Significance
Genome sequence analysis reveals that the zoonotic malaria parasite Plasmodium knowlesi consists of three highly divergent subpopulations. Two, commonly seen in sympatric human clinical infections in Malaysian Borneo, were identified in a previous study as corresponding to parasites seen in long-tailed and pig-tailed macaque hosts, respectively. A third type has been detected in a few laboratory-maintained isolates originally derived in the 1960s elsewhere in Southeast Asia. Divergence between the subpopulations varies significantly across the genome but overall is at a level indicating different subspecies. Analysis of the diversity within the most common type in human infections shows strong signatures of natural selection, including balancing selection and directional selection, on loci distinct from those under selection in endemic human malaria parasites.
Keywords: population genomics, Plasmodium diversity, reproductive isolation, zoonosis, adaptation
Abstract
Malaria cases caused by the zoonotic parasite Plasmodium knowlesi are being increasingly reported throughout Southeast Asia and in travelers returning from the region. To test for evidence of signatures of selection or unusual population structure in this parasite, we surveyed genome sequence diversity in 48 clinical isolates recently sampled from Malaysian Borneo and in five lines maintained in laboratory rhesus macaques after isolation in the 1960s from Peninsular Malaysia and the Philippines. Overall genomewide nucleotide diversity (π = 6.03 × 10−3) was much higher than has been seen in worldwide samples of either of the major endemic malaria parasite species Plasmodium falciparum and Plasmodium vivax. A remarkable substructure is revealed within P. knowlesi, consisting of two major sympatric clusters of the clinical isolates and a third cluster comprising the laboratory isolates. There was deep differentiation between the two clusters of clinical isolates [mean genomewide fixation index (FST) = 0.21, with 9,293 SNPs having fixed differences of FST = 1.0]. This differentiation showed marked heterogeneity across the genome, with mean FST values of different chromosomes ranging from 0.08 to 0.34 and with further significant variation across regions within several chromosomes. Analysis of the largest cluster (cluster 1, 38 isolates) indicated long-term population growth, with negatively skewed allele frequency distributions (genomewide average Tajima’s D = −1.35). Against this background there was evidence of balancing selection on particular genes, including the circumsporozoite protein (csp) gene, which had the top Tajima’s D value (1.57), and scans of haplotype homozygosity implicate several genomic regions as being under recent positive selection.
The zoonotic malaria parasite Plasmodium knowlesi is a significant cause of human malaria, with a wide spectrum of clinical outcomes including high parasitemia and death (1–4). Long known as a malaria parasite of long-tailed and pig-tailed macaques (5), the first large focus of human cases was described only in 2004 in the Kapit Division of Sarawak in Malaysian Borneo (6). Since then infections have been described from almost all countries in Southeast Asia (2, 7). Travelers to the region from Europe, North America, and Australasia also have recently acquired P. knowlesi malaria (7, 8). Until the application of molecular assays for specific detection, human P. knowlesi malaria was largely misdiagnosed as Plasmodium malariae, a morphologically similar but distantly related species (1, 6, 9, 10). Studies in the Kapit Division of Sarawak in Malaysian Borneo have indicated that P. knowlesi malaria is primarily a zoonosis with macaques as reservoir hosts (11) and that the forest-dwelling mosquito species Anopheles latens is the local vector for P. knowlesi (12). Other members of the Anopheles leucosphyrus group are vectors in different parts of Southeast Asia and may determine the geographical distribution of transmission (13, 14).
Although P. knowlesi malaria is regarded as an emerging infection, there clearly have been increased efforts in detection made since its existence as a significant zoonosis was discovered, and specific detection has been enhanced by the declining numbers of human cases caused by other malaria parasites in Southeast Asia (15). Aside from the first two human cases described several decades ago (5), there is direct evidence of human P. knowlesi infections from ∼20 y ago in Malaysian Borneo and Thailand obtained by retrospective molecular analysis of material from archived blood spots and slides (10, 16), and molecular population genetic evidence indicates the zoonosis has been in existence for a much longer time (11). The genetic diversity of P. knowlesi is high within humans as well as macaques, with sequence data on three loci [the circumsporozoite protein (csp) gene, 18S rRNA, and mtDNA genome] indicating extensive shared polymorphism and no fixed differences between P. knowlesi parasites from humans and monkeys sampled in the same area in Sarawak, Malaysian Borneo (6, 11). Analysis of samples from a smaller number of humans and monkeys in Thailand showed alleles of the P. knowlesi merozoite surface protein 1 (msp1) gene to be similarly diverse in both hosts (16), and there were shared polymorphisms of the csp gene in parasites from a few infections examined in humans and macaques in Singapore (17). Recent multilocus microsatellite analysis has indicated a deep population subdivision in P. knowlesi associated with long-tailed and pig-tailed macaques; both major types infect humans and occur sympatrically at most sites in Malaysia, but the two types show some additional geographical differentiation across sites (18).
Human populations have grown very rapidly in the Southeast Asian region and encroach on most of the wild macaque habitats, so it is vital to know if P. knowlesi parasites are adapting to human hosts or to anthropophilic mosquito vector species, either of which could cause human–mosquito–human transmission. Initial analysis of the P. knowlesi reference genome sequence (strain H) highlighted some unique features of the genome of this species (19), namely, schizont infected cell agglutination variant (SICAvar) and knowlesi interspersed repeat (KIR) variant antigen genes, which were widely dispersed instead of being predominantly localized in subtelomeric regions as seen in large gene families in Plasmodium falciparum and Plasmodium vivax (20). Here, we analyzed genomewide diversity in P. knowlesi and conducted scans for signatures of balancing and directional selection, revealing extremely high genetic diversity and significant structuring of this species into subpopulation clusters that appear to be reproductively isolated as well as loci that show evidence of recent strong selection.
Results
Sequencing and Nucleotide Diversity of Clinical and Laboratory P. knowlesi Isolates.
Paired-end short-read sequencing produced high-quality data for 53 P. knowlesi isolates (48 clinical infections and five laboratory-maintained lines), with genomewide average mapping depth to the P. knowlesi H strain reference genome (19) having a minimum and median fold coverage of 36× and 120×, respectively (Table S1). Another three clinical isolates had average fold coverage below 30× and were not used for further analysis. All isolates contained sequences that potentially encode amino acid motif groups that may be involved in molecular mimicry of macaque CD99, as previously seen in the P. knowlesi H strain reference genome (19).
