunctional platinum center containing only one labile site for coordination to DNA, rather than two, and coordinates DNA non-classically through adduct formation in the minor groove. This conjugate displays bifunctional, interdependent binding of mismatched DNA via metalloinsertion at a mismatch as well as covalent platinum binding. DNA sequencing experiments revealed that the preferred site of platinum coordination is not the traditional N7-guanine site in the major groove, but rather N3-adenine in the minor groove. The complex also displays enhanced cytotoxicity in mismatch repair-deficient and mismatch repair-proficient human colorectal carcinoma cell lines compared to the chemotherapeutic cisplatin, and triggers cell death via an apoptotic pathway, rather than the necrotic pathway induced by rhodium metalloinsertors.
INTRODUCTION
Platinum anticancer agents comprise an essential component in the current repertoire of chemotherapeutics. Cis-diammineplatinum(II) complexes such as cisplatin (Figure 1) and its derivatives have been extremely successful in the treatment of a variety of cancers but are also associated with a litany of severe side effects and resistance.1–5 These side effects arise primarily as a result of the mechanism by which the platinum complexes function biologically: slow displacement of cis-oriented labile leaving group ligands, such as chlorides or carboxylate groups, activates the platinum center for the formation of cytotoxic, covalent adducts with DNA.1,6 Although these complexes preferentially bind the nucleophilic N7 position of consecutive guanine residues to form what are known as 1,2-intrastrand crosslinks, the nature of the platinum binding is inherently nonspecific and can target the DNA of non-cancerous cells as well as malignant ones.2 Additionally, although DNA is widely considered to be the primary therapeutic target of cisplatin, platinum(II) complexes possess the ability to react with a number of biological ligands once inside the cell, including proteins. A major source of cisplatin resistance, for example, is the chelation and subsequent inactivation by sulfur-containing molecules, such as glutathione.7 Indeed, it is reported that only 1% of intracellular cisplatin reaches the genome.8 The ability to tune platinum therapeutics to target specific biomarkers of cancer would be invaluable in the development of next-generation platinum drugs.
Figure 1.
Chemical structures of complexes studied. [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl]3+ (left) is a bifunctional complex comprised of a trisheteroleptic rhodium metalloinsertor, which recognizes DNA mismatches, tethered to a cis-platinum (II) anticancer agent, which forms covalent adducts with DNA. [Rh(chrysi)(phen)(DPE)]2+ (center) is the rhodium metalloinsertor parent complex, which contains an unusual Rh—O axial coordination that contributes to its enhanced efficacy. Cis-dichlorodiammineplatinum (II) (right) is the FDA-approved chemotherapeutic known as cisplatin.
Our laboratory has focused largely on the development of octahedral rhodium(III) complexes for the targeted therapy of cisplatin-resistant cancers. These complexes selectively bind thermodynamically destabilized sites, such as base pair mismatches, in DNA.9 Mismatches, which arise naturally as a consequence of DNA replication, lead to cancerous mutations if left uncorrected by the complex of proteins known as the mismatch repair (MMR) machinery.10,11 As a result, deficiencies in the MMR pathway result in a buildup of these single base lesions in the genome, leading to several types of cancer. These malignancies are largely resistant to cisplatin and other classical chemotherapeutics, as MMR is involved in the recognition and processing of cisplatin-DNA lesions.12 As a result, cisplatin generally exhibits enhanced activity in MMR-proficient cells, leading to a buildup of resistance as malignant cells continue to proliferate.
Our rhodium complexes recognize DNA mismatches not through the formation of covalent adducts, as with cisplatin therapeutics, but rather through a non-covalent binding mode. These complexes bind DNA via insertion of a sterically expansive aromatic ligand, such as 5,6-chrysenequinone diimine (chrysi) (Figure 1) into the base stack of the duplex at the mismatched site. This event occurs from the minor groove, ejecting the thermodynamically destabilized, mismatched bases from the helix out towards the major groove.13–18 This binding mode, termed metalloinsertion, targets 80% of all mismatches with over 1000-fold specificity in all sequence contexts.13 More recently, we have demonstrated that these metalloinsertor complexes also target mismatches within the genome; metalloinsertors exhibit cytotoxicity preferentially in MMR-deficient cancer cells compared to isogenically-matched MMR-proficient cells, and this selectivity is correlated with localization to the nucleus rather than to the mitochondria.19–23
Rhodium metalloinsertors are a robust class of complexes that offer a promising alternative for targeting MMR-deficient cancers and circumventing resistance. New generations of metalloinsertors have exhibited increased potency surpassing that of cisplatin, while still maintaining selective targeting to MMR-deficiency.22,24 While these compounds are currently being explored as chemotherapeutic agents, they also hold promise as potential adjuvants that could confer their unique selectivity onto other therapeutic cargo. Recent efforts have focused on the development of bimetallic Rh-Pt complexes that bifunctionally target DNA through both metalloinsertion at mismatched sites as well as through the formation of covalent platinum crosslinks. Previous iterations of metalloinsertor-platinum complexes have included the conjugation of a platinum center to the rhodium complex through its inert amine ligand25 as well as the temporary attachment of the two metal centers via the labile platinum leaving group ligand.26
In our latest efforts to develop selective bifunctional conjugates, we turn to a new family of metalloinsertor complexes, developed and characterized only in the last two years. Each complex in this new generation of metalloinsertors contains an unusual ligand coordination involving an axial Rh—O bond. This coordination environment involves the bidentate N,O-chelation of a pyridylethanol ligand hinged by a quaternary carbon center, which can be functionalized with a variety of non-coordinating substituents. The coordination of anionic oxygen reduces the overall charge of the complex from [3+] to [2+], which raises the pKa of the complex; the protonated chrysi ligand thereby adopts a puckered rather than planar configuration when bound to DNA. These complexes also exhibit unprecedented potency in MMR-deficient cells, while maintaining high cell selectivity. It is purported that the unusual structure of these metalloinsertors alters their DNA binding mode; the buckled chrysi ligand potentially inserts into the base stack at a mismatch in a side-on configuration, producing a unique lesion in the genome that may be more readily recognized in MMR-deficient cells. Their biological properties as well as high functional group tolerance (with respect to the non-coordinating substituents on the N,O coordinating ligand) make these metalloinsertors promising new scaffolds for conjugate design.24 Here, we report the first-generation conjugate derived from this new family. We have synthesized a new bifunctional metalloinsertor complex, wherein a cisplatin group is attached to the previously characterized [Rh(chrysi)(phen)(DPE)]2+ (phen = 1,10-phenanthroline; DPE = 1,1-di(pyridin-2-yl)ethanol) via coordination to the extraneous pyridine. This conjugate, [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (Figure 1), thereby contains a monochloro platinum center with only one labile site available for coordination to DNA, distinguishing it from its doubly coordinating cis-platinum (II) predecessors. In this work, we demonstrate that [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ preferentially targets platinum to mismatched DNA in vitro and as a result forms unusual, nonclassical platinum adducts with adenine in the minor groove.
EXPERIMENTAL
Materials
Cisplatin and all organic reagents were purchased from Sigma-Aldrich unless otherwise noted. Commercially available chemicals were used as received without further purification. RhCl3 starting material was purchased from Pressure Chemical Co (Pittsburgh, PA). Sep-pak C18 solid-phase extraction (SPE) cartridges were purchased from Waters Chemical Co. (Milford, MA). Media and supplements were purchased from Invitrogen (Carlsbad, CA). BrdU, antibodies, and buffers were purchased in kit format from Roche Molecular Biochemical (Mannheim, Germany).
Oligonucleotides were ordered from Integrated DNA Technologies and purified by HPLC using a C18 reverse-phase column (Varian, Inc; Corona, CA). All HPLC purifications were carried out on a Hewlett-Packard 1100 HPLC. DNA purity was confirmed by MALDI-TOF mass spectrometry and quantified by UV-visible spectroscopy (UV-vis) using the extinction coefficients at 260 nm estimated for single-stranded DNA. UV-vis characterizations were performed on a Beckmann DU 7400 spectrophotometer. Radiolabeled [32P]-ATP was purchased from MP Biomedicals (Santa Ana, CA).
Synthesis
The syntheses of chrysene-5,6-dione (chrysi), 1,1-di(pyridin-2-yl)ethanol (DPE), and [Rh(chrysi)(phen)(DPE)]2+ were carried out according to published procedures.22,27,28
[Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]Cl3
A 250 ml round bottomed flask was charged with [Rh(chrysi)(phen)(DPE)]TFA2 (272 mg, 0.28 mmol) (prepared according to literature procedures) and cisplatin (305 mg, 1 mmol, 3.57 equiv) in 100 ml H2O. One drop of concentrated HCl was added, and the solution was stirred at reflux for an additional 48 h. The reaction was hot-filtered through a medium glass frit and purified by reverse-phase HPLC (85:15:0.1 to 40:60:0.1 H2O/MeCN/TFA gradient). Fractions were pooled and dried in vacuo to afford the bimetallic product as a red-brown solid. To obtain the complex as the chloride salt, [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl]TFA3 was redissolved in 50 mM HCl(aq) and freeze-dried under high vacuum. This process was repeated three times until the TFA counterion was eliminated. Yield: 60 mg (16% by HPLC). 1H NMR (500 MHz, D2O): δ 9.39 (d, J = 5.3 Hz, 1H), 8.95 (d, J = 8.0 Hz, 1H), 8.90 (d, J = 6.7 Hz, 2H), 8.87 – 8.68 (m, 1H), 8.49 – 8.35 (m, 1H), 8.34 – 8.27 (m, 1H), 8.24 – 8.18 (m, 1H), 8.14 – 8.11 (m, 1H), 8.07 (d, J = 10.3 Hz, 1H), 8.03 (s, 1H), 8.00 (d, J = 6.7 Hz, 2H), 7.96 (d, J = 8.0 Hz, 2H), 7.90 (d, J = 7.9 Hz, 1H), 7.81 (s, 1H), 7.72 (d, J = 7.5 Hz, 1H) 7.68 – 7.60 (m, 1H), 7.57 (s, 1H), 7.52 (t, J = 7.5 Hz, 1H), 7.41 (s, 2H), 7.32 (d, J = 6.8 Hz, 2H), 7.27 – 7.17 (m, 1H), 7.00 (d, J = 7.8 Hz, 1H), 3.66 – 3.59 (m, 3H), 3.54 (dd, J = 5.6, 3.5 Hz, 3H), 2.95 (s, 3H). ESI-MS (cation): m/z calc 1003.251, obs. 1001.8 (M – 2H+). UV-vis (H2O, pH 7.0): 270 nm (134,700 M−1 cm−1), 303 nm (72,400 M−1 cm−1), 442 nm (19,200 M−1 cm−1), 581 nm (10,600 M−1 cm−1).
