Abstract
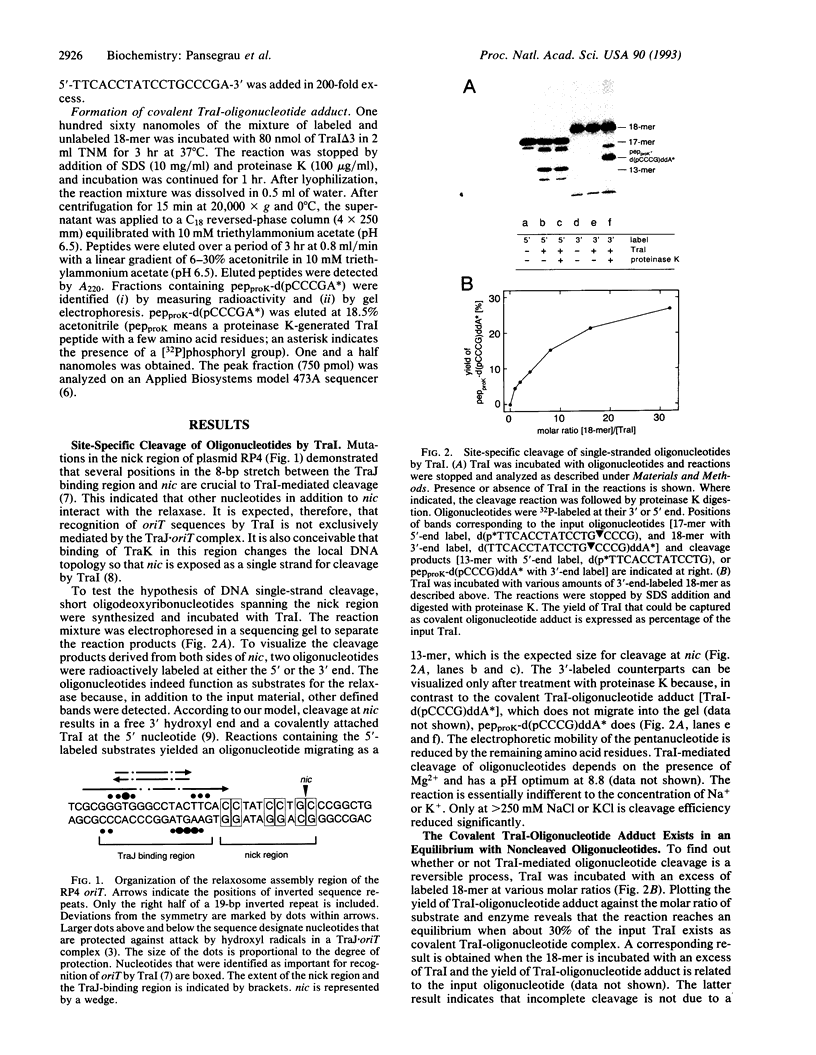
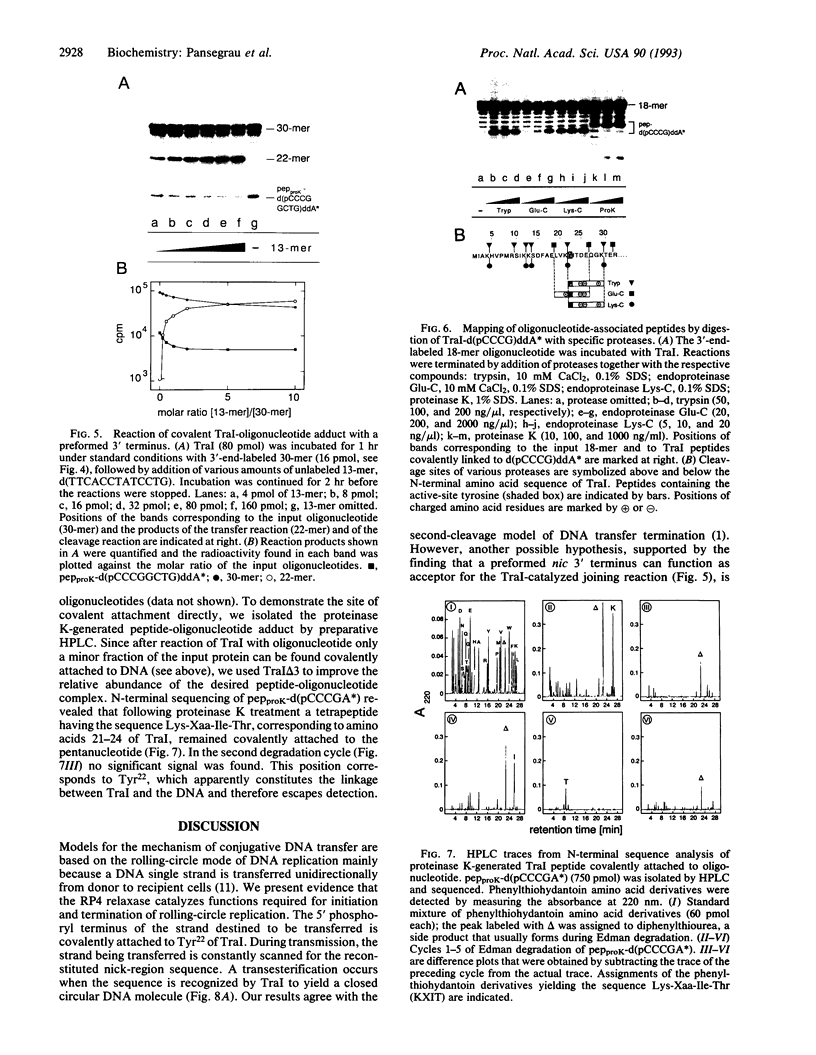
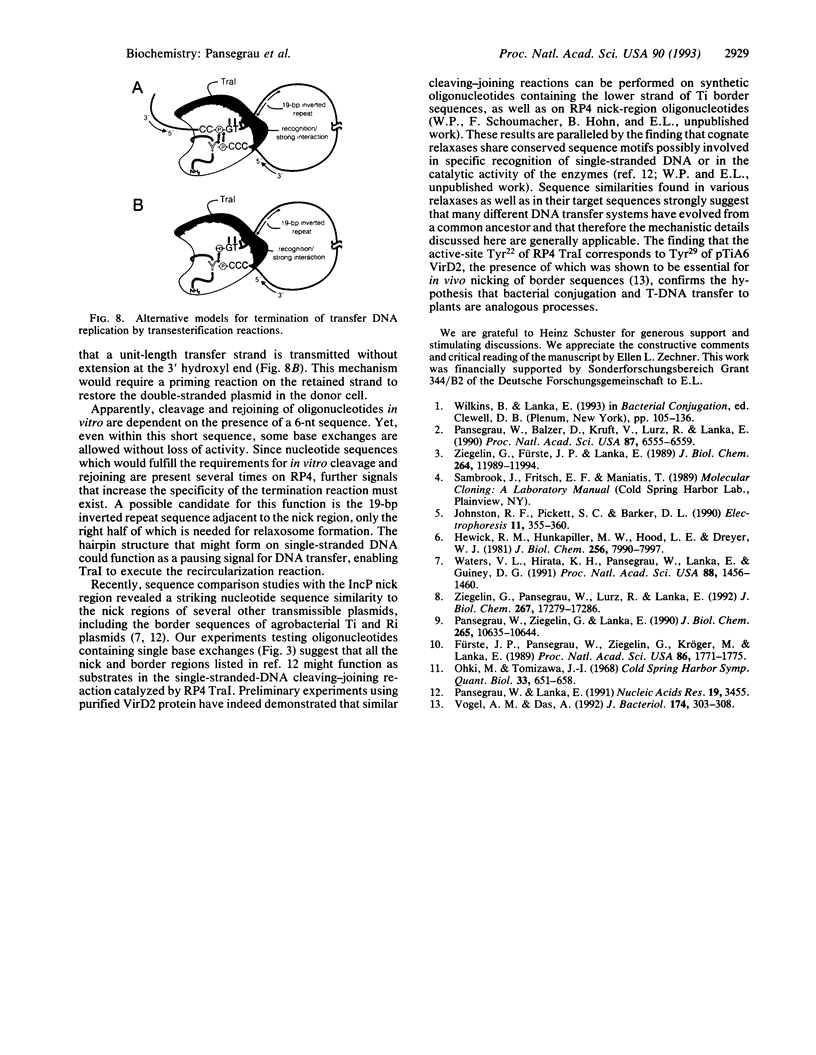
Conjugative DNA transfer of the self-transmissible broad-host-range plasmid RP4 is initiated by strand- and site-specific cleavage at the nick site (nic) of the transfer origin (oriT). Cleavage results in covalent attachment of the plasmid-encoded relaxase (TraI) to the 5'-terminal 2'-deoxycytidine residue at nic. We demonstrate that Tyr22 is the center of the catalytic site of TraI, mediating cleavage via formation of a phosphodiester between the DNA 5' phosphoryl and the aromatic hydroxyl group. The specificity of cleavage seen with form I oriT DNA was verified with short oligodeoxy-ribonucleotides embracing the nick region. The reaction requires TraI and Mg2+ but is independent of the relaxosome component TraJ. Cleavage produces one oligonucleotide fragment with a free 3' hydroxyl, the other part forms a covalent TraI-oligonucleotide adduct. Like nicking of form I oriT DNA, TraI-catalyzed oligonucleotide cleavage reaches an equilibrium when about 30% of the input TraI exists as a covalent protein-DNA complex. In the presence of two differently sized oligonucleotides, defined hybrid oligonucleotides are produced, demonstrating that TraI catalyzes