For analysis of polymorphism, we excluded subtelomeric and repeat regions (as defined in Materials and Methods) and members of large multigene families (SICAvar and KIR, n = 373 including fragments), allowing 92% of the genome to be analyzed (21.7 Mb of the 23.5-Mb reference genome), including 93% of all predicted protein-coding genes (4,860 genes). Overall, 975,642 biallelic high-quality SNPs were identified among the 53 isolates, indicating a very high level of nucleotide diversity [genomewide π = 6.03 × 10−3 (genic π = 4.84 × 10−3; noncoding π = 7.50 × 10−3)]. Almost half of the SNPs (423,160; 43.4% of the total) had a minor allele that occurred in only one isolate, and 21.8% (n = 213,375) had minor allele frequencies of above 10% in the overall sample of isolates.
All individual isolates were very distinct from one other (Fig. 1) except for two of the laboratory lines [labeled “H(AW)” and “Malayan”], which were virtually identical to each other and to the published H strain reference genome sequence (19). Only 251 SNPs were identified among these three sequences [numbers of pairwise differences: H reference vs. Malayan = 226; H reference vs. H(AW) = 221; Malayan vs. H(AW) = 57], 18 of which were in the coding regions of nine genes (Table S2) and two of which (PKH_112660 and PKH_133910) are members of the Apetala 2 (AP2) gene family of transcription factors. Interestingly, the SNP on one of the AP2 genes (PKH_133910) led to the only stop codon mutation identified in the laboratory isolates, in line H(AW). It is notable that the MR4-H strain sequence [obtained from the Malaria Research and Reference Reagent Resource Center (MR4) repository as supposedly the reference H strain genome] was very different from the published H strain reference sequence. This difference, together with the observation that the line called “Malayan” was virtually identical to the H strain reference sequence, suggests historical mislabeling or mix up of laboratory lines. It is likely that most or all of the SNPs differentiating the three nearly identical genome sequences (including those in the AP2 genes) were derived by mutations during laboratory passage in rhesus macaques or more recently in culture. For subsequent analysis of diversity between different sampled isolates, only one of these nearly identical H strain-like sequences (Malayan) was included.
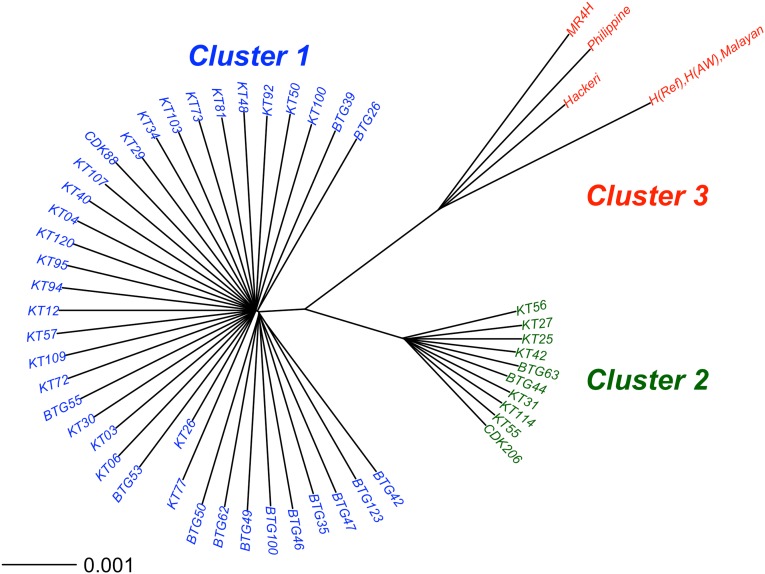
Fig. 1.
Deep genomic population substructure in P. knowlesi. Neighbor-joining tree based on pairwise nucleotide diversity (π) between isolates using high-quality SNPs from 53 samples (48 clinical isolates from human patients and five laboratory samples maintained in rhesus macaques). This tree shows three major clusters representing two subgroups of clinical isolates (cluster 1, n = 38; cluster 2, n = 10) and a third cluster of laboratory isolates (cluster 3, n = 5) together with the reference genome sequence H(Ref). Clusters 1 and 2 occurred sympatrically at both of the sampling sites (in Kapit, 25 and 8 in each cluster respectively; in Betong, 13 and 2 respectively). Two of the cluster 3 laboratory isolates, labeled “H(AW)” and “Malayan,” were nearly identical to each other and to the reference genome sequence. The isolate labeled here as “MR4H” was received from the MR4 reagent repository labeled as the “H” strain.
Major Substructure Within the P. knowlesi Species.
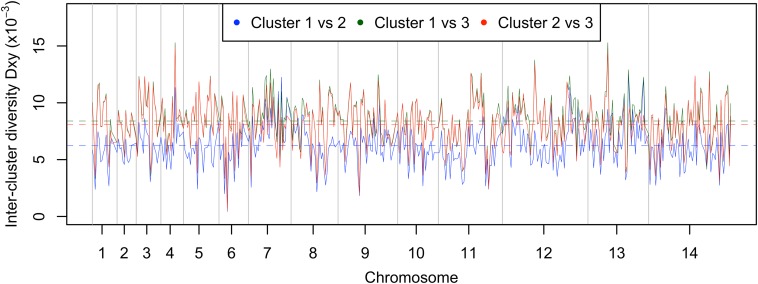
Analysis of pairwise nucleotide differences among all the isolates revealed three main clusters. The largest two represent subgroups of the clinical isolates (cluster 1, n = 38; cluster 2, n = 10), and a third comprises the laboratory isolates (cluster 3, n = 5 with n = 4 being distinct) (Fig. 1). Principal components analysis (PCA) confirmed the presence of the three groups, with the first two principal components accounting for 26% of the variation (Fig. S1). The nucleotide divergence between cluster 3 and the others [for clusters 1 vs. 3, genetic distance (DXY) = 8.4 × 10−3; for cluster 2 vs. 3 DXY = 8.1 × 10−3) was slightly higher than between the clinical isolate clusters 1 and 2 (DXY = 6.3 × 10−3) (Fig. S2). The assignment of the clinical isolates into two distinct clusters shown here also was compared with an independent assignment recently performed using a STRUCTURE analysis of 10-locus microsatellite genotype data for 40 of the 48 isolates (18). All isolates had the same cluster assignment by both methods, except for one cluster 1 isolate, which had an intermediate cluster assignment in the microsatellite analysis (Fig. S3).
Fig. S1.