Photocleavage Competition Titration
A 29mer DNA hairpin with the sequence 5’-GGCAGGCATGGCTTTTTGCCATCCCTGCC −3’ (underline denotes the mismatch) was labeled at the 5’-end with [32P]-ATP using polynucleotide kinase (PNK) at 37 °C for 2 h followed by purification using gel electrophoresis. A small amount of the labeled DNA (less than 1% of the total amount of DNA) was added to 2 µM DNA in 100 mM NaCl, 20 mM NaPi, pH 7.1 buffer. The DNA hairpin was annealed by heating at 90°C for 10 min and cooling slowly to ambient temperature over a period of 3 h. Racemic solutions of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ were prepared in Milli-Q water over a range of concentrations (100 nM – 50 µM). For each sample, 4 µM rac-[Rh(bpy)2chrysi]Cl3 (5 µl), which photocleaves DNA at mismatched sites, 2 µM annealed mismatched duplex DNA (10 µl), and the non-photocleaving competitor complex at various concentrations (5 µl) were combined to give 1 µM rac-[Rh(bpy)2chrysi]3+, 1 µM duplex DNA, and 50 mM NaCl(aq) as the final concentrations. Samples were irradiated on an Oriel (Darmstadt, Germany) 1000-W Hg/Xe solar simulator (340–440 nm) for 15 min, incubated at 37 °C for 10 min, and dried in vacuo. The irradiated samples were electrophoresed on a 20% denaturing polyacrylamide gel and exposed to a phosphor screen. The amounts of DNA in each band were analyzed by autoradiography and quantitated by phosphorimagery (ImageQuant).
Binding Constant Determination
To assess the binding of the rhodium subunit of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ at the CC mismatch, the fraction of cleaved DNA in each lane on the gel was quantified and expressed as a percentage of the total DNA in each lane and plotted against the log of the concentration of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+. The data from three independent titration experiments were each fit to a sigmoidal curve using OriginPro 8.5. The concentration of rhodium at the inflection point at the curve ([Rh50%]) was then used to solve simultaneous equilibria involving DNA, [Rh(bpy)2chrysi]Cl3, and [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ in Mathematica 8.0 to obtain the binding constant (KB).
Platinum Binding to Mismatched and Well-Matched DNA
A single-stranded DNA oligomer with the sequence 5*’-TTAGGATCATCCATATA-3’ (underline denotes the mismatch, asterisk denotes the radiolabel) was labeled at the 5’-end with [32P]-ATP and polynucleotide kinase (PNK) at 37 °C for 2 h. The radiolabeled DNA was purified by gel electrophoresis and annealed to either its mismatched complement (containing a CC mismatch) or a fully matched complement strand by heating to 90 °C in buffer (100 mM NaCl, 20 mM NaPi, pH 7.1), followed by slow cooling to ambient temperature over 2 h, to give a final concentration of 2 µM duplex DNA. Racemic solutions of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ were prepared in 50 mM NaCl(aq) over a range of concentrations (100 nM – 5 µM). For each sample, 2 µM annealed mismatched duplex DNA (10 µl) was mixed with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ at various concentrations (10 µl) to give 1 µM duplex DNA and 75 mM NaCl(aq) as the final concentrations. A “light” control, (ØRh, ØPt) consisting of 2 µM DNA mixed with 10 µl Milli-Q water, and a “dark” control (Ø hυ), containing the DNA mixed with the highest concentration of metalloinsertor without irradiation, were also prepared. The samples were incubated at 37 °C for periods of 1, 3, or 18h to promote the formation of the platinated DNA adducts. After the incubation period, samples were quenched with 50 µl of 0.1 M NaCl(aq) and cooled to 4 °C for 30 min. Except for the unirradiated controls, samples were irradiated on an Oriel (Darmstadt, Germany) 1000-W Hg/Xe solar simulator (340–440 nm) for 15 min and dried in vacuo. For DNA footprinting experiments, platinated DNA was precipitated with ethanol and subject to the appropriate sequencing method. The irradiated samples were electrophoresed on a 20% denaturing polyacrylamide gel and exposed to a phosphor screen. The amounts of DNA in each band were analyzed by autoradiography and quantitated by phosphorimagery (ImageQuant).
Dimethyl Sulfate Footprinting of Platinated DNA
DNA footprinting of guanine by dimethyl sulfate (DMS) was carried out according to literature procedures.29 Radiolabeled duplex DNA (well-matched or CC-mismatched; see above for sequence) was platinated with varying concentrations of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (0, 1, or 5 µM) or cisplatin (1 µM) as described above. The platination reaction was quenched via addition of 0.1 M NaCl(aq) (0.1 ml) followed by cooling to 4 °C for 30 min. Samples were purified by ethanol precipitation and dried in vacuo. The samples were taken up in 5 µl Milli-Q water, diluted with DMS buffer (50 mM sodium cacodylate, 1 mM EDTA, pH 7.5) (190 µl), and 2 mM calf-thymus DNA (4 µl) was added as a carrier DNA. Samples were cooled to 0 °C and treated with 5 µl DMS (10% v/v in EtOH, prepared immediately before use) for 5 min at 25 °C. The reaction was quenched via addition of the DMS stop solution (1.5 M NaOAc, 1 M β-mercaptoethanol, 250 µg/ml yeast tRNA) at 0 °C. Following ethanol precipitation of the DNA, samples were treated with 10% aqueous piperidine and heated to 90 °C for 30 min. The piperidine was removed in vacuo, and samples were electrophoresed on a 20% denaturing polyacrylamide gel and exposed to a phosphor screen. The amounts of DNA in each band were analyzed by autoradiography and quantitated by phosphorimagery (ImageQuant).
Methylation of Platinated DNA with Methyl Methanesulfonate (MMS)
Radiolabeled duplex DNA (well-matched or CC-mismatched; see above for sequence) was platinated with varying concentrations of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (0, 1, or 5 µM) or cisplatin (1 µM) as described above. The platination reaction was quenched via addition of 0.1 M NaCl(aq) (0.1 ml) followed by cooling to 4 °C for 30 min. Samples were purified by ethanol precipitation and dried in vacuo. The samples were taken up in 10 µl Milli-Q water, diluted with Tris-HCl buffer (10 mM Tris-HCl, 5 mM MMS, pH 7.8) (200 µl), and 2 mM calf-thymus DNA (4 µl) was added as a carrier DNA. The DNA methylation reaction was allowed to occur at ambient temperature for 16h, followed by ethanol precipitation. Strand breaks in the reacted DNA were generated by heating methylated DNA in 10 mM Tris-HCl buffer (0.1 ml) at 90 °C for 15 min to depurinate thermally labile adducts.30 Following precipitation with ethanol, DNA was then treated with 1 M piperidine at 90 °C for 30 min. Samples were then dried in vacuo, electrophoresed on a 20% denaturing polyacrylamide gel, and exposed to a phosphor screen. The amounts of DNA in each band were analyzed by autoradiography and quantitated by phosphorimagery (ImageQuant).
Analysis of Platinated DNA by Mass Spectrometry
Duplex DNA (1 µM well-matched or CC-mismatched; see above for sequence) was platinated with 5 µM [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ and incubated alongside unplatinated duplex DNA at 37 °C for 90 min. The reaction was quenched via incubation at 4 °C for 15 min, and the samples were irradiated for 15 min. The DNA was then treated with 1 M piperidine formate at 60 °C for 15 min, precipitated with ethanol at 4 °C, and depurinated with 1 M piperidine at 90 °C for 15 min. The piperidine was removed in vacuo and the dried DNA samples were analyzed by MALDI-TOF mass spectrometry.
Cell Culture
HCT116N (MMR-proficient) and HCT116O (MMR-deficient) cells were grown in RPMI medium 1640 supplemented with 10% fetal bovine serum, 400 µg/ml Geneticin (G418), 2 mM L-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 µg/ml streptomycin. Cells were grown in tissue culture flasks (Corning Costar, Acton, MA) at 37 °C under a humidified atmosphere (5% CO2).
MTT Cytotoxicity Assay
The cytotoxic effects of conjugate [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+, [Rh(chrysi)(phen)(DPE)]2+, and cisplatin were studied via MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay in the MMR-proficient HCT116N and MMR-deficient HCT116O cell lines.31 For biological experiments, [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ and cisplatin were prepared in saline solution (20 mM NaCl), and [Rh(chrysi)(phen)(DPE)]2+ was dissolved in deionized water. Cells were plated in 96-well plates at 50,000 cells/well and incubated with varying concentrations of metal complex for 72h under humidified atmosphere. After the incubation period, MTT was added, and the cells were incubated for an additional 4 h. The resulting formazan crystals were solubilized over a period of 24 h at 37 °C, 5% CO2. Formazan formation was quantified via electronic absorption at 550–600 nm with a reference wavelength of 690 nm. Cell viability is expressed as a function of formazan formation and normalized to that of untreated cells. Standard errors were calculated from 5 replicates.
