Abstract
IscR proteins are known as transcriptional regulators for Fe–S biogenesis. In the facultatively phototrophic bacterium, Rhodobacter sphaeroides IscR is the product of the first gene in the isc-suf operon. A major role of IscR in R. sphaeroides iron-dependent regulation was suggested in a bioinformatic study (Rodionov et al., PLoS Comput Biol 2:e163, 2006), which predicted a binding site in the upstream regions of several iron uptake genes, named Iron-Rhodo-box. Most known IscR proteins have Fe–S clusters featuring (Cys)3(His)1 ligation. However, IscR proteins from Rhodobacteraceae harbor only a single-Cys residue and it was considered unlikely that they can ligate an Fe–S cluster. In this study, the role of R. sphaeroides IscR as transcriptional regulator and sensor of the Fe–S cluster status of the cell was analyzed. A mutant lacking IscR is more impaired in growth under iron limitation than the wild-type and exhibits significantly increased ROS levels in iron-replete and iron-deplete conditions. Expression studies reveal that R. sphaeroides IscR in its cluster-bound form functions as transcriptional repressor of genes involved in iron metabolism by direct binding to the promoter region of genes preceded by the motif. A total of 110 genes are directly or indirectly affected by IscR. Furthermore, IscR possesses a unique Fe–S cluster ligation scheme with only a single cysteine involved.
Keywords: Fe–S proteins, iron, Iron-Rhodo-box, iron-sulfur cluster, IscR, Rhodobacter sphaeroides
Introduction
Iron-sulfur (Fe–S) clusters are ensembles of iron and sulfide centers. The most abundant types of Fe–S clusters found in nature are the rhombic [2Fe–2S] and the cubane [4Fe–4S] clusters which contain either ferrous (Fe2+) or ferric (Fe3+) iron and sulfide (S2−) (Drennan and Peters 2003; Johnson et al. 2005). These clusters are typically attached to their protein partners, called Fe–S proteins, via their iron atoms, which are preferentially bound to the sulfur atoms in the cysteine residues of the peptide backbone. However, an interesting feature of Fe–S clusters is the broad diversity of ligands that the iron atoms can ligate. There are several examples of nitrogen coordination, provided by histidine or arginine residues (Peters et al. 1998; Nicolet et al. 1999; Berkovitch et al. 2004), and oxygen coordination, from aspartate, glutamine or tyrosine (Calzolai et al. 1995; Dobritzsch et al. 2001). Furthermore, there are examples of coordination by exogenous ligands, such as water, enzyme substrates or cofactors (such as S-adenosylmethionine) (Flint and Allen 1996; Cheek and Broderick 2001; Berkovitch et al. 2004; Fontecave 2006).
Although Fe–S clusters are easily assembled in vitro with inorganic iron and sulfur sources in a reductive environment (Malkin and Rabinowitz 1966), the in vivo situation is more complex and requires Fe–S biogenesis systems. Three different types of bacterial Fe–S biogenesis systems, Isc (iron-sulfur cluster), Suf (sulfur formation) and Nif (nitrogen fixation) have been extensively characterized (Fontecave 2006; Py and Barras 2010). Such Fe–S assembly systems are essential for virtually all living organisms and are important for the activity of many enzymes involved in diverse cellular processes, including respiration, DNA synthesis and repair, gene regulation, RNA modification, and nitrogen and carbon metabolism (Py and Barras 2010). Thanks to their chemical versatility, Fe–S clusters can act as catalysts or redox sensors and are known or predicted to be used by a large number of proteins (e.g. over 150 in Escherichia coli; Py and Barras 2010). However, the increase in oxygen after the emergence of oxygenic photosynthesis created a threat to Fe–S proteins and, consequently, to the organisms relying on them (Imlay 2006). In particular, reactive oxygen species (ROS) cause destabilization of Fe–S cluster, leading to release of Fe2+ ions that in turn fuel ROS production by Fenton chemistry. Furthermore, iron switches from the soluble (0.1 mol/L at pH 7.0) ferrous state to the extremely insoluble (10−18 mol/L at pH 7.0) ferric form (Andrews et al. 2003). So, in oxic conditions, iron is both poorly available and potentially toxic. Therefore, bacteria have evolved mechanisms to maintain a precise intracellular iron concentration, including iron storage proteins and a general ferric iron buffering system (such as Fe–S cluster). Several regulators of iron metabolism have been extensively investigated in bacteria. In E. coli and in many other bacteria the Fur protein is a major regulator for iron-dependent gene expression (reviewed in e.g. Hantke 2001). Iron regulation in α-proteobacteria mainly occurs by regulators different from this type of Fur protein, namely by Irr, Fur/Mur, RirA, or IscR (Johnston et al. 2007; Peuser et al. 2012). In Rhodobacter capsulatus, the LysR-type transcriptional regulator HbrL is a crucial regulator for the control and coupling of heme synthesis with iron homeostasis (Zappa and Bauer 2013). While no RirA homolog is found in Rhodobacteraceae, previous data revealed that in Rhodobacter sphaeroides, neither Fur/Mur nor Irr appears as a master regulator of iron homeostasis (Peuser et al. 2011, 2012). Whether putative HbrL homologs play an important role in R. sphaeroides remains to be elucidated.
In Rodionov et al. (2006) identified a highly conserved 19-bp palindromic signal, which was termed Iron-Rhodo-box. This motif occurs in upstream regions of most iron uptake and storage genes in all 12 at that time available genome sequences of the Rhodobacteraceae group. The Iron-Rhodo-box motif has a significant similarity to the RirA-box motif in the Rhizobiales. However, the absence of a RirA homolog suggests that another transcription factor mediates the global control of iron transport genes. Rodionov et al. (2006) therefore hypothesized a potential major role of IscR in R. sphaeroides iron-dependent gene regulation. IscR belongs to the Rrf2 superfamily of transcriptional regulators and contains a helix-turn-helix DNA-binding domain. The Rrf2 family members are not well characterized in α-proteobacteria, but the presence of conserved Cys residues in several of them suggests that a subset of these proteins may ligate Fe–S clusters. In many organisms, IscR acts as both, a sensor of cellular Fe–S level and as a global transcriptional regulator for Fe–S biogenesis (Giel et al. 2006; Lee et al. 2008; Giel et al. 2013). Genome-wide transcript profiling showed that IscR regulates expression of at least 40 genes in E. coli, among which are the isc operon encoding IscR itself and the Fe–S cluster biogenesis genes (suf genes) (Schwartz et al. 2001; Giel et al. 2006). Other regulated genes encode both, Fe–S proteins as well as non-Fe–S proteins, suggesting an important role of IscR as global regulator (Giel et al. 2006; Loiseau et al. 2007; Angelini et al. 2008; Wu and Outten 2009). Furthermore, IscR acts as a sensor of the cellular demands for Fe–S cluster biogenesis (Fleischhacker et al. 2012). Most Fe–S proteins ligate Fe–S clusters via four Cys, while IscR proteins typically have Fe–S clusters featuring (Cys)3(His)1 ligation (Fleischhacker et al. 2012). In E. coli, IscR exists in two major forms: apo-IscR lacks the Fe–S cluster while the holo-protein is ligated to the Fe–S cluster. Apo-IscR signals that the cell is in need of Fe–S clusters and derepresses transcription of the isc operon. On the other hand, holo-IscR signals that other Fe–S proteins are momentarily oversaturated and blocks the transcription of the isc operon (Yeo et al. 2006). Since IscR only contains three Cys residues, IscR exhibits a decreased affinity for Fe–S cluster and is therefore only able to bind the clusters when other proteins do not require them (Schwartz et al. 2001). According to this, upon oxidative stress conditions, the Fe–S of IscR is likely one of the first clusters destroyed. Recent studies have shown that in E. coli all three conserved Cys residues are essential for the formation of the holo-protein (Yeo et al. 2006; Nesbit et al. 2009). Interestingly, the IscR proteins from Rhodobacteraceae harbor only a single-Cys residue. This difference in the primary structure of iscR raised the possibility that IscR proteins in Rhodobacteraceae cannot ligate an Fe–S cluster.
