Abstract
This study aimed to find candidate strains of Lactobacillus isolated from sheep dairy products (yogurt and ewe colostrum) with probiotic and anticancer activity. A total of 100 samples were randomly collected from yogurt and colostrum and 125 lactic acid bacteria were isolated. Of these, 17 Lactobacillus strains belonging to five species (L. delbrueckii, L. plantarum, L. rhamnosus, L. paracasei, and L. casei) were identified. L. plantarum 17C and 13C, which isolated from colostrums, demonstrated remarkable results such as resistant to low pH and high concentrations of bile salts, susceptible to some antibiotics and good antimicrobial activity that candidate them as potential probiotics. Seven strains (1C, 5C, 12C, 13C, 17C, 7M, and 40M), the most resistant to simulated digestion, were further investigated to evaluate their capability to adhere to human intestinal Caco-2 cells. L. plantarum 17C was the most adherent strain. The bioactivity assessment of L. plantarum 17C showed anticancer effects via the induction of apoptosis on HT-29 human cancer cells and negligible side effects on one human epithelial normal cell line (FHs 74). The metabolites produced by this strain can be used as alternative pharmaceutical compounds with promising therapeutic indices because they are not cytotoxic to normal mammalian cells.
Keywords: Colostrum, Lactobacillus, molecular method, probiotic
Introduction
Colostrum is defined as the first milk is secreted by the mammary gland in the initial 24–96 h of the postpartum period (Solomons 2002). The production varies depending on the animal species. This initial secreted milk is a rich source of nutrients (protein, vitamins, fat, lactose and minerals), antimicrobial substances and growth factors. It also contains a high diversity of probiotic bacteria such as Lactobacillus and Bifidobacterium strains (Soccol et al. 2010; De Dea Lindner et al. 2011), which have been used widely in functional or probiotic foods.
The majority of probiotic bacteria belong to the lactic acid bacteria (LAB) group. LAB exhibit fermentation activities and have been used in food preservation for thousands of years. The species of Lactobacillus genus are the most famous group of LAB that are recognized as probiotics (Haghshenas et al. 2014; Nami et al. 2014a). Lactobacillus strains exhibit health-promoting and preservation activities (Dubernet et al. 2002) and are used as a starter culture to enhance the texture, flavor, and nutrition value of some products, such as cheese, sourdough, wine, beer, silage, fermented plant, and meat (Singh et al. 2009; Giraffa et al. 2010). Specific probiotic strains have positive effects on atopic eczema (Bunselmeyer and Buddendick 2010), irritable bowel syndrome (Parkes 2010), diarrhea (Binns and Lee 2010), antibiotic-related diarrhea (Friedman 2012), vaginal infections (Reid and Bocking 2003), inflammatory bowel disease (Geier et al. 2007), and cancers (Nami et al. 2014b,c; Serban 2014; Haghshenas et al. 2015) by stimulating the immune mechanisms and balancing the human microbiota composition. Moreover, certain strains significantly affect the bio-availability of such nutrients as magnesium and calcium in the human body (Young and Huffman 2003).
Probiotic bacteria are resistant to gastrointestinal conditions (low pH and high concentrations of bile salts) (Sahadeva et al. 2011). Some LAB carry antibiotic resistance genes and thus exhibit high resistance against antibiotics (Temmerman et al. 2003).
For probiotics to be capable of inducing to promote effects on host health, they must tolerate environments with high concentrations of bile salts and low pH and display high antimicrobial activities (Biradar et al. 2005). Therefore, this study aimed to screen the yogurt and ewe colostrum microbiota to find new strains of the Lactobacillus genus with high probiotic capability and anticancer activity by employing suitable and effective biochemical and morphological assays.
Materials and Method
Sampling, isolation, and morphological/biochemical characterization
A total of 100 samples from traditional yogurt and ewe colostrum were used for the isolation of the probiotic bacteria. Colostrum samples were obtained from the first milking of the postpartum and the starter culture used for making yogurt was from sheep's traditional yogurt. For isolation of bacteria, 1 mL of each dairy sample was suspended in 2% w/v sodium citrate solution and homogenized by using a Stomacher 400 Circulator (2 min) (Seward, Inc., London, England). Then, 1 mL of the samples was added to 10 mL of MRS (de Man, Rogosa and Sharpe) broth (Difco Laboratories, Detroit, Mich., USA) as the specific growth medium for Lactobacillus strains. After 24 h of anaerobic growth (37°C), 0.02 mL of the diluted solutions was spread for 48 h on MRS agar media.
Colonies obtained on MRS agar were extracted and transferred to 10 mL of broth culture for the enrichment step. After 24 h of incubation (37°C), screening for isolates was performed through a primary morphological test with the use of a fluorescent microscope (BX61; Olympus) (Olympus, Tokyo, Japan) and biochemical tests, such as Gram staining and catalase tests. The isolates were stored in 30% (w/v) glycerol and 10% (w/v) skim milk at −70°C (Mirzaei and Barzgari 2012).
Amplification of 16S rRNA
The amplification of 16S rRNA of Lactobacillus strains was performed by using one primer pair (16F27 5′-AGAGTTTGATCMTGGCTCAG-3′ and 16R 5′-TACCTTGTTAGGACTTCACC-3′), as previously reported by Mirzaei and Barzgari (2012). These primers are genus-specific and can directly amplify the 16S rRNA (1500 bp) of Lactobacillus strains. The amplification was performed with a 25 μL final volume containing 0.4 μmol/L primer, 40 ng of chromosomal DNA, and master mix (Ampliqon, Herlev, Denmark). The PCR program cycles were set up as follows: denaturation at 95°C for 4 min, 32 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 95 sec, and a final extension at 72°C for 5 min.
Sequencing of amplified 16S rRNA
The amplified 16S rRNA were purified by using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and then sequenced by a Korean sequencing company (Macrogene). The isolates were then identified and discriminated by blasting their sequences with BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The strains were then compared with the sequences deposited in NCBI and GenBank.
Low pH and high concentrations of bile salts tolerance tests
Bacterial stocks were incubated in 5 mL of MRS growth medium at 37°C for 24 h. Then, 200 μL of bacterial media were incubated in 5 mL of fresh medium for another 24 h. Bacterial cells were suspended by centrifuging at 2000g for 15 min and then washed twice with PBS (Phosphate-buffered saline). Thereafter, the cells were re-suspended in adjusted PBS (1 mL) with pH 2.5 or media-THIO (thioglycollate media) broth (37°C) for 3 h. The media-THIO broth was prepared by mixing MRS broth with 0.3% (w/v) oxgall and 0.2% (w/v) sodium thioglycollate (Sigma, St. Louis, MO, USA). The resistant cells were isolated, and their survival rates were calculated by using the pour plate technique in agar media; the obtained rates were compared with those of strains incubated in normal PBS for 0 and 3 h (Walker and Gilliland 1993).
In the pour plate method, the isolates were incubated in MRS broth medium at 37°C for 48 h after undergoing maximum serial dilution. The single colonies were counted, and their survival rates were measured (Charteris et al. 1998). In this method, the mean data were calculated by conducting the experiment twice with three iterations for each run. The mean values were considered for each bacterial isolate.