MTT Caspase and PARP Inhibition Assays
The cytotoxic effects of conjugate [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ and cisplatin were studied via MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay in the HCT116O and HCT116N cell lines. Cells were plated in 96-well plates at 50,000 cells/well and incubated with 0 or 5 µM of metal complex. For caspase-inhibition assays, Z-VAD-FMK was added to a final concentration of 35 µM. For poly-ADP ribose polymerase (PARP) assays, the inhibitor 3,4-dihydro-5[4-(1-piperindinyl)butoxy]-1(2H)-isoquinoline (DPQ) was added to a final concentration of 50 µM. Controls wherein cells were treated with inhibitor alone in the absence of metal complex were included. Cells were incubated under humidified atmosphere for 72 h and labeled with MTT for an additional 4 h at 37 °C, 5% CO2. The ensuing formazan crystals were dissolved with a lysis buffer (10% SDS in 10 mM HCl) according to the manufacturer’s instructions. MTT reduction to formazan was quantified by electronic absorption at 570 nm (background: 690 nm), and percent viability was expressed as the amount of formazan in treated cells compared to that of the untreated controls.
RESULTS
DNA Binding Studies
The rhodium mismatch recognition and covalent platinum binding of DNA were analyzed with mismatched and well-matched DNA oligomers on 20% denaturing PAGE gels. Our laboratory has previously shown that rhodium metalloinsertors bearing N,O-coordinating ligands containing an axial Rh—O bond bind mismatches as both the Λ- and Δ-enantiomers with equal affinity in contrast to earlier chrysi complexes of Rh. As it has been demonstrated that [Rh(chrysi)(phen)(DPE)]2+ exhibits similar mismatch DNA binding affinities in both the racemic and enantiopure form, we carried out DNA binding experiments on racemic [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+. It is possible, however, that the addition of the platinum center increases preferential binding of one stereoisomer over the other. Mismatch recognition and platinum adduct formation can be visualized simultaneously under the same conditions (shown in Figure 2), however, platinum binding is optimally observed under saline conditions (75–100 mM NaCl(aq)). While this affords thermodynamic control over the DNA platination reaction, thereby enhancing selective platination of mismatched DNA, high salt concentrations make quantification of photocleavage at the mismatched site challenging. As a result, metalloinsertion at the mismatch was analyzed separately from platination of mismatched and well-matched DNA, under aqueous conditions.
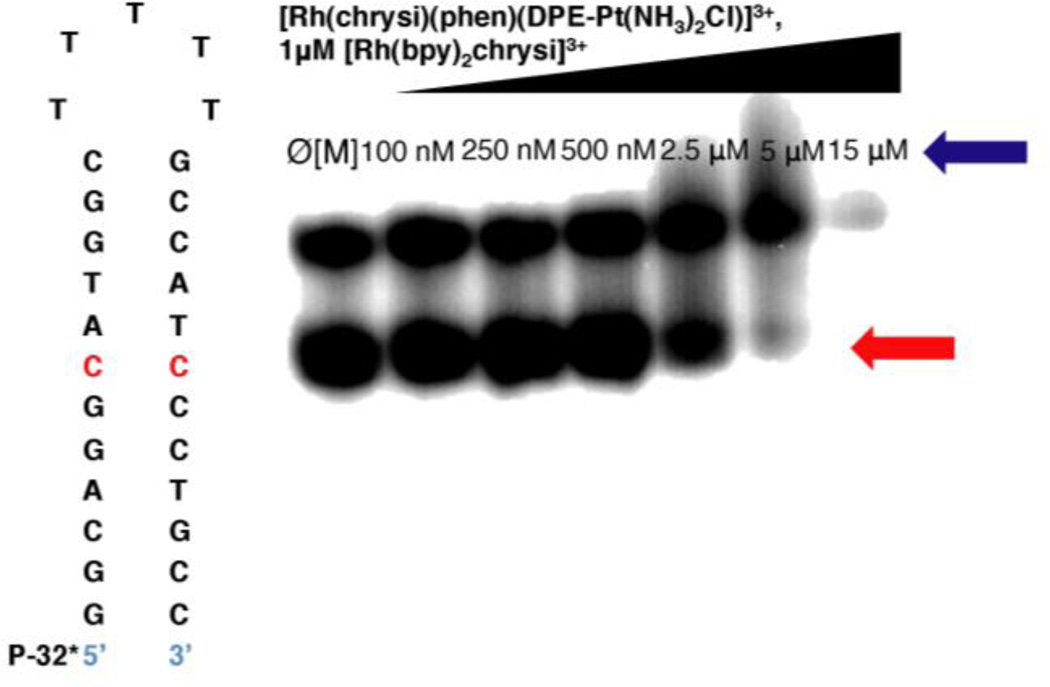
Figure 2.
Competition titration of increasing concentrations of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (0–15 µM) with 1 µM rac-[Rh(bpy)2chrysi]3+ on 1 µM 5’-[32P] labeled 29mer hairpin DNA of the sequence indicated containing a CC mismatch (denoted in red). Samples were irradiated (340–440 nm) for 15 min and electrophoresed on a 20% denaturing polyacrylamide gel. Controls without Rh were included (Ø[M]). [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ inhibits photocleavage by [Rh(bpy)2chrysi]3+ at the mismatched site. The site of photocleavage by [Rh(bpy)2chrysi]3+ at the mismatch is indicated by a red arrow at bands located below the unmodified parent band. Bands of reduced electrophoretic mobility, located above the unmodified parent DNA and indicated by a blue arrow, are indicative of covalent binding by the platinum subunit.
Binding Affinity of Rhodium at a CC Mismatch
In vitro DNA binding studies were performed with racemic aqueous solutions of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ and radiolabeled hairpin DNA containing a CC mismatch with the sequence 5*′-GGCAGGCATGGCTTTTTGCCATCCCTGCC-3′ (underline denotes the mismatch; asterisk denotes the radiolabel) Single-stranded DNA was labeled at the 5’-end with [32P]-ATP and polynucleotide kinase (PNK) at 37 °C for 2h as described above. The conjugate was bound with mismatched hairpin DNA at varying concentrations and irradiated (340–440 nm) for 15 min. Samples were then incubated at 37 °C for 10 min and electrophoresed on a 20% denaturing PAGE gel. As [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ does not cleave DNA upon irradiation, a competition titration was carried out using [Rh(bpy)2chrysi]3+, which does photocleave DNA at the site of a mismatch.9 The conjugate inhibits photocleavage by rac-[Rh(bpy)2chrysi]3+ at the mismatched site in a dose-dependent manner (Figure 2); this inhibition indicates that the complex binds specifically to the mismatch via metalloinsertion. Experimental conditions were carried out to minimize platinum adduct formation, thus limiting interference of covalent platinum binding on the equilibrium binding constant of the rhodium subunit at the mismatch. Nevertheless, some platinum binding is observed to occur simultaneously with mismatch binding, as indicated by the presence of slowly migrating bands located above the unmodified parent band. This result suggests that the complex is capable of binding mismatched DNA bifunctionally, through metalloinsertion at the mismatched site as well as the formation of covalent platinum adducts. The amount of photocleaved DNA was quantified and plotted against the logarithmic concentration of the complex (log[Rh(DPE)Pt]), and the KB value of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ was calculated by solving simultaneous equilibria at the inflection point of the titration curve (See Figure S2 of the Supporting Information). The binding affinity of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ for a CC mismatch, under conditions where platinum coordination is inhibited, was determined to be 4.8 × 106 M−1, comparable to that of monomeric metalloinsertors.20,22,23
Platination of Mismatched and Well-Matched DNA
The formation of platinum-DNA crosslinks was analyzed in vitro via denaturing polyacrylamide gel electrophoresis. Dissociation of the labile chloride ligand from the platinum center in solution enables the formation of covalent platinum adducts with DNA. The reaction between the conjugate and mismatched (CC) and well-matched duplex DNA oligomers was analyzed as a function of incubation time at 37 °C as well as complex concentration.
A time-course experiment was used to explore the formation of Pt-DNA adducts with radiolabeled duplex DNA of the sequence 5*’-TTAGGATCATCCATATA-3’ (underline denotes the site of a CC mismatch, asterisk denotes the radiolabel) annealed with either its mismatched or fully matched complement strands. Racemic mixtures of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (1 µM) and mismatched or well-matched DNA (1 µM) were incubated in buffer (75 mM NaCl, 10 mM NaPi, pH 7.1) at 37 °C for periods of either 1, 3, or 18 h. After the incubation period, samples were quenched with 0.1 M NaCl(aq), cooled to 4 °C, and electrophoresed on a 20% denaturing PAGE gel. Platination of the DNA is indicated by the appearance of bands with reduced electrophoretic mobility, located above the unmodified parent bands in the autoradiogram. The resulting autoradiogram is shown in Figure 3. The [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ conjugate exhibits a preference for mismatched DNA over fully matched oligomers after 1 and 3 h incubation periods. The 18 h incubations resulted in complete degradation of the DNA, and the bands could not be observed above background.
