Abstract
Multidrug-resistant Acinetobacter baumannii has recently emerged as an important pathogen in nosocomial infection; thus, effective antimicrobial regimens are urgently needed. Human antimicrobial peptides (AMPs) exhibit multiple functions and antimicrobial activities against bacteria and fungi and are proposed to be potential adjuvant therapeutic agents. This study examined the effect of the human cathelicidin-derived AMP LL-37 on A. baumannii and revealed the underlying mode of action. We found that LL-37 killed A. baumannii efficiently and reduced cell motility and adhesion. The bacteria-killing effect of LL-37 on A. baumannii was more efficient compared to other AMPs, including human ß–defensin 3 (hBD3) and histatin 5 (Hst5). Both flow cytometric analysis and immunofluorescence staining showed that LL-37 bound to A. baumannii cells. Moreover, far-western analysis demonstrated that LL-37 could bind to the A. baumannii OmpA (AbOmpA) protein. An ELISA assay indicated that biotin-labelled LL-37 (BA-LL37) bound to the AbOmpA74-84 peptide in a dose-dependent manner. Using BA-LL37 as a probe, the ~38 kDa OmpA signal was detected in the wild type but the ompA deletion strain did not show the protein, thereby validating the interaction. Finally, we found that the ompA deletion mutant was more sensitive to LL-37 and decreased cell adhesion by 32% compared to the wild type. However, ompA deletion mutant showed a greatly reduced adhesion defect after LL-37 treatment compared to the wild strain. Taken together, this study provides evidence that LL-37 affects A. baumannii through OmpA binding.
Introduction
Antimicrobial peptides (AMPs) are generated by a wide variety of organisms as a part of the host defense. In humans, AMPs can be produced by various cells and tissues and play a critical role in innate immunity [1,2]. AMPs are generally short (10–100 amino acids), positively charged (normally +2 to +9) and amphiphilic [3]. AMPs can be divided into three major classes based on their gross amino acid composition and certain structural features, including linear alpha-helical peptides (without cysteines), cysteine-containing peptides linked by disulfide bonds and peptides with a high ratio of specific amino acids [2]. For example, human defensins belong to the second class, and histatins are members of the third class. hCAP-18 (the only member of the cathelicidin AMP family in humans) contains an N-terminal domain, a cathelin domain and a C-terminal LL-37 domain [4]. LL-37 is extracellularly cleaved from hCAP-18 by proteinase 3 and belongs to the class of linear alpha peptides. LL-37 owes its name to the fact that it consists of 37 amino acids that begin with two leucine residues [5].
Different types of AMPs use different mechanisms to disrupt bacterial structures or inhibit cell growth [6,7]. For example, the amphipathic conformation change can help an AMP gain access or insert into the plasma membrane of bacteria to disrupt the cells [7]. However, AMPs not only attack membranes but also inhibit cell wall biosynthesis, protein folding, enzyme activity and even protein synthesis through DNA binding [6]. In addition to the direct killing of bacteria, AMPs also play an important role in immunomodulation [8]. AMPs activate the adaptive immune system by stimulating gene transcription to activate macrophages, inducing interleukin-8 in airway epithelial cells to recruit neutrophils, promoting histamine release to increase blood vessel permeability, activating fibroblast growth to facilitate wound healing and presenting chemotactic activity to recruit monocytes [1,9]. These multi-functional responses induced by AMP make it a promising candidate adjuvant therapeutic agent, especially against multidrug-resistant pathogens.
Human LL-37 is able to defend against various bacterial and fungal pathogens [10–12]. Recently, Acinetobacter baumannii has emerged as an important pathogen in nosocomial infections [13]. Infections and outbreaks caused by multidrug-resistant A. baumannii (MDRAB) are rapidly increasing [14]. Resistance to the last resort antibiotics for carbapenem-resistant A. baumannii, including tigecycline and colistin, has been reported [15,16]. A previous study reported a lipopolysaccharide (LPS)-deficient, colistin-resistant A. baumannii strain that showed reduced viability even at a low concentration of LL-37 [17]. Moreover, LL-37 and its fragments possess both antimicrobial and antibiofilm activities against MDRAB [18]. Therefore, human antimicrobial peptides (especially LL-37) may function as potential therapeutic alternatives or adjuvants to antibiotics.
The OmpA outer membrane protein of Escherichia coli and other enterobacteria is a multifaceted protein, which functions as an adhesin and invasin, participates in biofilm formation, acts as both an immune target and evasin, and serves as bacteriophage receptor [19]. The A. baumannii outer membrane protein A (AbOmpA) is a trimeric porin that is involved in solute transport and virulence [20]. The contributions of AbOmpA to pathogenesis include apoptosis, immunomodulation, cell adherence and invasion, biofilm formation and serum resistance. AbOmpA can induce dendritic cell death via targeting to the mitochondria [21]. Interaction of laryngeal epithelial cells with AbOmpA has a significant impact on the induction of innate immunity during the early stages of A. baumannii infection [22]. AbOmpA also plays a role in biofilm formation on abiotic surfaces [23]. Serum resistance to A. baumannii occurs through binding of factor H to outer membrane proteins (OMPs), including OmpA [24]. Because AbOmpA is multi-functional, we hypothesize that it may also bind to LL-37. Therefore, the aim of this study is to determine the effect of LL-37 on A. baumannii and to determine whether the effect was mediated via binding to OmpA.
Materials and Methods
Peptides, A. baumannii Strains, Media, and Growth Conditions
LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES), biotin-labeled LL-37 (BA-LL37), biotin-labeled human β-defensin-3 (BA-hBD3, QKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK), biotin-labeled histatin 5 (BA-Hst5, DSHAKRHHGYKRKFHEKHHSHRGY), OmpA164-181 (TYNADEEFWNYTALAGLN) and OmpA74-84 (GDVDGASAGAE) were synthesized by MDBio, Inc. (Taipei, Taiwan). The purity of these peptides was determined to be ≧85% by high performance liquid chromatography (HPLC) and mass spectrometry. The A. baumannii ATCC 17978 strain was used as the wild type. The media and growth condition were the same as described in our previous study [25].
Assays for LL-37 Anti-Acinetobacter Activity
Bacterial cells were grown overnight (for ~16 h) in LB broth and subcultured into 5 ml of fresh LB medium (initial OD600 ~0.27). Then, the cells were grown to an OD600 of 1.2 and harvested by centrifugation (6000 x g). Cell pellets were washed twice and re-suspended with phosphate-buffered saline (PBS). Different concentrations of LL-37, BA-LL37, BA-hBD3 or BA-Hst5 were incubated with the cells (1 X 107 cells/ml) in 750 μl of RPMI-1640 medium at 37°C with 5% CO2 for 30 min. After the incubation, the cells were serially diluted 10-fold with PBS, spotted onto LB agar plates (10 μl/per spot) and incubated at 37°C overnight. To determine colony forming units (CFUs), the cells were 10-fold serially diluted, and 100 μl of each sample was plated onto LB agar plates.
A. baumannii Adhesion Assay
The adhesion of A. baumannii was assessed as previously described with some modifications [26]. Briefly, bacterial cells were grown overnight in LB broth (~16 hr) and subcultured into 25 ml of fresh LB medium (initial OD600 ~0.35). The cells were grown to an OD600 of 1.2 and harvested by centrifugation (6000 x g). The cell pellets were washed twice and re-suspended with PBS. Different concentrations of LL-37 were incubated with the cells (4 X 108 cells/ml) in each well of a 96-well plate (NuncTM, Rochester, NY, USA) as previously described [26]. After incubation at 37°C for 1 hr with shaking (100 rpm), the non-adherent floating cells were discarded, and the adherent cells were washed three times with PBS. A total of 150 μl of crystal violet was added to each sample, and the plates were incubated at room temperature for 20 min. After removal of the crystal violent solution, each sample was washed three times with double-distilled water (ddH2O). The remaining crystal violet in each well was dissolved in 100 μl of 95% ethanol, and the absorbance at 595 nm was detected using an iMARK microplate reader (Bio-Rad Life Science, Hercules, CA, USA).