Rhodobacter sphaeroides is a facultative phototrophic bacterium that forms photosynthetic complexes at low oxygen tension or anoxic conditions. A global transcriptome analysis in the background of a ∆iscR strain revealed that IscR functions as transcriptional repressor of genes preceded by a specific DNA-binding motif (Iron-Rhodo-box). Furthermore, we confirmed that despite the marked differences in sequence IscR from R. sphaeroides coordinates an iron sulfur center and provide first hints to amino acids involved in this ligation.
Materials and Methods
All strains, plasmids, and oligonucleotides used in this study are listed in Tables S1–S3 of the supplementary data.
Bacterial strains and growth conditions
Escherichia coli strains were grown in Luria–Bertani medium at 37°C with shaking (180 rpm) or on solid growth medium, which contained 1.6% (w/v) agar. Rhodobacter sphaeroides strains were cultivated at 32°C in 50-mL Erlenmeyer flasks containing 40 mL malate minimal medium (Remes et al. 2014) with continuous shaking at 140 rpm, resulting in a constant dissolved oxygen concentration of ∼25–30 μmol/L during the exponential phase. Conditions of iron limitation were achieved as described previously (Peuser et al. 2011; Remes et al. 2014). When required, antibiotics were added to liquid or solid growth media at the following concentrations: spectinomycin (10 μg mL−1); kanamycin (25 μg mL−1); tetracycline (2 μg mL−1) (for R. sphaeroides); kanamycin (25 μg mL−1); and tetracycline (20 μg mL−1) (for E. coli).
Construction of a R. sphaeroides iscR deletion mutant
Rhodobacter sphaeroides strain 2.4.1ΔiscR was generated by transferring the suicide plasmid pPHU2.4.1ΔiscR:Sp into R. sphaeroides 2.4.1, and screening for insertion of the spectinomycin resistance cassette into the chromosome by homologous recombination. Briefly, parts of the iscR gene (RSP_0443) of R. sphaeroides, together with upstream and downstream sequences, were amplified by polymerase chain reaction (PCR) using oligonucleotides 0443_upA/0443_upB and 0443_downA/0443_downB. The amplified PCR fragments were cloned into the EcoRI-BamHI and BamHI-HindIII sites of the suicide plasmid pPHU281 (Hubner et al. 1991), generating the plasmid pPHU2.4.1ΔiscR. A 2.0 kb BamHI fragment containing the spectinomycin cassette from pHP45Ω (Prentki et al. 1991) was inserted into the BamHI site of pPHU2.4.1ΔiscR to generate pPHU2.4.1ΔiscR::Sp. This plasmid was transferred into E. coli strain S17-1 and diparentally conjugated into R. sphaeroides 2.4.1 wild-type strain. Conjugants were selected on malate minimal salt agar plates containing spectinomycin. By insertion of the spectinomycin cassette, 287 bp of the 468 bp R. sphaeroides iscR gene were deleted.
Complementation of the R. sphaeroides deletion mutant 2.4.1∆iscR
For complementation of the iscR deletion mutant of R. sphaeroides a 639 bp PCR fragment containing the entire iscR gene along with 104 bp of the upstream region and 74 bp of the downstream region was amplified by using the oligonucleotides IscR_complA and IscR_complB (Table S3). Following digestion with BamHI and XbaI, the fragment was cloned into the corresponding sites of pBBR1-MCS-2, resulting in plasmid pBBRiscR. To complement the iscR deletion in the wild-type strain 2.4.1, the plasmid pBBRiscR was transferred into E. coli S17-1 and conjugated into the 2.4.1∆iscR strain by biparental conjugation.
Cloning and expression of recombinant IscR protein from R. sphaeroides
Oligonucleotides IscR-His_fwd and IscR-His_rev (Table S3) were used for amplifying the coding region of iscR. The purified 497-bp PCR product was digested with BamHI and HindIII and ligated into the pQE30 cloning vector (Qiagen, Hilden, Germany) to yield plasmid pQE30::iscR. This plasmid was transformed into E. coli M15 cells and overexpressed as described earlier (Rische and Klug 2012). Aliquots of the fractions were separated by 10% sodiumdodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After visualization with silver staining, the protein was determined as pure, since there was only one major band visible.
For site-directed mutagenesis of residues in IscR the plasmid pQE30::iscR was used as PCR template. Mutations were inserted with the following primers by inverse PCR: IscR_H93A_A, IscR_H93A_B, IscR_H121A_A, IscR_H121A_B, IscR_H127A_A, IscR_H127A_B, IscR_C142A_A, IscR_C142A_B, IscR_P143A_A, and IscR_P143A _B (Table S3). IscR_H93A carries a mutation at position 277–278 (CA to GC), IscR_H121A carries a mutation at position 361–362 (CA to GC), IscR_H127A carries a mutation at position 389–390 (CA to GC), IscR_C142A carries a mutation at position 424–425 (TG to GC), and IscR_P143A carries a mutation at position 427 (C to G). The mutated clones were selected and confirmed for mutations by nucleotide sequencing. After transformation with E. coli S17-1, the resulting plasmids were transferred to R. sphaeroides by diparental conjugation, yielding the corresponding reporter strains (Table S1).
Reconstitution of iron-sulfur clusters in IscR
The chemical reconstitution of IscR was performed after Ni-NTA purification with a 100-μmol/L protein solution under strictly anaerobic conditions in a Coy anaerobic chamber. Reconstitution was performed as described elsewhere (Fluhe et al. 2012). Incubation overnight in ammonium iron citrate and lithium sulfide resulted in a dark brown solution.
Construction of a cfp reporter system
For construction of the fluorescence based in vivo reporter system we used a vector system designed for fluorescence based in vivo localization studies. Therefore we originally inserted the multiple cloning site (MCS) of pET28(a) (Novagen, Darmstadt, Germany), by use of the restriction sites XbaI and XhoI, into the broad host range vector pBBR1MCS2 (Kovach et al. 1995) using the same restriction sites. Thereby a ribosomal-binding site (RBS), His-tag and thrombin cleavage site were transferred from pET28(a) to pBBR1MCS2 and the derived vectors were renamed to pBE (B = pBBR1MCS2, E = pET28). A regulative DNA element for strong constitutive expression (RSP_4352 upstream sequence, see Mank et al. 2012) was inserted with XbaI, 5′to the RBS (pBE4352). A first eCFP DNA fragment without stop codon was inserted with the help of NdeI and EcoRI restriction sites (pBE4352::eCFP), a second eCFP DNA fragment with stop codon was inserted by EcoRI and HindIII 3′to the RBS (pBE4352::eCFP:eCFP). The exchange of one of the present eCFP fragments with a sequence of interest, the exchange of the constitutive promoter by a promoter of interest, or a constitutive over expression of N- or C-terminally tagged eCFP fusions are possible.
The promoter region of IscR (PiscR) was used for cfp fusion on plasmid pBE4352::eCFP:eCFP. A DNA fragment with a length of 169 bp was amplified using primers iscR_repA and iscR_repB. The resulting fragment contains positions −169 to −1with respect to the start codon and includes a predicted IscR interaction site (−119 to −100). Primers (Table S3) generated XbaI/BamHI restriction sites in the corresponding PCR product, which were sub-cloned into the pJET1.2 cloning vector (Thermo Fisher Scientific, Waltham, MA, USA) and after digestion with XbaI/BamHI inserted into pBE4352::eCFP:eCFP. The resulting reporter plasmid pBE::PiscR::eCFP was used for transformation with E. coli S17-1 and subsequently transferred to R. sphaeroides by diparental conjugation, yielding the corresponding reporter strain (Table S1).
UV-visible spectroscopy analysis and fluorescence measurements
The UV-visible spectroscopy analyses of the IscR variants were recorded on a Spectral photometer SPECORD 50 (Analytic Jena, Jena, Germany). Equal amount of proteins were analyzed immediately after protein purification to avoid the oxidation of the Fe–S cluster of the IscR proteins.