Identifying the population structure using PCA. A PCA of SNPs from all 53 sequenced P. knowlesi samples reveals three main subgroups representing two clusters of the clinical isolates and a third cluster of laboratory samples. These subgroups correspond exactly to the three clusters shown in Fig. 1. Samples from Kapit (n = 33) and Betong (n = 15) are both distributed evenly in the two major clusters (overlapping points on the plot have obscured the visibility of two isolates from Betong within cluster 2). The first two principal components shown here account for a large proportion (26%) of the total variation in the data.
Fig. S2.
Divergence (DXY) between each of the three major P. knowlesi clusters in sliding windows of 50 kb (with a step size of 25 kb) across the genome. The average DXY values (differences per nucleotide × 10−3) for the 14 chromosomes were 5.44, 5.61, 6.25, 6.38, 5.71, 5.54, 7.64, 6.25, 6.20, 5.83, 5.62, 6.64, 7.08, and 5.87, respectively.
Fig. S3.
The concordance of cluster assignment based on sequence data of 40 clinical isolates analyzed here with the previous assignment of these isolates based on a previously published STRUCTURE analysis of 10-locus microsatellite genotypes (18).
The average pairwise nucleotide diversity was higher among the cluster 1 clinical isolates (π = 5.28 × 10−3) and among the cluster 3 laboratory isolates (π = 5.67 × 10−3, n = 4 excluding nearly identical isolates) than among the cluster 2 clinical isolates (π = 3.15 × 10−3). This result is not caused by genetic complexity within infections. Generally, there are low levels of complexity per infection in clinical P. knowlesi isolates, most containing one predominant haploid genotype each (Table S3). The estimates of within-infection genomic complexity (1 − FWS) were similar for cluster 1 and cluster 2 isolates [mean within-infection fixation index (FWS) values of 0.94 and 0.95 respectively; Mann–Whitney test; P = 0.40] and correlated highly with indices independently derived by counting the number of distinct alleles in a previously performed 10-locus microsatellite genotypic analysis (Pearson’s r2 = 0.76, P = 6.4 × 10−10) (Table S3) (18).
Despite the differences in overall nucleotide diversity in the different clusters, the relative proportions of synonymous, nonsynonymous, intronic, and intergenic SNPs observed were similar in each (respectively 14%, 16%, 13%, and 57% for cluster 1; 14%, 17%, 13%, and 56% for cluster 2; and 13%, 18%, 14%, and 55% for cluster 3). A genomewide scan shows that lower nucleotide diversity among the cluster 2 isolates is observed in most chromosomes; exceptions with similar levels of diversity include chromosome 5 and regions of chromosomes 8, 10, and 11 (Fig. 2). In contrast, clusters 1 and 3 show similar variation in levels of diversity across the genome, so it is clear that cluster 2 differs in this particular respect.
Fig. 2.
Distribution of the average nucleotide diversity (π) for sliding windows of 50-kb regions within the three main P. knowlesi subpopulation clusters: cluster 1 (n = 38, blue line), cluster 2 (n = 10, green line), and cluster 3 (n = 4, red line). The dotted lines represent the genomewide mean values for the three respective clusters. The solid black line represents the overall nucleotide diversity across all samples.
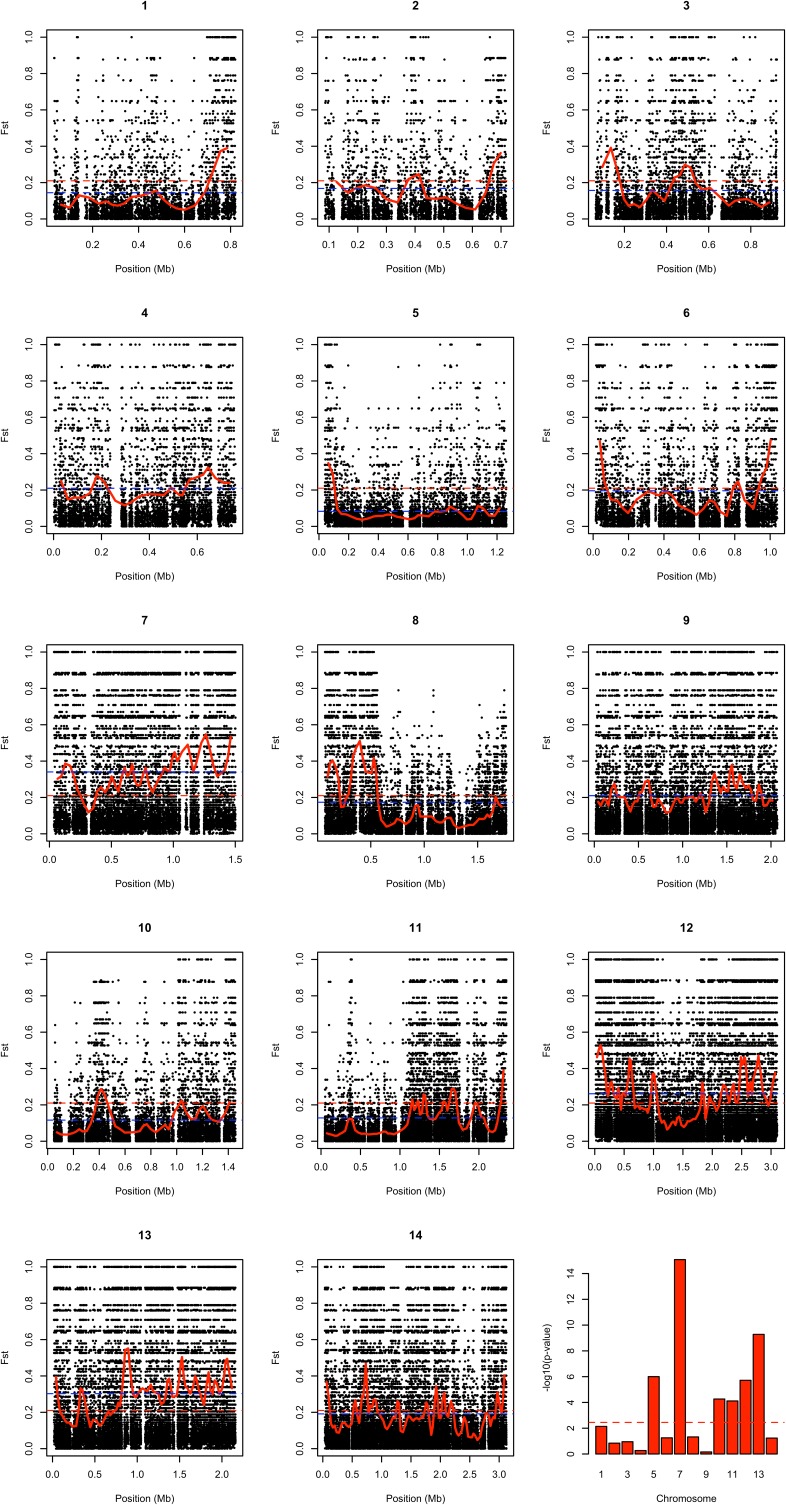
Comparison of cluster 1 and 2 clinical isolates revealed a very high level of differentiation across the genome [genomewide average fixation index (FST) = 0.21] (Fig. 3). Moreover, a large number of SNP positions (n = 9,293) showed complete fixation of alternative alleles between these clusters (FST = 1.0), and these SNP positions with complete fixation occurred on all chromosomes (Fig. 3B and Fig. S4). However, there was marked heterogeneity in the level of differentiation between clusters 1 and 2 across the genome, with chromosomal mean FST scores ranging from 0.08 (chromosome 5) to 0.34 (chromosome 7) (Fig. 3 and Fig. S4). Analysis of windows of 500 consecutive SNPs revealed that chromosomes 5, 10, and 11 had the most windows below the genomewide average FST (Fisher’s exact tests, P = 3.6 × 10−7, 1.4 × 10−5, and 2.5 × 10−5, respectively), whereas chromosomes 7, 12, and 13 had the most windows above the genomewide average (3.7 × 10−16, 4.0 × 10−6, and 2.1 × 10−11, respectively). Chromosomes 7, 12, and 13 also had high DXY cluster 1 versus cluster 2 divergence values of 7.64 × 10−3, 6.64 × 10−3, and 7.08 × 10−3, respectively (Fig. S2), indicating that the elevated FST values are not caused entirely by the lower diversity within cluster 2. There also was heterogeneity across different regions within chromosomes (Fig. 3 and Fig. S4), with window mean FST values ranging from <0.05 to >0.40. Regions with high FST values contained many SNPs that had completely fixed differences between the clusters (FST = 1.0) (Fig. 3B and Fig. S4). Application of Moran’s I test of autocorrelation indicated that chromosomes 7, 8, 11, 12, and 13 had particularly significant nonrandom intrachromosomal clustering of FST (Fig. S4).