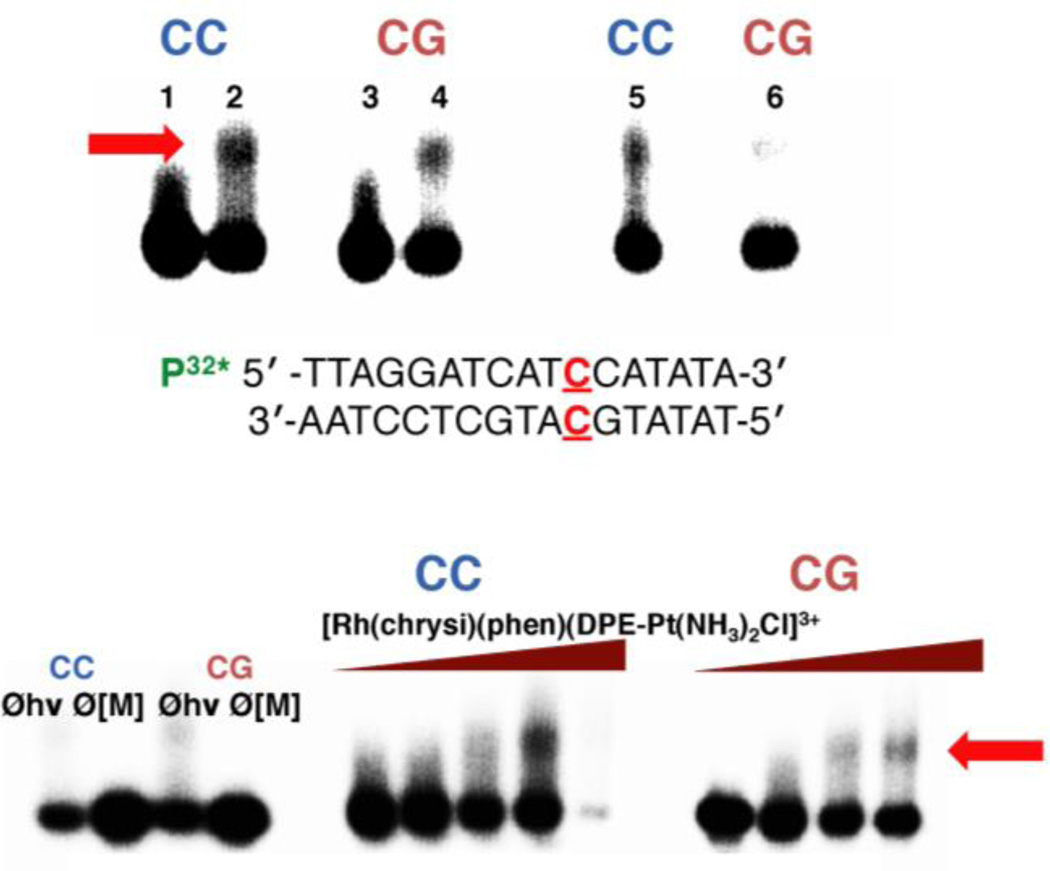
Figure 3.
Platinum binding to mismatched and well-matched DNA. Samples were irradiated for 15 min and electrophoresed on a 20% denaturing PAGE gel. Platinum crosslinking of 5’-32P-labelled DNA is indicated by the appearance of slow-moving bands located above the unmodified parent DNA; platinated DNA is denoted by a red arrow. (Top) Autoradiogram showing the formation of covalent platinum adducts with mismatched and well-matched DNA duplexes (1 µM ) as a function of time. [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (1 µM ) was incubated with 5’end radiolabeled duplex DNA of the sequence indicated (bottom; the site of the CC mismatch is denoted in red) as well as the corresponding well-matched duplex in buffer (75 mM NaCl, 10 mM NaPi, pH 7.1) at 37 °C for 1, 3, or 18h (left to right). Lanes: (1) untreated duplex DNA containing a CC mismatch; (2) mismatched DNA incubated with metal complex for 1h; (3) untreated well-matched DNA; (4) well-matched DNA incubated with metal complex for 1h; (5) mismatched DNA treated with metal complex for 3h; (6) well-matched DNA treated with metal complex for 3h. Samples treated with metal complex for 18h were degraded on the gel and are not visible in the autoradiogram. (Bottom) Autoradiogram showing the formation of covalent platinum adducts with mismatched and well-matched DNA duplexes (1 µM ) as a function of metalloinsertor concentration. Controls without irradiation (Øhν) and without metal complex (Ø[M]) were included for each type of DNA (mismatched DNA is denoted by “CC” in blue; well-matched DNA is denoted by “CG” in red) and are depicted on the left.
DNA platination was also analyzed in a dose-dependent manner, as can be seen in Figure 3. Racemic mixtures of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (0.1 – 5 µM) and mismatched or well-matched DNA (1 µM) were incubated at 37 °C for 2h and electrophoresed on a 20% denaturing polyacrylamide gel. The platinum-DNA bands were quantified by autoradiography, (See Figure S3 of the Supporting Information), revealing a preference for mismatched DNA at low concentrations (0.1 – 1 µM) of conjugate. At 500 nM [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+, 41 ± 5.4% of mismatched duplex contains platinum adducts versus 25 ± 5.3% of well-matched DNA. Optimal selectivity is achieved at stoichiometric Pt:DNA (1 µM), with 52 ± 5.1% platinated mismatched DNA versus 36 ± 5.7% (p < 0.05 by unpaired two-tailed t test). Not surprisingly, this differential platinum binding diminishes at high concentrations of the complex, where mismatched and well-matched DNA are platinated equally; at 5 µM complex, 72% and 70% platination of mismatched and well-matched DNA is observed, respectively. It would appear as though the formation of platinum crosslinks is guided at least in part by mismatch recognition by the rhodium subunit.
Determination of Platinum Binding Site
Given the preferential platination of mismatched DNA over well-matched sequences, we performed DNA sequencing reactions to probe the site of covalent binding within the duplex. Typically, the preferential DNA binding site of platinum(II) complexes is the N7 position of guanine, a major groove adduct. Methylation of N7-guanine by dimethyl sulfate (DMS) promotes site-selective depurination of these residues.29 The degree of DMS-induced guanine cleavage indicates whether platinum is coordinated; uncoordinated guanines will incur relatively high levels of cleavage upon DMS treatment, while platinated sites will be protected. Duplex DNA (1 µM) containing a single CC mismatch, as well as a similarly well-matched sequence was radiolabeled at the 5’-end with [32P] and incubated with either cisplatin (1 µM) or [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (1 or 5 µM) for 90 min at 37 °C to promote the formation of Pt-DNA adducts; untreated controls of mismatched and well-matched DNA were also included. The DNA was then purified and subjected to treatment with 10% DMS, followed by cleavage by piperidine (1 M) and denaturing gel electrophoresis (20% polyacrylamide).
The resulting autoradiogram is shown in Figure 4. The cleavage products of the two guanine residues in the radiolabeled strand are indicated by bands of high electrophoretic mobility located below the unmodified parent bands. For both mismatched and well-matched DNA, treatment with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ conjugate does not confer protection of the guanine residues from DMS methylation and cleavage. In fact, a marked increase in guanine cleavage product is observed with conjugate-bound DNA at both 1 and 5 µM treatment, compared to untreated and cisplatin-treated DNA. Furthermore, this increase is observed for both guanine residues, which occur consecutively in the sequence. However, the conjugate clearly forms covalent adducts, as is indicated by the presence of slow-migrating bands located above the unmodified parent bands. The [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ complex does not coordinate at the expected guanine sites; rather, the alternative platinum binding site likely results in a conformational change to the DNA that enhances the accessibility of both guanine residues to methylation by DMS.
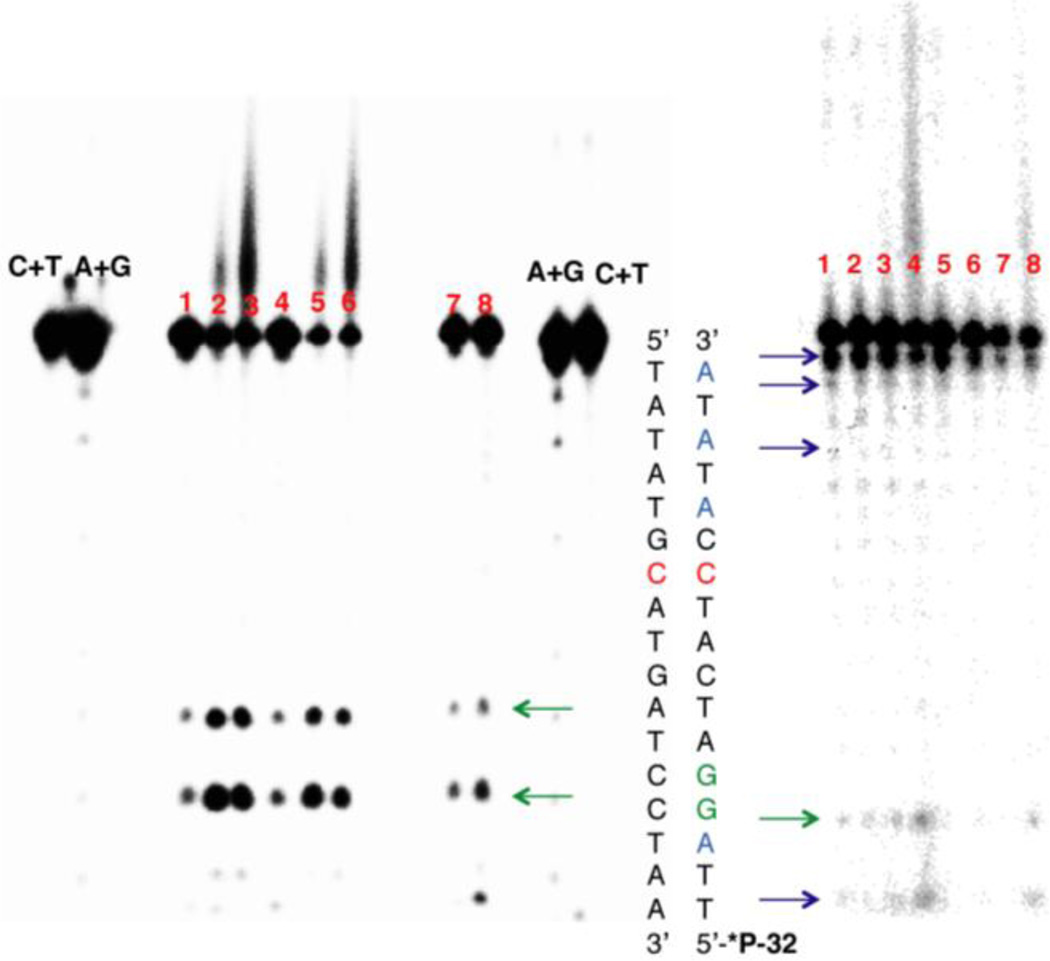
Figure 4.