Antimicrobial activity assay
A modified agar diffusion method described by Bauer et al. (1966) was used to determine antimicrobial activity. After overnight incubation in MRS broth medium at 37°C, the isolates were centrifuged for 10 min at 14,000g. The supernatants were filtered by using a 0.2 μm filter. Then, 50 μL of filtered of neutralized supernatant were added to 7 mm diameter wells created on a MRS agar plate preinoculated with indicator pathogens. The agar was incubated at 37°C overnight. The clear zone formation indicated a positive antimicrobial activity of isolated metabolites on the pathogens (Bauer et al. 1966). This experiment was performed against some clinically important human pathogens including Salmonella typhimurium (14028), Staphylococcus aureus (ATCC 25923), Escherichia coli (026), Bacillus cereus (ATCC 11778), Listeria monocytogenes (PTCC 1163), Klebsiella pneumoniae (ATCC 10031), and Shigella flexneri (PTCC 1234).
Antibiotic susceptibility assay
The disk diffusion method was used to determine bacterial susceptibility against clinically important antibiotics including chloramphenicol (30 μg), tetracycline (30 μg), erythromycin (15 μg), ampicillin (10 μg), gentamycin (10 μg), clindamycin (2 μg), sulfamethoxazol (15 μg), penicillin (10 μg), and vancomycin (30 μg). The isolates were completely incubated in Mueller-Hinton agar plates, and the antibiotic disks were placed on plates with the use of sterilized forceps and then incubated at 37°C for 18–24 h. Diameters of clear zones around disks were measured by using a digital caliper. According to the results and disk producer guidelines (NCCLS, 2007), the isolates were classified into sensitive, intermediate, and resistant groups.
Survival in simulated in vitro digestion
To assess in vitro digestion, the method previously described by Seiquer et al. (2001) was used with some modifications. To recreate the gastric digestion, pepsin with a final concentration of 5% (w/v) was added to the samples, the pH values of which were adjusted to 3.0. The samples were incubated for 120 min at 37°C with gentle agitation at 110 rpm. To simulate intestinal digestion, the samples were adjusted to pH 6.0, and solutions of bile salts and pancreatin were added at final concentrations of 0.3% and 0.1% (w/v), respectively. The samples were incubated at 37°C for 180 min with gentle agitation at 110 rpm. To determine cell count, the samples were removed before and after gastric and intestinal digestion, and the aliquots were serially diluted and plated in triplicate on MRS agar. The plates were incubated for 48 h under anaerobic conditions.
Adhesion of LAB to Caco-2 cells
Bacteria were evaluated for their adhesion capability to the human colon carcinoma cell line Caco-2. The cells were cultured in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin–streptomycin mixture. Cells were cultured on 24-well tissue culture plates and incubated at 37°C in 5% CO2 under a relatively humidified atmosphere until a confluent monolayer was formed.
Before the adhesion assay, the media in the wells containing a Caco-2 cell monolayer were removed and replaced once with fresh antibiotic-free RPMI. Thereafter, 1 × 107 cfu/mL of bacteria was added to each well with a total volume of 1 mL and then incubated for 3 h at 37°C under an atmosphere of 5% (v/v) CO2. To remove nonattached bacterial cells, the wells were washed thrice with a sterile prewarmed PBS solution. To detach the bacteria cells from the wells, 1 mL of 1% (v/v) Triton X-100 was added to each well, and the mixture was stirred for 10 min. To measure the viable cell count, the cell suspension was plated onto MRS agar and incubated at 37°C under anaerobic conditions. This assay was performed in triplicate.
MTT assay
The cytotoxicity of the isolated Lactobacillus strains to tumor/normal cells was evaluated by using the microculture tetrazolium [MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. Briefly, HT-29 and FHs 74 cells (1.2 × 104 cell/well) were seeded into each well of a 96-well microplate with RPMI growth medium. After reaching 50% confluence, 24 h after seeding, the cells were treated with the filtered supernatant of the isolated strain at a time point of 24 h. After treatment, the medium was replaced with 200 μL of fresh medium containing 50 μL of MTT solution (2 mg/mL in PBS) and then incubated for an additional 4 h at 37°C. After incubation, the MTT mixture was carefully removed, and 200 μL of dimethyl sulfoxide and 25 μL of Sorenson's glycine buffer (0.1 mol/L glycine, 0.1 mol/L NaCl at pH 10.5) were added to each well. The mixture was then incubated for 30 min. Finally, the absorbance of each well was measured after 30 sec of shaking by using a microplate reader (ELx 800; Biotek, Winooski, VT, USA) at 570 nm. The cells were treated with MRS (bacterial culture medium) and Taxol (Tocris Cookson Ltd., Bristol, UK) (anticancer drug as a reference) as negative and positive controls, respectively.
Apoptotic cell detection by fluorescent microscopy
To detect apoptotic cells, sterile cover slip slides were placed into each well of a six-well culture plate. Then, 2 mL of HeLa cancer cells (1.2 × 104 cells/mL) were added to each well and then incubated at 37°C under 5% CO2. After reaching 50–60% confluence, 500 μL of the cell-free filtered supernatant of selected strains were added to each well and incubated for 24 h at same condition. The treated cells were washed with prewarmed tissue culture media 24 h after seeding and were carefully removed and replaced with freshly prepared fixative solution (prewarmed RPMI containing 4% formaldehyde).After the plates were incubated for 5 min at 37°C, the cells were fixed, washed twice with PBS, and then permeabilized with PBS containing 0.1% Triton X-100 for 5 min at 37°C. The cells were then stained with 50 μL per well of DAPI 4′6-diamidino-2-phenylindole (Sigma, St. Louis, MO, USA) (1:2000 dilution) for 3 min of incubation at room temperature. Finally, the slides were washed with PBS and then assessed by using fluorescent microscopy.
Statistical analysis
All data were analyzed by one-way ANOVA. Significant differences in means (P < 0.05) were then compared by Duncan's test using the SPSS (SPSS Inc, Chicago, IL, USA) 19.0 software. All graphs were prepared by using Microsoft Office Excel.
Results and Discussion
Sampling, isolation, and morphological/biochemical characterization
The isolated single colonies in the MRS agar growth medium were hemispherically round with white or yellow color. By performing morphological and biochemical tests, the rod- or spherical-shaped gram-positive and catalase-negative single colonies that could belong to the Lactobacillus genus were isolated. A total of 125 bacteria were isolated from yogurt and colostrum. Of these, only 17 isolates belonged to Lactobacillus genus (Table1).
Table 1.