Fig. 3.
Genomewide FST scans between the cluster 1 and 2 subpopulations of P. knowlesi clinical isolates. (A) Sliding window plot of mean FST scores for windows of 500 consecutive SNPs shows within-chromosome variation of FST scores. The blue and gray shades represent alternating chromosomes. The dashed red lines show the genomewide mean FST value of 0.21. (B) Heterogeneity within individual chromosomes illustrated by plots of FST values for chromosomes 8, 11, and 12. Black dots show values for individual SNPs, and red lines show mean FST values for consecutive windows of 500 SNPs. Dashed blue lines represent chromosome-wide mean FST values, and dashed red lines show the genomewide mean FST value of 0.21.
Fig. S4.
Plots of FST values indicating differentiation in SNP allele frequencies between P. knowlesi cluster 1 (38 isolates) and cluster 2 (10 isolates) throughout the genome. The panels show widespread distribution of high-FST SNPs (black dots) as well as extended low-FST regions. The solid red lines show mean FST values for sliding windows of 500 consecutive SNPs. The dashed red lines represent the genomewide average FST score of 0.21, and the dashed blue lines represent the mean FST score for each chromosome (chromosomes 1–14 having values of 0.14, 0.17, 0.16, 0.21, 0.08, 0.2, 0.34, 0.17, 0.21, 0.12, 0.13, 0.26, 0.3, and 0.19, respectively). The bar plot in the bottom right panel shows significance values as −log10 P values of Fisher’s exact test statistics for the 14 chromosomes. In testing the number of windows with mean FST values greater or less than the genomewide mean, chromosomes 5, 10, and 11 were found to have a significantly greater number of low-FST windows, and high-FST windows were significantly overrepresented on chromosomes 7, 12, and 13. In addition, positive Moran’s I indices indicated nonrandom intrachromosomal clustering of FST values, particularly for chromosomes 7, 8, 11, 12, and 13 (with Moran’s I indices of 0.41, 0.48, 0.39, 0.42, and 0.40, respectively).
Scan for Genes Showing Signatures of Balancing Selection.
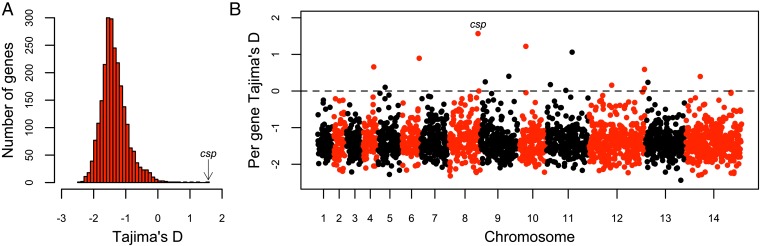
Because of the overall deep population structuring and the small numbers of isolates belonging to clusters 2 and 3, scans for signatures of selection were focused on the main cluster 1 of clinical isolates (n = 38). A total of 759,409 biallelic SNPs were identified within cluster 1, 230,398 (30%) of which were found in coding regions (2,399 genes). A genomewide scan of allele frequency distributions conducted on genes that had a minimum of three SNPs (2,381 genes) showed a negatively skewed distribution, with a mean Tajima’s D value of −1.35 (Fig. 4A). Only 16 of the 2,381 genes (0.67%) had a positive Tajima’s D value (Fig. 4 and Dataset S1), with the highest being for the csp gene (Tajima’s D = 1.57). Most other genes close to the top of the list encode putative proteins that have not been studied previously, although orthologs of particular antigen genes previously shown to be under balancing selection in other malaria parasite species (21) have quite high values here (e.g., the trap and msp1 genes, with ranks of 24 and 61, respectively). However not all antigen genes have such high values; for example, the P. knowlesi apical membrane antigen 1 (ama1) and DBP-α genes had Tajima’s D values of −1.68 and −1.31, respectively, close to the genomewide average and in contrast with the elevated values for their orthologs in population studies of other species (21).
Fig. 4.
Scan of Tajima’s D values for 2,381 genes with a minimum of three SNPs within the major P. knowlesi subpopulation cluster 1 (n = 38 isolates). (A) Frequency distribution of Tajima’s D values shows a highly negative skew genomewide and only a minority of genes with positive values (the gene with the highest value was csp, encoding the circumsporozoite protein). (B) Tajima’s D values for 2,381 genes with a minimum of three SNPs plotted according to their chromosomal positions (black and red colors indicate consecutive chromosomes numbered from the smallest upwards). Tajima’s D values for each of the individual genes are listed in Dataset S1.