DNA sequencing gels to determine the site of platinum binding. (Left) Dimethyl sulfate (DMS) footprinting of 5’-end radiolabeled duplex DNA containing a CC mismatch (denoted in red) and a d(GpG) site (denoted in green). Samples were incubated with platinum and treated with 10% DMS, followed by piperidine cleavage. Samples were electrophoresed on a 20% denaturing PAGE gel. Lanes: (1) CC-mismatched DNA in the absence of platinum; (2) mismatched DNA with 1 µM conjugate; (3) mismatched DNA with 5 µM conjugate; (4) well-matched DNA in the absence of platinum; (5) well-matched DNA with 1 µM conjugate; (6) well-matched DNA with 5 µM conjugate; (7) mismatched DNA with 1 µM cisplatin; (8) well-matched DNA with 1 µM cisplatin; Maxam-Gilbert sequencing lanes (C+T; A+G) are indicated. Bands of high electrophoretic mobility below the unmodified parent bands represent sites of guanine cleavage and are indicated by the green arrows. (Right) Methyl methanemonosulfate (MMS) footprinting of adenine residues (shown in blue) in the same sequence. Samples were incubated with platinum, treated with 5 mM MMS for 18h, and depurinated by neutral thermal hydrolysis followed by piperidine cleavage. Samples were electrophoresed on a 20% denaturing polyacrylamide gel. Sites of adenine cleavage are denoted by the blue arrows; site of guanine cleavage is denoted by the green arrow Lanes: (1) CC-mismatched DNA in the absence of platinum; (2) mismatched DNA with 1 µM cisplatin; (3) mismatched DNA with 1 µM conjugate; (4) mismatched DNA with 5 µM conjugate; (5) well-matched DNA in the absence of platinum; (6) well-matched DNA with 1 µM cisplatin; (7) well-matched DNA with 1 µM conjugate; (8) well-matched DNA with 5 µM conjugate.
As the metalloinsertion binding mode occurs from the minor groove, we considered the possibility that the platinum subunit forms minor groove adducts with DNA. Specifically, we explored platination of the N3 position of adenine, a common binding site for alkylating agents that target the minor groove due to its nucleophilicity and its relative accessibility.32 Treatment of double stranded DNA with methyl methanesulfonate (MMS) leads to methylation of N3-adenine as well as, to a lesser extent, N7-guanine in the major groove.30 As with DMS treatment, a decrease in cleavage at methylation sites is indicative of protection by platinum coordination. Mismatched and well-matched DNA duplexes were treated with either cisplatin (1 µM) or [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (1 or 5 µM) as described above and exposed to MMS (5 mM in 10 mM Tris-HCl buffer, pH 7.8) for 18 h at ambient temperature. Alkylated DNA was cleaved by heating at 90 °C followed by treatment with hot piperidine (1 M), and samples were electrophoresed on a 20% denaturing polyacrylamide gel.
Figure 4 displays the resulting autoradiogram. Note that depurination due to MMS methylation appears to occur more readily at residues near the ends of the DNA strand and less so in the middle of the sequence, likely due to accessibility. Quantitated PAGE data are shown in Figure S4 of the Supporting Information. Near the 3’-end of the radiolabeled strand, a decrease in cleavage of adenine residues occurs concurrently with the appearance of slow-migrating Pt-DNA bands. This decrease suggests that platinum coordination confers protection of these residues from methylation. At the 5’-end of the labeled strand of the mismatched sequence, an increase in cleavage is observed with increasing conjugate concentration at the adenine and guanine residues located the farthest away from the mismatched site. This effect is less pronounced for the well-matched sequence (See Figure S4 of the Supporting Information), suggesting that the duplex distortions that promote methylation by MMS at these sites in mismatched DNA are perhaps less severe when the complex is bound to well-matched DNA.
As a corollary to denaturing PAGE experiments, we also analyzed platinated DNA by mass spectrometry. Mismatched and well-matched sequences were platinated as described previously and exposed to 1 M piperidine formate at 60 °C for 15 min to depurinate the DNA. Following purification by ethanol precipitation, samples were cleaved at purine residues via treatment with 1 M piperidine at 90 °C. Samples were analyzed by MALDI-TOF mass spectrometry, and the results are summarized in Table 1. The corresponding spectra are shown in the Supporting Information. Whereas MMS footprinting primarily labeled purine residues located near the ends of the nucleic acid sequence, mass spectrometry analysis herein favors cleavage towards the middle of the sequence. Nevertheless, fragments corresponding to platinated adenine sites were observed for both mismatched and well-matched sequences.
Table 1.
MALDI-TOF Analysis of Platinated DNA Sequences
| Fragmenta | m/z Calculatedb | m/z Observedc |
|---|---|---|
| 5’-TTAGGATCATCCATATA-3’ (mismatched, parent) |
5168.4 | 5164.3 |
| 5’-TTAGGATCA-Pt-3’ (mismatched) | 3705.7 | 3696.3 |
| 5’-Pt-ATA-3’ (mismatched) | 1836.4 | 1838.8 |
| 5’-5’-TTAGGA-Pt-3’ (mismatched) | 2799.1 | 2815.8 |
| 5’- TTAGGATCATGCATATA-3’ (well-matched, parent) |
5208.5 | 5199.9 |
| 5’-Pt-ATATA-3’ (well-matched) | 2453.9 | 2456.5 |
| 5’-TTA-Pt-3’ (well-matched) | 1827.4 | 1816.0 |
Double stranded DNA comprising the sequence 5’-TTAGGATCAXCATATA-3’ and its fully matched or CC-mismatched (at site X) complement was platinated with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+, cleaved at purine residues, and analyzed by mass spectrometry. After each parent strand, fragments are presented in order of decreasing abundance for each type of DNA (mismatched or well-matched). Cleavage at platinated adenine residues is denoted in bold.
Calculated masses are derived from the mass of each DNA fragment plus the mass of the coordinated [Rh(chrysi)(phen)(DPE-Pt(NH3)2]3+ (m/z = 967.8 amu).
Mass accuracy (defined as the difference between the calculated and observed mass divided by the calculated mass) ranged from 0.08–0.62%.
MTT Cytotoxicity Assay
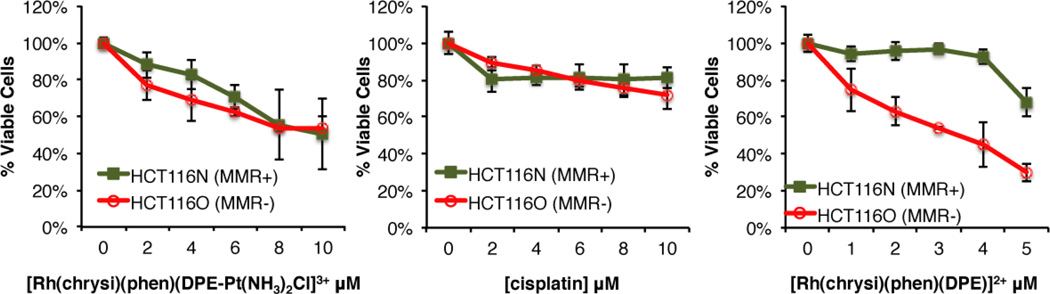
The cytotoxic effects of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ were probed via MTT assay (MTT = (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide). Metabolically active cells reduce MTT to formazan, which has a characteristic absorbance at 570 nm. Quantification of formazan by electronic absorption indicates the amount of viable cells in each sample.31 The isogenically matched human colorectal carcinoma cell lines HCT116N (MMR-proficient) and HCT116O (MMR-deficient) cells were plated in 96-well plates at 5.0 × 105 cells/well and treated with varying concentrations of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+. Cells were also treated with each parent subunit, [Rh(chrysi)(phen)(DPE)]2+ and cisplatin, for 72h under humidified atmosphere. Percent viability is defined as the ratio of the amount of formazan in treated cells to that of untreated cells. The cytotoxic effects of the complexes in the HCT116N and HCT116O cell lines are shown in Figure 5.
Figure 5.
MTT cytotoxicity assay of HCT116N (MMR-proficient) and HCT116O (MMR-deficient) cells treated with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (left), cisplatin (center), and [Rh(chrysi)(phen)(DPE)]2+ (right). Cells were incubated with each complex at the concentrations indicated for 72h. After the incubation period, cells were treated with the MTT reagent for 4 h, and the resulting formazan crystals were solubilized with acidified SDS. Percent cell viability is defined as the percentage of formazan normalized to that of untreated cells. Standard errors were calculated from 5 replicates.