For fluorescence measurements a plate reader from the Tecan Infinity M200 series (Tecan, Männedorf, Switzerland) was used. ROS generation was measured using an oxidation-sensitive fluorescent probe, 2,7-dihydrodichlorofluorescein diacetate (DCFH-DA; Thermo Fisher Scientific, Waltham, MA, USA). Exponential cells were incubated with the probe at a final concentration of 10 μmol/L for 30 min. The excitation wavelength was 492 nm, emission wavelength was 525 nm. For cfp-based fluorescence measurements the excitation wavelength was 434 nm, the emission wavelength was 480 nm. For both methods the reading mode was top with 0 μsec lag time, 20 μsec integration time, 25 reads, and 0 msec settle time.
EMSA with dsDNA substrates and IscR protein
For electromobility shifts (EMSAs) the putative regulatory regions (about 200–300 bp) of the respective genes were amplified using the pairs of oligonucleotides shown in Table S3 and were end labeled with [γ32P]-ATP using T4 polynucleotide kinase (Fermentas). Binding reactions were carried out in a final volume of 15 μL and contained the indicated amount of protein, ∼10 fmol [γ32P]ATP-labeled DNA probe (10,000 c.p.m.), salmon sperm DNA (1 mg), and 7.5 μL of a binding buffer as described elsewhere (Wu and Outten 2009). The binding reaction was performed at 4°C for 30 min. Samples were loaded on a 6% (w/v) nondenaturing polyacrylamide gel in TBE buffer (22 mmol/L Tris-HCl, 22 mmol/L boric acid, 0.5 mmol/L EDTA (ethylenediaminetetraacetic acid), pH 8.3). Electrophoresis was done at 4°C at 200 V for 4 h.
RNA isolation and quality assignment
Rhodobacter sphaeroides cultures were grown in the presence or absence of iron in triplicate cultures inoculated separately from three independent starter cultures. RNA isolation for quantitative real-time RT-PCR or microarray analysis was performed as previously described (Remes et al. 2014).
Microarray analysis
Microarray analysis was performed as described before (Peuser et al. 2011). In brief, 2 μg of total RNA of three independent experiments of strains wild-type 2.4.1 and 2.4.1∆iscR was chemically labeled with Cy3 and Cy5, respectively. Transcriptome profiles were analyzed on two arrays including six biological replicates. Differentially labeled RNA samples were mixed and competitively hybridized to microarrays. Hybridizations and scanning were performed according to the specifications from Agilent (Böblingen, Germany). Multiarray analysis and normalization according to LOESS were accomplished with the Bioconductor package Limma for R and performed as described elsewhere (Smyth and Speed 2003; Ritchie et al. 2007). On the basis of calculated MA plots, genes were considered reliable if the average signal intensity [A-value: 1/2 log2 (Cy3 × Cy5)] was ≥12. Fold changes were calculated using MS Excel (Microsoft Corp. Redmond, WA, USA). The data shown in this study represent the results from two individual microarrays (biological replicates), each containing a pool of three independent experiments for each sample. The microarray data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE65537 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65537).
Quantitative real-time RT-PCR
The One-Step Brilliant III QRT-PCR Master Mix Kit (Agilent) was used for reverse transcription followed by PCR as described in the manufacturer's manual. RT-PCR samples containing 4 ng of total RNA per μL were run in a Rotor-Gene 3000 real-time PCR cycler (Qiagen, Hilden, Germany) for relative quantification of mRNAs in each of the three independent experiments. Primers are listed in Table S3. Crossing points (Cp) with a fluorescence threshold of 0.002 were visualized with the Rotor-Gene software 6.0 (Corbett Research). The relative mRNA levels were normalized to the housekeeping gene rpoZ and calculated according to Pfaffl (Pfaffl 2001).
Homology modeling
The sequence of R. sphaeroides IscR was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto 2000). IscR has 155 amino acid residues and the accession number is RSP_0443. Comparative modelling was done by searching the protein data bank (PDB) for known protein structures using the sequence of IscR as the query with the program MODELLER (Sali and Blundell 1993). The search was executed using BLASTp, and the results provided three PDB-related potential templates for modelling. The templates are 2Y75, CymR, the global cysteine regulator of Bacillus subtilis (Shepard et al. 2011), 4CIC, the transcriptional regulator from Thermincola potens (Santos et al. 2014) and 4HF1, HTH-type transcriptional regulator IscR of E. coli (Rajagopalan et al. 2013).
Results
Organization of isc and suf genes in a single operon in R. sphaeroides
In contrast to the situation in E. coli, the isc and suf genes that code for Fe–S cluster assembly proteins are in R. sphaeroides colocalized on the chromosome. These genes encode a regulator (IscR), two cysteine desulfurases (IscS and SufS), an iron-regulated ABC transporter (SufB), a hypothetical protein (RSP_0439), an ATPase (SufC), an Fe–S cluster assembly protein (SufD), and a putative sulfate transporter (composed of RSP_0433 and RSP_0432) (Fig.1). In a differential RNAseq approach (Sharma et al. 2010) under iron limitation we identified two transcriptional start sites (TSS), one in front of the iscR gene, a second within the gene encoding iscS (Fig.1) (Remes et al. 2014). This motivated us to test if all genes are part of an operon and are transcribed together. Selected noncoding regions (RSP_0443-0442; RSP_0442-0440; RSP_0440-0439; RSP_0432-0431) were amplified together with upstream and downstream coding regions via RT-PCR. All RT-PCR reactions showed transcripts with the expected size (Fig. S1). To investigate the role of the internal TSS, two sets of primers were used. In the first primer set one primer hybridizes to the 3′ end of iscS upstream of the internal TSS, whereas the forward primer of the second primer set hybridizes to the 3′ end of iscS downstream of the internal TSS. In both sets the second primer hybridizes to the 5′ end of sufB. The amplification products (Fig. S1) revealed the existence of an RNA extending from the 5′ part of iscS into sufB, strongly implying that transcripts initiating at the TSS in front of iscR also comprise the coding regions downstream of sufB. We therefore assume that the whole gene cluster is transcribed as one operon, albeit the two promoters may lead to differential expression.
Figure 1.
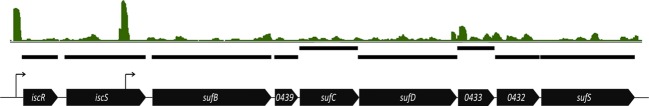
Schematic representation and RNA-seq read coverage of the isc-suf operon in Rhodobacter sphaeroides. Top: Read coverage of the isc-suf operon in R. sphaeroides visualized by the Integrated Genome Browser. Bottom: Schematic representation of the operon. Genes are represented by black boxes and transcriptional start sites are shown by black arrows. The direction of the arrow indicates the direction of transcription.
An iscR mutant shows elevated ROS levels and an increased sensitivity to iron limitation
To elucidate the role of the predicted regulator encoded by the first gene of the operon, an iscR deletion strain was constructed as described in Materials and Methods. The growth of the ΔiscR mutant was similar to that of the wild-type in iron-replete conditions, whereas growth of the mutant was more severely impeded than that of the wild-type in iron-limiting conditions (Fig.2). To investigate the impact of oxidative stress on this growth behavior, ROS levels were monitored in both strains. As previously described, exposure to iron starvation cause a strongly increased ROS accumulation in the wild-type cells (Peuser et al. 2011; Remes et al. 2014). An iscR mutant showed in comparison to the wild-type significantly increased ROS production irrespective of iron availability (Fig.3).
Figure 2.
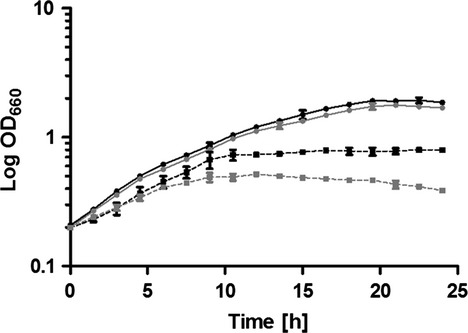
An iscR mutant is more sensitive to iron limitation. Growth curves of wild-type Rhodobacter sphaeroides (black) and the ∆iscR mutant (gray) was performed in iron-replete (continuous line) or iron-limiting (dashed line) conditions. The optical density at 660 nm (OD660) was determined over time. The data represent the mean of at least three independent experiments, and the error bars indicate the standard deviation.