Informative tests of allele frequency distributions by Tajima’s D require larger numbers of isolates than were sequenced for clusters 2 and 3. To explore the allele frequency distribution in the isolates in cluster 2 (n = 10), Tajima’s D values were calculated for 2,117 genes with three or more SNPs, yielding an overall mean value of −0.26, which was much less negative than seen for the larger sample of cluster 1 isolates. To test if this difference from cluster 1 was an artifact of the small sample size, Tajima’s D values were computed from randomly generated subsamples of 10 isolates from cluster 1, yielding a mean value of −0.22, which was not significantly different from that of cluster 2. This result illustrates the low information content of this index with very small numbers of samples and the need for more extensive sampling of clusters 2 and 3 to enable allele frequency-based tests for selection within each of these clusters.
Evidence of Recent Positive Directional Selection.
A genomewide scan using both coding and noncoding SNPs from 38 isolates of the main cluster 1 identified 235 positions with integrated haplotype score (|iHS|) values of >4.89 (Fig. 5), which clustered into 57 windows of contiguous elevated haplotype homozygosity covering a total of 1,978 kb. To focus on the most significant regions, we identified nine of these windows that had at least one SNP with |iHS| >6.0 and that did not have gaps of more than 20 kb between consecutive genotyped SNPs, covering a combined total of 593 kb. Individual window sizes ranged from 10–161 kb (Table S4). These windows contained 146 genes (Table S5), 75 (51%) of which encoded conserved Plasmodium proteins of unknown function and 11 (8%) of which are annotated as members of the species-specific SICAvar gene family. Because SNPs in SICAvar genes were excluded from analysis, the five windows that contained SICAvars are caused by signals from SNPs in the surrounding regions. Other genes within elevated iHS regions include csp and two P. knowlesi-specific proteins (PKH_083491 and PKH_112405) with no reported orthologs in other Plasmodium species. As expected, given the assumptions that P. knowlesi cases in humans are primarily zoonotic and that the reservoir parasite population is not under antimalarial drug selection, no signals of positive selection were observed around five orthologs of known P. falciparum drug-resistance genes: the chloroquine resistance transporter (crt, PKH_010710), multidrug resistance-1 (mdr1, PKH_100920), dihydrofolate reductase (dhfr, PKH_052130), dihydropteroate synthase (dhps, PKH_142780), and kelch K13 (PKH_121080).
Fig. 5.
Scan for evidence of recent positive selection in the main P. knowlesi subpopulation cluster 1. Plot of genomewide |iHS| scores shows regions of the genome that have windows of elevated values, consistent with the operation of recent positive directional selection. The dashed lines represent values of 4.89 (blue) and 6 (red), used to define nine windows containing SNPs with overlapping regions of extended haplotype homozygosity, as described in Materials and Methods. The coordinates of these windows and the genes within them are listed in Tables S4 and S5.
Discussion
This whole-genome population study of zoonotic malaria reveals much higher diversity in P. knowlesi than in either of the major endemic human malaria parasite species, P. falciparum or P. vivax (22–25). Consistent with previous analyses of diversity of mitochondrial genome sequences (11) and microsatellite genotyping (18), the estimate of genomic complexity here confirms that most human infections are dominated by single genotypes. A very deep population substructure was uncovered within this zoonotic species. The mean genomewide differentiation between the two main clusters of clinical isolates (FST = 0.21) was similar to that indicated by a recent study using microsatellite markers, which indicated that these clusters were associated with long-tailed and pig-tailed macaque reservoir hosts, respectively (18).
A third cluster comprised the small number of lines that have been maintained in laboratory rhesus macaques for many years. It is possible that the divergence of this latter cluster relates to geographical origins, because the isolates originally were obtained in the 1960s from peninsular Malaysia and the Philippines, although sequence analysis here indicates uncertainty in the true identity of some of these isolates. A recent study has indicated dimorphism in whole-genome sequences of six clinical isolates (three belonging to each of two divergent types) from Sarikei in Sarawak (26), where our previous microsatellite analysis identified both cluster 1 and cluster 2 as occurring at approximately equal frequencies (18).
Although it is likely that strong reproductive isolation is maintained between these sympatric subpopulations, mosaicism in the level of differentiation across the genome is apparent. Further sampling and population genetic analyses may address whether there has been occasional hybridization leading to introgression of parts of the genome between the different subpopulations, potentially enabling parasites to establish infections in different host species, including humans. Introgression between subspecies has been associated with gain of virulence in another human eukaryotic pathogen, Trypanosoma brucei (27), and was reported potentially to have enhanced growth rates and infectivity in hybrids between schistosome populations from cattle and human hosts (28). Such adaptive roles of introgression have been implicated in other cases, including adaptation of pigs to hot and cold climates (29). Although differences in FST have been used to scan for genomic islands related to speciation in other taxa, such measures can be misleading if values are inflated by a lack of diversity in particular populations (30). Because in the present study there were fewer isolates in cluster 2 than in cluster 1, it will be valuable to analyze a larger number of cluster 2 isolates in future to investigate further the genomewide variation in divergence.
Because of the pronounced population substructure, the scan for signatures of selection in this study was focused on the majority of the clinical isolates that appeared to belong to a single, undivided population, the 38 isolates in cluster 1. The overall negative skew of allele frequency distributions as shown by the Tajima’s D values suggests previous long-term population growth of this parasite. It is interesting that the csp gene appears to be under stronger balancing selection in P. knowlesi (having the highest Tajima’s D value genomewide) than in P. falciparum (in which its ortholog has a weaker positive value), whereas the ama1 gene did not have an elevated value in P. knowlesi although it is consistently identified as under balancing selection in other species (21). To investigate whether these or other loci show evidence of selection within clusters 2 and 3, further sampling is required to obtain sufficient numbers of isolate sequences of these rarer clusters.
The high integrated haplotype scores observed in multiple chromosomal regions suggests current or recent occurrence of strong positive selection on a number of different genes. In P. falciparum population studies, such strong selection signals were reported around drug-resistance loci where selective sweeps recently have increased the frequency of resistance-associated haplotypes (31). The absence of any signature of positive selection on the P. knowlesi orthologs of drug-resistance genes is consistent with the hypothesis that most selection on these parasites occurs in macaque rather than human hosts (11), specifically in long-tailed macaques for the cluster 1 subpopulation (18). Interestingly, the csp gene was located within an elevated window of haplotype homozygosity on chromosome 8, suggesting that it could be a target of both balancing and directional selection or that it might have hitchhiked to intermediate allele frequencies by a linked locus under selection within the cluster 1 subpopulation. The relatively small number of isolate sequences of cluster 2 precluded comparable tests of selection for that subpopulation, although, given the different population structure and association with pig-tailed macaque reservoir hosts (18), it will be important to investigate selection in cluster 2 by further sampling.