Sequencing results for 17 representative isolates and survival rate of 17 representative isolates in simulated digestion condition and their capacity to adhere to Caco-2 cells
| Isolates | Sequencing results | Dairy origen | Digestion survival (%) | Adhesion to Caco-2 (cfu/mL) |
|---|---|---|---|---|
| 11C | Lactobacillus delbrueckii | Colostrum | 0 | 0 |
| 17C | Lactobacillus plantarum | Colostrum | 54 | 2.9 × 105 |
| 13C | Lactobacillus plantarum | Colostrum | 51 | 2.6 × 105 |
| 12C | Lactobacillus rhamnosus | Colostrum | 47 | 4.9 × 104 |
| 5C | Lactobacillus rhamnosus | Colostrum | 45 | 4.1 × 104 |
| 1C | Lactobacillus rhamnosus | Colostrum | 37 | 3.2 × 104 |
| 15C | Lactobacillus casei | Colostrum | 0 | 0 |
| 25C | Lactobacillus casei | Colostrum | 0 | 0 |
| 36C | Lactobacillus casei | Colostrum | 0 | 0 |
| 14C | Lactobacillus paracasei | Colostrum | 0 | 0 |
| 32C | Lactobacillus paracasei | Colostrum | 0 | 0 |
| 17M | Lactobacillus casei | Yogurt | 0 | 0 |
| 20M | Lactobacillus casei | Yogurt | 0 | 0 |
| 19M | Lactobacillus paracasei | Yogurt | 0 | 0 |
| 13M | Lactobacillus paracasei | Yogurt | 0 | 0 |
| 40M | Lactobacillus plantarum | Yogurt | 32 | 2.8 × 104 |
| 7M | Lactobacillus rhamnosus | Yogurt | 48 | 2.6 × 104 |
Amplification and sequencing of 16S rRNA
The presence of Lactobacillus strains in the samples was confirmed by amplification of 1500 bp fragments of 16S rRNA gene with genus-specific primers. Among 17 strains, 11 were isolated from colostrum, whereas six were isolated from yogurt. According to results and FAO/WHO guidelines, identification of microorganisms by 16S rRNA patterns can be considered as a more suitable technique than other costly and time-consuming molecular techniques (Ben Amor et al. 2007). This technique has been effectively used for analyzing and isolating lactic and acetic acid bacteria from fermented dairy products (Pogačić et al. 2013; Tulini et al. 2013). The result confirmed that this method can be used as an accessible, low-cost, and suitable technique for isolating Lactobacillus strains from traditional dairy products.
The 1500 bp fragments of 16S rRNA gene were sequenced. By using BLAST software in NCBI site and comparing the results with deposited sequences in GenBank, we identified the isolates with 98–100% homology. The threshold value for taxonomical studies was approximately 97%. Although, 16S rRNA sequencing was a valid and accurate technique for phylogenetic clustering (Deng et al. 2008), the sequencing results should be compared with other molecular methods like GTG-PCR, ERIC-PCR, and ARDRA outcomes.
The strains belonged to five species of Lactobacillus genus (L. delbrueckii, L. plantarum, L. rhamnosus, L. paracasei, and L. casei). The results of our study and those of previous studies imply that the homology levels between some Lactobacillus strains reached more than 99% (Ritchie et al. 2010). Hence, this technique is valid for discrimination at the species level but is invalid and insufficient for discrimination at the strain levels. Sequencing results were comparable with those of other molecular methods, such as GTG-PCR, ERIC-PCR, and ARDRA. The biodiversity of Lactobacillus species in fermented dairy products is variable and region specific. In traditional Spanish cheese (Armada cheese), the predominant Lactobacilli are L. casei subsp. casei and L. brevis (Herreros et al. 2003). Meanwhile, in Greek goat cheese (Batzos cheese), L. paracasei and L. sakei are the dominant species (Psoni et al. 2003), whereas in Brazilian fresh cheese (Minas Frescal cheese) the predominant species is L. acidophilus (Lollo et al. 2012). It revealed that our results are quite different from those in terms of species and prevalence.
Low pH and high concentrations of bile salts tolerance tests
Most probiotic bacteria are delivered through food. Therefore, these bacteria must survive for a minimum 90 min under low pH conditions or in the presence of secreted high concentrations of bile salts in the digestive system before colonization in the gastrointestinal tract and before displaying health-promoting effects. These defense mechanisms destroy the entering microorganisms, such that the assessment of resistance under harsh conditions is important (Sahadeva et al. 2011).
The in vitro low pH and high concentrations of bile salts tolerance experiments show the same cell viability results. The assessment of the harsh condition tolerance of probiotic bacteria can be performed by using an in vitro method with the same pH (pH 2.5) or concentration of oxgall [0.3% (w/v)] in the gastrointestinal tract (Ben Salah et al. 2012).
The survival rates for 17 isolated Lactobacillus strains after 3 h of incubation at pH 2.5 and the substance at 0.3% (w/v) high concentrations of bile salts are shown in Table2. All isolates displayed high growth at low pH and high concentrations of bile salts.
Table 2.
The survival rates of strains after 3 h incubation at pH 2.5 and 0.3% bile salts
| Isolates | Final counts (log CFU/mL) after incubation at | Final counts (log CFU/mL) after incubation at | ||
|---|---|---|---|---|
| 0 h | 3 h | 0 h | 3 h | |
| 11C | 9.02 ± 0.02 | 5.86 ± 0.04 | 8.92 ± 0.03 | 6.96 ± 0.02 |
| 17C | 8.96 ± 0.03 | 7.17 ± 0.03 | 9.05 ± 0.02 | 7.96 ± 0.03 |
| 13C | 8.12 ± 0.02 | 6.17 ± 0.01 | 9.17 ± 0.05 | 7.98 ± 0.04 |
| 12C | 8.47 ± 0.04 | 6.69 ± 0.02 | 8.99 ± 0.03 | 8.00 ± 0.03 |
| 5C | 8.51 ± 0.05 | 6.47 ± 0.03 | 8.93 ± 0.02 | 8.04 ± 0.03 |
| 1C | 8.77 ± 0.03 | 7.19 ± 0.03 | 9.07 ± 0.05 | 8.07 ± 0.02 |
| 15C | 8.94 ± 0.01 | 5.54 ± 0.03 | 9.11 ± 0.05 | 6.65 ± 0.03 |
| 25C | 9.07 ± 0.02 | 5.44 ± 0.05 | 8.88 ± 0.03 | 6.39 ± 0.05 |
| 36C | 9.38 ± 0.04 | 6.00 ± 0.04 | 9.18 ± 0.04 | 6.98 ± 0.02 |
| 14C | 8.99 ± 0.02 | 5.21 ± 0.02 | 9.09 ± 0.03 | 6.45 ± 0.03 |
| 32C | 8.82 ± 0.04 | 5.29 ± 0.03 | 9.15 ± 0.03 | 6.50 ± 0.05 |
| 17M | 8.69 ± 0.05 | 5.39 ± 0.05 | 8.93 ± 0.02 | 6.70 ± 0.02 |
| 20M | 8.74 ± 0.05 | 5.68 ± 0.04 | 8.97 ± 0.03 | 6.55 ± 0.04 |
| 19M | 8.78 ± 0.04 | 5.00 ± 0.04 | 8.77 ± 0.04 | 6.05 ± 0.03 |
| 13M | 9.20 ± 0.03 | 5.80 ± 0.02 | 9.13 ± 0.04 | 6.57 ± 0.02 |
| 40M | 9.15 ± 0.02 | 7.14 ± 0.03 | 9.17 ± 0.05 | 7.79 ± 0.02 |
| 7M | 9.03 ± 0.04 | 7.22 ± 0.01 | 9.00 ± 0.02 | 8.19 ± 0.02 |
The isolates showed high survival rates of more than 57% of that of other strains under low pH conditions. The survival rates of the L. plantarum and L. rhamnosus strains (76–82%) were also significantly higher than those of the other strains (L. delbrueckii, L. casei, and L. paracasei), which ranged from 57% to 65%, at a 0.05 significance level. These results are in accordance with those of other studies, which showed that some Lactobacillus strains, such as L. plantarum, tolerate low pH conditions better than other strains (Abriouel et al. 2012).