Figure 3.
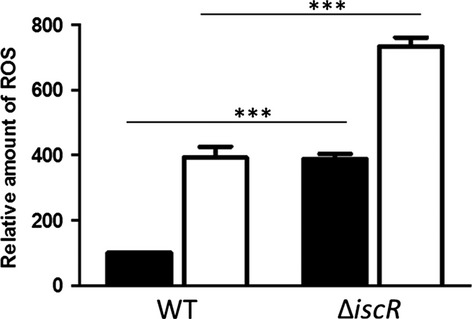
The iscR deletion strain shows significantly increased ROS levels. Determination of intracellular levels of ROS in wild-type Rhodobacter sphaeroides and the ∆iscR mutant strain. Cultures were grown under iron-replete (black) and iron-limiting (white) conditions. ROS generated by the cells were analyzed after reaction with 10 mmol/L 2,7-dihydrodichlorofluorescein diacetate. The fluorescence intensity was normalized to the optical densities of the samples. The resulting values are presented in arbitrary units. The data represent the mean of three independent experiments, and the error bars indicate the standard deviation. A P-value was computed using the student's t test. ***Significant at P ≤ 0.001.
IscR regulates genes involved in iron and sulfur metabolism
A high-density oligonucleotide microarray was applied for comparing gene expression in strain ∆iscR and its parental wild-type strain. A total of 110 protein coding genes are differently expressed (log2 fold change ≥0.6 or ≤ −0.6) in the absence of iscR (Table S4). Table1 gives an overview of differently expressed genes involved in iron homeostasis as well as genes preceded by an Iron-Rhodo-box motif. The iscR deletion strain carries an antibiotic resistance cassette within the iscR gene, which also abolishes expression of iscS, while three genes (sufCD, RSP_0439) of the isc-suf operon were strongly expressed. Indeed, besides the promoter upstream of iscR an additional promoter element was identified by an RNAseq approach upstream of sufB (Fig.1). Thus, transcription of the sufBCD genes in the iscR mutant seems to originate from this additional, iscR-independent promoter. Nevertheless, as mentioned above, we proved the existence of an RNA extending from the 5′ part of iscS into sufB (Fig. S1). According to this, the suf transcripts are at least partly initiated at the iscR promoter in the wild-type situation.
Table 1.
Selection of IscR-responsive genes in Rhodobacter sphaeroides as determined by microarray analysis
| RSP no. | Gene | log2 FC | Description |
|---|---|---|---|
| RSP_0437 | sufC | 0.68 | Suf C, ATPase |
| RSP_0439 | 0.67 | Hypothetical protein | |
| RSP_0440 | sufB | 0.67 | Putative SufB |
| RSP_0442 | iscS | −1.00 | Putative aminotransferase |
| RSP_0443 | iscR | −1.15 | Rrf2 family transcriptional regulator |
| RSP_0920 | exbB | 1.98 | Biopolymer transport protein, ExbB |
| RSP_0921 | exbD | 1.59 | Biopolymer transport protein, ExbD |
| RSP_0922 | tonB | 1.59 | Putative TonB protein |
| RSP_1438 | 0.62 | ABC Fe hydroxamate (ferrichrome) transporter, fused inner membrane subunits | |
| RSP_1440 | 0.93 | TonB dependent, hydroxamate-type ferrisiderophore, outer membrane receptor | |
| RSP_1543 | 0.72 | Hypothetical protein | |
| RSP_1544 | 0.92 | Hypothetical signal peptide protein | |
| RSP_1545 | (0.59) | Probable thiol oxidoreductase with 2 cytochrome c heme-binding sites | |
| RSP_1546 | bfr | 1.50 | Bacterioferritin |
| RSP_1547 | bfd | 2.30 | Probable bacterioferritin-associated ferredoxin |
| RSP_1548 | 2.53 | Putative iron-regulated protein | |
| RSP_1818 | feoB | 0.86 | Fe2 transport system protein B |
| RSP_1819 | feoA1 | 0.87 | Ferrous iron transport protein A |
| RSP_1949 | 0.65 | FeS assembly SUF system protein | |
| RSP_2913 | afuA | 1.93 | ABC Fe siderophore transporter, periplasmic substrate-binding protein |
| RSP_3076 | 0.75 | Hypothetical protein | |
| RSP_3077 | 0.95 | Hypothetical protein | |
| RSP_3078 | 1.01 | Hypothetical protein | |
| RSP_3079 | 0.66 | ABC Fe siderophore transporter, periplasmic substrate-binding protein | |
| RSP_3223 | 0.85 | TonB-dependent receptor precursor | |
| RSP_3416 | (0.07) | ABC Fesiderophore transporter, periplasmic-binding protein | |
| RSP_3417 | (0.00) | TonB-dependent outer membrane ferrichrome-iron receptor | |
| RSP_3678 | 0.77 | Siderophore-interacting protein | |
| RSP_4275 | fecI | (0.32) | sigma24, FecI |
| RSP_6006 | 2.53 | Hypothetical protein | |
| RSP_6020 | feoA2 | 1.15 | Ferrous iron transport protein A |
Significant expression changes (log2 fold change ≥0.6 or ≤0.6) of selected genes (2.4.1∆iscR vs. wild-type) are shown. Genes with RSP numbers in boldface type are located in operons preceded by an Iron-Rhodo-box motif. Numbers in parentheses did not pass our selection criteria (log2 FC ≥0.6 or ≤ −0.6).
Many genes with predicted functions in ferric/ferrous iron uptake or iron storage showed a higher expression in the mutant strain, including exbBD-tonB, RSP_1438-1440 encoding a ferrichrome transporter, bfd-bfr encoding a bacterioferritin, irpA encoding an iron-regulated protein, feoAB encoding a ferrous transport system, genes encoding Fe3+-siderophore transporters (afuA, RSP_3079, RSP_3678) and hemP encoding an iron uptake protein. Furthermore, several genes encoding proteins involved in adaptation to cold stress (RSP_1952, RSP_3260/21), oxidative stress (rpoE, RSP_1091, phrA), glycolysis (RSP_2968), pyruvate decarboxylation (pdhAB), flagellum biosynthesis (fli, flg) and chemotaxis (che) had significantly lower expression levels in the mutant compared to the wild-type (Table S4).
To validate the microarray data real-time RT-PCR was used for the quantification of mRNAs transcribed from several selected genes. In addition to genes involved in iron homeostasis one gene of the flagellum biosynthesis (fliS), the ABC zinc transporter gene znuA, a gene for bacteriochlorophyll synthesis (bchL), and for a structural protein of the reaction center (pufL) were selected for validation. Increase or decrease of expression levels as revealed by microarray analysis were confirmed by real-time RT-PCR (Fig. S2). Nevertheless for some genes, the extent of change varied between the two approaches and was mostly more pronounced in the real-time RT-PCR data set. As a consequence some genes showed a clear difference in expression between the two strains only in the RT-PCR analysis (e.g. RSP_3416 and RSP_3417).
Rhodobacter sphaeroides IscR exhibits a new type of Fe–S coordination
Apo-IscR typically ligates an Fe–S cluster in its C-terminal part by three Cys residues and one histidine residue. This motif is conserved in many proteobacteria (Fleischhacker et al. 2012), but only a single Cys residue is present in the C-terminal part of Rhodobacteraceae IscR at a position not matching the Fe–S coordinating Cys residues of other IscR proteins (Fig. S3). We used the oxidized apo-form of IscR for chemical reconstitution under strictly anaerobic conditions, yielding a brown protein solution. The reconstituted protein showed the typical absorption shoulder at 410–420 nm, which is characteristic for iron-sulfur proteins (Fig.4A) (Kulzer et al. 1998; Zeng et al. 2008). This absorption pattern disappeared after cluster reduction in aerobic conditions (Fig.4A).
Figure 4.