recombination of two single strands at nic. This finding shows that TraI possesses cleaving-joining activity resembling that of a type I topoisomerase. Reactions are dependent on the sequence of the 3'-terminal 6 nucleotides adjacent to nic. Only certain base changes in a few positions are tolerated, whereas the sequence of the 5' terminal nucleotides apparently is irrelevant for recognition by TraI. The reactions described here further support the hypothesis that DNA transfer via conjugation involves a rolling circle-like mechanism which generates the immigrant single strand while DNA-bound TraI protein scans for the occurrence of a second cleavage site at the donor-recipient interface.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fürste J. P., Pansegrau W., Ziegelin G., Kröger M., Lanka E. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Johnston R. F., Pickett S. C., Barker D. L. Autoradiography using storage phosphor technology. Electrophoresis. 1990 May;11(5):355–360. doi: 10.1002/elps.1150110503. [DOI] [PubMed] [Google Scholar]
- Ohki M., Tomizawa J. Asymmetric transfer of DNA strands in bacterial conjugation. Cold Spring Harb Symp Quant Biol. 1968;33:651–658. doi: 10.1101/sqb.1968.033.01.074. [DOI] [PubMed] [Google Scholar]
- Pansegrau W., Balzer D., Kruft V., Lurz R., Lanka E. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6555–6559. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991 Jun 25;19(12):3455–3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Ziegelin G., Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5'-terminal nucleotide at the relaxation nick site. J Biol Chem. 1990 Jun 25;265(18):10637–10644. [PubMed] [Google Scholar]
- Vogel A. M., Das A. Mutational analysis of Agrobacterium tumefaciens virD2: tyrosine 29 is essential for endonuclease activity. J Bacteriol. 1992 Jan;174(1):303–308. doi: 10.1128/jb.174.1.303-308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters V. L., Hirata K. H., Pansegrau W., Lanka E., Guiney D. G. Sequence identity in the nick regions of IncP plasmid transfer origins and T-DNA borders of Agrobacterium Ti plasmids. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1456–1460. doi: 10.1073/pnas.88.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelin G., Fürste J. P., Lanka E. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J Biol Chem. 1989 Jul 15;264(20):11989–11994. [PubMed] [Google Scholar]
- Ziegelin G., Pansegrau W., Lurz R., Lanka E. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J Biol Chem. 1992 Aug 25;267(24):17279–17286. [PubMed] [Google Scholar]