A. baumannii Motility Assay
The motility assay was performed as previously described [27]. Cells from an overnight culture were subcultured into 5 ml of fresh LB medium and grown to an OD600 of 1.2. The cells were harvested by centrifugation (6000 x g), and a 5 μl cell suspension (~1 X 109 cells) was spotted onto motility agar (1% tryptone, 0.5% NaCl and 0.4% agarose) and incubated at 37°C for 10 hr.
Flow Cytometric Analysis
Cells from an overnight culture were inoculated into 5 ml of fresh LB medium (initial OD600 0.27) and grown to an OD600 of 1.2. Cell pellets were harvested by centrifugation (6000 x g), washed twice with PBS and re-suspended with 750 μl of ice-cold PBS (containing 5 X 106 cells). Then, the cells were incubated overnight at 4°C with or without different concentrations of BA-LL37, BA-LL37, BA-hBD3, or BA-Hst5. Binding of the BA-AMPs to the bacterial cells was assessed by flow cytometry based on SA-4,6-dichlorortriazinyl aminofluorescein (SA-DTAF) detection. Three microliters of SA-DTAF was used in each reaction (Jackson ImmunoResearch, West Grove, PA, USA). Reactions were quantified using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) according to a previously described method [28]. Fluorescence data for 2 X 104 cells were acquired per experiment.
Immunofluorescence Staining
Bacterial cells from overnight cultures were subcultured into 5 ml of fresh LB medium (initial OD600 ~0.27). Cells were grown to an OD600 of 1.2 and harvested by centrifugation (6000 x g). The cells were re-suspended with PBS to an OD600 of 1, mixed with or without BA-LL37 (20 μg/ml) to a final volume of 750 μl, and grown overnight at 4°C. Next, the cell pellets were harvested by centrifugation (6000 x g), washed twice with ice-cold PBS and re-suspended in 750 μl of ice-cold PBS containing SA-DTAF (3 μg/reaction). The mixture was incubated at 4°C for 2 hr. Finally, the cell pellets were collected by centrifugation and re-suspended in 40 μl of ice-cold PBS. The cell suspension was then transferred to a cover slip at 4°C for 20 min. The samples were examined with a Carl Zeiss AXIO IMAGER A1 Microscope.
Extraction of A. baumannii Outer Membrane Proteins (AbOMPs)
Extraction of AbOMPs was performed as previously described [29] with some modifications. Briefly, A. baumannii cells were grown overnight, subcultured into 100 ml of fresh LB medium and incubated at 37°C with shaking (220 rpm) for 2 hr. The cell pellets were harvested by centrifugation (6000 x g at room temperature) and washed twice with PBS. Then, the cells were re-suspended in 20 ml of RPMI-1640 medium and incubated at 37°C with shaking (100 rpm) for 1 hr. The cell pellets were washed twice with PBS and re-suspended in 20 ml of 10 mM phosphate buffer (pH7.2) supplemented with phenylmethanesulfonylfluoride (PMSF) at a final concentration of 1 mM. The cells were disrupted by sonication for 12 min on ice (10 sec sonication at intervals of 10 sec). The cell debris was discarded by centrifugation (3000 x g), and the supernatant was subjected to centrifugation at 13,700 x g (4°C, 45 min). Then, the supernatant was discarded, and the extracted proteins were solubilized at room temperature using 2% sodium lauryl sarcosinate (Sarkosyl) in 10 mM phosphate buffer for 30 min. Finally, the AbOMPs were collected by centrifugation at 13,700 x g (4°C, 45 min), re-suspended in 62.6 mM Tris-HCl buffer, and stored at -20°C.
Western and Far-Western Analysis
AbOMP extracts were mixed with sample buffer and heated at 100°C for 10 min. OMP samples were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride (PVDF) membrane (Pall Corporation, Port Washington, NY, USA) using a TE77 ECL Semi-Dry Transfer Unit (Amersham Biosciences). The membrane was blocked with 3% non-fat milk in PBST (PBS with 1% Tween 20) at room temperature for 1 hr and washed with PBST. Then, the membrane was hybridized overnight at 4°C with anti-OmpA (1:5000; a kind gift from Luis A. Actis, Miami University, OH, USA) in PBST containing 1% BSA. The blotted membrane was washed with PBST for 25 min and probed with anti-rabbit IgG (1:5000; GeneTex) at room temperature for 40 min. The membrane was again washed with PBST for 25 min. The Western Lightning Plus-ECL reagent (PerkinElmer Life Science, SC-2004) and an ImageQuantTM LAS 4000 mini system (GE Healthcare Science) were used to detect proteins immobilized on the membrane according to the manufacturer’s instructions.
For the far-western analysis, outer membrane extraction and separation were performed as described above for the western analysis. The membrane was blocked with 3% bovine serum albumin in PBST at room temperature for 2 hr and washed with PBST. Then, the membrane was hybridized overnight with 10 μg/ml of BA-LL37 (in PBST) at 4°C. Proteins were detected using the Western Lightning Plus-ECL reagent and an ImageQuantTM LAS 4000 mini system.
Coomassie Blue Staining
After SDS-PAGE, the gel was soaked overnight in the fixing buffer (35% ethanol and 2% phosphoric acid) at room temperature. Next, the gel was washed with water for 90 min at room temperature, with water changes every 30 min. After washing, the gel was soaked in 50 ml of staining buffer (34% methanol, 17% (NH4)2SO4, and 3% phosphoric acid) for 1 hr at room temperature. Finally, 25 mg of Coomassie Blue G-250 was dissolved into 50 ml of the staining buffer. The gel was stained in the solution until blue colored bands appeared.
Prediction of LL-37 Binding Sites within the AbOmpA Protein and ELISA Assay to Investigate LL-37 Binding to the AbOmpA74-84 and AbOpmA164-181 Peptides
Based on our previous report [26], ΦHWXΦXΦXΦ (Φ: a hydrophobic amino acid residue; X: any amino acid residue) is a consensus sequence derived from different peptides that can bind LL-37. This sequence was used to blast search for possible LL-37 binding sites within the AbOmpA protein. Members of the OmpA family of bacteria are known to commonly contain four surface-exposed loop structures [30]. Amino acid sequences representing these structures were identified within the AbOmpA protein. We found that the amino acid residues between 74 to 84 and 164 to 181 of the AbOmpA protein matched the consensus sequences for LL-37 binding and were possibly located on two independent loops exposed towards the outside of the outer membrane. Therefore, the two peptides AbOmpA74-84 and AbOmpA164-181 were used to facilitate the following experiments.
Both AbOmpA164-181 (TYNADEEFWNYTALAGLN) and AbOmpA74-84 (GDVDGASAGAE) were synthesized by MDBio, Inc. (Taipei, Taiwan). The peptides were mixed with carbonate-bicarbonate coating buffer (30 mM Na2CO3 and 69 mM NaHCO3, pH 9.6) as previously described [31]. A total of 5~10 μg of AbOmpA74-81 or AbOmpA164-181 in 100 μl of carbonate-bicarbonate coating buffer was transferred into a Nunc Maxisorp 96-well plate (NuncTM, Rochester, NY, USA) and immobilized at 4°C overnight. After peptide immobilization, each well was washed three times with PBST to remove non-attached peptides. For blocking, 100 μl PBST and 2% BSA were added to each well; the mixture was incubated at room temperature for 1 hr, followed by removal of the blocking reagents. Each well was washed three times with PBST. Different concentrations of BA-LL37 (in 100 μl of PBS) were added to each well and incubated at room temperature for 1 hr. Unbound BA-LL37 was removed, and the wells were washed three times with PBST. Finally, SA-HRP (1:200 in PBST) was added to each well and incubated at room temperature for 30 min. After removing the SA-HRP solution, each well was washed three times with PBST. For signal detection, 100 μl of 3,3',5,5'-tetramethylbenzidine (TMB) was added to each well until the blue color began to appear. Then, 100 μl of 1N H2SO4 was added to stop the reaction, and the signal was detected at OD450 using an iMARK ELISA reader (BioRad).