To understand further the signatures of selection and to test for evidence of host-switching and possible adaptation to humans, it is also a priority to obtain whole-genome sequence data from P. knowlesi parasites in natural infections of macaque reservoir hosts. Although these data are difficult to obtain because of the generally low densities of parasites within the blood of these animals and because most macaques are infected with multiple species of Plasmodium (11, 32), a promising approach may be flow cytometric sorting of individual parasites for single-cell sequencing, as recently applied to human malaria parasites (33). In addition, future culture adaptation of P. knowlesi clones from the divergent genotypic clusters and the application of high-efficiency transfection technologies to study gene functions systematically could enable the investigation of phenotypic differences. These studies could include attempts at genetic crosses by feeding mixed cultures to vector mosquitoes to identify competent vectors for each of the clusters and test for the possible existence of reproductive isolation mechanisms maintaining the differences between the clusters.
Materials and Methods
Peripheral blood samples were collected from patients with P. knowlesi malaria at Betong and Kapit Hospitals in Sarawak, Malaysian Borneo, following informed consent and with the approval of the Medical Research and Ethics Committee of the Ministry of Health, Malaysia, the Ethics Committee of the London School of Hygiene and Tropical Medicine, and the Institutional Biosafety and BioEthics Committee of King Abdullah University of Science and Technology, Saudi Arabia. P. knowlesi genomic DNA was prepared after depletion of human leukocytes (34) and processed for Illumina paired-end short-read sequencing. DNA samples from five laboratory strains of P. knowlesi, isolated in the 1960s and subsequently maintained in laboratory rhesus macaques, were similarly sequenced. Three of the laboratory strains (labeled “Hackeri,” “Malayan,” and “H”) were originally isolated from peninsular Malaysia (35, 36), and one (labeled as “Philippine”) was from the Philippines (37). They were obtained through the American Type Culture Collection (ATCC) and MR4 repository, and, along with a previously in vitro adapted line of the H strain (38, 39), here termed “H(AW),” were culture-adapted to rhesus macaque erythrocytes in vitro, cloned by limiting dilution and grown for 1–2 wk before DNA extraction.
For additional information on patient sampling and laboratory strains, DNA library preparation and genome sequencing, quality control and processing of sequence data (40, 41), and population genetic and comparative sequence analyses (24, 42–46), see SI Materials and Methods.
SI Materials and Methods
Sampling and Initial Processing of Parasites from Human P. knowlesi Cases.
Peripheral blood samples (10 mL) were collected in EDTA tubes from P. knowlesi malaria cases admitted to Betong and Kapit Hospitals in Sarawak, Malaysian Borneo, from 2012 to 2014 after informed consent had been obtained. A total of 51 clinical samples were processed for preparation of P. knowlesi genomic DNA after removal of leukocytes by CF11 cellulose column filtration (34).
DNA Library Preparation and Sequencing of P. knowlesi Samples.
DNA was extracted using Qiagen Midi extraction kits (Qiagen), and 500 ng of DNA from each sample was sheared and prepared for sequencing using Illumina’s HiSeq 2000 platform. Briefly, after shearing the DNA on Covaris, paired-end libraries were made on the IP-Star automated platform (Diagenode) using the protocol “Illumina library prep” as per the manufacturer’s protocol from end repair until adapter ligation and purification. After the ligation step, gel size selection and PCR amplification using the standard Illumina PCR cycle (Kapa high-fidelity master mix) were done manually. The samples then were run on a bioanalyzer and were quantified on Qubit (Thermo Fisher). They then were normalized and pooled for sequencing (pools with read lengths of 93 and 101 bp; insert size of 400 bp) and submitted for HiSeq 2000 sequencing at the Bioscience Core Laboratory of King Abdullah University of Science and Technology.
In addition, five strains of P. knowlesi that were maintained in rhesus macaques underwent whole-genome sequencing using the Illumina platform at the Broad Institute of MIT and Harvard. Vials of four of these strains, labeled as P. knowlesi “Hackeri,” “Philippine,” “Malayan,” and “MR4-H,” respectively, were obtained through MR4 and ATCC as part of the Biodefense and Emerging Infections Research Resources Repository, National Institute of Allergy and Infectious Diseases, NIH. The MR4-H strain (MRA-456) is the H strain that was isolated in 1961 from a natural human infection (35), and no information is available from MR4 about the original contributor. The P. knowlesi line Hackeri (MRA-547) was contributed by W. E. Collins and was originally isolated in 1961 from an Anopheles hackeri mosquito in Peninsular Malaysia (36). The P. knowlesi Philippine line (MRA-457) was contributed by W. E. Collins and had been isolated from a naturally infected long-tailed macaque (Macaca fascicularis) from the southern Philippines in January 1964 (37). The P. knowlesi Malayan strain (MRA-487) was deposited by W. E. Collins and was isolated from an Anopheles hackeri mosquito and established in the NIH in 1960. These strains, along with the culture-adapted laboratory H strain provided by A. W. Thomas [Biomedical Primate Research Center (BPRC), Rijswijk, The Netherlands], here termed “H(AW),” were culture-adapted to rhesus macaque erythrocytes in vitro, cloned by limiting dilution and grown for a period of 1–2 wk; then DNA was extracted. The H(AW) strain was previously in vitro adapted, initially at the BPRC (38) and subsequently at the Department of Immunology and Infectious Diseases, Harvard T. H. Chan School of Public Health (39).
Quality Control and Processing of Sequence Data.
Short reads were aligned to version 1.0 of the reference P. knowlesi genome strain H (19) (accessed at plasmodb.org/plasmo/), using the BWA alignment tool (40) (bwa mem, default parameters). Short reads with poor quality toward the end were trimmed before mapping using the software Trimmomatic (41). Average fold coverage of properly aligned reads was used to assess the yield for each isolate. Properly aligned reads are defined as post-quality control paired reads that aligned to the reference genome with the correct orientation and within the expected insert size distribution. An average fold coverage of 30× was used as a threshold for inclusion of isolates, resulting in a total of 53 samples (48 clinical isolates and five laboratory lines).