The isolates displayed higher tolerance of high concentrations of bile salts conditions (≥69%). The high concentrations of bile salts tolerance capability was strain specific, similar to the case under acidic conditions. The effects of high concentrations of bile salts on bacterial cells can be distinguished from the acidic effects, but some combined results can be observed. Stress adaptation mechanisms caused the resistance to high concentrations of bile salts to be higher than that under acidic conditions (Sahadeva et al. 2011). The high tolerance capability of Lactobacillus strains under high concentrations of bile salts has been reported by other researchers (Pan et al. 2009).
Similar to those under acidic conditions, the survival rates for L. plantarum and L. rhamnosus strains (85–91%) were higher than those of the other strains (L. delbrueckii, L. casei, and L. paracasei) at a range of 69–78% at a 0.05 significance level.
Survival in simulated in vitro digestion and adhesion assay to Caco-2 cells
One of the most desirable features of probiotics is their capability to remain alive in the gastrointestinal tract. A total of 17 isolated Lactobacillus were tested to evaluate the strains further through a simulated digestion test. Results showed that seven strains have considerable digestion survivability. The resistant strains were three L. plantarum and four L. rhamnosus. Five of these resistant strains were isolated from colostrum, whereas two were isolated from yogurt. The highest percentage of survivability was observed for L. plantarum 17C and 13C (both isolated from colostrum), with survivability values of 54% and 51%, respectively (Table2). Similar to our results, high survival rates were reported for three commercial probiotic strains including L. casei subsp. shirota, L. casei subsp. immunitas, and L. acidophilus subsp. Johnsonii in simulated digestion condition (Lo Curto et al. 2011).
These seven digestion-resistant LAB strains were examined for their capability to adhere to Caco-2 cells. The results showed that L. plantarum 17C and 13C were the most adherent strains, with adhesion values of 2.9 × 105 (≈3 bacteria per Caco-2 cell) and 2.6 × 105 cfu/mL (≈2.5 bacteria per Caco-2 cell), respectively (Table1). These results are in accordance with those of other studies, which showed that some L. plantarum strains, such as L. plantarum L2, L. plantarum CH3 and CH41, could adhere to Caco-2 cells better than other Lactobacilli (Wang et al. 2009; Ramos et al. 2013).
A requirement for bacteria to be recognized as a probiotic is their capability to remain alive while passing through the upper digestive tract to reach the large intestine, where their useful actions are expected. To be colonized in the intestine, probiotic bacteria have to adhere to the intestinal mucosa to avoid being removed from the colon by peristalsis. Seventeen “dairy isolates” were examined for this test. Among these isolates, only seven strains survived exposure to the simulated digestion conditions of the stomach. The capability to adhere to Caco-2 cells was investigated for further analysis. We found that the strains L. plantarum 17C and 13C isolated from colostrum were the most resistant strains to digestion conditions and were the best strains in terms of adherence to Caco-2 cells. This result showed that these strains could be new potential probiotic candidates and that colostrum could be a suitable source of probiotics.
Antimicrobial activity assay
The inhibitory characteristics of probiotics against pathogens can be considered as their most important health-promoting properties (Cizeikiene et al. 2013; Gerez et al. 2013). According to the diameter of the inhibition zone, the antipathogen activity was classified into strong (diameter ≥ 20 mm), moderate (20 mm > diameter > 10 mm), and weak (diameter ≤ 10 mm) (Lim 2010).
Among the 17 isolates, 15 isolates displayed moderate antimicrobial activities, whereas two isolates (L. paracasei 19M and L. casei 25C) did not exhibit any antagonistic activities. In contrast, gram-negative pathogens, such as S. typhimurium and E. coli (026), were more sensitive to extracted metabolites than gram-positive pathogens, such as B. cereus and L. monocytogenes. This finding can be linked to the thin cell walls of gram-negative pathogens and their susceptibility to acidic metabolites (Table3).
Table 3.
Antimicrobial activity of isolates against the pathogenic bacteria
| Diameter of inhibition zone (mm) | |||||||
|---|---|---|---|---|---|---|---|
| Bacteria | Salmonella typhimurium | Staphylococcus aureus | Escherichia coli (026) | Bacillus cereus | Listeria monocytogenes | Klebsiella pneumoniae | Shigella flexneri |
| Lactobacillus delbrueckii 11C | 11.8 ± 0.4a | 0.0 ± 0.0b | 12.0 ± 0.6a | 0.0 ± 0.0b | 12.7 ± 1.2a | 0.0 ± 0.0b | 12.3 ± 0.3a |
| Lactobacillus plantarum 17C | 12.3 ± 1.2b | 11.7 ± 0.3b | 12.3 ± 0.7b | 10.0 ± 0.0c | 13.7 ± 0.9a | 12.0 ± 0.6b | 12.0 ± 0.0b |
| Lactobacillus plantarum 13C | 12.3 ± 0.7bc | 12.0 ± 0.6c | 12.0 ± 0.0c | 11.7 ± 1.2c | 14.0 ± 0.6a | 13.3 ± 0.3ab | 12.7 ± 0.0bc |
| Lactobacillus plantarum 40M | 14.0 ± 1.0b | 10.3 ± 0.3d | 15.7 ± 0.3a | 12.0 ± 0.0c | 0.0 ± 0.0e | 0.0 ± 0.0e | 12.0 ± 1.0c |
| Lactobacillus rhamnosus 12C | 15.3 ± 0.7a | 0.0 ± 0.0d | 13.0 ± 0.6c | 0.0 ± 0.0d | 14.0 ± 0.6b | 0.0 ± 0.0d | 0.0 ± 0.0d |
| Lactobacillus rhamnosus 5C | 13.3 ± 0.3c | 13.0 ± 0.0c | 15.7 ± 0.3b | 0.0 ± 0.0d | 17.0 ± 0.0a | 13.3 ± 1.2c | 14.0 ± 0.6c |
| Lactobacillus rhamnosus 1C | 16.7 ± 0.3a | 13.0 ± 0.0bc | 14.0 ± 1.0b | 12.3 ± 1.2c | 0.0 ± 0.0d | 0.0 ± 0.0d | 13.3 ± 0.3bc |
| Lactobacillus rhamnosus 7M | 12.8 ± 0.8a | 0.0 ± 0.0b | 12.3 ± 1.2a | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b | 12.0 ± 0.6a |
| Lactobacillus casei 15C | 0.0 ± 0.0c | 0.0 ± 0.0c | 0.0 ± 0.0c | 0.0 ± 0.0c | 0.0 ± 0.0c | 13.3 ± 0.7a | 11.0 ± 0.0b |
| Lactobacillus casei 25C | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a |
| Lactobacillus casei 36C | 0.0 ± 0.0c | 0.0 ± 0.0c | 13.7 ± 1.2a | 0.0 ± 0.0c | 0.0 ± 0.0c | 12.7 ± 0.3b | 0.0 ± 0.0c |
| Lactobacillus casei 17M | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b | 12.0 ± 0.0a | 0.0 ± 0.0b |
| Lactobacillus casei 20M | 15.0 ± 0.6a | 0.0 ± 0.0d | 11.0 ± 0.0c | 0.0 ± 0.0d | 0.0 ± 0.0d | 12.3 ± 1.2b | 12.0 ± 0.0b |
| Lactobacillus paracasei 14C | 0.0 ± 0.0c | 0.0 ± 0.0c | 0.0 ± 0.0c | 0.0 ± 0.0c | 0.0 ± 0.0c | 12.7 ± 1.2a | 11.0 ± 0.0b |
| Lactobacillus paracasei 32C | 12.0 ± 1.0a | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b |
| Lactobacillus paracasei 19M | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.0a |
| Lactobacillus paracasei 13M | 0.0 ± 0.0c | 13.7 ± 1.2a | 0.0 ± 0.0c | 0.0 ± 0.0c | 0.0 ± 0.0c | 13.0 ± 1.0a | 11.3 ± 0.7b |
Salmonella typhimurium (14028); Staphylococcus aureus (ATCC 25923); Escherichia coli (026) (native strain); Bacillus cereus (ATCC 11778); Listeria monocytogenes (PTCC 1163); Klebsiella pneumoniae (ATCC 10031); Shigella flexneri (PTCC 1234) values are mean ± standard error mean (SEM). All incubations were performed in triplicate. Within a row, values with different superscripts are significantly different at P ≤ 0.05.