As expected, the [Rh(chrysi)(phen)(DPE)]2+ parent complex displays cell-selective cytotoxicity in the MMR-deficient HCT116O line, with an IC50 value of approximately 3.5 µM. Cisplatin exhibits no effect in either cell line, possibly due to being administered from saline solution, to provide an adequate control for the [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ complex, which is also prepared in aqueous NaCl (20 mM). The conjugate displays intermediary cytotoxic effects compared to its monomeric rhodium and platinum subunits: the cell selectivity of the rhodium subunit is abolished, as both MMR-proficient and MMR-deficient cell lines are targeted equally. However, the conjugate exhibits enhanced potency compared to its platinum subunit (IC50 ≈ 10 µM), signifying that conjugation to rhodium does play some role in enhancing the efficacy of the cisplatin parent complex, either through increased cellular uptake or DNA targeting. The potency of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ is also comparable to the earlier-generation metalloinsertor-oxaliplatin conjugate, which has an IC50 value of 9 µM in the HCT116O cell line.26
Caspase and PARP Inhibition Assays
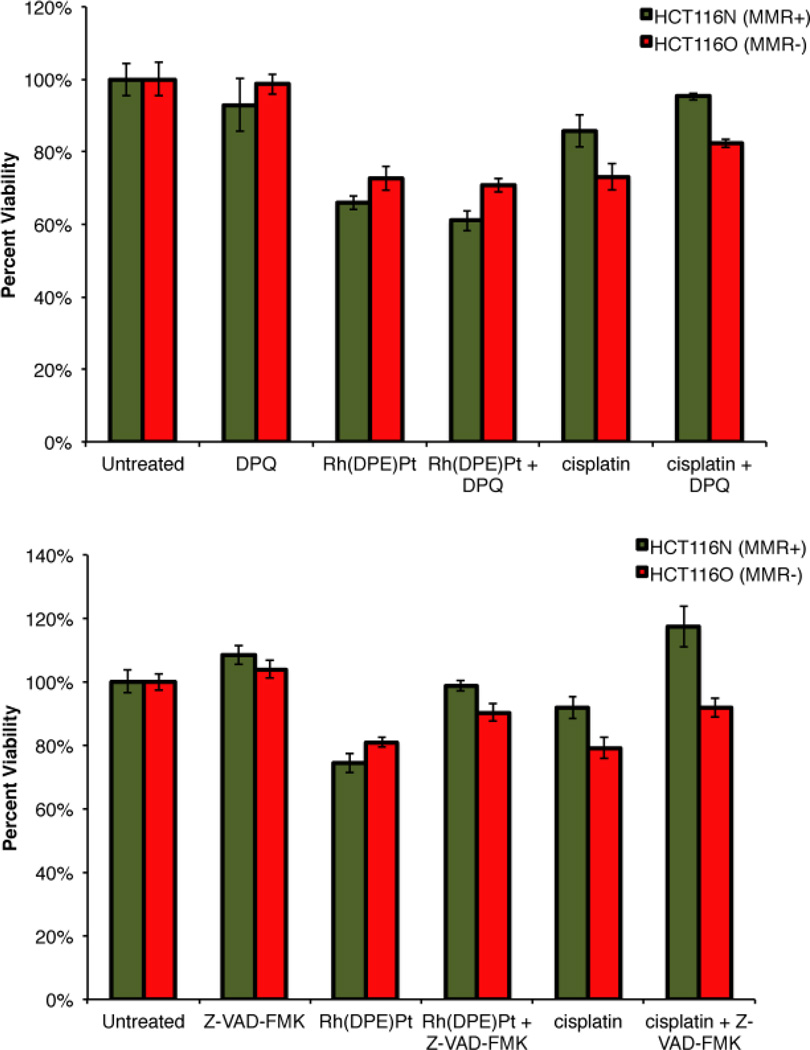
Characterization of a previous metalloinsertor-platinum conjugate revealed that the cytotoxic effects arose not from the necrotic cell death mechanism induced by monomeric metalloinsertors,21 but rather through an apoptotic pathway more characteristic of cis-platinum complexes.26,33,34 Here, we examined whether [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ also triggers apoptosis, which may account for its lack of cell-selectivity. HCT116N and HCT116O cells were treated with conjugate (5 µM) and poly-ADP ribose polymerase (PARP) inhibitor 3,4-dihydro-5[4-(1-piperindinyl)butoxy]-1(2H)-isoquinoline (“DPQ,” 50 µM)35 for 72h, and cell viability was assayed by MTT. Cells were treated similarly with DPQ (50 µM) and cisplatin (5 µM) as a control. The addition of PARP inhibitor DPQ protects cells from necrotic death, as PARP mediates this pathway through severe depletion of cellular ATP.36 As can be seen in Figure 6, treatment of both cell lines with DPQ alone effects no change in viability. Similarly, DPQ has no effect on the viability of cells treated with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+. HCT116N cells exhibit 66 ± 2.0% cell viability in the presence of conjugate alone, and 61 ± 2.8% viability with metal complex administered in combination with PARP inhibitor. For HCT116O cells, viability is 73 ± 3.4% and 71 ± 2.0% in the presence of the conjugate alone and the combination treatment, respectively. These results indicate that the cytotoxic effects of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ in HCT116 cells are independent of the PARP pathway and therefore do not proceed via necrosis.
Figure 6.
MTT cytotoxicity assay with PARP and caspase inhibitors. Viability is normalized to untreated controls. (Top) Cell viability in HCT116N (green, MMR-proficient) and HCT116O (red, MMR-deficient) cells after 72h treatment with PARP inhibitor DPQ. Treatment with DPQ (50 µM) alone has no effect on cell viability. Likewise, DPQ does not increase the viability of cells treated with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl]3+ (“Rh(DPE)Pt,” 5 µM). A modest increase in viability is observed when cells are exposed to DPQ in combination with cisplatin (5 µM). (Bottom) Cell viability in HCT116N (green, MMR-proficient) and HCT116O (red, MMR-deficient) cells after 72h treatment with caspase inhibitor Z-VAD-FMK. Treatment with Z-VAD-FMK (35 µM ) alone has no effect on cell viability. When administered in combination with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl]3+ (“Rh(DPE)Pt,” 5 µM), a statistically significant increase in viability is observed in both cell lines. A similar result is observed when capase inhibitor is added in combination with cisplatin (5 µM). These results signify caspase-dependent apoptosis (p < 0.0001 by unpaired two-tailed t-test).
Curiously, co-treatment of cells with cisplatin (5 µM) and DPQ (50 µM) results in a statistically significant increase (p < 0.0001 by unpaired two-tailed t test) in cell viability compared to treatment with cisplatin alone: the percentage of viable HCT116N cells increases from 86 ± 4.3% to 95 ± 1.0% upon the addition of PARP inhibitor, and the fraction of viable HCT116O cells increases from 73 ± 3.6% to 82 ± 1.1%. While these are modest changes overall, these results suggest that cisplatin induces necrosis in these cell lines to some degree.
The experiment was also performed in the presence of a pan-caspase inhibitor, Z-VAD-FMK. By irreversibly binding to the active site of caspases, Z-VAD-FMK inhibits apoptosis.37 Previously, it has been shown that appendage of a platinum moiety to a metalloinsertor triggers caspase-dependent cell death, signifying apoptosis rather than necrosis.26 Here, treatment of HCT116N and HCT116O cells with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ in combination with caspase inhibitor results in a similar outcome. Cells were treated with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (5 µM) or cisplatin (5 µM) in combination with Z-VAD-FMK (35 µM) for 72h, and cell viability was determined by MTT cytotoxicity assay (Figure 6).
A clear increase in cell viability upon addition of the caspase inhibitor is observed for both cell lines treated with conjugate. In fact, caspase inhibition almost completely abolishes the cytotoxic effects of the conjugate: the percentage of viable HCT116N cells increases from 74 ± 3.0% with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ alone to 99 ± 1.6% upon addition of Z-VAD-FMK, and the percentage of viable HCT116O cells is similarly enhanced from 81 ± 1.5% to 90 ± 2.7%. For both cell lines, these differences were determined to be statistically significant by unpaired two-tailed t-test (p < 0.0001). These results, in combination with the results of the MTT assay in combination with PARP inhibitor, signify that the cytotoxicity of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ is caspase-dependent and PARP-independent. [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ induces an apoptotic mode of cell death in both HCT116N and HCT116O cell lines.
The cisplatin-treated cells display similar results upon addition of Z-VAD-FMK: cell viability increases 25 ± 2.9% and 13 ± 0.3% for HCT116N and HCT116O cells, respectively, compared to treatment with cisplatin alone (p < 0.0001 by unpaired two-tailed t test). Exposure of cisplatin-treated cells to caspase inhibitor results in a markedly more dramatic increase in cell viability compared to treatment with PARP inhibitor, suggesting that while some cells may be undergoing necrotic death, the apoptotic pathway is likely the major mechanism of cisplatin cytotoxicity.
DISCUSSION
Synthesis of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+
We have synthesized a new bimetallic Rh-Pt metalloinsertor derived from a recently characterized family of complexes bearing axial Rh—O bonds.24 Metalloinsertors containing these ligands, which coordinate through a five-membered pyridylethanol ring, have been shown to exhibit enhanced potency and cell-selectivity in MMR-deficient cells.22,24 Furthermore, these complexes can accommodate a wide variety of functional groups incorporated into the N,O-coordinating ligand without sacrificing DNA binding ability or biological activity, making this class of complexes an attractive scaffold for the development of next-generation bifunctional metalloinsertor conjugates.