SNPs initially were identified using SAMTools/bcftools (mpileup -B -Q 23 -d 2000 -C 50 –ugf; varFilter -d 10 -D 2000). A second step of SNP calling and quality control was used as follows. First, a high-quality list of potential variant positions was obtained from the 53 samples (Q >30) and was used for a second round of strict SNP calling using a read depth-based approach as described previously in a study of P. falciparum (24). Briefly, using the initially identified list of SNP positions, we identified the major genotype, as determined by the highest read count, for each potential variant site across all isolates (minimum read depth of 10×). Second, a mapping quality (mq) and base quality (bq) check were used to remove positions with an excess of low-quality reads that align to multiple locations. A ratio of read depth values at high-quality (mq = 26, bq = 23) and low-quality (mq = 0, bq = 0) thresholds was computed for each isolate and was used to exclude positions where the ratio fell below 0.5 even though the minimum coverage requirement of 10× was achieved. Additional filtering excluded positions where the reference allele was unknown (i.e., ambiguous positions with Ns on the reference genome) and singleton and triallelic sites. Multigene families (SICAVars and KIRs) and subtelomeric regions also were excluded to minimize potential false-positive calls caused by erroneous alignment and variant calling. Subtelomeric regions were defined by aligning each chromosome of P. knowlesi to the P. vivax homolog (BlastN -F F). A visual analysis of syntenic regions toward chromosome ends was used to mark subtelomeric regions. The last conserved telomeric protein-coding gene was used to define boundaries of subtelomeric regions, with the core genome defined as all non-subtelomeric regions and excluding all sequences of SICAvar and KIR gene families.
Population Genomic and Comparative Sequence Analyses.
A pairwise nucleotide diversity index (π) was computed for all samples and individual clusters using customized Perl scripts. Heterogeneity of nucleotide diversity within chromosomes was visualized by taking sliding windows of 50 kb (as shown in Fig. 2). Similarly, the pairwise nucleotide diversity between clusters was estimated using the DXY statistic, and sliding windows of 50 kb were used to look at regions of the genome with high and low intercluster diversity.
The genotypic mixedness within each P. knowlesi clinical isolate was estimated using the within-infection fixation index (FWS) as described previously for analyses of P. falciparum (24, 42, 43), which has a potential range from 0 to 1. The estimated level of mixedness is 1 − FWS, and isolates with FWS scores of >0.95 (or 1 − FWS <0.05) were considered to contain a predominant single clone. Customized R scripts were used for the analyses of FWS indices, the overall allele frequency spectrum, Tajima’s D indices (44), FST, and PCA. A neighbor-joining tree (Ape package in R-statistical environment, type = “unrooted”) was used to visualize potential substructures from all 53 samples using a pairwise nucleotide diversity (π) matrix. PCA was performed using 213,375 SNPs without missing data and a minor allele frequency of >0.1.
A sliding window analysis (window size = 500 SNPs, slide by 250 SNPs) was used on genomewide FST scores between the two main clusters (cluster 1, n = 38 and cluster 2, n = 10 using SNPs with a minor allele frequency >0.1). Mean FST values were computed for each window and were used to test for overrepresentation of high- or low-FST windows (i.e., windows with mean FST above or below the genomewide mean, 0.21) using Fisher’s exact test. Chromosomes with P values <0.003571 (cutoff adjusted to account for 14 tests) were found to contain a significant proportion of high- or low-FST windows compared with the rest of the genome.
The rehh package (45) was used to compute standardized |iHS| values (46) on 38 samples of the main cluster using biallelic SNPs without missing calls and a minor allele frequency of >0.1. For SNPs with high |iHS| scores (top 0.0001%; |iHS| >4.892), putative selection windows were defined by merging overlapping extended haplotype homozygosity windows (defined from a core SNP until the frequency of extended haplotypes is below 0.05). A final set of windows was defined by focusing on windows that contained at least one SNP with |iHS| >6 and windows that did not contain excessive gaps (i.e., the largest distance between consecutive SNPs). Excessive gaps within windows were defined as gaps that accounted for more than 60% of the window. Information on ortholog groups was obtained from OrthoMCL (version 5, orthomcl.org/orthomcl/) and were used to identify orthologous genes in P. falciparum 3D7 and P. vivax Savador I.
A scan for reads that contained a CD99 motif was conducted initially using a customized C++ script that looked for exact matches of motifs (word size/kmer = 10 aa) in a given Fastq file. Reads were translated in all of the six reading frames during the matching process. A total of nine motifs were obtained from the genome paper (19) and were grouped into five motifs as follows: Motif 1 = ATPKKPSAGDDFDLGDAVVDGG; Motif 2 = GGSDGGGSHRKEGEE; Motif 3.1 = KPNPNPNQAGSSGSFS; Motif 3.2 = KPNPNPNQAGSSGSFSDA, Motif 4.1 = PKKPSAGDDFDLGDAVVDG; Motif 4.2 = PKKPSAGDDFDLGDAVVDGGN; Motif 4.3 = PKKPSAGDDFDLGDAVVDGGNE; Motif 5.1 = VSGGEGKGGSDGGGS; and Motif 5.2 = VSGGEGKGGSDGGGSHR. Short reads that contained a motif were extracted as FASTA files and were used for a more exhaustive matching using blast (amino acid motifs as a database and nucleotide FASTA sequences as query; −F F –m 8; minimum length 10 aa; minimum identity 99%). Reads that mapped to the P. knowlesi reference genome and that contained a motif then were counted for each motif group to determine whether KIR genes from a given sample have sequence similarities to CD99, suggesting a molecular mimicry as postulated in the reference genome.
Supplementary Material
Acknowledgments
We thank all patients, nurses, doctors, and laboratory technicians at Kapit and Betong Hospitals and core staff in our research institutes for their assistance; Hifzur Rahman Ansari (King Abdullah University of Science and Technology) for submission of the sequences to European Nucleotide Archive; the Malaria Research and Reference Reagent Resource Center for providing the P. knowlesi strains contributed by William E. Collins; and the Director General of Health in Malaysia for permission to publish this paper. This study was supported by Universiti Malaysia Sarawak Grants E14054/F05/54PK1/09/2012(01) and 01(TD03)/1003/2013(01); UK Medical Research Council Grants MRC G1100123, MR/K000551/1, and MR/M01360X/1; European Research Council Advanced Award AdG-2011-294428; NIH Grant 5R01AI091787; Bill and Melinda Gates Foundation Grant OPP1023594; a postgraduate scholarship from the Ministry of Education in Malaysia; and faculty baseline funding from King Abdullah University of Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. X.S. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive (accession no. PRJEB10288) and NCBI database (accession no. SRP063014).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509534112/-/DCSupplemental.