The antimicrobial activities of Lactobacillus strains can be attributed to the secretion of different antipathogen substances, such as bacteriocins, biosurfactants, H2O2, and organic acids (hydrochloric, lactic, and acetic acids) (Giraffa et al. 2010).
Among the treated isolates, L. plantarum strains, particularly L. plantarum 17C and L. plantarum 13C, displayed good antagonistic activities, as reflected by the inhibition of all indicator pathogens. Similar to our results, high antagonistic activity was reported for L. plantarum strains isolated from fermented food and dairy products against different pathogens (Gupta and Srivastava 2014; Ryu et al. 2014).
Antibiotic susceptibility assay
Antibiotic overuse has caused the antibiotic resistance genes to spread across regions and transfer onto probiotic societies, such that the sensitivity of probiotics to conventional antibiotics is a fundamental health-promoting characteristic which causes to avoid resistant genes flow to other pathogenic/nonpathogenic bacteria especially intestine microflora (Temmerman et al. 2003).
The antibiotic susceptibility of 17 isolates against the high consumption of clinically important antibiotics was assessed on the basis of the formation of inhibition zones (Table4). The results were expressed in terms of resistance, moderate susceptibility, or susceptibility by comparing with the interpretative zone diameters given by the performance standards for antimicrobial disk susceptibility tests (NCCLS, 2007).
Table 4.
Antibiotic susceptibility of isolated bacteria against the high consumption antibiotics performed by disk diffusion assay
| Diameter of inhibition zone (mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolates | C | TE | E | AM | GE | CC | SMZ | P | V |
| Lactobacillus delbrueckii 11C | 28S | 30S | 28S | 18S | 13S | 29S | 22S | 22S | 28S |
| Lactobacillus plantarum 17C | 25S | 22S | 22I | 30S | 28S | 24S | 24S | 30S | 0R |
| Lactobacillus plantarum 13C | 30S | 28S | 21I | 38S | 12S | 20S | 22S | 40S | 0R |
| Lactobacillus plantarum 40M | 22S | 26S | 26S | 26S | 30S | 28S | 0R | 28S | 0R |
| Lactobacillus rhamnosus 12C | 14S | 20S | 25S | 29S | 16S | 29S | 0R | 15I | 26S |
| Lactobacillus rhamnosus 5C | 14I | 22S | 0R | 11R | 14S | 30S | 22S | 13I | 0R |
| Lactobacillus rhamnosus 1C | 13I | 14I | 26S | 22S | 0R | 17I | 0R | 26S | 13I |
| Lactobacillus rhamnosus 7M | 36S | 28S | 0R | 0R | 22S | 26S | 20S | 0R | 0R |
| Lactobacillus casei 15C | 28S | 30S | 26S | 28S | 30S | 30S | 0R | 32S | 0R |
| Lactobacillus casei 25C | 20S | 20S | 20I | 26S | 18S | 28S | 15I | 28S | 0R |
| Lactobacillus casei 36C | 22S | 30S | 0R | 30S | 22S | 22S | 30S | 30S | 0R |
| Lactobacillus casei 17M | 0R | 28S | 28S | 28S | 0R | 28S | 26S | 0R | 0R |
| Lactobacillus casei 20M | 28S | 22S | 25S | 26S | 24S | 30S | 32S | 0R | 0R |
| Lactobacillus paracasei 14C | 22S | 30S | 0R | 30S | 18S | 22S | 20S | 22S | 20S |
| Lactobacillus paracasei 32C | 30S | 20S | 30S | 26S | 30S | 26S | 0R | 26S | 26S |
| Lactobacillus paracasei 19M | 22S | 14I | 26S | 28S | 20S | 30S | 0R | 30S | 22S |
| Lactobacillus paracasei 13M | 20S | 20S | 28S | 28S | 22S | 25S | 26S | 20S | 18S |
Erythromycin results based on R < 13 mm; I: 13–23 mm; S > 23 mm. Gentamycin results based on R ≤ 6 mm; I: 7–9 mm; S ≥ 10 mm. Vancomycin results based on R < 12 mm; I: 12–13 mm; S > 13 mm. Chloramphenicol, Tetracycline, Ampicillin, Clindamycin, Penicillin, and Sulfamethoxazol based on I: intermediate (zone diameter, 12.5–17.4 mm); R: resistant (zone diameter, ≤12.4 mm); S: susceptible (zone diameter, ≥17.5) (NCCLS, 2007). C, chloramphenicol (30 μg); TE, tetracycline (30 μg); E, erythromycin (15 μg); AM, ampicillin (10 μg); GE, gentamycin (10 μg); CC, clindamycin (2 μg); SMZ, sulfamethoxazol (15 μg); P, penicillin (10 μg); V, vancomycin (30 μg).
Isolated Lactobacillus strains displayed high sensitivity to treated antibiotics. Meanwhile, resistance to tetracycline, cefixime, pefloxacin, neomycin, enoxacin, sulfamethoxazol, lincosamide, cloxacillin, penicillin G, streptomycin, gentamycin, erythromycin, and chloramphenicol was reported among Lactobacillus strains isolated from dairy products (Bernardeau et al. 2008).