Genomic DNA Extraction, RNA Isolation and Reverse Transcription (RT)-PCR
Genomic DNA extraction, RNA isolation and RT-PCR were performed as described in our previous work [25]. For genomic DNA extraction, cells from an overnight culture (1 ml) were harvested by centrifugation (6000 x g) for 1 min and then mixed with 600 μl of lysis solution (200 mM Tris-HCl [pH 8.5], 100 mM EDTA [pH 8.0], and 35 mM SDS). Cells were lysed in an 80°C water bath for 10 min; then, 200 μl of 10 M NH4OAC was added, and the samples were vortexed vigorously for 20 sec. The mixture was centrifuged (16500 x g, 4°C) for 5 min, and the supernatant (650 μl) was mixed with an equal volume of ice-cold PCIA (phenol [pH 7.0]-chloroform-isoamyl alcohol [25:24:1, v/v]). After centrifugation (16500 x g, 4°C) for 5 min, 500 μl of the supernatant was mixed with an equal volume of isopropanol to precipitate the DNA. DNA pellets were collected by centrifugation (16,500 x g, 4°C) for 5 min, washed with 75% ethanol and centrifuged again. The pellets were re-suspended with 400 μl of water containing 75 μg/ml RNase A and incubated at 37°C for 15 min. Then, the sample was mixed with two volumes of 99.5% ethanol and a 1/10 volume of 3 M ammonium acetate to precipitate the DNA. The sample was centrifuged at 16,500 x g at 4°C for 5 min, and the DNA pellets were re-suspended with 100 μl of water.
To extract total RNA, cells were grown in LB broth to the mid-log phase and harvested by centrifugation at 4°C. Cell pellets were re-suspended in 200 μl of ice-cold RNA extraction buffer (0.1 M Tris-HCl [pH 7.5], 0.1 M LiCl, 0.01 M EDTA [pH 8.0], 5% SDS, and 2% β-mercaptoethanol) and 200 μl of ice-cold PCIA (pH 4.5). The extraction was repeated three times, and the extracts were collected by centrifugation. Two volumes of ethanol (pre-cooled at -20°C) and 0.1 volumes of 3 M NaOAc were added to precipitate the RNA overnight at -80°C. RNA was pelleted by centrifugation at maximum speed (5 min) and re-suspended in 25–100 μl of DEPC-treated water. DNA contaminants were removed using Ambion® TURBO™ DNase. cDNAs were synthesized using the High-Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems).
Construction of an ompA Deletion (ΔompA) Mutant of A. baumannii
To determine the involvement of OmpA in LL-37 binding, the ompA gene (A1S_2840) was deleted and replaced with a kanamycin resistance (Kan R) gene using the pEX18Tc plasmid [25]. Briefly, 1000 bp flanking sequences upstream or downstream of the ompA gene were independently PCR-amplified from the genomic DNA of the A. baumannii ATCC 17978 strain. The primer pairs used for amplification of the ompA-upstream sequences were ompA5’F (the SalI site is underlined) and ompA5’R (the BamHI site is double underlined; see S1 Table). The primer pairs used for amplification of the ompA-downstream sequences were ompA3’F (the KpnI site is underlined) and ompA3’R (the SacI site is double underlined). The PCR products containing the ompA upstream and downstream flanking regions were digested with SalI/BamHI and KpnI/SacI and independently cloned into pEX18Tc, generating pEX18Tc-ompAUD. The Kan R gene was obtained by PCR amplification using the TOPO® vector as a template [32] and the primer pairs kanF (the BamHI site is underlined) and kanR (the KpnI site is double underlined). The PCR product carrying Kan R was digested with BamHI and KpnI and cloned into the BamHI/KpnI sites of pEX18Tc-ompAUD to generate pEX18Tc-OmpAUD-Kan. To construct the ompA deletion mutant, pEX18Tc-ompAUD-Kan was transformed into the E. coli S17-1 λ-pir strain. The E. coli transformant and A. baumannii ATCC 17978 strain were grown separately overnight and mixed (1:3) in 1 ml of fresh LB medium. The cell mixture was further incubated at 37°C for 1 hr. Then, 20 μl of the cell mixture was spotted onto an LB agar plate and incubated overnight at 37°C to obtain cells with a single gene crossover. To eliminate E. coli from the mixed culture, the mixture was grown overnight in 5 ml of LB medium containing 100 μg/ml ampicillin. To obtain cells with two gene crossovers, the mixed culture was pelleted and plated onto LB agar plate containing 10% sucrose and kanamycin (50 μg/ml) at 37°C for 16 hr. The sucrose-resistant colonies were picked, and the ompA deletion was verified by PCR and validated by RT-PCR and SDS-PAGE.
Results
LL-37 Kills A. baumannii in a Dose-Dependent Manner
To assess the anti-Acinetobacter activity of LL-37, bacterial cells were treated with different concentrations of LL-37; viable cells were counted and represented as the number of CFUs. In Fig 1, the results indicated that LL-37 harbored dose-dependent bactericidal activity against A. baumannii. Approximately 32%, 80%, and 99% of the cells died following treatment with 2.5, 5 and 7.5 μg/ml of LL-37, respectively. Moreover, no viable cells were apparent after treatment with 20 μg/ml of LL-37 (Fig 1). The bacterial killing activity of LL-37 on two clinical isolates of A. baumannii was also performed using spot assay, which showed the anti-bacterial effect augmented with LL-37 concentrations increasing (S1 Fig). These results indicate that LL-37 exhibits bactericidal activity against A. baumannii.
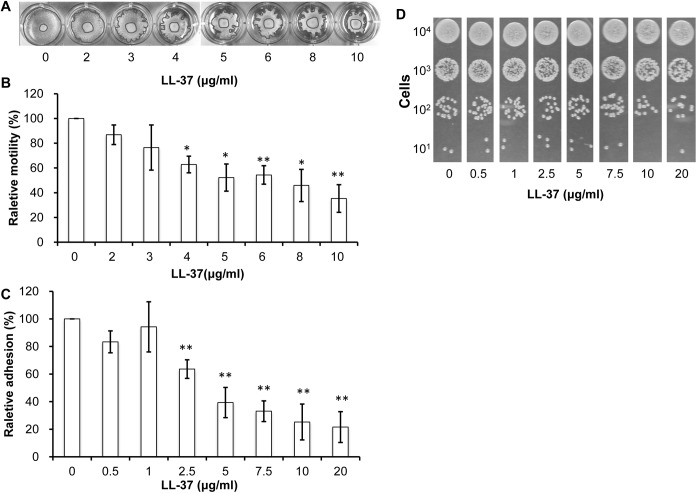
Fig 1. Anti-Acinetobacter activity of LL-37.
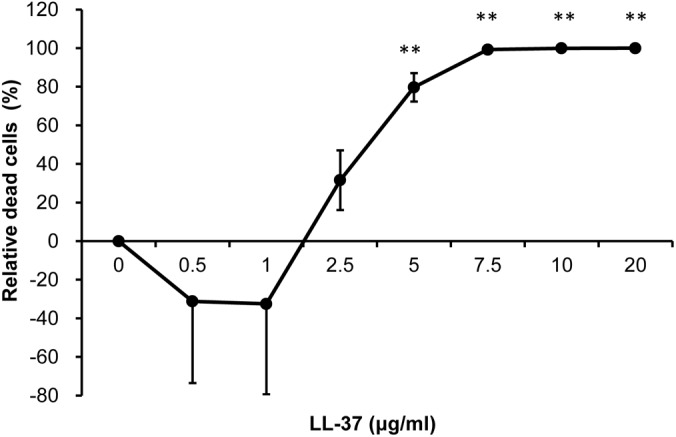
Anti-Acinetobacter activity of LL-37 was determined by counting CFUs. The CFU counts of cells (1 X 107 cells/ml) in 750 μl of RPMI-1640 medium treated with different concentrations of LL-37 were normalized to the control without LL-37 treatment and presented as a percentage. The result indicated that LL-37 exhibited a bactericidal effect on A. baumannii. These experiments were performed in triplicates (N = 3). The Student’s t-test was used to determine (**, p<0.01) the statistical significance of the experimental data.
LL-37 Inhibits A. baumannii Motility and Adhesion
Although A. baumannii is generally considered to be “non-motile”, several studies indicated that the A. baumannii ATCC 17978 strain had the ability to migrate under certain conditions [27,33]. Moreover, motility was recently identified as an A. baumannii virulence factor [27]. To test the effect of different concentrations of LL-37 on A. baumannii motility, cells were grown to the exponential phase and spotted onto motility agar plates (Fig 2A). A reduction in A. baumannii motility was observed concomitant with treatment with increasing concentrations of LL-37 (Fig 2B).