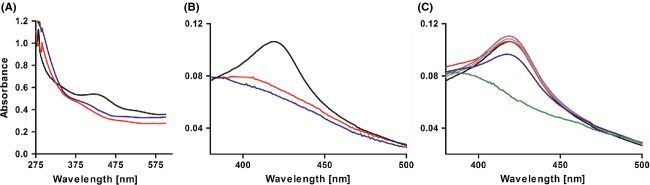
UV-visible absorption spectra of various IscR samples in native state. (A) UV-vis spectra of purified IscR (∼60 μmol/L). After Ni-NTA purification, His-tagged IscR was first oxidized (red), then reconstituted in strictly anaerobic conditions (black), and finally oxidized again (Blue). (B) UV-Vis spectra of His-tagged IscR (∼10 μmol/L) purified under reduced conditions (black), iron-limiting conditions (red), or treated with 1 mmol/L H2O2 (blue), respectively. (C) UV-Vis spectra of His-tagged IscR (∼10 μmol/L) (black) and its single-site mutants H93A (red), H121A (gray), H127A (purple), C142A (green), P143A (yellow), and the triple-site mutant H93A/H121A/H127A (blue), respectively.
For further analysis, we measured absorption spectra of reduced IscR, which showed a similar absorption pattern with a peak at 420 nm (Fig.4B). By contrast, IscR grown and purified under iron-limiting conditions showed nearly no absorption at 420 nm relative to the control, indicating a strongly reduced amount of Fe–S clusters ligated to the protein (Fig.4B). To determine the effect of ROS on Fe–S cluster integrity, purified His-tagged IscR was incubated with H2O2 (1 mmol/L) for 2 min prior to UV-visible spectroscopy. The results showed a total loss in IscR absorbance at 420 nm (Fig.4B), suggesting that in the presence of oxidative stress, the Fe–S clusters ligated to IscR were targets for H2O2 -mediated oxidation that resulted in destabilization of Fe–S bound to the protein.
Since other IscR proteins coordinate the Fe–S cluster via 3 Cys and one His residue we replaced the single Cys and the three His residues in the C-terminal part of R. sphaeroides IscR. His-tagged IscR and IscR variants carrying these amino acid substitutions (H93A, H121A, H127A, and C142A) were purified and equal amounts of the proteins were then subjected to UV-visible spectroscopy. The IscR variant C142A had nearly no absorption at 420 nm indicating that binding of Fe–S cluster to this protein is severely impaired (Fig.4C). In contrast, the three mutated IscR variants H93A, H121A and H127A and also the triple mutant H93A/H121A/H127A showed the typical IscR absorption at 420 nm.
Furthermore, R. sphaeroides IscR harbors the so-called “heme regulatory motif” (HRM) in its C-terminal region. HRMs are heme-binding sequences that are found in proteins involved in many aspects of heme and iron metabolism. The consensus sequence comprises a stretch of residues where only a Cys-Pro dipeptide is absolutely conserved and in most cases flanked by hydrophobic amino acids (Lathrop and Timko 1993; Zhang and Guarente 1995). Thus, the Pro141-Cys-Pro-Ala-Val145 sequence reflects a typical HRM found in proteins that bind heme. To exclude that the observed absorption peak at 420 nm is due to a heme bound to the HRM rather than to an Fe–S cluster, we mutated the essential amino acids in the conserved HRM. While a total loss in absorption was observed for the IscR variant C142A, IscR variant P143A shows the same absorption at 420 nm as the wild-type (Fig.4C).
IscR requires an Fe–S cluster to repress target genes with an Iron-Rhodo-box motif
In the Rhodobacteraceae, a highly conserved 19-bp palindromic signal, termed Iron-Rhodo-box, is located in the regulatory regions upstream of genes encoding iron uptake and storage proteins, and was predicted for an unknown regulator or for IscR (Rodionov et al. 2006). Expression analyses revealed that most of these genes are induced in response to iron starvation in wild-type cells (Peuser et al. 2011; Remes et al. 2014), and all genes showed higher expression in iron replete conditions in the background of ∆iscR (Fig. S4A). However, deletion of iscR resulted in an abolished expression of the downstream gene iscS. In the complemented strain ∆iscR_pBBRiscR, the expression of the target genes reached wild-type-like levels (Fig. S4B), while the transcript level of iscS is unchanged in comparison to the iscR mutant (data not shown). Thus, the repression of its target genes is solely IscR dependent. However, no difference or even lower expression levels were observed in the mutant compared to the parental wild-type strain in iron-limiting conditions (Fig. S4A). To further investigate a potential Fe–S cluster requirement for the repressor function of IscR to its own promoter (PiscR), we measured the activity of PiscR in both, iron-deplete and iron-replete conditions. For this approach, the target promoter region PiscR was transcriptionally fused to the cfp gene on plasmid pBE4352: eCFP: eCFP and transformed in both, wild-type and iscR deletion strain. In the presence of iron, deletion of iscR results in a significant induction of PiscR activity (Fig.5). However, under iron-limiting conditions, IscR exhibited a severe defect in PiscR repression (Fig.5). Thus, IscR negatively regulates its own transcription only in the presence of iron.
Figure 5.
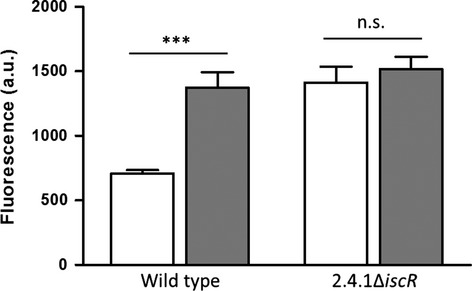
IscR negatively regulates expression of its own promoter (PiscR). Relative fluorescence was measured from the wild-type strain and deletion strain ∆iscR, both containing the eCFP:PiscR reporter fusions on plasmid pBE. Strains were grown in iron-replete (white bars) or iron-limiting (gray bars) conditions. The data represent the average activity of three independently isolated strains. A P-value was computed using the student's t test. Variations were considered statistically significant when the P-value was ≤0.05. ***Significant at P ≤ 0.001.
To study the DNA-binding function of IscR with predicted iscR-controlled promoters, we chose the iscR and hemP promoter regions for binding assays. His-tagged IscR was purified after heterologous overexpression and radiolabeled DNA probes containing the predicted binding sites were co-incubated and separated on a nondenaturing polyacrylamide gel. With increasing IscR concentrations, formation of a retarded DNA protein complex was observed for both fragments (Fig. S5, lane 2–5). Addition of an up to 100-fold molar excess of an unlabeled nonspecific probe containing the regulatory region of znuA did not interfere with the complex formation, confirming specificity of binding (Fig. S5, lane 6–7). The znuA gene is IscR-dependently regulated (Table S4) but does not carry the Iron-Rhodo-box motif. However, almost a complete loss of the retardation was observed when 10-fold molar excess of the specific unlabeled DNA probe was used to compete out the labeled probe (Fig. S5, lane 8–9). This result indicates that genes preceded by the Iron-Rhodo-box motifs are under direct control of IscR.
Figure6 illustrates the effect of IscR on the R. sphaeroides genes with predicted Iron-Rhodo-boxes. Based on previous dRNAseq analyses (Remes et al. 2014), we also show the position of the Iron-Rhodo-box motif in relation to the TSS. RSP_3417 was only very weakly expressed under the chosen conditions and no TSS could be identified. No TSS could be identified directly upstream of RSP_1545. The dRNAseq data strongly suggest that RSP_1545-1543 are cotranscribed together with hemP-irpA-bfd-bfr. This suggests one IscR-binding site directly in the hemP promoter and an additional binding site within the operon. All other IscR-binding sites including that upstream of hemP completely or partially overlap the −35/−10 region or are positioned very close to it as in the case of the afuA promoter. The consensus sequence of these motifs matches well to the consensus sequence for the Iron-Rhodo-box motif of Rhodobacteraceae as published by Rodionov et al. (2006).
Figure 6.