All isolates were susceptible or intermediate to tetracycline and clindamycin. The high resistant strains to tetracycline among the probiotic bacteria were reported by other researchers (Chang et al. 2011). Despite limited reports on clindamycin resistance genes among probiotics (Casado Muñoz et al. 2014), these resistance genes can be easily transferred to pathogenic strains, such as S. aureus, and thus cause outbreaks of bacteremia (Dubey et al. 2013). The sensitivity to these antibiotics can probably be attributed to the limited use of antibiotics in the rural area of Iran (Kermanshah province), such that isolated probiotics can safely be consumed after antibiotic (tetracycline and clindamycin) therapy.
The maximum resistance was observed for vancomycin, more than half of the strains (10 strains) displayed high resistance. The strict homo-fermentative Lactobacillus species, such as L. paracasei strains, are sensitive to vancomycin, which agrees with our results. Other Lactobacillus strains, such as L. casei and L. plantarum, which are similar to our isolates, carry the vancomycin resistance genes (Bernardeau et al. 2008).
L. delbrueckii 11C and L. paracasei 13M displayed the best results and were sensitive or semisensitive to all antibiotics, whereas L. rhamnosus 7M and L. casei 17M, were the most resistant strains among isolates on the basis of their resistance to four antibiotics.
Cell viability assay
The MTT method is commonly used to scrutinize the cytotoxicity and viability of living cells in a 96-well plate format. This evaluation was performed by assessing L. plantarum secretion metabolites on cancerous/normal human cell lines to determine their capability for growth inhibition using the MTT assay. Paclitaxel, a standard commercial drug for treating different cancers, was utilized as a positive control. The MTT assay is a colorimetric method based on the metabolic capability of cells to decrease the yellow tetrazolium salt 3-[4, 5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide or MTT to a blue crystalline formazan product. This method is widely used to analyze the cytotoxicity and cell viability of living cells in a 96-well plate.
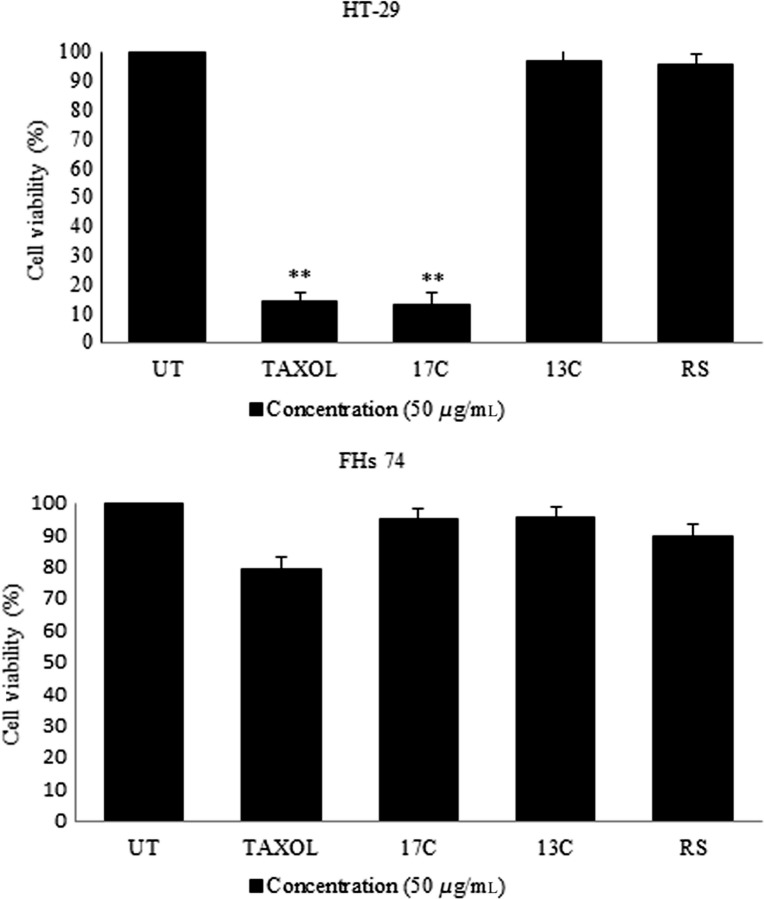
Figure1A illustrates the cytotoxicity potential of L. plantarum 17C and 13C supernatant on HT-29 cancer cell. Lactobacillus plantarum ATCC 8014 was used as a reference strain for comparison. The viability of HT-29 carcinoma cells was inhibited by L. plantarum 17C secretions after 24 h of incubation, and the viable cells of HT-29 comprised 13% after 24 h of incubation. Our findings revealed that the antiproliferative effect of L. plantarum 17C secretion metabolites on HT-29 cancer cells had significant differences with that of the L. plantarum 13C and L. plantarum ATCC 8014 as control. We also used normal FHs 74 to determine the effect of this strain on normal cells (Fig.1B). No significant cytotoxic effects for these cells were observed, and approximately 94% of the normal cells grew well.
Figure 1.
Effect of Lactobacillus plantarum17C and 13C supernatant on the viability of HT-29 cancer cells and FHs 74 normal cell by 50 μg/mL concentration and 24 h incubation. Data are expressed as mean viability ratio ± SD. Asterisks denote statistically significant differences (**p ≤ 0.01; *p ≤ 0.05).All incubations were carried out in triplicate. UT means untreated and RS means reference strain L. plantarum ATCC 8014 to compare their effects.
Apoptosis detection by DAPI staining
Apoptosis is the primary means of programmed cell death and serves a significant function in regulating tissue development and homeostasis. Hence, the induction of apoptotic cell death is a favorable emerging scheme for the inhibition and treatment of cancer. Morphological changes offer the most direct criteria for distinguishing the apoptotic process. Thus, fluorescence microscopy was used to detect apoptosis on the basis of changes in cellular morphology, condensation and fragmentation of nuclei, as well as cell shrinkage and membrane blebbing.
To scrutinize the effect of L. plantarum 17C on HT-29 cell viability, HT-29 cells were exposed to the supernatant of the late stationary phase growth of L. plantarum 17C and analyzed by fluorescent microscopy (Olympus BX61). None of apoptotic-related signals were observed in the nontreated HT-29 cells (Fig.2A). Significant numbers of apoptotic cells were found in the cells after 24 h of incubation with 50 μg/mL of L. plantarum 17C supernatant. The number of apoptotic cells with condensed and fragmented nuclei was significantly higher than that of blue intact normal cells. The treated HT-29 cells showed distinctive signs of apoptosis, including micronucleus formation, membrane blebbing, cell shrinkage, apoptotic bodies, and nuclear fragmentation, after 24 h of exposure (Fig.2B). Membrane blebbing and micronucleus formation were prominent apoptotic features of the HT-29 cells treated with L. plantarum 17C.
Figure 2.
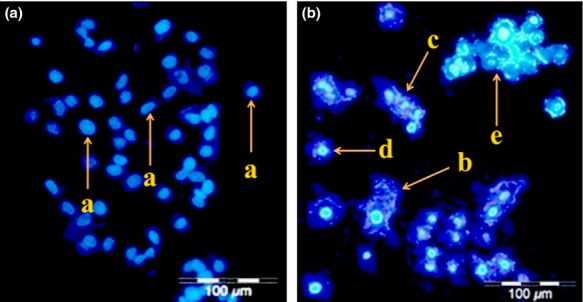
Detection of normal and apoptotic cells of HT-29 cancer cells without treating by Lactobacillus plantarum supernatant (A) and with treating by L. plantarum supernatant (B) after 24 h incubation. a: blue intact normal cell; b: membrane blebbing; c: nucleus fragmentation; d: cell shrinkage; e: apoptotic bodies.