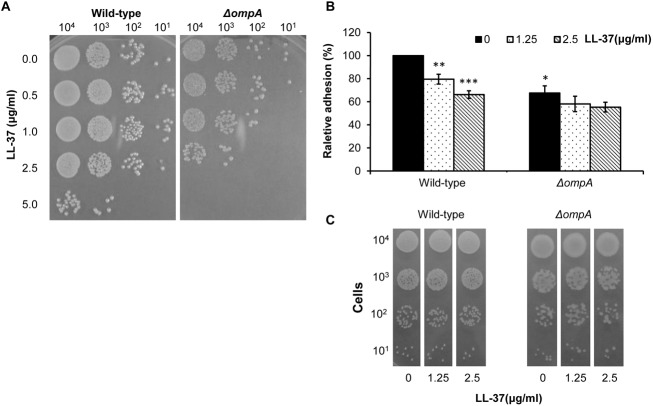
Fig 2. Inhibition of A. baumannii adhesion and motility by LL-37.
(A) Motility of A. baumannii on semisolid agar. Three microliters of cells from mid-log phase were spotted onto the middle of semisolid agar containing different concentrations of LL-37 and incubated at 37°C for 7 hr. (B) Cell motility was quantified using the Image J program (http://imagej.nih.gov/ij/). The whole area of the entire colony and the original spot was measured. Cell motility was obtained by subtracting the area of the original spot from the area of the entire colony. The relative percentage of motility was obtained by comparing the motility of cells treated with LL-37 to of the motility of the control without LL-37 treatment. The results of both (A) and (B) showed that A. baunmannii motility was reduced by increasing concentrations of LL-37. (C) Cell adhesion to a polystyrene surface. Cells were mixed with different concentrations of LL-37 in RPMI-1640 and incubated at 37°C for 1 hr. Adherent cells were detected by crystal violet staining, and relative adhesion was represented as a percentage. (D) Non-adherent floating cells were collected by centrifugation, serially diluted and spotted onto an LB agar plate. The results indicated that A. baumannii cell attachment was decreased with treatment of increasing concentrations of LL-37 and that most non-adherent cells were alive. Both adhesion and motility experiments were performed in triplicates (N = 3). The Student’s t-test (**, p<0.01) was used to determine the statistical significance of the experimental data.
Adhesion is another A. baumannii virulence factor [34]. To determine the effects of LL-37 on A. baumannii adhesion, cells were grown in a 96-well microplate and adherent cells were quantified by crystal violet staining. We found that A. baumannii cell attachment was decreased with increasing concentrations of LL-37 (Fig 2C). To determine whether the inhibition of bacterial adhesion by LL-37 was due to bacterial cell death, the floating non-adherent cells were serially diluted and spotted onto LB agar plates. As shown in Fig 2D, bacterial growth was not significantly affected among the floating cells treated with different concentrations of LL-37. Therefore, LL-37-mediated inhibition of adhesion was not a consequence of LL-37-induced cell death.
LL-37 Binds to A. baumannii Cells
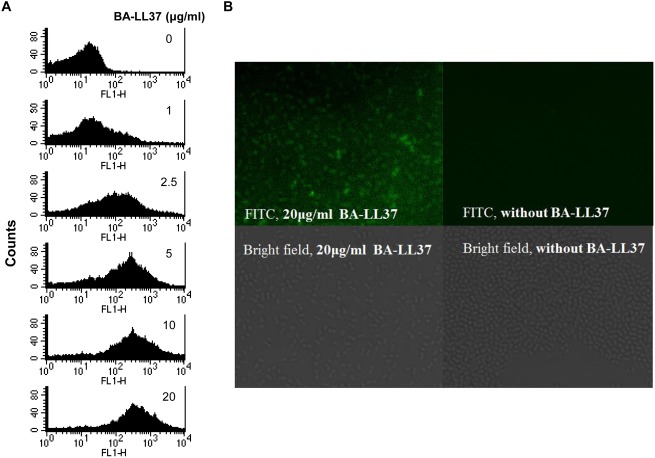
In many cases, the first step in the AMP bacterial killing ability is to bind to the bacterial cell surface [6]. Therefore, we hypothesized that LL-37 might directly bind to the cell surface. To test this hypothesis, we used a flow cytometric assay based on SA-DTAF detection (as described in the Materials and Methods). An increase in the fluorescence intensity was correlated with increasing concentrations of BA-LL-37, indicating that LL-37 could directly bind to A. baumannii cells (Fig 3A). Immunofluorescence staining was also performed. A. baumannii cells were incubated with BA-LL37 overnight and stained with SA-DTAF. In the presence of 20 μg/ml LL-37, increased fluorescence was observed compared to the control cells without BA-LL37 treatment (Fig 3B). Therefore, these studies showed that LL-37 bound to A. baumannii cells.
Fig 3. BA-LL37 binding to A. baumannii.
(A) LL-37 binding to A. baumannii cells was determined by flow cytometry. Cells were incubated with different concentrations of BA-LL37 at 4°C in ice-cold PBS overnight. The binding between BA-LL37 and A. baumannii was detected by adding SA-DTAF. The increase of signal was detected and correlated to cells treated with increasing concentrations of LL-37, indicating that LL-37 binds directly to A. baumannii. These data are representative of three independent experiments with similar results. FL1-H indicates the extent of the fluorescence intensity. (B) An immunofluorescence staining assay was performed to verify LL-37 binding to A. baumannii. In the presence of 20 μg/ml LL-37, fluorescence was observed compared to the control (without BA-LL37 treatment). Samples were observed using a Carl Zeiss AXIO IMAGER A1 microscope.
LL-37 Is Highly Efficient against A. baumannii Compared to Other AMPs
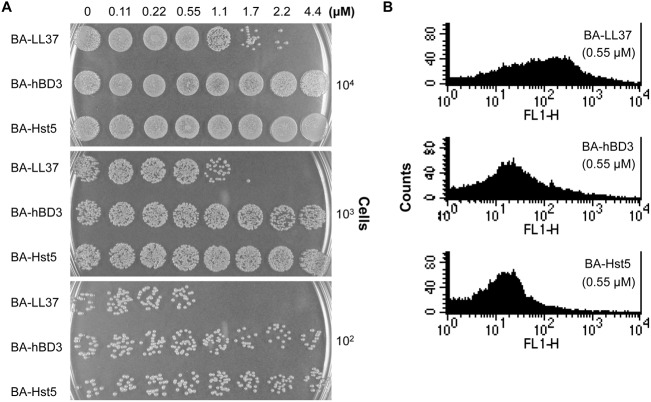
There are various AMPs in the human immune system, including LL-37, hBD3 and Hst5 [2]. To compare their anti-Acinetobacter activity, cells were treated with different concentrations of these three AMPs and spotted onto LB agar plates. Fig 4A showed that LL-37 killed A. baumannii cells more efficiently than hBD3 or Hst5. Additionally, the ability of hBD3 and Hst5 to bind to A. baumannii was determined by flow cytometric analysis. Compared to LL-37, hBD3 and Hst5 exhibited much poorer binding to A. baumannii when the same concentration of each AMP was used (Fig 4B). We concluded that the effects of LL-37 on A. baumannii were more efficient compared to hBD3 and Hst5.
Fig 4. Efficient action of LL-37 on A. baumannii.
(A) A spot assay was performed to compare the anti-Acinetobacter activities of three different AMPs. The cells were treated with different concentrations of BA-LL37, BA-hBD3 and BA-Hst5 for 30 min, ten-fold serially diluted and then spotted onto LB agar plates. BA-LL37 showed a more efficient killing of A. baumannii than BA-hBD3 or BA-Hst5. The data are representative of three independent experiments with similar results. (B) Binding of different AMPs to A. baumannii was examined using flow cytometry. Cells were incubated with 0.55 μM of each BA-AMP (in ice-cold PBS) at 4°C overnight, and the attachment of BA-AMP to the cells was analyzed by flow cytometry based on SA-DTAF detection. FL1-H represents the extent of the fluorescence intensity. Compared to BA-LL37, BA-hBD3 and BA-Hst5 showed poor binding to A. baumannii. The image is representative of three independent experiments with similar results.