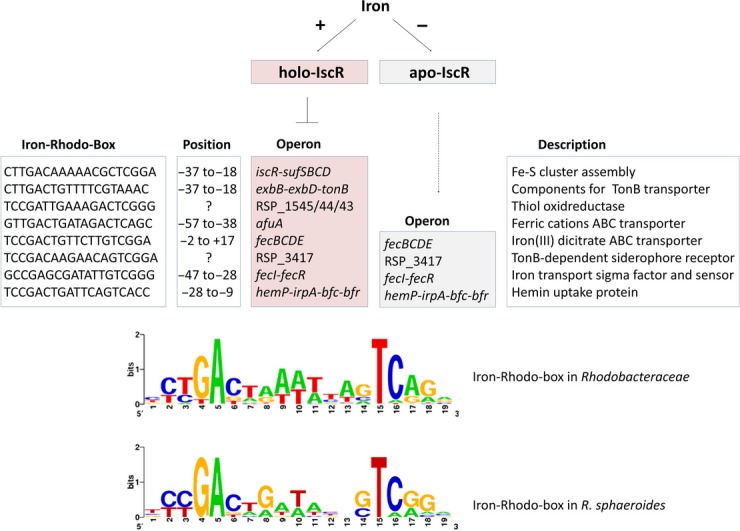
Model of the IscR regulon in Rhodobacter sphaeroides. Fe–S cluster-containing IscR inhibits transcription of all genes preceded by an Iron-Rhodo-box motif, while apo-IscR activates transcription of some of them. The position relative to the transcriptional start site as determined by differential RNAseq (Remes et al. 2014) is shown. A consensus logo of the Iron-Rhodo-box motif for the Rhodobacteraceae (Rodionov et al. 2006) and for R. sphaeroides was drawn, using the WebLogo package version 2.8.2 (http://weblogo.berkeley.edu).
Discussion
E. coli IscR acts as a sensor of the cellular demands for Fe–S cluster biogenesis by binding an Fe–S cluster with (Cys)3(His)1 (Fleischhacker et al. 2012). IscR thereby acts as global regulator involved in a homeostatic mechanism controlling Fe–S cluster biogenesis (Giel et al. 2006; Lee et al. 2008; Wu and Outten 2009; Giel et al. 2013). In E. coli, holo-IscR directly represses the Isc Fe–S biogenesis pathway, while as an apo-protein, it activates an alternative Fe–S biogenesis pathway, encoded by the suf genes (Schwartz et al. 2001; Giel et al. 2006; Yeo et al. 2006; Lee et al. 2008). An important question was therefore, whether the unusual R. sphaeroides IscR serves a similar function as its E. coli homolog and is able to ligate an Fe–S cluster.
In contrast to all other IscR proteins from proteobacteria, IscR proteins from Rhodobacteraceae lack the three conserved Cys residues, which are essential for Fe–S cluster ligation (Rodionov et al. 2006; Yeo et al. 2006; Nesbit et al. 2009). A phylogenetic tree revealed that IscR proteins from Rhodobacteraceae form a distinct cluster within the group of alpha proteobacteria (Rodionov et al. 2006) (Fig. S6). There is only a single-Cys residue present in the C-terminal region and it was therefore proposed that Rhodobacteraceae IscR is unlikely to ligate an Fe–S cluster (Rodionov et al. 2006). However, our data confirm that R. sphaeroides IscR contains an Fe–S cluster and that the single Cys residue is essential for this ligation (Fig.4).
A homology model based on the comparative modelling method from known PDB-related three-dimensional structures shows that this Cys residue is surface-exposed (Fig. S7). It is therefore conceivable that apo-IscR coordinates one Fe–S cluster by Cys residues of distinct IscR protomers. Similar results were observed for Aquifex aeolicus IscU and Thermosynechococcus elongates IscA, which coordinate one Fe–S cluster by two or three conformationally distinct protomers, respectively (Morimoto et al. 2006; Shimomura et al. 2008). Although, both proteins contain the three-conserved Cys residues. To analyze if R. sphaeroides IscR forms an oligomeric complex, the purified protein was subjected to gel filtration on a Superdex 200 16/60 column. The elution profile indicated that IscR even under aerobic conditions forms dimers and tetramers (data not shown). In Rieske proteins, the Fe–S cluster is ligated by an unusual (Cys)2(His)2, but no His residues are involved in R. sphaeroides IscR Fe–S cluster binding (Fig.4C). However, it is conceivable that the cluster is coordinated by three additional noncysteinyl ligands. Further biochemical analysis and structural characterization is required to establish a clear picture of the Fe–S cluster ligation and to identify the precise composition of Fe–S clusters bound to IscR.
In contrast to the situation in E. coli, the isc and suf genes in Rhodobacteraceae are co-localized on the chromosome and in R. sphaeroides transcribed as one operon (Fig. S1). Several genes of the operon are highly expressed under various oxidative stresses or iron-limiting conditions to compensate the decreased Fe–S availability (Zeller et al. 2007; Peuser et al. 2011; Berghoff et al. 2013; Remes et al. 2014). Regulation of the R. sphaeroides isc-suf operon by oxidative stress is under control of the global regulator OxyR, which senses H2O2 (Zeller et al. 2007; Remes et al. 2014). An oxyR mutant showed increased ROS production and impaired growth under iron limitation (Remes et al. 2014). The same phenotype was observed for the iscR deletion strain of R. sphaeroides (Figs.2, 3) and of Pseudomonas aeruginosa (Romsang et al. 2014). It is likely that the lack of IscR prevents activation of iron uptake genes, leading to lower intracellular iron concentrations than in the wild-type, and consequently to an impaired growth behavior.
IscR has recently emerged as a pleiotropic regulator that influences the expression of ∼40 genes in E. coli or 67 genes in Vibrio vulnificus (Giel et al. 2006; Lim and Choi 2014). Our transcriptome study identified an R. sphaeroides IscR regulon comprising ∼110 protein coding genes. Deletion of iscR also abolished expression of iscS. Although this had no influence on genes with predicted Iron-Rhodo-boxes (Fig. S4B), we cannot completely rule out that some genes of the regulon are affected by the deletion of iscS. The IscR regulons of the three organisms E. coli, V. vulnificus, and R. sphaeroides share important functions, including iron homeostasis, motility or oxidative stress response. For most of the R. sphaeroides regulated genes, a similar expression pattern was previously observed for the wild-type strain in response to iron limitation in oxic conditions, potentially due to elevated ROS levels (Peuser et al. 2011; Remes et al. 2014). In agreement with this, of the 110 IscR-dependent genes 33 are affected by iron levels, 28 by hydrogen peroxide, and 44 by singlet oxygen stress (Zeller et al. 2005; Peuser et al. 2011; Berghoff et al. 2013). The increased ROS levels in strain ∆iscR compared to the wild-type in iron-replete and iron-deplete conditions (Fig.3) support an indirect regulation for those genes in response to oxidative stress.
Oxidative stress and iron limitation result in a decrease of holo-IscR (Fig.4B), but in an increased iscR mRNA level (Zeller et al. 2005; Peuser et al. 2011; Remes et al. 2014), Since the repressor function of IscR in R. sphaeroides was no longer present under iron-limiting conditions (Figs.5, S4A), the cluster coordination is a necessary feature for IscR to repress its target promoters. In E. coli transcriptional regulators, such as IscR and SoxR, bind their DNA targets independently of an Fe–S cluster, while transcription of target genes is cofactor dependent (Hidalgo et al. 1998; Nesbit et al. 2009; Rajagopalan et al. 2013). Binding of R. sphaeroides IscR to the iscR and hemP promoter region did also not require the Fe–S cluster, but IscR only represses both genes in its holo-form. However, IscR also activates some genes as apo-protein (Fig. S4A), maximizing the cellular capacity for iron uptake, Fe–S cluster assembly and maintenance.
Since no IscU scaffold protein is present in R. sphaeroides, it was proposed that a complex of SufBCD could function as both, scaffold protein, and transporter for Fe–S cluster to apo-IscR (Wollers et al. 2010; Vinella et al. 2013). Therefore, the SufBCD proteins have to be able to distinguish between IscR and other apo-protein targets. While the details of this putative target specificity remain disputable, the ligation scheme with only one Cys may differentiate IscR from other apo-proteins. Due to this ligation scheme R. sphaeroides IscR exhibits probably a decreased affinity for Fe–S cluster and is only able to bind the clusters if no other proteins require them. We propose that sufficient amounts of Fe–S cluster are sensed by forming holo-IscR, while in turn PiscR gets repressed by a negative feedback loop to keep appropriate levels of cellular Fe–S cluster formation and delivery.