In conclusion, traditional yogurt and colostrum were preliminary screened because variable animal feeding and rare use of antibiotics can introduce novel and promising probiotic bacteria. Our findings indicated that L. plantarum 17C and L. plantarum 13C strains, which were isolated from colostrum, displayed a desirable tolerance to low pH and high concentrations of bile salts, favorable antipathogen activity, and acceptable antibiotic susceptibility. Thus, these two strains can be considered as potential probiotics. In addition, L. plantarum 17C showed significant antiproliferative effects on HT-29 human colon cancer cell line.
Acknowledgments
The financial support from the University Putra Malaysia, the Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran, and the moral patronage of Abolfazl Barzegari and Simin Sharifi are gratefully acknowledged.
Conflict of Interest
None declared.
References
- Abriouel H, Benomar N, Cobo A, Caballero N, Fernandez Fuentes MA, Perez-Pulido R, et al. Characterization of lactic acid bacteria from naturally-fermented Manzanilla Aloreña green table olives. Food Microbiol. 2012;32:308–316. doi: 10.1016/j.fm.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Bauer AW, Kirby WM, Sherris JC. Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- Ben Amor K, Vaughan EE. de Vos WM. Advanced molecular tools for the identification of lactic acid bacteria. J. Nutr. 2007;137:741S–747S. doi: 10.1093/jn/137.3.741S. [DOI] [PubMed] [Google Scholar]
- Ben Salah R, Trabelsi I, Ben Mansour R, Lassoued S, Chouayekh H. Bejar S. A new Lactobacillus plantarum strain, TN8, from the gastro intestinal tract of poultry induces high cytokine production. Anaerobe. 2012;18:436–444. doi: 10.1016/j.anaerobe.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Bernardeau M, Vernoux JP, Henri-Dubernet S. Gueguen M. Safety assessment of dairy microorganisms: the Lactobacillus genus. Int. J. Food Microbiol. 2008;126:278–285. doi: 10.1016/j.ijfoodmicro.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Binns C. Lee MK. The use of probiotics to prevent diarrhea in young children attending child care centers: a review. J. Exp. Clin. Med. 2010;2:269–273. [Google Scholar]
- Biradar SS, Bahagvati ST. Shegunshi B. Probiotics and antibiotics: a brief overview. Int. J. Nutr. Wellness. 2005 , and http://ispub.com/IJNW/2/1/7058. [Google Scholar]
- Bunselmeyer B. Buddendick K. Chapter 19 - Probiotics and prebiotics-prevention and therapy in atopic eczema. In: Watson RR, Preedy VR, editors; Bioactive Foods in Promoting Health. Boston: Academic Press; 2010. pp. 279–292. , eds., and. [Google Scholar]
- Casado Muñoz MDC, Benomar N, Lerma LL, Gálvez A. Abriouel H. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented Aloreña table olives throughout fermentation process. Int. J. Food Microbiol. 2014;172:110–118. doi: 10.1016/j.ijfoodmicro.2013.11.025. [DOI] [PubMed] [Google Scholar]
- Chang YH, Cui SW, Roberts KT, Ng PKW. Wang Q. Evaluation of extrusion-modified fenugreek gum. Food Hydrocol. 2011;25:1296–1301. [Google Scholar]
- Charteris WP, Kelly PM, Morelli L. Collins JK. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 1998;84:759–768. doi: 10.1046/j.1365-2672.1998.00407.x. [DOI] [PubMed] [Google Scholar]
- Cizeikiene D, Juodeikiene G, Paskevicius A. Bartkiene E. Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Con. 2013;31:539–545. [Google Scholar]
- De Dea Lindner J, Santarelli M, Tiemi Yamaguishi C, Ricardo Soccol C. Neviani E. Recovery and identification of bovine colostrum microflora using traditional and molecular approaches. Food Tech. Biotech. 2011;49:364–368. [Google Scholar]
- Deng W, Xi D, Mao H. Wanapat M. The use of molecular techniques based on ribosomal RNA and DNA for rumen microbial ecosystem studies: a review. Mol. Biol. Rep. 2008;35:265–274. doi: 10.1007/s11033-007-9079-1. [DOI] [PubMed] [Google Scholar]
- Dubernet S, Desmasures N. Guéguen M. A PCR-based method for identification of lactobacilli at the genus level. FEMS Microbiol. Lett. 2002;214:271–275. doi: 10.1111/j.1574-6968.2002.tb11358.x. [DOI] [PubMed] [Google Scholar]
- Dubey D, Rath S, Sahu MC, Rout S, Debata NK. Padhy RN. A report on infection dynamics of inducible clindamycin resistance of Staphylococcus aureus isolated from a teaching hospital in India. Asian Pacific J. Trop. Biomed. 2013;3:148–153. doi: 10.1016/S2221-1691(13)60040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman G. The Role of probiotics in the prevention and treatment of antibiotic-associated diarrhea and Clostridium difficile colitis. Gastroenterol. Clin. North Am. 2012;41:763–779. doi: 10.1016/j.gtc.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Geier MS, Butler RN. Howarth GS. Inflammatory bowel disease: current insights into pathogenesis and new therapeutic options; probiotics, prebiotics and synbiotics. Int. J. Food Microbiol. 2007;115:1–11. doi: 10.1016/j.ijfoodmicro.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Gerez CL, Torres MJ, Font de Valdez G. Rollán G. Control of spoilage fungi by lactic acid bacteria. Biol. Con. 2013;64:231–237. [Google Scholar]
- Giraffa G, Chanishvili N. Widyastuti Y. Importance of lactobacilli in food and feed biotechnology. Res. Microbiol. 2010;161:480–487. doi: 10.1016/j.resmic.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Gupta R. Srivastava S. Antifungal effect of antimicrobial peptides (AMPs LR14) derived from Lactobacillus plantarum strain LR/14 and their applications in prevention of grain spoilage. Food Microbiol. 2014;42:1–7. doi: 10.1016/j.fm.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Haghshenas B, Abdullah N, Nami Y, Radiah D, Rosli R. Yari Khosroushahi A. Different effects of two newly-isolated probiotic Lactobacillus plantarum 15HN and Lactobacillus lactis subsp. lactis 44Lac strains from traditional dairy products on cancer cell lines. Anaerobe. 2014;30:51–59. doi: 10.1016/j.anaerobe.2014.08.009. [DOI] [PubMed] [Google Scholar]
- Haghshenas B, Nami Y, Abdullah N, Radiah D, Rosli R. Yari Khosroushahi A. Anticancer impacts of potentially probiotic acetic acid bacteria isolated from traditional dairy microbiota. LWD- Food Sci. Technol. 2015;60:690–697. [Google Scholar]
- Herreros MA, Fresno JM, González Prieto MJ. Tornadijo ME. Technological characterization of lactic acid bacteria isolated from Armada cheese (a Spanish goats’ milk cheese) Int. Dairy J. 2003;13:469–479. [Google Scholar]
- Lim YS.2010. Probiotic Characteristics of Bacteriocinogenic Lactobacillus Plantarum Strains Isolated from Malaysian Foods PhD thesis, Universiti Putra Malaysia. 192–208.