AbOmpA Is a Binding Target of LL-37
Because LL-37 bound to A. baumannii cells, we were interested in identifying potential target(s) for LL-37 on the cell surface, and particularly on the A. baumannii outer membrane. OMPs were isolated from A. baumannii [29], immediately subjected to SDS-PAGE, and transferred onto a PVDF membrane (Fig 5, Lane 1). Far western analysis was performed using BA-LL37 as a probe. Several OMPs bound BA-LL37, with a protein with a molecular mass of ~38 kDa showing the strongest signal (Fig 5, Lane 2). The molecular mass of 38 kDa is close to that of AbOmpA, an A. baumannii outer membrane porin protein. To determine whether the protein was indeed AbOmpA, western blotting was performed using an anti-AbOmpA antibody. As shown in lane 3 of Fig 5, a protein of ~38 kDa was detected. These results suggest that AbOmpA is an LL-37 binding target.
Fig 5. LL-37 binding to A. baumannii OmpA.
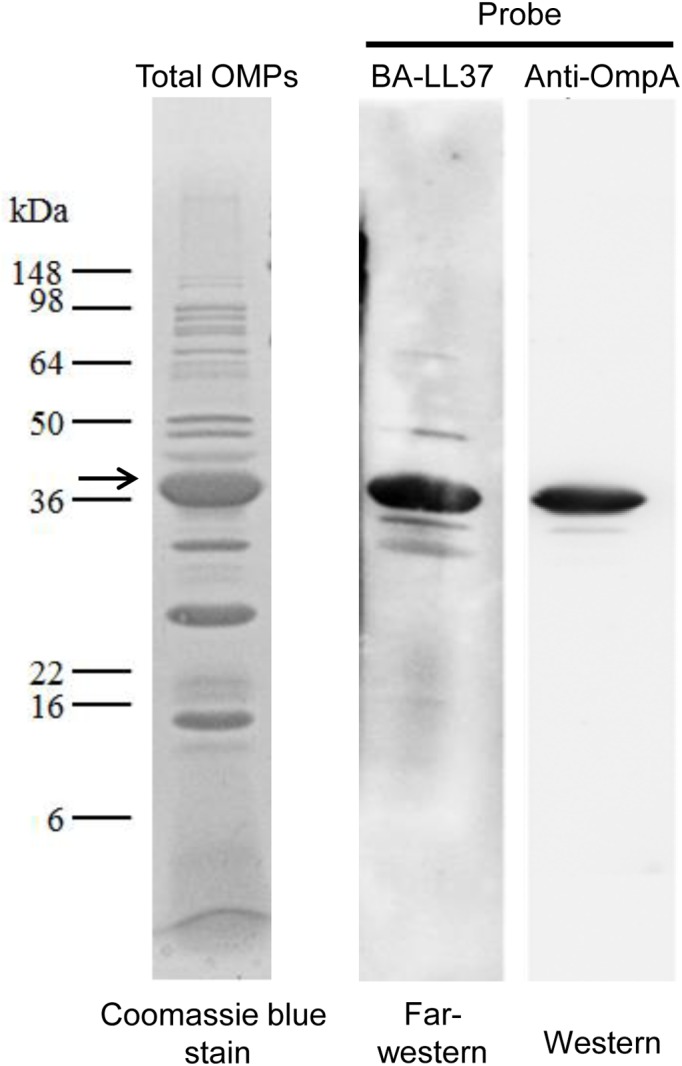
Total OMPs of A. baumannii were isolated, subjected to 12% SDS-PAGE and detected by western blotting. After electrophoresis, OMPs were stained with Coomassie blue (Lane 1). OMPs in another gel were transferred onto a PVDF membrane using a semi-dry transfer system and blotted with SA-HRP (Lane 2) or an anti-OmpA antibody (Lane 3). The signals were detected using enhanced chemiluminescence (ECL).
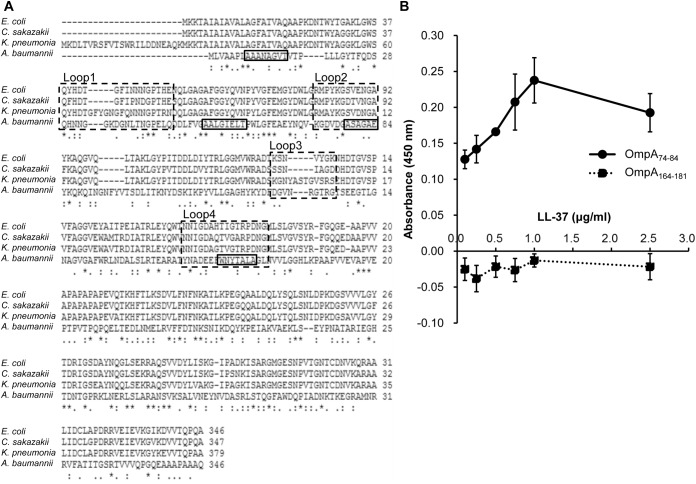
LL-37 Associates with Certain Regions of AbOmpA
Using phage display screening of a linear dodeca-peptide library, several peptide sequences associated with LL-37 were identified in a previous study from our laboratory [26]. From these identified LL-37-binding peptides, a conserved sequence of ΦHWXΦXΦXΦ (Φ: a hydrophobic residue; X: any residues) was proposed. Additionally, sequence analysis of AbOmpA revealed 4 loops that were highly conserved in the OmpA protein family of Gram-negative bacteria (Loops 1 to 4, boxed in dashed lines, Fig 6A) [30]. To determine whether AbOmpA was truly an LL-37-binding target, the consensus peptide sequence was used to blast search against the entire protein sequence of AbOmpA. The conserved sequence derived from the phage display assay aligned with four regions of the AbOmpA protein (boxed in solid lines, Fig 6A). Two of the regions (AbOmpA amino acid residues 74–84 and 164–181) were located within Loops 2 and 4, respectively (Fig 6A). Therefore, two peptides (AbOmpA74-84 and AbOmpA164-181) were synthesized and used in an ELISA assay to examine their association with LL-37. The results indicated that BA-LL37 could bind to the AbOmpA74-84 peptide in a dose-dependent manner (Fig 6B). However, the AbOmpA164-181 peptide did not bind to BA-LL37. These results raise the possibility that LL-37 binding to AbOmpA may be dependent on the recognition of specific region(s).
Fig 6. Detection of binding between OmpA peptides and LL-37.
(A) Amino acid sequence alignment of OmpA from E. coli, Klebsiella pneumoniae, Cronobacter sakazakii and A. baumannii. The alignment was performed using Clustal W2. The conserved Loop 1 to Loop 4 were boxed in dashed lines, and four potential LL-37-binding sequences were boxed in solid lines. (B) ELISA assay for binding of the AbOmpA74-84 and AbOmpA164-181 peptides to LL-37. Each peptide was mixed with carbonate coating buffer and immobilized on each well of a microplate at 4°C overnight and then washed with PBS. After blocking, various concentrations of BA-LL37 were added to the immobilized peptides and the binding activity was detected with SA-HRP. The results indicated that BA-LL37 could bind to the AbOmpA74-84 peptide, but not the AbOmpA164-181 peptide, in a dose-dependent manner. All assays were performed in triplicate and repeated three times.
LL-37-AbOmpA Interaction
To verify the finding that LL-37 bound to AbOmpA, we constructed an ompA deletion mutant by gene displacement. The successful construction of the mutant was confirmed by RT-PCR and western blot analysis (S2 Fig). Then, a far western assay was performed to compare BA-LL37 binding to the wild type and ompA deletion strain. Using BA-LL37 as a probe, the ~38 kDa OmpA was detected in the wild type but absent in the ΔompA strain (Fig 7), thereby confirming that LL-37 indeed bound to AbOmpA.
Fig 7. LL-37 and AbOmpA interaction.