The metalloinsertor parent complex, [Rh(chrysi)(phen)(DPE)]2+ (Figure 1), contains a non-coordinating pyridine functionality within the pyridyl-ethanol ligand scaffold. This extraneous pyridine serves as the site of coordination for cisplatin. Simple reflux of commercially available cisplatin with the rhodium parent complex under acidic conditions displaces one of the labile chloride ligands on the platinum center, affording [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (Figure 1) in a single step in reasonable yield. This conjugate, then, contains a platinum center with only a single labilization site at the remaining chloride, and is therefore expected to form “monofunctional” platinum adducts – that is, the platinum will only coordinate a single nucleobase on the DNA, rather than binding two nearby residues and forming the classical 1,2- or 1,3-intrastrand crosslinks characteristic of the cisplatin parent complex.
Monofunctional platinum anticancer complexes, particularly those with the general structure cis-[Pt(NH3)2(L)Cl]+ (where L is an N-heterocycle), have been heavily investigated by Lippard and others.38–41 Long considered to be clinically uninteresting owing to the lack of activity of the first studied monofunctional compounds, [Pt(dien)Cl]+ (dien = diethylenetriamine) and [Pt(NH3)3Cl]+,42–44 interest in this class of complexes has been renewed in recent years with the development of more active analogues, such as pyriplatin (cis-[Pt(NH3)2(pyridine)Cl]2+)45 and the highly potent phenanthriplatin (cis-[Pt(NH3)2(phenanthridine)Cl](NO3)), which is being investigated as a new chemotherapeutic agent.46,47 These complexes have been shown to form monofunctional adducts with single bases on DNA, usually at the N7 position of guanine.38,45 Monofunctional adducts distort the DNA in a manner that is structurally distinct from that of cisplatin and other doubly-coordinating cis-platinum (II) complexes, resulting in considerably less bending and unwinding of the DNA.48–51 These complexes thus exert their anticancer activity via different biological mechanisms, providing orthogonality in the treatment of cisplatin-resistant cancers.48 In addition to the distinctive DNA binding exhibited by cis-[Pt(NH3)2(L)Cl]+ complexes, the presence of the bulky N-heterocycle protects the metal center from deactivating protein thiols as well as recognition by nucleotide excision repair proteins, which repair Pt-DNA adducts and lead to resistance.38,45,52 As a result, monofunctional, cationic platinum (II) complexes are a growing class of platinum-based drugs that can be effective against cisplatin-resistant cancers. Here, the synthesis and characterization of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ represents a monofunctional platinum complex conjugated to a rhodium metalloinsertor, as well as an example of a bifunctional conjugate developed from the Rh—O metalloinsertor family.
DNA Binding Behavior
The [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ complex was designed as were previous iterations of Rh-Pt metalloinsertor conjugates, comprising a rhodium(III) subunit coordinated to a 5,6-chrysenequinone diimine ligand for base pair mismatch recognition and a thermally activated platinum subunit for covalent DNA binding. The complex was analyzed for both mismatch binding and platinum coordination on hairpin and duplex radiolabeled DNA containing a CC mismatch. The conjugate is capable of simultaneous metalloinsertion at a mismatch and platinum adduct formation with hairpin DNA (Figure 2). Additionally, platinum binding was explored with mismatched and well-matched duplex DNA, and it was revealed that the complex preferentially binds mismatched DNA over well-matched sequences.
The preferential platination of mismatched DNA over well-matched in vitro likely results from the ability of the complex to target mismatched sites in DNA by metalloinsertion. This behavior has been shown previously in our laboratory with a metalloinsertor-cisplatin conjugate.25 However, for this earlier complex, preferential binding was found to be highly dependent on the presence and location of a d(GpG) site (the preferred binding site of cisplatin); if there was no d(GpG) site, or if it was inaccessible to the platinum center due to limitations in the length and flexibility of the alkyl tether (i.e., situated too closely to the site of the mismatch), then minimal platination occurred. Likewise, there was no preference for mismatched DNA in these scenarios. Selective DNA platination, then, is highly sequence-dependent for this complex.
The structural limitations of the first-generation metalloinsertor-platinum conjugate do not appear to be present for [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+, despite the fact that the platinum subunit is considerably more constricted in its coordination to the DPE ligand. The complex has been shown to platinate mismatched (and, to a lesser extent, well-matched) DNA in both hairpin and duplex sequences, including variation in the sequence context surrounding the site of the CC mismatch. Furthermore, in the case of the duplex DNA sequence, the d(GpG) site is located six and seven base pairs away from the mismatch – an unreachable distance for simultaneous metalloinsertion and guanine platination by a complex with virtually no separation between the subunits.
We considered the possibility that the simultaneous mismatch binding and adduct formation occur independently of one another – one equivalent binds at the mismatch, which in turn stabilizes the duplex for coordination of a second equivalent at the distal d(GpG) site. However, DNA sequencing of the guanine residues by DMS and MMS footprinting revealed that [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ does not form covalent adducts with either guanine on the radiolabeled strand. In fact, the binding of the conjugate results in an increase in the efficiency of guanine N7 methylation by alkylating agents (rather than the decrease that would be expected for platinum bound at that site), implying that the site of platination potentially distorts and/or unwinds the DNA helix in a manner that leaves the guanines more accessible to methylation.
DNA sequencing experiments with MMS revealed that [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ appears to form covalent adducts with the N3 position of adenosine residues in the minor groove, indicated by the lack of depurination at these sites in platinated DNA compared to untreated sequences. For well-matched DNA bound with [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+, a pronounced decrease in adenine cleavage, compared to untreated or cisplatin-bound DNA, is observed for the two residues located near the 3’-end. In contrast, shielding of these residues is less dramatic for mismatched DNA treated with the conjugate; however, mass spectrometry analysis suggests that platination of internal adenine residues, located closer to the mismatch but not visible in the MMS footprinting experiments, may occur more readily in this sequence. These potential differences in binding sites for mismatched versus well-matched sequences may explain the preference of the conjugate for platination of mismatched DNA: metalloinsertion by the rhodium center at the CC mismatch favorably drives the association of the platinum center with proximal adenines residue in the minor groove. Notably, an increase in depurination of the 5’-end adenine (N3) and guanine (N7) residues, located 7 and 8 base pairs away from the mismatch, respectively, is observed with increasing platinum binding. This increase is similar to the result observed with DMS treatment; platinum coordination appears to make these sites more sensitive to methylation and cleavage, possibly through distortions in the duplex that improve their accessibility to alkylating agents.
It is interesting that the conjugate appears to favor the minor groove N3-adenine adduct over the major groove N7-guanine adduct even in the absence of a mismatch (and hence in the absence of metalloinsertion in the minor groove). It is possible that the presence of the bulky rhodium metalloinsertor alters the ability of the platinum center to access the major groove in a manner that promotes coordination. Possibly, the coordination environment of the monochloro-platinum center could reduce the electrophilicity of the complex, limiting the kinetic factors that favor guanine coordination. Additionally, while the lack of guanine coordination is unusual for a DNA platinating agent, minor groove coordination of N3-adenine is not uncommon for DNA alkylating agents that are tethered to groove-specific binders. For instance, mustard-type nucleophiles conjugated with the 9-anilinoacridine intercalator favor N3-adenine adducts.53 Additionally, minor groove binder distamycin directs alkylation of adenine when tethered to methylsulfonate esters.54 Most significantly, the platinum complex PT-ACRAMTU is another monochloro cis-platinum (II) intercalator conjugate that exclusively coordinates N3-adenine. Here, the presence of a 9-aminoacridine intercalator directs the platinum center to the minor groove, where bifunctional binding occurs.32,55 Thus, for [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+, the preference of both the rhodium and platinum centers for minor groove binding of mismatched DNA represents an unusual but not unprecedented binding mode.
Characterization in Cell Tissue Culture
Encouraged by the mismatch specificity exhibited by [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ in vitro, we sought to examine whether this translated to cell-selective cytotoxicity in cancer cells deficient in mismatch repair. The cytotoxic effects of the conjugate were explored in the isogenic human colorectal carcinoma cell lines HCT116N (MMR-proficient) and HCT116O (MMR-deficient). While the conjugate does not display the potency or cell-selective targeting of HCT116O cells exhibited by its [Rh(chrysi)(phen)(DPE)]2+ parent complex, it outperforms its other parent complex, the FDA-approved chemotherapeutic cisplatin, in both cell lines. The lack of selective targeting of MMR-deficient cells is attributed to the complex triggering an apoptotic, rather than necrotic, mode of cell death as determined by cytotoxicity assays performed with caspase and PARP inhibitors. As has been seen previously, the appendage of a platinum(II) functionality circumvents the biological response to genomic mismatch recognition by metalloinsertors, resulting in a toxic but nonspecific apoptotic response.26
Although the cell-specific biological activity of the [Rh(chrysi)(phen)(DPE)]2+ metalloinsertor could not be transferred to a cytotoxic platinum subunit, [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ is quite efficacious for a monofunctional cis-platinum(II) compound. Early examples of monofunctional platinum complexes, such as [Pt(dien)Cl]+ and [Pt(NH3)3Cl]+, display no cytotoxicity in cellulo.42–44 Pyriplatin, the exploratory lead compound for monofunctional platinum complexes and a close structural analogue of the platinum subunit of [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+, also exhibits limited potency, with reported IC50 values surpassing 200 µM for HCT116 cells as well as a spectrum of various cancer cell lines.56 Indeed, aside from phenanthriplatin, there are few examples of monofunctional platinum(II) complexes that surpass cisplatin in potency, despite the evaluation of many derivations of these complexes in a variety of cancer cell lines.46 It is presently unclear precisely how the rhodium subunit enhances the efficacy of the monofunctional platinum center in this conjugate; it is possible that properties such as increased lipophilicity and charge afforded by attachment of the metalloinsertor enhances cellular uptake, as was seen for the previously reported metalloinsertor-oxaliplatin complex.26 Perhaps the bulky rhodium center shields the platinum moiety from deactivating proteins or creates a bulky lesion that blocks DNA synthesis, as is observed with phenanthriplatin.52 Future studies may probe the underlying biological mechanisms of this unusual complex.