Acknowledgments
This work was supported by the DFG (Kl563/25 and the IRTG program GRK 1384 “Enzymes and multi-enzyme complexes acting on nucleic acids”)
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. The Rhodobacter sphaeroides isc-suf operon is cotranscribed as determined by RT-PCR. Predicted RT-PCR products are represented by lines under the genes and predicted RT-PCR product sizes are shown in parentheses. RT-PCR was performed with total RNA in the presence (+) or absence (−) of reverse transcriptase or with genomic DNA (G).
Figure S2. Validation of microarray data by real-time RT-PCR. Quantified log2 fold changes of iron-responsive genes in 2.4.1ΔiscR compared to the wild-type was determined by real-time RT-PCR. Values are normalized to that of the housekeeping gene rpoZ. The data presented are the means of at least three experiments, and the standard deviations of the means are indicated (error bars). Numbers in parentheses show the fold change of the respective genes as determined by microarray analysis.
Figure S3. Similarity of Rhodobacter sphaeroides IscR to Escherichia coli IscR. Amino acid sequence alignment of the two proteins generated with “align” showed 43% identical residues (*) and 23% similar residues (:). Solid lines denote the helix-turn-helix DNA-binding domains; the three Cys residues (C92, C98, and C104) of E. coli IscR that coordinate the Fe–S cluster are framed.
Figure S4. Functional analysis of IscR repressor activity via real-time RT PCR. (A) Relative gene expression in strain 2.4.1ΔiscR in comparison to that of the wild-type under iron-replete conditions (black) or under iron limitation (gray). (B) Relative gene expression of the mutant 2.4.1ΔiscR (black) or the complemented mutant ΔiscR_pBBRiscR (white) in comparison to the wild-type under iron-replete conditions. Values are normalized to that of the housekeeping gene rpoZ. The data represent the mean of at least three independent experiments. A P-value was computed using the student's t test. Variations were considered statistically significant when the P-value was ≤0.05. *Significant at P ≤ 0.05; **significant at P ≤ 0.01; ***significant at P ≤ 0.001.
Figure S5. Binding of purified IscR to the promoter regions of iscR (A) and hemP (B) as determined by EMSAs. All reactions contain the same amount of 32P end-labeled DNA fragment (∼5 fmol/lane) comprising the promoter sequence, including the Iron-Rhodo-box motif. Lane 1 contains no IscR; lanes 2–5 contain increasing amounts of IscR (100–500 ng); lanes 6–9 contain 500 ng IscR; lanes 6 and 7 contain excess amounts of an unlabeled nonspecific probe containing the regulatory region of znuA. Lanes 8–9 contain excess amounts of the respective unlabeled DNA probe as competitor.
Figure S6. (A) Phylogenetic Tree of IscR from proteobacteria (modified from Rodionov et al. 2006; Fig. 3B). The genome abbreviations are listed in (B).
Figure S7. Homology model of the IscR monomer calculated using the MODELLER program. The cartoon representation in blue of the IscR protein shows a helix-turn-helix fold. The surface accessible residue Cys-142 is represented as a stick model in magenta.
Table S1. Rhodobacter sphaeroides and Escherichia coli strains used in this study.
Table S2. Plasmids used in this study.
Table S3. Oligodeoxynucleotide used in this study.
Table S4. Gene expression changes in 2.4.1ΔiscR as determined by microarray analysis.
References
- Andrews SC, Robinson AK. Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Angelini S, Gerez C, Ollagnier-De Choudens S, Sanakis Y, Fontecave M, Barras F, et al. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 2008;283:14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- Berghoff BA, Konzer A, Mank NN, Looso M, Rische T, Forstner KU, et al. Integrative “omics”-approach discovers dynamic and regulatory features of bacterial stress responses. PLoS Genet. 2013;9:e1003576. doi: 10.1371/journal.pgen.1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovitch F, Nicolet Y, Wan JT, Jarrett JT. Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethi-onine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolai L, Gorst CM, Zhao ZH, Teng Q, Adams MW. la Mar GN. 1H NMR investigation of the electronic and molecular structure of the four-iron cluster ferredoxin from the hyperthermophile Pyrococcus furiosus. Identification of Asp 14 as a cluster ligand in each of the four redox states. Biochemistry. 1995;34:11373–11384. doi: 10.1021/bi00036a010. [DOI] [PubMed] [Google Scholar]
- Cheek J. Broderick JB. Adenosylmethionine-dependent iron-sulfur enzymes: versatile clusters in a radical new role. J. Biol. Inorg. Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- Dobritzsch D, Schneider G, Schnackerz KD. Lindqvist Y. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001;20:650–660. doi: 10.1093/emboj/20.4.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan CL. Peters JW. Surprising cofactors in metalloenzymes. Curr. Opin. Struct. Biol. 2003;13:220–226. doi: 10.1016/s0959-440x(03)00038-1. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M. Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, et al. Characterization of the [2Fe–2S] cluster of Escherichia coli transcription factor IscR. Biochemistry. 2012;51:4453–4462. doi: 10.1021/bi3003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DH. Allen RM. Ironminus sign Sulfur proteins with nonredox functions. Chem. Rev. 1996;96:2315–2334. doi: 10.1021/cr950041r. [DOI] [PubMed] [Google Scholar]
- Fluhe L, Knappe TA, Gattner MJ, Schafer A, Burghaus O, Linne U, et al. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 2012;8:350–357. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- Fontecave M. Iron-sulfur clusters: ever-expanding roles. Nat. Chem. Biol. 2006;2:171–174. doi: 10.1038/nchembio0406-171. [DOI] [PubMed] [Google Scholar]
- Giel JL, Rodionov D, Liu M, Blattner FR. Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- Giel JL, Nesbit AD, Mettert EL, Fleischhacker AS, Wanta BT. Kiley PJ. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe–2S]-IscR in Escherichia coli. Mol. Microbiol. 2013;87:478–492. doi: 10.1111/mmi.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Hidalgo E, Leautaud V. Demple B. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 1998;17:2629–2636. doi: 10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner P, Willison JC, Vignais PM. Bickle TA. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 1991;173:2993–2999. doi: 10.1128/jb.173.9.2993-2999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD. Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Johnston AW, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, Gelfand MS, et al. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals. 2007;20:501–511. doi: 10.1007/s10534-007-9085-8. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Kulzer R, Pils T, Kappl R, Huttermann J. Knappe J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J. Biol. Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- Lathrop JT. Timko MP. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science. 1993;259:522–525. doi: 10.1126/science.8424176. [DOI] [PubMed] [Google Scholar]
- Lee KC, Yeo WS. Roe JH. Oxidant-responsive induction of the suf operon, encoding a Fe–S assembly system, through Fur and IscR in Escherichia coli. J. Bacteriol. 2008;190:8244–8247. doi: 10.1128/JB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JG. Choi SH. IscR is a global regulator essential for pathogenesis of Vibrio vulnificus and induced by host cells. Infect. Immun. 2014;82:569–578. doi: 10.1128/IAI.01141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau L, Gerez C, Bekker M, Ollagnier-De Choudens S, Py B, Sanakis Y, et al. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2007;104:13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin R. Rabinowitz JC. The reconstitution of clostridial ferredoxin. Biochem. Biophys. Res. Commun. 1966;23:822–827. doi: 10.1016/0006-291x(66)90561-4. [DOI] [PubMed] [Google Scholar]
- Mank NN, Berghoff BA, Hermanns YN. Klug G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc. Natl. Acad. Sci. USA. 2012;109:16306–16311. doi: 10.1073/pnas.1207067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K, Yamashita E, Kondou Y, Lee SJ, Arisaka F, Tsukihara T, et al. The asymmetric IscA homodimer with an exposed [2Fe–2S] cluster suggests the structural basis of the Fe–S cluster biosynthetic scaffold. J. Mol. Biol. 2006;360:117–132. doi: 10.1016/j.jmb.2006.04.067. [DOI] [PubMed] [Google Scholar]