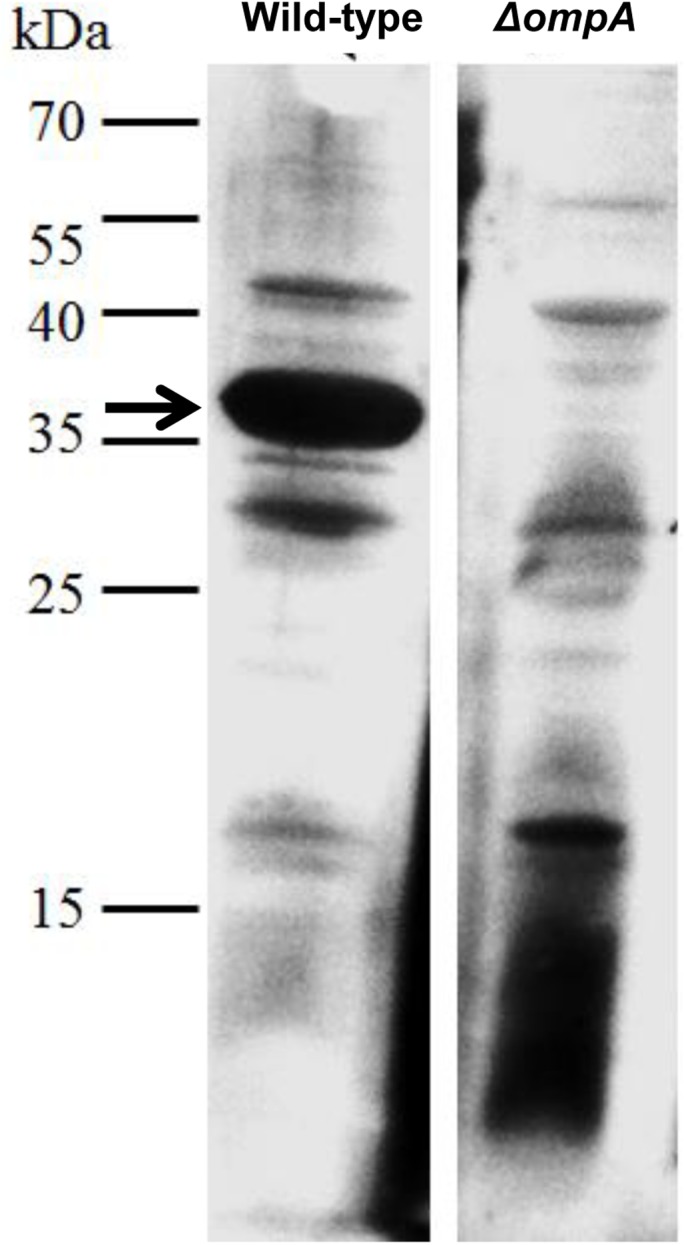
Binding of LL-37 to the wild type and ΔompA strains was determined by far-western analysis. OMPs were isolated and subjected to SDS-PAGE. The proteins were transferred onto a PVDF membrane, followed by probing with an anti-OmpA antibody. The ~38 kDa OmpA was detected in the wild type strain but was absent in the ΔompA mutant.
AbOmpA Influences LL-37’s Effect on Bacterial Killing and Bacterial Adhesion
To determine the influence of AbOmpA on LL-37-mediated bacteria-killing, the sensitivity of the ΔompA strain to LL-37 was examined using a spot assay. The result showed that the ΔompA strain was more sensitive to LL-37 than the wild type strain (Fig 8A). This result was unexpected. Because bacterial OmpA interacts with lipopolysaccharide (LPS) and both OmpA and LPS are major components of the outer membrane [35,36], we used the spot assay to compare LL-37 susceptibility between the wild type strain and an LPS-defective mutant (S3 Fig). The result indicated that the LPS-defective mutant had a better tolerance to LL-37 than the wild type. Moreover, the adhesion ability of the wild type and ΔompA strains was compared with or without LL-37 treatment. The ΔompA strain showed a decrease in adhesion of 32% compared to the wild type strain without LL-37 treatment (Fig 8B). After ompA deletion, the effect of LL-37 on adhesion was greatly reduced compared to the wild strain. Bacterial growth was not significantly different between the floating cells treated with 1.25 and 2.5 μg/ml of LL-37 (Fig 8C). This result implies that LL-37 may impair the adhesion of A. baumannii through binding to OmpA.
Fig 8. Comparison of LL-37 sensitivity and adhesion ability between the wild type and ΔompA strains.
(A) Sensitivity of the wild-type and ΔompA strains to LL-37 killing was examined using the spot assay. Cells were incubated with different concentrations of LL-37 for 1 hr. Then, the cells were 10-fold serially diluted and spotted onto LB agar plates. The result showed that the ΔompA mutant was more sensitive to LL-37 than the wild type. (B) Cell adhesion of the wild type and ΔompA strains was compared. The wild type and ΔompA cells were attached to polystyrene for 1 hr. Then, non-adherent cells were removed by centrifugation and the adherent cells were stained with crystal violet. The ΔompA mutant showed a decrease in adhesion of 32% compared to the wild type. The adhesion defect in the ΔompA strain was not augmented as obviously as that of the wild type after the addition of LL-37. (C) The spot assay demonstrated that the difference in bacterial adhesion induced by LL-37 was not due to bacterial cell death. The Student’s t-test (**p <0.01) was used to determine the statistical significance of the experimental data.
Discussion
LL-37 is an important component of the human innate immune defense [5,37]. LL-37 not only plays a critical role in bacterial clearance but also regulates host activities related to the immune response, including chemotactic migration and wound healing. The cationic, α-helical peptide LL-37 efficiently kills both Gram-positive and Gram-negative bacteria. Besides, the discovery of LL-37-inducing components, such as butyrate and vitamin D(3), has opened new avenues to prevent or treat infections by boosting innate immune response [38,39]. Although different approaches (e.g., bulk assay, model membrane assay and minimum inhibitory concentration test) have been used and different mechanisms have been proposed for the activity of LL-37 against bacteria [5,40], the detailed mechanism underlying LL-37 killing of A. baumannii is mostly unknown.
In this study, we examined the effects of LL-37 on A. baumannii and found that the effects were exerted in a LL-37 dose-dependent manner and related with the cell concentrations tested. When the cells (1 X 107 cells/ml) in 750 μl of RPMI-1640 medium were used to examine the anti-bacterial effect, LL-37 above 5 μg/ml could effectively kill the cells (Fig 1). However, the cell adhesion (Fig 2A and 2B) and cell motility (Fig 2C and 2D) data are still valid beyond 5 μg/ml LL-37 concentration because a higher cell density (4 X 108 cells/ml) and cell count (~1 X 109 cells) were used in the adhesion and motility tests, respectively. Although LL-37-mediated inhibition of adhesion and biofilm formation in bacteria has been reported previously [41,42], this study is the first to report an inhibitory effect of LL-37 on bacterial motility. LL-37 is commonly secreted at mucosal surfaces at a concentration ranging between 2 to 5 mg/ml [43,44]. The effects of LL-37 on A. baumannii in our study were observed at the physiological concentrations in humans. Moreover, our study showed that BA-LL37 exhibited the most efficient killing of A. baumannii among the three tested AMPs (Fig 4A and 4B). The charges of LL-37, hBD3 and Hst5 in the physiological environment are +6, +11 and +12, respectively [45]. Both LL-37 and Hst5 have random coil conformations in hydrophilic environments and α-helical structures under hydrophobic conditions. In contrast, hBD3 has a β-sheet structure due to the presence of three intra-molecular disulfide bridges. How these structural differences among the three AMPs influence their ability to kill A. baumannii deserves further study.
Most studies suggest that AMPs act on Gram-negative bacteria through their surface LPS molecules [46–48]. However, several studies have emphasized the interaction between AMPs and OMPs. Outer membrane protein I (OprI) of Pseudomonas aeruginosa has been shown to be the target of cationic AMP [49]; this AMP can also interact with OmpF from E. coli [50]. In this study, far-western analysis revealed there were several LL-37-binding candidates; among them, OmpA was confirmed using an anti-OmpA antibody (Fig 5). This finding adds a pluripotent function to A. baumannii OmpA in addition to its roles in cytotoxicity, cell adhesion and immunomodulation. OmpA is an outer membrane porin protein and has an amino acid sequence that is highly conserved among Gram-negative bacteria [30]. In our study, BA-LL37 bound to the AbOmpA74-84 peptide (loop 2) but not the loop 4 peptide in a dose-dependent manner (Fig 6B). A hypothetical arrangement of the OmpA protein suggests that it repeatedly traverses the outer membrane in a cross-β structure, exposing the four loops to the outside [51]. Of these four loops, loop 2 appears to interact with the core carbohydrates of LPS. As shown in S3 Fig, an LPS defect was able to influence the bacteria-killing ability of LL-37, which implicates LPS as another target of LL-37. These results suggest the possible association and involvement of specific regions of OmpA and LPS as the mode of action of LL-37 against A. baumannii.