CONCLUSIONS
Here we report the synthesis, in vitro characterization, and biological evaluation of a bimetallic Rh(III)-Pt(II) metalloinsertor conjugate that incorporates both the unusual ligand coordination of a recently characterized family of metalloinsertors as well as a monofunctional cis-[Pt(NH3)2(N-heterocycle)Cl]+ subunit. While not cell-selective, the conjugate displays increased potency compared to FDA-approved cisplatin in both cell lines studied. Moreover, the complex exhibits enhanced platination of mismatched over well-matched DNA in vitro, which arises from uncharacteristic minor-groove coordination to adenine preferentially over guanine by platinum in addition to mismatch recognition by the rhodium subunit. The results thus confirm that rhodium metalloinsertors containing axial Rh—O bonds can be developed as scaffolds for conjugation, resulting in selective targeting of their cargo towards mismatched DNA. This work also provides the foundation for exploration of non-classical platinum complexes that deviate from traditional structure-activity rules as potential mismatch-targeting agents.
Supplementary Material
Synopsis.
We have synthesized a bimetallic Rh(III)/Pt(II) DNA binding complex capable of selectively targeting mismatched DNA through both metalloinsertion of the rhodium center at the mismatched site as well as through platinum adduct formation. The platinum center forms non-classical monofunctional adducts with DNA in the minor groove, preferentially coordinating to the N3-position of adenine rather than the N7 position of guanine.
Acknowledgements
Financial support for this work from the NIH (GM033309) is gratefully acknowledged. We also thank the NIH for a training grant (NIH/NRSA 5T32GM7616-33) to A.G.W.
Footnotes
Supporting Information
Synthesis and supporting figures. This material is available free of charge via the internet at http://www.pubs.acs.org
REFERENCES
- 1.Wang D, Lippard SJ. Nature Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 2.Jamieson ER, Lippard SJ. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 3.Decatris MP, Sundar S, O’Byrne KJ. Cancer Treat. Rev. 2004;30:53–81. doi: 10.1016/S0305-7372(03)00139-7. [DOI] [PubMed] [Google Scholar]
- 4.Fink D, Aebi S, Howell SB. Clin. Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- 5.Aebi S, Fink D, Gordon R, Kim HK, Zheng H, Fink JL, Howell SB. Clin. Cancer Res. 1997;3:1763–1767. [PubMed] [Google Scholar]
- 6.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 7.Kehe K, Szinicz L. Toxicology. 2005;214:198–209. doi: 10.1016/j.tox.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Reedijk J. Chem. Rev. 1999;99:2499–2510. doi: 10.1021/cr980422f. [DOI] [PubMed] [Google Scholar]
- 9.Jackson BA, Barton JK. J. Am. Chem. Soc. 1997;119:12986–12987. [Google Scholar]
- 10.Loeb LA. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- 11.Bhattacharya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carethers JM, Hawn MT, Chauhan DP, Luce MC, Marra G, Koi M, Boland CR. J. Clin. Invest. 1996;98:199–206. doi: 10.1172/JCI118767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson BA, Barton JK. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 14.Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 15.Pierre VC, Kaiser JT, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2007;104:429. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordier C, Pierre VC, Barton JK. J. Am. Chem. Soc. 2007;129:12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeglis BM, Pierre VC, Kaiser JT, Barton JK. Biochemistry. 2009;48:4247. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song H, Kaiser JT, Barton JK. Nature Chem. 2012;4:615–620. doi: 10.1038/nchem.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart JR, Glebov O, Ernst RJ, Kirsch IR, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst RJ, Song H, Barton JK. J. Am. Chem. Soc. 2009;131:2359–2366. doi: 10.1021/ja8081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst RJ, Komor AC, Barton JK. Biochemistry. 2011;50:10919–10928. doi: 10.1021/bi2015822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komor AC, Schneider CJ, Weidmann AG, Barton JK. J. Am. Chem. Soc. 2012;134:19223–19233. doi: 10.1021/ja3090687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidmann AG, Komor AC, Barton JK. Philos. Trans. R. Soc. A. 2013;371:20120117. doi: 10.1098/rsta.2012.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komor AC, Barton JK. J. Am. Chem. Soc. 2014;136:14160–14172. doi: 10.1021/ja5072064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petitjean A, Barton JK. J. Am. Chem. Soc. 2004;126:14728–14729. doi: 10.1021/ja047235l. [DOI] [PubMed] [Google Scholar]
- 26.Weidmann AG, Barton JK. Inorg. Chem. 2014;53:7812–7814. doi: 10.1021/ic501509x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu A, Bhaduri S, Sapre NY, Jones PG. J. Chem. Soc., Chem. Commun. 1987;22:1724–1725. [Google Scholar]
- 28.Muerner H, Jackson BA, Barton JK. Inorg. Chem. 1998;37:3007–3012. [Google Scholar]
- 29.Brabec V, Leng M. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5345–5349. doi: 10.1073/pnas.90.11.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly JD, Shah D, Chen F-X, Wurdeman R, Gold B. Chem. Res. Toxicol. 1998;11:1481–1486. doi: 10.1021/tx9801763. [DOI] [PubMed] [Google Scholar]
- 31.Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 32.Guddneppanavar R, Bierbach U. Anticancer Agents Med. Chem. 2007;7:125–138. doi: 10.2174/187152007779313991. [DOI] [PubMed] [Google Scholar]
- 33.Siddik ZH. Oncogene. 2003;22:7265. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 34.Arango D, Wilson AJ, Shi Q, Corner GA, Arañes MJ, Nicholas C, Lesser M, Mariadason JM, Augenlicht LH. Br. J. Cancer. 2004;91:1931. doi: 10.1038/sj.bjc.6602215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costantino G, Macchiarulo A, Camaioni E, Pellicciari R. J. Med. Chem. 2001;44:3786–3794. doi: 10.1021/jm010116l. [DOI] [PubMed] [Google Scholar]
- 36.Ha HC, Snyder SH. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandenabeele P, Vanden Berghe T, Festjens N. Sci. STKE. 2006;358:e44. doi: 10.1126/stke.3582006pe44. [DOI] [PubMed] [Google Scholar]
- 38.Johnstone TC, Wilson JJ, Lippard SJ. Inorg. Chem. 2013;52:12234–12249. doi: 10.1021/ic400538c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollis LS, Amundsen AR, Stern EW. J. Med. Chem. 1989;32:128–136. doi: 10.1021/jm00121a024. [DOI] [PubMed] [Google Scholar]
- 40.Baird CL, Griffitts AE, Baffic S, Bryant P, Wolf B, Lutton J, Berardini M, Arvanitis GM. Inorg. Chim. Acta. 1997;256:253–262. [Google Scholar]
- 41.Sundquist WI, Bancroft DP, Lippard SJ. J. Am. Chem. Soc. 1990;112:1590–1596. [Google Scholar]
- 42.Macquet JP, Butour JL. J. Natl. Cancer Inst. 1983;70:899–905. [PubMed] [Google Scholar]
- 43.Brabec V, Reedijk J, Leng M. Biochemistry. 1992;31:12397–12402. doi: 10.1021/bi00164a014. [DOI] [PubMed] [Google Scholar]
- 44.Bursova V, Kasparkov J, Hofr C, Brabec V. Biophys. J. 2005;88:1207–1214. doi: 10.1529/biophysj.104.051771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovejoy KS, Todd RC, Zhang S, McCormick MS, D’Aquino JA, Reardon JT, Sancar A, Giacomini KM, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8902–8907. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park GY, Wilson JJ, Song Y, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11987–11992. doi: 10.1073/pnas.1207670109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnstone TJ, Park GY, Lippard SJ. Anticancer Res. 2014;34:471–476. [PMC free article] [PubMed] [Google Scholar]
- 48.Lempers ELM, Bloemink MJ, Brouwer J, Kidani Y, Reedijk J. J. Inorg. Biochem. 1990;40:23–25. doi: 10.1016/0162-0134(90)80037-x. [DOI] [PubMed] [Google Scholar]
- 49.Bellon SF, Lippard SJ. Biophys. Chem. 1990;35:179–188. doi: 10.1016/0301-4622(90)80007-t. [DOI] [PubMed] [Google Scholar]
- 50.Chottard JC, Ledner JA, Bauer WA, Ushay HM, Caravana C, Lippard SJ. J. Am. Chem. Soc. 1980;102:2487–2488. [Google Scholar]
- 51.Keck MV, Lippard SJ. J. Am. Chem. Soc. 1992;114:3386–3390. [Google Scholar]
- 52.Wang D, Zhu G, Huang X, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9584–9589. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan JY, Ohms SJ, Boyd M, Denny WA. Chem. Res. Toxicol. 1999;12:1166. doi: 10.1021/tx990089q. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Chen FX, Mehta P, Gold B. Biochemistry. 1993;32:7954. doi: 10.1021/bi00082a017. [DOI] [PubMed] [Google Scholar]
- 55.Rao L, West TK, Saluta G, Kucera GL, Bierback U. Chem. Res. Toxicol. 2010;23:1148–1150. doi: 10.1021/tx100170p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lovejoy KS, Serova M, Bieche I, Emami S, D’Incalci M, Broggini M, Erba E, Gespach C, Cvitkovic E, Faivre S, Raymond E, Lippard SJ. Mol. Cancer Ther. 2011;10:1709–1719. doi: 10.1158/1535-7163.MCT-11-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.