- Lo Curto A, Pitino I, Mandalari G, Dainty JR, Faulks RM. John Wickham MS. Survival of probiotic lactobacilli in the upper gastrointestinal tract using an in vitro gastric model of digestion. Food Microbiol. 2011;28:1359–1366. doi: 10.1016/j.fm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Lollo PCB, Cruz AG, Morato PN, Moura CS, Carvalho-Silva LB, Oliveira CAF, et al. Probiotic cheese attenuates exercise-induced immune suppression in Wistar rats. J. Dairy Sci. 2012;95:3549–3558. doi: 10.3168/jds.2011-5124. [DOI] [PubMed] [Google Scholar]
- Mirzaei H. Barzgari A. Isolation and molecular study of potentially probiotic lactobacilli in traditional white cheese of Tabriz in Iran. Ann. Biol. Res. 2012;3:2213–2216. [Google Scholar]
- Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R. Khosroushahi AY. Assessment of probiotic potential and anticancer activity of newly isolated vaginal bacterium Lactobacillus plantarum 5BL. Microbiol. Immunol. 2014a;58:492–502. doi: 10.1111/1348-0421.12175. [DOI] [PubMed] [Google Scholar]
- Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R. Yari Khosroushahi A. Probiotic potential and biotherapeutic effects of newly isolated vaginal Lactobacillus acidophilus 36YL strain on cancer cells. Anaerobe. 2014b;28:29–36. doi: 10.1016/j.anaerobe.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R. Yari Khosroushahi A. A newly isolated probiotic Enterococcus faecalis strain from vagina microbiota enhances apoptosis of human cancer cells. J. Appl. Microbiol. 2014c;117:498–508. doi: 10.1111/jam.12531. [DOI] [PubMed] [Google Scholar]
- NCCLS. 2007. Performance standards for antimicrobial susceptibility testing, seventeenth informational supplement, M100-S1727.
- Pan X, Chen F, Wu T, Tang H. Zhao Z. The acid, bile tolerance and antimicrobial property of Lactobacillus acidophilus NIT. Food Con. 2009;20:598–602. [Google Scholar]
- Parkes GC. Chapter 30 - The Role of Probiotics in the Treatment of Irritable Bowel Syndrome. In: Ronald Ross W, Victor RP, editors. Bioactive Foods in Promoting Health. Boston: Academic Press; 2010. pp. 513–528. [Google Scholar]
- Pogačić T, Mancini A, Santarelli M, Bottari B, Lazzi C, Neviani E, et al. Diversity and dynamic of lactic acid bacteria strains during aging of a long ripened hard cheese produced from raw milk and undefined natural starter. Food Microbiol. 2013;36:207–215. doi: 10.1016/j.fm.2013.05.009. [DOI] [PubMed] [Google Scholar]
- Psoni L, Tzanetakis N. Litopoulou-Tzanetaki E. Microbiological characteristics of Batzos, a traditional Greek cheese from raw goat's milk. Food Microbiol. 2003;20:575–582. [Google Scholar]
- Ramos CL, Thorsen L, Schwan RF. Jespersen L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013;36:22–29. doi: 10.1016/j.fm.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Reid G. Bocking A. The potential for probiotics to prevent bacterial vaginosis and preterm labor. Am. J. Obs. Gynecol. 2003;189:1202–1208. doi: 10.1067/s0002-9378(03)00495-2. [DOI] [PubMed] [Google Scholar]
- Ritchie LE, Burke KF, Garcia-Mazcorro JF, Steiner JM. Suchodolski JS. Characterization of fecal microbiota in cats using universal 16S rRNA gene and group-specific primers for Lactobacillus and Bifidobacterium spp. Vet. Microbiol. 2010;144:140–146. doi: 10.1016/j.vetmic.2009.12.045. [DOI] [PubMed] [Google Scholar]
- Ryu EH, Yang EJ, Woo ER. Chang HC. Purification and characterization of antifungal compounds from Lactobacillus plantarum HD1 isolated from kimchi. Food Microbiol. 2014;41:19–26. doi: 10.1016/j.fm.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Sahadeva RPK, Leong SF, Chua KH, Tan CH, Chan HY, Tong EV, et al. Survival of commercial probiotic strains to pH and bile. Int. Food Res. J. 2011;18:1515–1522. [Google Scholar]
- Seiquer I, Aspe T, Vaquero P. Navarro P. Effects of heat treatment of casein in the presence of reducing sugars on calcium bioavailability: In vitro and in vivo assays. J. Agric. Food Chem. 2001;49:1049–1055. doi: 10.1021/jf001008v. [DOI] [PubMed] [Google Scholar]
- Serban DE. Gastrointestinal cancers: influence of gut microbiota, probiotics and prebiotics. Cancer Lett. 2014;10:258–270. doi: 10.1016/j.canlet.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Singh S, Goswami P, Singh R. Heller KJ. Application of molecular identification tools for Lactobacillus, with a focus on discrimination between closely related species: a review. LWT - Food Sci. Technol. 2009;42:448–457. [Google Scholar]
- Soccol CR, Porto SVL, Rigon SM, Bianchi PMA, Tiemi Yamaguishi C, De Dea Lindner J, et al. The Potential of probiotics: a review. Food Tech. Biotech. 2010;48:413–434. [Google Scholar]
- Solomons NW. Modulation of the immune system and the response against pathogens with bovine colostrum concentrates. Eur. J. Clin. Nutr. 2002;56:24–28. doi: 10.1038/sj.ejcn.1601480. [DOI] [PubMed] [Google Scholar]
- Temmerman R, Pot B, Huys G. Swings J. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 2003;81:1–10. doi: 10.1016/s0168-1605(02)00162-9. [DOI] [PubMed] [Google Scholar]
- Tulini FL, Winkelströter LK. De Martinis ECP. Identification and evaluation of the probiotic potential of Lactobacillus paraplantarum FT259, a bacteriocinogenic strain isolated from Brazilian semi-hard artisanal cheese. Anaerobe. 2013;22:57–63. doi: 10.1016/j.anaerobe.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Walker DK. Gilliland SE. Relationships among bile tolerance, bile salt deconjugation, and assimilation of cholesterol by Lactobacillus acidophilus. J. Dairy Sci. 1993;76:956–961. doi: 10.3168/jds.s0022-0302(93)77422-6. [DOI] [PubMed] [Google Scholar]
- Wang B, Li J, Li Q, Zhang H. Li N. Isolation of adhesive strains and evaluation of the colonization and immune response by Lactobacillus plantarum L2 in the rat gastrointestinal tract. Int. J. Food Microbiol. 2009;132:59–66. doi: 10.1016/j.ijfoodmicro.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Young RJ. Huffman S. Probiotic use in children. J. Pediatric Health Care. 2003;17:277–283. doi: 10.1016/s0891-5245(03)00070-1. [DOI] [PubMed] [Google Scholar]