References
- 1.William T, et al. Severe Plasmodium knowlesi malaria in a tertiary care hospital, Sabah, Malaysia. Emerg Infect Dis. 2011;17(7):1248–1255. doi: 10.3201/eid.1707.101017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 2013;26(2):165–184. doi: 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daneshvar C, et al. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin Infect Dis. 2009;49(6):852–860. doi: 10.1086/605439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox-Singh J, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46(2):165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garnham P. Malaria Parasites and Other Haemosporidia. Blackwell Scientific Publications Ltd.; Oxford, UK: 1966. [Google Scholar]
- 6.Singh B, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 7.Tyagi RK, Das MK, Singh SS, Sharma YD. Discordance in drug resistance-associated mutation patterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. J Antimicrob Chemother. 2013;68(5):1081–1088. doi: 10.1093/jac/dks508. [DOI] [PubMed] [Google Scholar]
- 8.Müller M, Schlagenhauf P. Plasmodium knowlesi in travellers, update 2014. Int J Infect Dis. 2014;22:55–64. doi: 10.1016/j.ijid.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 9.Lee KS, Cox-Singh J, Singh B. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J. 2009;8:73. doi: 10.1186/1475-2875-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KS, Cox-Singh J, Brooke G, Matusop A, Singh B. Plasmodium knowlesi from archival blood films: Further evidence that human infections are widely distributed and not newly emergent in Malaysian Borneo. Int J Parasitol. 2009;39(10):1125–1128. doi: 10.1016/j.ijpara.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KS, et al. Plasmodium knowlesi: Reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7(4):e1002015. doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan CH, Vythilingam I, Matusop A, Chan ST, Singh B. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar J. 2008;7:52. doi: 10.1186/1475-2875-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vythilingam I, et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit Vectors. 2008;1(1):26. doi: 10.1186/1756-3305-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins WE. Plasmodium knowlesi: A malaria parasite of monkeys and humans. Annu Rev Entomol. 2012;57:107–121. doi: 10.1146/annurev-ento-121510-133540. [DOI] [PubMed] [Google Scholar]
- 15.William T, et al. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax Malaria in Sabah, Malaysia. PLoS Negl Trop Dis. 2013;7(1):e2026. doi: 10.1371/journal.pntd.0002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jongwutiwes S, et al. Plasmodium knowlesi Malaria in humans and macaques, Thailand. Emerg Infect Dis. 2011;17(10):1799–1806. doi: 10.3201/eid1710.110349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeslyn WP, et al. Molecular epidemiological investigation of Plasmodium knowlesi in humans and macaques in Singapore. Vector Borne Zoonotic Dis. 2011;11(2):131–135. doi: 10.1089/vbz.2010.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Divis PCS, et al. Admixture in humans of two divergent Plasmodium knowlesi populations associated with different macaque host species. PLoS Pathog. 2015;11(5):e1004888. doi: 10.1371/journal.ppat.1004888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pain A, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455(7214):799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conway DJ. Paths to a malaria vaccine illuminated by parasite genomics. Trends Genet. 2015;31(2):97–107. doi: 10.1016/j.tig.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39(1):113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 23.Neafsey DE, et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 2012;44(9):1046–1050. doi: 10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487(7407):375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39(1):126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 26.Pinheiro MM, et al. Plasmodium knowlesi genome sequences from clinical isolates reveal extensive genomic dimorphism. PLoS One. 2015;10(4):e0121303. doi: 10.1371/journal.pone.0121303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodhead I, et al. Whole-genome sequencing of Trypanosoma brucei reveals introgression between subspecies that is associated with virulence. MBio. 2013;4(4):e00197-13. doi: 10.1128/mBio.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huyse T, et al. Bidirectional introgressive hybridization between a cattle and human schistosome species. PLoS Pathog. 2009;5:e1000571. doi: 10.1371/journal.ppat.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai H, et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 2015;47:217–225. doi: 10.1038/ng.3199. [DOI] [PubMed] [Google Scholar]
- 30.Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Molecular Ecology. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- 31.Nwakanma DC, et al. Changes in malaria parasite drug resistance in an endemic population over a 25-year period with resulting genomic evidence of selection. J Infect Dis. 2014;209(7):1126–1135. doi: 10.1093/infdis/jit618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muehlenbein MP, et al. Accelerated diversification of nonhuman primate malarias in Southeast Asia: Adaptive radiation or geographic speciation? Mol Biol Evol. 2015;32(2):422–439. doi: 10.1093/molbev/msu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair S, et al. Single-cell genomics for dissection of complex malaria infections. Genome Res. 2014;24(6):1028–1038. doi: 10.1101/gr.168286.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkatesan M, et al. Using CF11 cellulose columns to inexpensively and effectively remove human DNA from Plasmodium falciparum-infected whole blood samples. Malar J. 2012;11:41. doi: 10.1186/1475-2875-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin W, Contacos PG, Coatney GR, Kimball HR. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science. 1965;149(3686):865. doi: 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- 36.Wharton RH, Eyles DE. Anopheles hackeri, a vector of Plasmodium knowlesi in Malaya. Science. 1961;134(3474):279–280. doi: 10.1126/science.134.3474.279. [DOI] [PubMed] [Google Scholar]
- 37.Collins WE, Contacos PG, Chin W. Infection of the squirrel monkey Saimiri sciureus, with Plasmodium knowlesi. Trans R Soc Trop Med Hyg. 1978;72(6):662–663. doi: 10.1016/0035-9203(78)90030-5. [DOI] [PubMed] [Google Scholar]
- 38.Kocken CH, et al. Plasmodium knowlesi provides a rapid in vitro and in vivo transfection system that enables double-crossover gene knockout studies. Infect Immun. 2002;70(2):655–660. doi: 10.1128/IAI.70.2.655-660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim C, et al. Expansion of host cellular niche can drive adaptation of a zoonotic malaria parasite to humans. Nat Commun. 2013;4:1638. doi: 10.1038/ncomms2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auburn S, et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS One. 2012;7(2):e32891. doi: 10.1371/journal.pone.0032891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mobegi VA, et al. Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Mol Biol Evol. 2014;31(6):1490–1499. doi: 10.1093/molbev/msu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gautier M, Vitalis R. rehh: An R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28(8):1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 46.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.