- Nesbit AD, Giel JL, Rose JC. Kiley PJ. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe–S cluster ligation. J. Mol. Biol. 2009;387:28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet Y, Piras C, Legrand P, Hatchikian CE. Fontecilla-Camps JC. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- Peters JW, Lanzilotta WN, Lemon BJ. Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- Peuser V, Metz S. Klug G. Response of the photosynthetic bacterium Rhodobacter sphaeroides to iron limitation and the role of a Fur orthologue in this response. Environ. Microbiol. Rep. 2011;3:397–404. doi: 10.1111/j.1758-2229.2011.00245.x. [DOI] [PubMed] [Google Scholar]
- Peuser V, Remes B. Klug G. Role of the Irr protein in the regulation of iron metabolism in Rhodobacter sphaeroides. PLoS One. 2012;7:e42231. doi: 10.1371/journal.pone.0042231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P, Binda A. Epstein A. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene. 1991;103:17–23. doi: 10.1016/0378-1119(91)90385-o. [DOI] [PubMed] [Google Scholar]
- Py B. Barras F. Building Fe–S proteins: bacterial strategies. Nat. Rev. Microbiol. 2010;8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Teter SJ, Zwart PH, Brennan RG, Phillips KJ. Kiley PJ. Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat. Struct. Mol. Biol. 2013;20:740–747. doi: 10.1038/nsmb.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remes B, Berghoff BA, Forstner KU. Klug G. Role of oxygen and the OxyR protein in the response to iron limitation in Rhodobacter sphaeroides. BMC Genom. 2014;15:794. doi: 10.1186/1471-2164-15-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rische T. Klug G. The ordered processing of intervening sequences in 23S rRNA of Rhodobacter sphaeroides requires RNase J. RNA Biol. 2012;9:343–350. doi: 10.4161/rna.19433. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS, Todd JD, Curson AR. Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comput. Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romsang A, Duang-Nkern J, Leesukon P, Saninjuk K, Vattanaviboon P. Mongkolsuk S. The iron-sulphur cluster biosynthesis regulator IscR contributes to iron homeostasis and resistance to oxidants in Pseudomonas aeruginosa. PLoS One. 2014;9:e86763. doi: 10.1371/journal.pone.0086763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A. Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Santos JA, Alonso-Garcia N, Macedo-Ribeiro S. Pereira PJ. The unique regulation of iron-sulfur cluster biogenesis in a Gram-positive bacterium. Proc. Natl. Acad. Sci. USA. 2014;111:E2251–E2260. doi: 10.1073/pnas.1322728111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, et al. IscR, an Fe–S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe–S cluster assembly proteins. Proc. Natl. Acad. Sci. USA. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Shepard W, Soutourina O, Courtois E, England P, Haouz A. Martin-Verstraete I. Insights into the Rrf2 repressor family – the structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J. 2011;278:2689–2701. doi: 10.1111/j.1742-4658.2011.08195.x. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Wada K, Fukuyama K. Takahashi Y. The asymmetric trimeric architecture of [2Fe–2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 2008;383:133–143. doi: 10.1016/j.jmb.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Vinella D, Loiseau L, Ollagnier De Choudens S, Fontecave M. Barras F. In vivo [Fe–S] cluster acquisition by IscR and NsrR, two stress regulators in Escherichia coli. Mol. Microbiol. 2013;87:493–508. doi: 10.1111/mmi.12135. [DOI] [PubMed] [Google Scholar]
- Wollers S, Layer G, Garcia-Serres R, Signor L, Clemancey M, Latour JM, et al. Iron-sulfur (Fe–S) cluster assembly: the SufBCD complex is a new type of Fe–S scaffold with a flavin redox cofactor. J. Biol. Chem. 2010;285:23331–23341. doi: 10.1074/jbc.M110.127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. Outten FW. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J. Bacteriol. 2009;191:1248–1257. doi: 10.1128/JB.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo WS, Lee JH, Lee KC. Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe–S assembly proteins. Mol. Microbiol. 2006;61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- Zappa S. Bauer CE. The LysR-type transcription factor HbrL is a global regulator of iron homeostasis and porphyrin synthesis in Rhodobacter capsulatus. Mol. Microbiol. 2013;90:1277–1292. doi: 10.1111/mmi.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Moskvin OV, Li K, Klug G. Gomelsky M. Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides. J. Bacteriol. 2005;187:7232–7242. doi: 10.1128/JB.187.21.7232-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Mraheil MA, Moskvin OV, Li K, Gomelsky M. Klug G. Regulation of hydrogen peroxide-dependent gene expression in Rhodobacter sphaeroides: regulatory functions of OxyR. J. Bacteriol. 2007;189:3784–3792. doi: 10.1128/JB.01795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Zhang K, Liu J. Qiu G. Expression, purification, and characterization of iron-sulfur cluster assembly regulator IscR from Acidithiobacillus ferrooxidans. J. Microbiol. Biotechnol. 2008;18:1672–1677. [PubMed] [Google Scholar]
- Zhang L. Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The Rhodobacter sphaeroides isc-suf operon is cotranscribed as determined by RT-PCR. Predicted RT-PCR products are represented by lines under the genes and predicted RT-PCR product sizes are shown in parentheses. RT-PCR was performed with total RNA in the presence (+) or absence (−) of reverse transcriptase or with genomic DNA (G).
Figure S2. Validation of microarray data by real-time RT-PCR. Quantified log2 fold changes of iron-responsive genes in 2.4.1ΔiscR compared to the wild-type was determined by real-time RT-PCR. Values are normalized to that of the housekeeping gene rpoZ. The data presented are the means of at least three experiments, and the standard deviations of the means are indicated (error bars). Numbers in parentheses show the fold change of the respective genes as determined by microarray analysis.
Figure S3. Similarity of Rhodobacter sphaeroides IscR to Escherichia coli IscR. Amino acid sequence alignment of the two proteins generated with “align” showed 43% identical residues (*) and 23% similar residues (:). Solid lines denote the helix-turn-helix DNA-binding domains; the three Cys residues (C92, C98, and C104) of E. coli IscR that coordinate the Fe–S cluster are framed.
Figure S4. Functional analysis of IscR repressor activity via real-time RT PCR. (A) Relative gene expression in strain 2.4.1ΔiscR in comparison to that of the wild-type under iron-replete conditions (black) or under iron limitation (gray). (B) Relative gene expression of the mutant 2.4.1ΔiscR (black) or the complemented mutant ΔiscR_pBBRiscR (white) in comparison to the wild-type under iron-replete conditions. Values are normalized to that of the housekeeping gene rpoZ. The data represent the mean of at least three independent experiments. A P-value was computed using the student's t test. Variations were considered statistically significant when the P-value was ≤0.05. *Significant at P ≤ 0.05; **significant at P ≤ 0.01; ***significant at P ≤ 0.001.
Figure S5. Binding of purified IscR to the promoter regions of iscR (A) and hemP (B) as determined by EMSAs. All reactions contain the same amount of 32P end-labeled DNA fragment (∼5 fmol/lane) comprising the promoter sequence, including the Iron-Rhodo-box motif. Lane 1 contains no IscR; lanes 2–5 contain increasing amounts of IscR (100–500 ng); lanes 6–9 contain 500 ng IscR; lanes 6 and 7 contain excess amounts of an unlabeled nonspecific probe containing the regulatory region of znuA. Lanes 8–9 contain excess amounts of the respective unlabeled DNA probe as competitor.
Figure S6. (A) Phylogenetic Tree of IscR from proteobacteria (modified from Rodionov et al. 2006; Fig. 3B). The genome abbreviations are listed in (B).
Figure S7. Homology model of the IscR monomer calculated using the MODELLER program. The cartoon representation in blue of the IscR protein shows a helix-turn-helix fold. The surface accessible residue Cys-142 is represented as a stick model in magenta.
Table S1. Rhodobacter sphaeroides and Escherichia coli strains used in this study.
Table S2. Plasmids used in this study.
Table S3. Oligodeoxynucleotide used in this study.
Table S4. Gene expression changes in 2.4.1ΔiscR as determined by microarray analysis.