LL-37 not only exerts its antimicrobial effect by the formation of membrane pores leading to membrane disruption [6], but can also cross lipid membranes of host cells, resulting in gene/protein stimulation or a block of gene/protein expression [3]. In this study, the ompA deletion strain was more sensitive to LL-37 than the wild type strain (Fig 8A). Although the LL-37-binding target AbOmpA was absent, other OMPs might be bound by LL-37 (represented in Fig 5). Because OmpA exhibits low pore-forming function and permeability, with a pore size approximately 2 nm in diameter [52], LL-37 might bind to other OMPs with higher pore-forming abilities and permeabilities in the ompA deletion strain, resulting in increased cell death. Moreover, AbOmpA plays an important role in adhesion and biofilm formation in A. baumannii [23]. Fig 8B showed that the adhesion defect in the ΔompA strain was not augmented as obviously as the defect in the wild type strain by the addition of LL-37. Therefore, we suggest that the LL-37-mediated adhesion defect may be explained by interference with AbOmpA.
LL-37 may bind to LPS with high affinity, but its bactericidal activity is not LPS-dependent. The increased sensitivity of LPS-deficient colistin-resistant A. baumannii to LL-37 has been demonstrated and ascribed to increased membrane permeability [17]. However, colistin-resistant isolates (due to mutations in the PmrB domains post-colistin treatment) induced cross-resistance to LL-37 [53]. Hence, we speculated that the LPS-defective strain with increased resistance to LL-37 (S3 Fig) might be a result of a PmrB mutation. While studying P. aeruginosa, Lin et al. proposed a model that suggested that the associated LPS and fatty acids of OprI were eliminated by AMP hRNase 7 treatment, followed by subsequent internalization of OprI with the invading hRNase 7. According to the results of our experiment, it is possible that LL-37 exerts its action on A. baumannii via OmpA binding in a manner that is similar to the results reported for P. aeruginosa.
There were some limitations to this study. First, the adhesion assay was performed on abiotic polystyrene plate whose characteristics were completely different from the cell surface in vivo. Second, the effects of LL-37 on A. baumannii were not demonstrated in human cell platforms. Finally, the contribution of LPS to LL-37 action in A. baumannii deserved further investigation. In conclusion, our study demonstrated that the human antimicrobial peptide LL-37 affected A. baumannii via binding to OmpA. We hope that this study can serve as a starting point to understand the complete mechanism underlying the effect of LL-37 on A. baumannii.
Supporting Information
The bacterial killing activity of LL-37 on two clinical isolates of A. baumannii was performed using spot assay, which also showed the anti-bacterial effect augmented with LL-37 concentrations increasing. The clinical strains are from our previous study [54].
(TIF)
(A) RT-PCR was performed to detect ompA expression. Total RNAs were isolated from wild type and the ΔompA mutant, cDNAs were synthesized and RT-PCR was performed. The absence of the ompA transcript (504 bp) was observed in the mutant strain.(B) OmpA protein expression was detected by Coomassie blue staining and western blot. OMPs of the wild-type and ΔompA strains were extracted and subjected to SDS-PAGE. After transferring the proteins onto PVDF membrane, gel was stained by Coomassie blue and the membrane was blotted by anti-OmpA antibody. No OmpA protein expressed was detected in the ΔompA mutant.
(TIF)
(A) LPS of the wild type and the LPS-defective mutant was visualized by silver staining. LPS was isolated, subjected to a polyacryamide gel and analyzed by electrophoresis. Lane 1 was the LPS control from E. coli. (B) Sensitivity of the wild type and a LPS-defective strain to LL-37 was examined by spot assay. The wild type and LPS-defect strains were mixed with different concentrations of LL-37 for 1 hr, 10-fold serially diluted, and spotted on LB agar plate. The LPS-defect strain had better tolerance to LL-37 compared to the wild type.
(TIF)
(PDF)
Acknowledgments
We are grateful to Luis A. Actis (Miami University, OH, USA), Kai-Chih Chang (Tzu Chi University, Taiwan) and M.-L. Liou (Yuanpei University, Taiwan) for providing antibodies and bacterial strains.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grant MOST104-2622-B-007-001-CC2 (to CYL) from the Ministry of Science and Technology (Taiwan).
References
- 1. Hancock RE, Sahl HG (2006) Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 24: 1551–1557. [DOI] [PubMed] [Google Scholar]
- 2. Bals R (2000) Epithelial antimicrobial peptides in host defense against infection. Respir Res 1: 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jenssen H, Hamill P, Hancock RE (2006) Peptide antimicrobial agents. Clin Microbiol Rev 19: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zaiou M, Nizet V, Gallo RL (2003) Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J Invest Dermatol 120: 810–816. [DOI] [PubMed] [Google Scholar]
- 5. Durr UH, Sudheendra US, Ramamoorthy A (2006) LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta 1758: 1408–1425. [DOI] [PubMed] [Google Scholar]
- 6. Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3: 238–250. [DOI] [PubMed] [Google Scholar]
- 7. Nguyen LT, Haney EF, Vogel HJ (2011) The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol 29: 464–472. 10.1016/j.tibtech.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 8. Lai Y, Gallo RL (2009) AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol 30: 131–141. 10.1016/j.it.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gudmundsson GH, Agerberth B (1999) Neutrophil antibacterial peptides, multifunctional effector molecules in the mammalian immune system. J Immunol Methods 232: 45–54. [DOI] [PubMed] [Google Scholar]
- 10. Cirioni O, Giacometti A, Ghiselli R, Bergnach C, Orlando F, Silvestri C, et al. (2006) LL-37 protects rats against lethal sepsis caused by gram-negative bacteria. Antimicrob Agents Chemother 50: 1672–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chromek M, Arvidsson I, Karpman D (2012) The antimicrobial peptide cathelicidin protects mice from Escherichia coli O157:H7-mediated disease. PLoS One 7: e46476 10.1371/journal.pone.0046476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsai PW, Cheng YL, Hsieh WP, Lan CY (2014) Responses of Candida albicans to the human antimicrobial peptide LL-37. J Microbiol 52: 581–589. 10.1007/s12275-014-3630-2 [DOI] [PubMed] [Google Scholar]
- 13. Lin MF, Lan CY (2014) Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J Clin Cases 2: 787–814. 10.12998/wjcc.v2.i12.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA (2007) Global challenge of multidrug-resistant Acinetobacter baumannii . Antimicrob Agents Chemother 51: 3471–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Navon-Venezia S, Leavitt A, Carmeli Y (2007) High tigecycline resistance in multidrug-resistant Acinetobacter baumannii . J Antimicrob Chemother 59: 772–774. [DOI] [PubMed] [Google Scholar]
- 16. Cai Y, Chai D, Wang R, Liang B, Bai N (2012) Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67: 1607–1615. 10.1093/jac/dks084 [DOI] [PubMed] [Google Scholar]
- 17. Moffatt JH, Harper M, Mansell A, Crane B, Fitzsimons TC, Nation RL, et al. (2013) Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect Immun 81: 684–689. 10.1128/IAI.01362-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng X, Sambanthamoorthy K, Palys T, Paranavitana C (2013) The human antimicrobial peptide LL-37 and its fragments possess both antimicrobial and antibiofilm activities against multidrug-resistant Acinetobacter baumannii . Peptides 49: 131–137. 10.1016/j.peptides.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 19. Smith SG, Mahon V, Lambert MA, Fagan RP (2007) A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett 273: 1–11. [DOI] [PubMed] [Google Scholar]
- 20. Mortensen BL, Skaar EP (2012) Host-microbe interactions that shape the pathogenesis of Acinetobacter baumannii infection. Cell Microbiol 14: 1336–1344. 10.1111/j.1462-5822.2012.01817.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee JS, Choi CH, Kim JW, Lee JC (2010) Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting. J Microbiol 48: 387–392. 10.1007/s12275-010-0155-1 [DOI] [PubMed] [Google Scholar]
- 22. Kim SA, Yoo SM, Hyun SH, Choi CH, Yang SY, Kim HJ, et al. (2008) Global gene expression patterns and induction of innate immune response in human laryngeal epithelial cells in response to Acinetobacter baumannii outer membrane protein A. FEMS Immunol Med Microbiol 54: 45–52. 10.1111/j.1574-695X.2008.00446.x [DOI] [PubMed] [Google Scholar]
- 23. Gaddy JA, Tomaras AP, Actis LA (2009) The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77: 3150–3160. 10.1128/IAI.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim SW, Choi CH, Moon DC, Jin JS, Lee JH, Shin JH, et al. (2009) Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol Lett 301: 224–231. 10.1111/j.1574-6968.2009.01820.x [DOI] [PubMed] [Google Scholar]
- 25. Lin MF, Lin YY, Yeh HW, Lan CY (2014) Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol 14: 119 10.1186/1471-2180-14-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsai PW, Yang CY, Chang HT, Lan CY (2011) Human antimicrobial peptide LL-37 inhibits adhesion of Candida albicans by interacting with yeast cell-wall carbohydrates. PLoS One 6: e17755 10.1371/journal.pone.0017755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, et al. (2010) The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 192: 6336–6345. 10.1128/JB.00917-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loehfelm TW, Luke NR, Campagnari AA (2008) Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol 190: 1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuenca FF, Pascual A, Martinez Marinez L, Conejo MC, Perea EJ (2003) Evaluation of SDS-polyacrylamide gel systems for the study of outer membrane protein profiles of clinical strains of Acinetobacter baumannii . J Basic Microbiol 43: 194–201. [DOI] [PubMed] [Google Scholar]
- 30. Krishnan S, Prasadarao NV (2012) Outer membrane protein A and OprF: versatile roles in Gram-negative bacterial infections. Febs j 279: 919–931. 10.1111/j.1742-4658.2012.08482.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krauel K, Weber C, Brandt S, Zahringer U, Mamat U, Greinacher A, et al. (2012) Platelet factor 4 binding to lipid A of Gram-negative bacteria exposes PF4/heparin-like epitopes. Blood 120: 3345–3352. 10.1182/blood-2012-06-434985 [DOI] [PubMed] [Google Scholar]
- 32. Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, et al. (2010) A rapid and simple method for constructing stable mutants of Acinetobacter baumannii . BMC Microbiol 10: 279 10.1186/1471-2180-10-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gohl O, Friedrich A, Hoppert M, Averhoff B (2006) The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl Environ Microbiol 72: 1394–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH (2011) Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett 323: 44–51. 10.1111/j.1574-6968.2011.02362.x [DOI] [PubMed] [Google Scholar]
- 35. Puspurs A, Medon P, Corless C, Hackett J, Reeves P (1983) A class of ompA mutants of Escherichia coli K12 affected in the interaction of OmpA protein and the core region of lipopolysaccharide. Mol Gen Genet 189: 162–165. [DOI] [PubMed] [Google Scholar]
- 36. Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS, et al. (2012) Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol 50: 155–160. 10.1007/s12275-012-1589-4 [DOI] [PubMed] [Google Scholar]
- 37. Doss M, White MR, Tecle T, Hartshorn KL (2010) Human defensins and LL-37 in mucosal immunity. J Leukoc Biol 87: 79–92. 10.1189/jlb.0609382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu PT, Stenger S, Tang DH, Modlin RL (2007) Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol 179: 2060–2063. [DOI] [PubMed] [Google Scholar]
- 39. van der Does AM, Bergman P, Agerberth B, Lindbom L (2012) Induction of the human cathelicidin LL-37 as a novel treatment against bacterial infections. J Leukoc Biol 92: 735–742. 10.1189/jlb.0412178 [DOI] [PubMed] [Google Scholar]
- 40. Sochacki KA, Barns KJ, Bucki R, Weisshaar JC (2011) Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc Natl Acad Sci U S A 108: E77–81. 10.1073/pnas.1101130108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hell E, Giske CG, Nelson A, Romling U, Marchini G (2010) Human cathelicidin peptide LL37 inhibits both attachment capability and biofilm formation of Staphylococcus epidermidis . Lett Appl Microbiol 50: 211–215. 10.1111/j.1472-765X.2009.02778.x [DOI] [PubMed] [Google Scholar]
- 42. Overhage J, Campisano A, Bains M, Torfs EC, Rehm BH, Hancock RE (2008) Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun 76: 4176–4182. 10.1128/IAI.00318-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bowdish DM, Davidson DJ, Lau YE, Lee K, Scott MG, Hancock RE (2005) Impact of LL-37 on anti-infective immunity. J Leukoc Biol 77: 451–459. [DOI] [PubMed] [Google Scholar]
- 44. Bowdish DM, Davidson DJ, Scott MG, Hancock RE (2005) Immunomodulatory activities of small host defense peptides. Antimicrob Agents Chemother 49: 1727–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. De Smet K, Contreras R (2005) Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol Lett 27: 1337–1347. [DOI] [PubMed] [Google Scholar]
- 46. Ding L, Yang L, Weiss TM, Waring AJ, Lehrer RI, Huang HW (2003) Interaction of antimicrobial peptides with lipopolysaccharides. Biochemistry 42: 12251–12259. [DOI] [PubMed] [Google Scholar]
- 47. Rosenfeld Y, Shai Y (2006) Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: Role in bacterial resistance and prevention of sepsis. Biochimica et Biophysica Acta (BBA)—Biomembranes 1758: 1513–1522. [DOI] [PubMed] [Google Scholar]
- 48. Chai H, Allen WE, Hicks RP (2014) Synthetic Antimicrobial Peptides Exhibit Two Different Binding Mechanisms to the Lipopolysaccharides Isolated from Pseudomonas aeruginosa and Klebsiella pneumoniae. Int J Med Chem 2014: 809283 10.1155/2014/809283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin YM, Wu SJ, Chang TW, Wang CF, Suen CS, Hwang MJ, et al. (2010) Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein. J Biol Chem 285: 8985–8994. 10.1074/jbc.M109.078725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Apetrei A, Asandei A, Park Y, Hahm KS, Winterhalter M, Luchian T (2010) Unimolecular study of the interaction between the outer membrane protein OmpF from E. coli and an analogue of the HP(2–20) antimicrobial peptide. J Bioenerg Biomembr 42: 173–180. 10.1007/s10863-010-9273-z [DOI] [PubMed] [Google Scholar]
- 51. Morona R, Klose M, Henning U (1984) Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J Bacteriol 159: 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sugawara E, Nikaido H (2012) OmpA is the principal nonspecific slow porin of Acinetobacter baumannii . J Bacteriol 194: 4089–4096. 10.1128/JB.00435-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Napier BA, Burd EM, Satola SW, Cagle SM, Ray SM, McGann P, et al. (2013) Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii . MBio 4: e00021–00013. 10.1128/mBio.00021-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin MF, Lin YY, Tu CC, Lan CY (2015) Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J Microbiol Immunol Infect. In press. 10.1016/j.jmii.2015.04.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The bacterial killing activity of LL-37 on two clinical isolates of A. baumannii was performed using spot assay, which also showed the anti-bacterial effect augmented with LL-37 concentrations increasing. The clinical strains are from our previous study [54].
(TIF)
(A) RT-PCR was performed to detect ompA expression. Total RNAs were isolated from wild type and the ΔompA mutant, cDNAs were synthesized and RT-PCR was performed. The absence of the ompA transcript (504 bp) was observed in the mutant strain.(B) OmpA protein expression was detected by Coomassie blue staining and western blot. OMPs of the wild-type and ΔompA strains were extracted and subjected to SDS-PAGE. After transferring the proteins onto PVDF membrane, gel was stained by Coomassie blue and the membrane was blotted by anti-OmpA antibody. No OmpA protein expressed was detected in the ΔompA mutant.
(TIF)
(A) LPS of the wild type and the LPS-defective mutant was visualized by silver staining. LPS was isolated, subjected to a polyacryamide gel and analyzed by electrophoresis. Lane 1 was the LPS control from E. coli. (B) Sensitivity of the wild type and a LPS-defective strain to LL-37 was examined by spot assay. The wild type and LPS-defect strains were mixed with different concentrations of LL-37 for 1 hr, 10-fold serially diluted, and spotted on LB agar plate. The LPS-defect strain had better tolerance to LL-37 compared to the wild type.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper.