Abstract
The presence or absence of specific transcription factors, chromatin remodeling machineries, chromatin modification enzymes, post-translational histone modifications and histone variants all play crucial roles in the regulation of pathogenicity genes. Chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-seq) provides an important tool to study genome-wide protein-DNA interactions to help understand gene regulation in the context of native chromatin. ChIP-seq is a convenient in vivo technique to identify, map and characterize occupancy of specific DNA fragments with proteins against which specific antibodies exist or which can be epitope-tagged in vivo. We optimized existing ChIP protocols for use in the wheat pathogen Zymoseptoria tritici and closely related sister species. Here, we provide a detailed method, underscoring which aspects of the technique are organism-specific. Library preparation for Illumina sequencing is described, as this is currently the most widely used ChIP-seq method. One approach for the analysis and visualization of representative sequence is described; improved tools for these analyses are constantly being developed. Using ChIP-seq with antibodies against H3K4me2, which is considered a mark for euchromatin, and H3K9me3, which is considered a mark for heterochromatin, the overall distribution of euchromatin and heterochromatin in the genome of Z. tritici can be determined. Our ChIP-seq protocol was also successfully applied to Z. tritici strains with high levels of melanization or aberrant colony morphology, and to different species of the genus (Z. ardabiliae and Z. pseudotritici), suggesting that our technique is robust. The methods described here provide a powerful framework to study new aspects of chromatin biology and gene regulation in this prominent wheat pathogen.
Keywords: histone modifications, protein-DNA interactions, chromatin immunoprecipitation, ChIP, genome-wide histone modification maps
1. Introduction
The genome of Zymoseptoria tritici is one of the best-assembled eukaryotic genomes (Goodwin et al., 2011). The reference isolate IPO323 has a total of 21 chromosomes of which eight are accessory chromosomes (Wittenberg et al., 2009; Goodwin et al., 2011). The 39 Mb genome of IPO323 has an overall relatively high content of repetitive DNA, 18%. On the 13 core chromosomes few expressed genes are found in “islands” of repetitive DNA. The accessory chromosomes, however, are enriched with repetitive DNA and contain fewer expressed and predicted genes (Dhillon et al., 2014). This well-assembled genome provides an excellent resource for comparative genomics (Stukenbrock et al., 2011), analyses of genome-wide transcription patterns (Kellner et al., 2014; Yang et al., 2013), and studies of protein-DNA interaction and chromatin structure. So far, protein-DNA interactions and chromatin have been little studied in this species. Here, we describe how methods based on chromatin immunoprecipitation provide a powerful framework for the identification of protein binding sites and protein distribution across a small eukaryotic genome.
Chromatin is a complex of histones and non-histone proteins packing the DNA within the nuclei (van Holde, 1989). The basic unit of chromatin is the nucleosome consisting of a ~150 bp segment of double-stranded DNA wound around a core octamer of the histone proteins, H2A, H2B, H3 and H4. Tails of histones carry a wide range of post-translational modifications that can influence their interaction with the associated DNA and determine chromatin condensation state to generate two cytologically recognizable conformations, euchromatin and heterochromatin (Jenuwein and Allis, 2001; Strahl and Allis, 2000; van Holde, 1989). In contrast to euchromatin, which is decondensed during interphase, constitutive heterochromatin remains condensed throughout the cell cycle. Posttranslational histone modifications are thought to affect these “open” or “closed” chromatin states and thus mediate a wide range of molecular processes, including transcription of genes by direct or indirect changes in chromatin structure or recruitment of protein complexes, resulting in gene activity or silencing (Grewal and Jia, 2007; Kouzarides, 2007). Over the past 15 years, correlations of certain histone modifications with euchromatin and active transcription or with heterochromatin and silencing have been noted (Bernstein et al., 2002; Bernstein et al., 2006; Mikkelsen et al., 2007). In filamentous fungi, three different modifications of histone H3 have been most widely analyzed to distinguish euchromatin and heterochromatin. These are dimethylation of the lysine 4 of the tail of histone H3 (H3K4me2), which is normally associated with euchromatin, and trimethylation of lysine 9 or 27 on H3 (H3K9me3 and H3K27me3), which is usually associated with heterochromatin (Chujo and Scott, 2014; Connolly et al., 2013; Reyes-Dominguez et al., 2010; Tamaru and Selker, 2001; Tamaru et al., 2003; Smith et al., 2011; Soyer et al., 2014).
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq; Fig. 1) is an exceedingly powerful method for the generation of genome-wide maps of interactions between proteins and DNA, including histone modifications, transcription factor binding sites, occupancy with chromatin remodeling factors and mapping of histone variants (Johnson et al., 2007; Meyer and Liu, 2014; Park, 2009; Smith et al., 2011). In filamentous fungi, ChIP-based approaches have been widely used to study chromatin modification and centromere structure of Neurospora crassa (Honda et al., 2010; Jamieson et al., 2013; Lewis et al., 2009; 2012; Smith et al., 2008; 2011; Tamaru and Selker, 2001; Tamaru at al., 2003). Recently, ChIP has also been applied in studies of host-pathogen interactions. In the rice blast fungus Magnaporthe oryzae, ChIP allowed the identification of binding sites of a transcription factor (TF) crucial for the regulation of virulence-related genes (Kim et al., 2010). In Fusarium graminearum and Leptosphaeria maculans, the roles of chromatin modifications in regulation of secondary metabolite and effector-encoding genes were likewise demonstrated using ChIP-seq and ChIP-qPCR with antibodies against euchromatic and heterochromatic marks (Connolly et al., 2013; Soyer et al., 2014). These studies demonstrated the importance of DNA-protein interactions in the regulation of pathogenicity determinants and highlighted the potential of ChIP-based approaches in studies of chromatin structures and their modifications in fungal pathogens. We adapted and improved existing protocols for studies of protein-DNA interactions and chromatin modifications for the use in Z. tritici and related species of the same genus (Z. ardabiliae and Z. pseudotritici). ChIP-seq allows us to address new questions related to key biological processes, such as gene regulation, pathogenicity, population genomics and evolution of important plant pathogenic fungi.
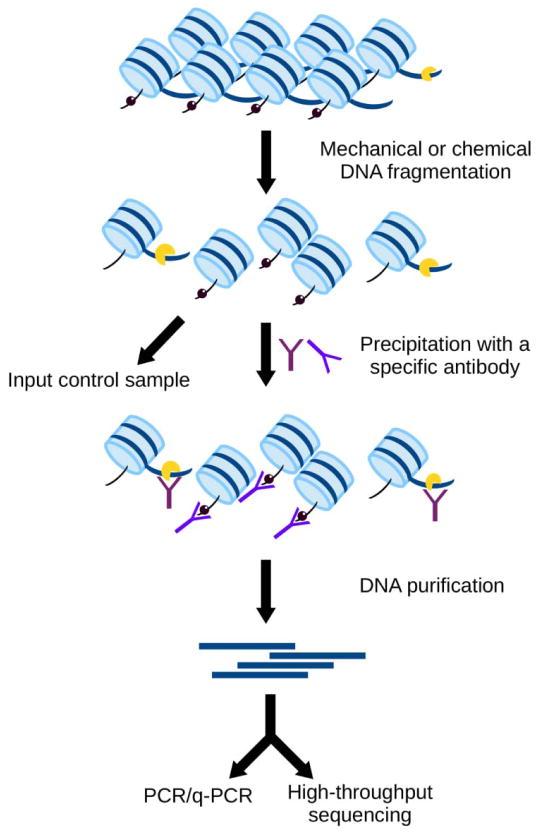
Fig. 1.
General overview of a chromatin immunoprecipitation (ChIP) experiment. The genomic DNA associated with a specific histone modification or a non-histone protein (e.g. a transcription factor) can be detected in vivo by ChIP. DNA is fragmented by mechanical or enzymatic sheering. Antibodies (dark and light purple marks) against a particular post-translational histone modification (black spheres) or non-histone protein (yellow) are used for the immunoprecipitation of DNA associated with the targeted proteins. Purified DNA is amplified and sequenced allowing identification of DNA associated with the protein of interest.
2. Methods
2.1. Overview of ChIP
We focus on ChIP followed by high-throughput sequencing (ChIP-seq) as perhaps the most powerful method for the analysis of chromatin states, and supply a step-for-step method optimized for Z. tritici in the supplementary information (Supplementary Method) and data obtained with this method with Z. tritici (Figs. 2 and 3). The purpose of the following sections is to provide an outline of the method and to discuss important aspects or variations and why specific methods are preferred over others, always with a view to the specifics of Zymoseptoria compared to other species of filamentous fungi. ChIP is an in vivo method to identify genomic regions associated with a particular DNA binding protein such as a TF (Orlando et al., 1997; 1998) or to identify regions enriched with a given histone modification (Hecht and Grunstein, 1999; Wu and Grunstein, 2000). Briefly, chromatin fragments associated with proteins of interest, for example specific TFs, are cross-linked, typically with formaldehyde (crosslinking, or “X-Chip”), sheared by sonication or digested with enzymes down to mononucleosome size, and captured by reactions with protein-specific antibodies. This is the “experimental” sample, whereas the “input” control is not treated with antibodies. DNA from immunoprecipitated samples or input control chromatin fragments is isolated by precipitation after proteins have been degraded with proteinases. Thus, the DNA serves as the readout for the original pattern of protein occupancy (Fig. 1). Because of the relatively strong interactions between histone octamers and DNA, chromatin can sometimes also be used in its native state (Native-Chip”) (Turner, 2001; see Cosseau et al., 2009, and Soyer et al., 2014, for methods that can be applied to fungi). This method can be useful when treatment with formaldehytde “over-crosslinks” chromatin, resulting in an essentially irreversible state; even with prolonged shearing chromatin cannot be broken into small fragments after over-crosslinking (Orlando et al., 1997)
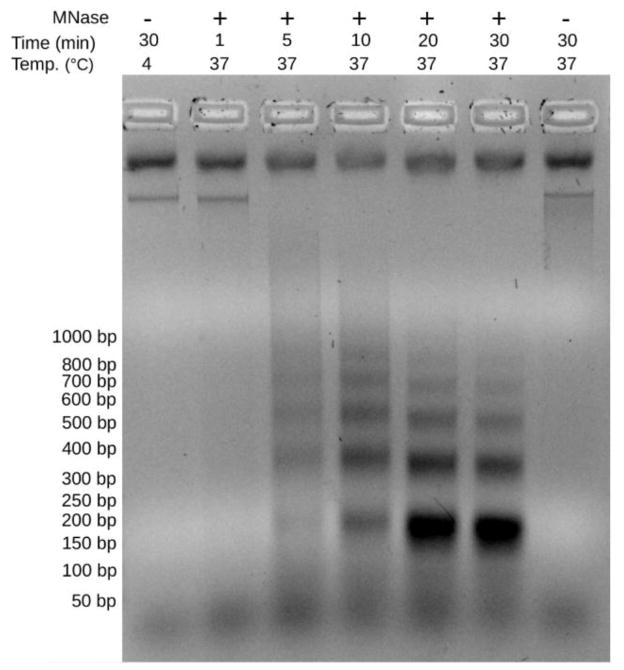
Fig. 2.
Optimization of micrococcal nuclease (MNase) digestion. Many different conditions were tested for the three Zymoseptoria species, the best being 20 minutes at 37°C. Shown here is MNase digestion of Z. ardabiliae chromatin with either 1000 units of MNase (“+”; New England Biolabs) or no enzyme (“−”) at 4°C and 37°C. The digestion time was optimized to release mostly mononucleosomes (~150 bp). Digestion profiles for Z. tritici and Z. pseudotritici revealed similar optimal conditions (data not shown). Optimal digestion can vary for different species and strains and should be done and re-checked for each different set of strains.
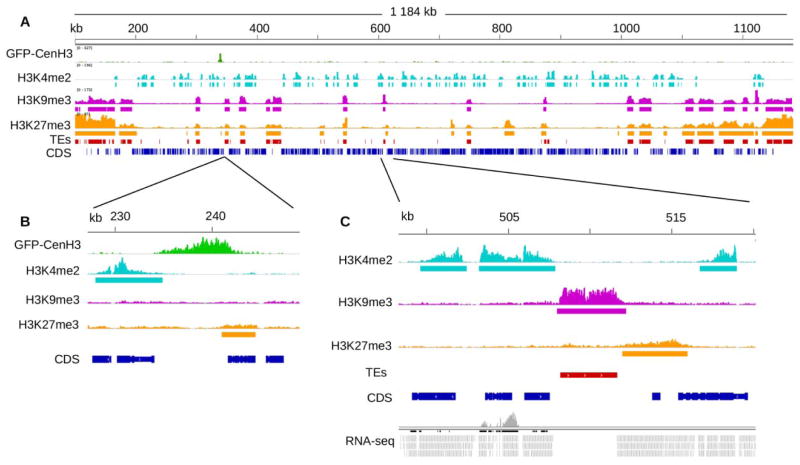
Fig. 3.
Centromere location and enrichment of histone modifications identified by ChIP-seq in Z. tritici. (A) View of chromosome 13 of Z. tritici IPO323. Reads obtained from ChIP-seq with GFP-CenH3 (green) identify the centromere, and regions enriched with H3K4me2 (light blue), H3K9me3 (purple), H3K27me3 (orange) identify euchromatin, and heterochromatin, respectively. ChIP-seq results are compared to the location of genomic features; shown are coding sequences (dark blue; according to a new annotation based on RNA-seq; Grandaubert et al., in review) and transposable elements (TEs, red). Tracks were visualized with the Integrative Genome Viewer (IGV; http://www.broadinstitute.org/software/igv; Thorvaldsdóttir et al., 2013). (B) Detailed view of the centromeric region. (C) Detailed view of a smaller region of chromosome 13 to show histone modifications and mapping of RNA-seq reads (grey). Statistically enriched regions predicted by analyses with RSEG (Song and Smith, 2011) appear as rectangles under the called peak for each modification.
2.2. Experimental outline and important considerations
2.2.1. Strains and culture conditions
Strains of Z. tritici are sensitive to environmental conditions, for example a small change in temperature or medium composition may cause dramatic changes in cell or colony morphology. As always, biological replicates are essential and necessary to capture the natural variation within and among strains. We use a minimum of two biological replicates for each growth experiment and repeat experiments with different strains at least twice. We have assessed presence of centromere proteins and various histone modifications of different isolates grown in rich (yeast-malt-sucrose) medium. Yeast-like cells of different Z. tritici isolates were grown at 18°C in a shaking incubator (200 rpm) until a cell density of approximately 107 cells/ml was reached (Kellner et al., 2014). Chromatin was prepared according to the detailed method described in the Supplementary Method. This protocol should work to study chromatin while the fungus is growing in isolation or within its plant host. The challenge will be to obtain sufficient amounts of material but this can be achieved by pooling several parallel samples. Results obtained with an endophyte, Epichloë (Chujo and Scott, 2014), suggest that this will be possible in the near future.
2.2.2. Methods to break cells for chromatin isolation
Originally, ChIP was carried out in Drosophila embryos and other animal cells and thus no special breakage procedure was used. Whole embryos or Schneider cells were cross-linked and directly sheared by sonication (Orlando et al., 1997; 1998). Tougher yeast cells were ground under liquid nitrogen or subjected to bead-beating (Hecht and Grunstein, 1999). Initial protocols for N. crassa also relied on bead-beating (Tamaru et al., 2003) but in recent years two different sonication steps have been used to first break the cells and then shear chromatin (Honda et al., 2012). We attempted to break Z. tritici cells by sonication alone, as this speeds up the procedure, especially when processing multiple samples, a protocol that works well for F. graminearum (Connolly et al., 2013) and F. fujikuroi (Studt et al., 2013; Wiemann et al., 2013). We found, however, that this yielded insufficient chromatin shearing and amounts of DNA obtained after ChIP were too low for subsequent analyses. Thus, we now apply grinding under liquid nitrogen, optimized for the isolation of RNA and protein from fungi in plant material (S. Poppe, P. Happel and E.H. Stukenbrock, in review; Kellner et al., 2014). We encountered similar difficulties with other species; for example, numerous strains of Trichoderma reesei, F. solani, F. oxysporum, and L. maculans. In these cases we either employ bead-beating (T. reesei, Karimi-Aghcheh et al., 2013; Seiboth et al., 2012) or grinding under liquid nitrogen followed by native ChIP (L. maculans, Soyer et al., 2014).
2.2.3. Methods to fragment chromatin
The goal of chromatin fragmentation is to generate mono- or dinucleosomal fragments for precise mapping of protein occupancy. Depending on protein occupancy or regional chromatin structure, resistance of DNA within chromatin to enzymatic digestion or mechanical shearing can vary, resulting in bias. Micrococcal nuclease (MNase) digests linker DNA until it encounters a physical barrier, such as a TF binding a regulatory element or a nucleosome. MNase digestion appears not to induce significant bias in the subsequent analysis (Allan et al., 2012). Overall, MNase digestion is more reproducible than shearing with a single tube tip sonicator. However, if a Covaris sonication device is used, sonication is preferable over enzymatic digestion, because of improved reproducibility of shearing. Our studies with Z. tritici suggest that fragmentation of chromatin by MNase digestion and sonication results in similar patterns of the distribution of the centromere-specific H3, CenH3 (Fig. 3 and K. Schotanus, L.R. Connolly, M. Freitag and E.H. Stukenbrock, unpublished data). In other filamentous fungi we found this to be an important variable. While chromatin can be fragmented equally well by sonication and MNase digestion in Zymoseptoria, N. crassa, F. graminearum, F. verticillioides and F. fujikuroi, reproducible sonication with high chromatin yields proved so far difficult with various isolates of F. solani, F. oxysporum and L. maculans, especially from crosslinked samples. In these cases, MNase digestion, sometimes in the absence of formaldehyde crosslinking has proved more successful (Soyer et al., 2014; L.R. Connolly and M. Freitag, unpublished data). Other considerations favor the use of MNase as well. Mechanical shearing requires a tip sonicator or Covaris machine, both expensive, and optimizing parameters for crosslinking and shearing is time-consuming. In contrast, MNase digestion is easy to optimize, reproducible and does not require specific equipment. For all fragmentation methods, the optimal time for digestion to mono- and di-nucleosomes, the amount of input material and the concentration of the enzyme have to be determined. Over-digestion can result in degradation of DNA below ~150 bp fragments. The procedure should be optimized even for species of the same genus or strains of the same species. We optimized the MNase digestion conditions for three different Zymoseptoria species (Fig. 2 shows Z. ardabiliae).
2.2.4. Choice of antibodies
Proteins of interest are selectively immunoprecipitated with antibodies raised against the whole or fragments of the protein. Alternatively, an epitope tag can be included in the DNA sequence encoding the protein of interest to generate a translational fusion. There are advantages and disadvantages with both approaches. Tagged versions of proteins must be carefully tested and ideally should complement all defects of a deletion mutant. The advantage, however, is that commercially available antibodies have been thoroughly tested in other organisms (Table 1). Raising antibodies to parts or all of a protein of interest can be expensive and may not work, even after repeated trials. Usually, however, protein-specific antibodies work well for ChIP-seq, especially if they have not been excessively purified (Smith et al., 2010). Not all antibodies are ChIP-grade, even if they work well for westerns or co-immunoprecipitation experiments; all should be carefully checked (Cosseau et al., 2009).
Table 1.
List of commercially available antibodies used in analyses of protein-DNA interactions in filamentous fungi.
| Company | Epitope | Catalog # | type | beads | ChIP | Western |
|---|---|---|---|---|---|---|
| abcam | GFP | ab290 | rabbit polyclonal | protein A | excellent | yes |
| Active Motif | H3K9acetyl | 39137 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | H3K14acetyl | 39127 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | H3K27acetyl | 39133 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | H3 C-terminus | 39163 | rabbit polyclonal | protein A | no | yes |
| Active Motif | H3S10,28phospho | 39147 | rabbit polyclonal | protein A | no | yes |
| Active Motif | H3K4trimethyl | 39159 | rabbit polyclonal | protein A | excellent | yes |
| Active Motif | H3K9dimethyl | 39240 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | H3K9trimethyl | 39161 | rabbit polyclonal | protein A | excellent | yes |
| Active Motif | H3K27trimethyl | 39535 | mouse-monoclonal | protein G | excellent | yes |
| Active Motif | H4K16acetyl | 39167 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | H4K20trimethyl | 39181 | rabbit polyclonal | protein A | yes | yes |
| Sigma | FLAG | F3165 | mouse-monoclonal | protein G | ok | yes |
| Roche | HA | 11 867 423 001 | rat monoclonal | protein G | no | yes |
| abcam | H3K9acetyl | ab4441 | rabbit polyclonal | protein A | yes | yes |
| Millipore | H3K4dimethyl | 07–030 | rabbit polyclonal | protein A | excellent | yes |
| Active Motif | H3K27trimethyl | 39155 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | H3K9acetyl | 39137 | rabbit polyclonal | protein A | yes | yes |
| Millipore | H3K14acetyl | 07–353 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | H3K27ac | 39133 | rabbit polyclonal | protein A | yes | yes |
| Millipore | H3K27me2 | 07–452 | rabbit polyclonal | protein A | yes | yes |
| Abcam | H3K9trimethyl | ab8898 | rabbit polyclonal | protein A | ok | no |
| Millipore | H3S10ph | 04–817 | rabbit polyclonal | protein A | no | yes |
| Millipore | H4 | 05–858 | rabbit polyclonal | protein A | no | yes |
| Millipore | H3 | 07–690 | rabbit polyclonal | protein A | no | yes |
| abcam | H3K36me3 | ab9050 | rabbit polyclonal | protein A | yes | yes |
| Active Motif | RNA PolII CTD S5ph | 39233 | rabbit polyclonal | protein A | yes | yes |
| abcam | V5 | ab9116 | rabbit polyclonal | protein A | yes | yes |
| Invitrogen | V5 | R960–25 | rabbit polyclonal | protein A | excellent | yes |
2.2.5. Quantification of ChIP results
After immunoprecipitation, cross-links between chromatin components are reversed, proteins digested and DNA precipitated. The amount of DNA obtained from a single ChIP sample varies but is usually between 5 and 50 ng in a 30 to 50 μl volume. While the minimum amount of DNA required for successful Illumina sequencing is very low, between 5–10 ng per sample for most fungi, slightly more is required for other organisms (Park, 2009). It is sometimes advisable to pool several ChIP samples to avoid having to overamplify ChIP-seq libraries at later steps in the protocol. Using too many amplification steps results in bias towards over- or underrepresentation of AT-rich regions (L.R. Connolly, P.A. Phatale, S. Friedman, J.M. Galazka and M. Freitag, unpublished data); the same bias has been observed in analyses of methylated DNA (Ji et al., 2014).
There are several ways to quantify the relative enrichment of precipitated DNA. Quantification can be targeted to particular loci of interest by using region-specific primers (multiplex ChIP-PCR or ChIP-qPCR). Multiplex PCR involves one PCR with labeled nucleotides and at least two primer pairs, one amplifying an experimental locus (presumably enriched with the protein binding to the isolated DNA), the other amplifying a control region (presumably not enriched). The internal control allows calculating ratios of relative enrichment in a very straightforward manner (Smith et al., 2008; 2011; Tamaru et al., 2003) but primer pairs and PCR conditions must be carefully selected to assure a linear relationship between number of PCR cycles and product amplified. Real-time quantitative PCR (qRT-PCR) has been used extensively to replace multiplex PCR with ChIP-qPCR (e.g. Soyer et al., 2014) but there are no internal controls, primer pairs must be even more carefully selected and the method is quite labor-intensive and expensive. Thus, high-throughput sequencing of a representative portion of all isolated DNA (ChIP-seq) has become the standard qualitative and quantitative method for ChIP analyses in filamentous fungi. Assaying individual regions is mostly done as validation of whole-genome ChIP-seq data or for quantitative approaches in well-defined genomes regions.
2.2.6. ChIP-seq library preparation
Before the DNA can be subjected to ChIP-seq by next generation sequencing, specific adapters must be ligated to the immunoprecipitated DNA fragments. During these steps, the DNA is size-selected, purified and enriched. We recently published a detailed protocol for Illumina library preparation, including tips for troubleshooting (Pomraning et al., 2012). In recent years, many core sequencing facilities or companies started offering library preparation services, there are commercial kits available and, lastly, for different sequencing platforms different library methods are required. There are no steps in which the biological origin of the DNA matters significantly; thus, we will not discuss library preparation in great detail here. Our most recent protocol, relying largely on Illumina TruSeq methods is part of the Z. tritici-specific supplementary information. One aspect needs to be considered, however. We, and others (Ji et al., 2014), have noticed that heterochromatic sequences, which are usually AT-rich, can be depleted by Illumina protocols, likely during PCR overamplification. Thus, special considerations need to be made when studying heterochromatin, for example keeping the number of final library amplification PCR cycles below ten.
2.2.7. Controls
The most frequently used strategy to generate a control for ChIP consists of preserving a fraction of each sheared chromatin sample that will not undergo immunoprecipitation, the so-called “input control” (Park, 2009; Tamaru et al., 2003). In Chip-seq, libraries are constructed with this input DNA to distinguish enriched regions from the background noise of sequencing and to limit sequencing bias from many sources (local chromatin structure, repetitive DNA, library preparation) (Chen et al., 2012; Teytelman et al., 2009). However, quantitative analyses of enriched regions showed that input controls also can be highly variable between biological replicates (Cheung et al., 2011; de Boer et al., 2014). Deep sequencing of the precipitated sample DNA from several replicates and different conditions (for example, “induced” versus “uninduced” expression of a tagged TF) limits false negatives of poorly enriched and false positives or spuriously enriched regions. Specialized analyses tools have been developed to analyze sample data without the need of an input control. In the past we compared both methods; we used input controls and inducible protein expression to quantify occupancy of TFs but comparisons between several independent samples need to be made to assess presence of histone modifications.
2.3. Analysis of ChIP-seq data: considerations and tools for Z. tritici
The output of ChIP is DNA associated with the immunoprecipitated protein of interest. Sequencing of this DNA followed by mapping to a reference genome allows us to identify the genomic locus of the DNA and thus the specific binding sites for proteins, ideally down to short palindromic binding sites, e.g. for TFs (Johnson et al., 2007; Smith et al., 2010). For ChIP-seq, 50-nt short-read sequencing is sufficient. On an Illumina Hi-Seq2000 machine approximately 250 million reads can be produced in a single lane, translating to about 12.5 Gb of sequence, more than 300-fold of the typical Zymoseptoria genome content. Data will include both sequence reads that are true signal and non-specific background or “noise”. Alignment (or mapping) of Chip-seq reads to the reference genome generates a histogram of stacked reads on chromosome coordinates (x axis). On the y axis the number of times a specific nucleotide was found as part of a read is shown. Alignment will thus result in “enriched” regions or “peaks” that represent the DNA to which the protein or protein complex was bound. In ChIP-seq with fungal genomes there is almost always background noise because coverage is very deep. The y axis value will rarely go to “0”; this is usually the case when a sequence that is present in the reference genome is missing from a specific sample, e.g. by deletion or translocation of chromosome fragments.
2.3.1. Sequencing coverage and data quality
As with de novo sequencing approaches, the appropriate depth of ChIP-seq depends largely on the size of the genome. Excellent or adequate ChIP-seq datasets for most filamentous fungi consist, after filtering and quality control, of 4 to 20 million mapped reads (Connolly et al., 2013; Jamieson et al., 2013; Karimi-Aghcheh et al., 2013; Smith et al., 2010; 2011), overall fewer than for other eukaryotes (Pepke et al., 2009). In our experiments with Z. tritici we obtained variable numbers of 50-nt reads with antibodies targeting different proteins or protein modifications (Fig. 3). For ChIP-seq with the centromere-specific CenH3 protein tagged with GFP we obtained 15,270,094 reads, compared to 7,349,409 reads from H3K4me2; 9,750,281 reads from H3K9me3 and 7,263,505 reads from H3K27me3 corresponding to a calculated genome coverage of 9–19X. This level of depth is usually sufficient to call peaks and calculate statistically significant enrichment above background levels (Fig. 3). Coverage alone is not a good predictor of how well peaks, or -in the case of histones- regions of enrichment can be called, because background noise tends to increase with sequencing depth as well, though not in a linear fashion. For TF ChIP, peak calling using two different conditions (“induced” or “uninduced”) can help to localize true peaks.
In our experiments, quality control of ChIP-seq reads was done with grooming, trimming, filtering and masking tools from the Galaxy server (www.galaxyproject.org, Blankenberg et al., 2010; Giardine et al., 2005; Goecks et al., 2010;). In brief, reads with an overall quality score below 20 and reads with less than 25% of nucleotides with a quality score above 20 were discarded. For the remaining reads, nucleotides with quality scores below 20 were masked with Ns.
2.3.2. Mapping against a reference genome and data visualization
The choice of appropriate mapping software depends on the type of sequence data. For ChIP-seq data, software developed for mapping of small reads, such as Bowtie2 or SOAP2 is suitable (Langmead, 2010; Langmead and Salzberg, 2012; Li et al., 2009). Both methods implement algorithms that allow the rapid and efficient mapping of millions of reads to a reference genome. They generate a summary report with mapping statistics, including unmapped reads, reads which only mapped once and reads that map to multiple positions. We almost exclusively use Bowtie2 for mapping of ChIP-seq reads to the Z. tritici reference genome of IPO323 (Goodwin et al., 2011) or those of other filamentous fungi. We allowed reads with multiple “hits” (mostly short identical regions in stretches of repetitive DNA, for example transposons or rDNA) to be mapped one by one to different identical matching regions in the genome; thus, each region receives almost the same number of reads but the number is an integer. An alternative choice is to calculate the number of identical (“clonal”) reads and assign the average to each of a discrete number of identical matching positions in the genome.
Multiple genome viewers are available for visualization of mapping data. A genome viewer provides an important tool to evaluate genome-wide distribution of reads, helps to spot chromosome rearrangements or deletions, assess data quality and consistency between replicates. Genome browsers also provide the only way to show different types of data such as tracks for genes, transcripts, repetitive sequences, ChIP-seq and transcript levels in a comprehensive manner. In studies with Z. tritici, we use two browsers extensively. One, the Z. tritici gbrowse site, is currently housed at OSU and provides stable genome-wide access to published genome, updated transcript and now ChIP-seq data; access to the site is password-protected but as soon as data are published it is made publicly available. The gbrowse viewer is excellently suited for genomes that are completely assembled. Although most tracks give qualitative, not quantitative information, “browser glancing” is usually the first way to develop new hypothesis based on high-throughput sequencing data. Installation and upkeep of a gbrowse site is somewhat labor-intensive. Thus we employ another, perhaps more nimble, genome viewer for our work, the Integrative Genome Viewer (IGV) (http://www.broadinstitute.org/software/igv; Thorvaldsdóttir et al., 2013). IGV also supports large datasets and allows the parallel visualization of different tracks, such as annotations and ChIP-seq or RNA-seq at chromosome scale as well as local scales (e.g. down to the nucleotide level). Both browsers allow upload of tracks that convey quantitative evaluations of data, e.g. peaks that show statistically significant enrichment or quantitative changes in gene expression. In addition, there are several other genome browsers available (Argo, Artemis), but IGV and gbrowse are the two most commonly used by researchers who work on filamentous fungi.
2.3.3. Peak calling analysis and peak annotation
Numerous peak-callers have been developed, optimized for different types of experiments such as the identification of TF binding sites or histone modifications (for examples, see reviews by Bailey et al., 2013; Pepke et al., 2009). Peak calling software addresses background “noise” and normalization in different ways; every operator needs to be knowledgeable on this and the field is changing quickly, so that specific recommendations are not very useful. Peak calling analysis is applied to identify those genomic regions, which statistically are enriched with sequencing reads over the background noise of reads (Park, 2009). The methodological approaches and challenges of peak-calling analyses have been reviewed and discussed only recently (Meyer and Liu, 2014). It is usually a good idea to support conclusions from one analysis (i.e. detection and quantification of enriched sites or regions) by analyses with a second software tool (Meyer and Liu, 2014). Most tools are dedicated to the quantification of TF binding sites in specific regions. More recently, the most widely used program, MACS (Feng et al., 2012; Zhang et al., 2008), has been updated to allow for the quantification of the broader regions of histone enrichment, which can extend over a few to dozens of nucleosomes, (i.e. from 200 to thousands of base pairs; Bailey et al., 2013). Examples from our previous work with N. crassa and F. graminearum illustrate these differences (Connolly et al., 2013; Smith et al., 2010; 2011).
For Z. tritici we applied two different peak-calling algorithms: RSEG (Song and Smith, 2011), which was originally developed to identify genome-wide regions of histone modifications, and MACS2, an updated version of MACS that now combines detection of TF binding sites and broader regions of enrichment (Feng et al., 2012). Both RSEG and MACS can be run without the input control that used to be preferred in earlier studies. MACS models background signals by using a Poisson distribution. The user predefines a p-value for significantly enriched domains or peaks (the default p-value is <10−5). RSEG relies on a Hidden Markov Model approach to provide the coordinates of enriched domains. In both cases the coordinates for statistically significant peaks from ChIP-seq can be correlated to gene and transcript coordinates from RNA-seq (Fig. 3). For Z. tritici data we called enriched domains with RSEG and MACS, and found a high degree of overlap between peak coordinates; thus, both peak callers appear to yield similar results. We also analyzed the overlap between biological replicates by assessing the distribution of H3K4me2, H3K9me3 and H3K27me3 and across chromosomes and comparing the output by correlation analyses (Fig. 3).
The goal of ChIP-seq analysis is not only to locate significantly enriched regions across the genome but ideally also to help quantify the enrichment, for example of different histone modifications in different samples and to correlate these enriched regions with gene expression under various conditions (Connolly et al., 2013). One also seeks to determine the association of different peaks with genomic features such as coding sequences and transposable elements. Combining ChIP-seq data of histone modifications with RNA-seq data from Z. tritici allows detailed analyses of gene transcription across the genome (Fig. 3). This correlation provides the functional definition for heterochromatic (transcriptionally silent) and euchromatic (transcriptionally active) regions.
3. Concluding remarks
In spite of the economic importance of Z. tritici as a worldwide wheat pathogen, the biology of this fungus is still poorly understood. During host infection, Z. tritici undergoes a life-style transition from biotrophic to necrotrophic growth. This hemibiotrophic infection pattern must entail a finely tuned regulation of gene expression likely involving changes at the chromatin level. Chromatin-mediated regulation of pathogenicity-related genes has recently been shown in other phytopathogenic fungi, including Epichloe festucae, F. graminearum, F. fujikuroi and L. maculans (Chujo and Scott, 2014; Connolly et al., 2013; Studt et al., 2013; Soyer et al., 2014; Wiemann et al., 2013), supporting this hypothesis. Here we describe a ChIP-seq based approach to study chromatin-based gene regulation in Z. tritici during in vitro growth. We defined euchromatic and heterochromatic regions by mapping reads obtained by ChIP with various antibodies against modified histones to the reference genome of the IPO323 strain (see Fig. 3), and to correlate the average chromatin structure with genome annotation and transcriptomic data (K. Schotanus, M. Möller, J. Soyer, J. Grandaubert, L.R. Connolly, M. Freitag, E.H. Stukenbrock, in preparation). The next logical step, already mentioned above, will be to further develop existing ChIP methodology for in planta experiments, something that has not been done yet for any phytopathogenic fungus. As histone modifications are highly conserved, antibodies used with fungi will also detect plant histone modifications. Thus, either ChIP-seq will need to be carried out at great depth or more specific antibodies may need to be developed to increase specificity of the isolated DNA. In E. festucae, ChIP-qPCR with specific primers was used on in planta material for comparison of chromatin structures during in vitro and in planta growth, demonstrating induction of secondary metabolite cluster genes during host infection (Chujo and Scott, 2014). The best approach for the future will be to generate ChIP-seq and RNA-seq data for both organisms in one experiment to get a more complete view of gene regulation during infection. As sequencing and reagent costs decrease, this will be the goal of our future studies.
Supplementary Material
Highlights.
Chromatin structures and modifications important for gene regulation
Chromatin immunoprecipitation and NGS sequencing (ChIP--seq) to study histone modifications
Optimized ChIPseq based methods for the wheat pathogen ymoseptoria tritici
Genome--wide maps of chromatin modifications
Heterochromatin and euchromatin distribution in Z. tritici
Acknowledgments
Chromatin research in the group of EHS is supported by the Max Planck Society. JLS is funded by a Young Scientist Fellowship from INRA. KS acknowledges the EMBO for a short-term fellowship to visit Oregon State University. Chromatin work in the Freitag lab is supported by funds from the NIH (R01 GM097637) to MF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Appendix A, Supplementary Method file
The Supplementary information provides a detailed annotated protocol of the ChIP assay and library preparation as used for Z. tritici.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allan J, Fraser RM, Owen-Hughes T, Keszenman-Pereyra D. Micrococcal nuclease does not substantially bias nucleosome mapping. J Mol Biol. 2012;417:152–164. doi: 10.1016/j.jmb.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T, Krajewski P, Ladunga I, Lefebvre C, Li Q, Liu T, Madrigal P, Taslim C, Zhang J. Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput Biol. 2013;9:e1003326. doi: 10.1371/journal.pcbi.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Kuster GV, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;89:19.10:19.10.1–19.10.21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Negre N, Li Q, Mieczkowska JO, Slattery M, Liu T, Zhang Y, Kim TK, He HH, Zieba J, Ruan Y, Bickel PJ, Myers RM, Wold BJ, White KP, Lieb JD, Liu XS. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods. 2012;9:609–614. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MS, Down TA, Latorre I, Ahringer J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res. 2011;39:e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Scott B. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte plant symbiosis. Mol Microbiol. 2014;92:413–434. doi: 10.1111/mmi.12567. [DOI] [PubMed] [Google Scholar]
- Connolly LR, Smith KM, Freitag M. The Fusarium graminearum histone H3K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 2013;9:e1003916. doi: 10.1371/journal.pgen.1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosseau C, Azzi A, Smith K, Freitag M, Mitta G, Grunau C. Native chromatin immunoprecipitation (N-ChIP) and ChIP-Seq of Schistosoma mansoni: Critical experimental parameters. Mol Biochem Parasitol. 2009;166:70–76. doi: 10.1016/j.molbiopara.2009.02.015. [DOI] [PubMed] [Google Scholar]
- de Boer BA, van Duijvenboden K, van den Boogaard M, Christoffels VM, Barnett P, Ruijter JM. OccuPeak: ChIP-Seq peak calling based on internal background modelling. PLoS One. 2014;9:e99844. doi: 10.1371/journal.pone.0099844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon B, Gill N, Hamelin RC, Goodwin SB. The landscape of transposable elements in the finished genome of the fungal wheat pathogen Mycosphaerella graminicola. BMC Genomics. 2014;15:1132. doi: 10.1186/1471-2164-15-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SB, M’barek SB, Dhillon B, Wittenberg AH, Crane CF, Hane JK, Foster AJ, Van der Lee TA, Grimwood J, Aerts A, Antoniw J, Bailey A, Bluhm B, Bowler J, Bristow J, van der Burgt A, Canto-Canché B, Churchill AC, Conde-Ferràez L, Cools HJ, Coutinho PM, Csukai M, Dehal P, De Wit P, Donzelli B, van de Geest HC, van Ham RC, Hammond-Kosack KE, Henrissat B, Kilian A, Kobayashi AK, Koopmann E, Kourmpetis Y, Kuzniar A, Lindquist E, Lombard V, Maliepaard C, Martins N, Mehrabi R, Nap JP, Ponomarenko A, Rudd JJ, Salamov A, Schmutz J, Schouten HJ, Shapiro H, Stergiopoulos I, Torriani SF, Tu H, de Vries RP, Waalwijk C, Ware SB, Wiebenga A, Zwiers LH, Oliver RP, Grigoriev IV, Kema GH. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011;7:e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- Honda S, Lewis ZA, Huarte M, Cho LY, David LL, Shi Y, Selker EU. The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa. Genes Dev. 2010;24:443–454. doi: 10.1101/gad.1893210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Lewis ZA, Shimada K, Fischle W, Sack R, Selker EU. Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation. Nat Struct Mol Biol. 2012;19:471–477. doi: 10.1038/nsmb.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson K, Rountree MR, Lewis ZA, Stajich JE, Selker EU. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc Natl Acad Sci USA. 2013;110:6027–6032. doi: 10.1073/pnas.1303750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ji L, Sasaki T, Sun X, Ma P, Lewis ZA, Schmitz RJ. Methylated DNA is over-represented in whole-genome bisulfite sequencing data. Front Genet. 2014;5:341. doi: 10.3389/fgene.2014.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Karimi-Aghcheh R, Bok JW, Phatale PA, Smith KM, Baker SE, Lichius A, Omann M, Zeilinger S, Seiboth B, Rhee C, Keller NP, Freitag M, Kubicek CP. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 (Bethesda) 2013;3:369–378. doi: 10.1534/g3.112.005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner R, Bhattacharyya A, Poppe S, Hsu TY, Brem RB, Stukenbrock EH. Expression profiling of the wheat pathogen Zymoseptoria tritici reveals genomic patterns of transcription and host-specific regulatory programs. Genome Biol Evol. 2014;6:1353–1365. doi: 10.1093/gbe/evu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Hu J, Oh Y, Park J, Choi J, Lee YH, Dean RA, Mitchell TK. Combining ChIP-chip and expression profiling to model the MoCRZ1 mediated circuit for Ca/calcineurin signaling in the rice blast fungus. PLoS Pathog. 2010;6:e1000909. doi: 10.1371/journal.ppat.1000909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010;11:11–7. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schübeler D, Selker EU. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1997. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Meyer CA, Liu XS. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet. 2014;15:709–721. doi: 10.1038/nrg3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6:S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Bredeweg EL, Connolly LR, Phatale PA, Freitag M. Library preparation and data analysis packages for rapid genome sequencing. Methods Mol Biol. 2012;944:1–22. doi: 10.1007/978-1-62703-122-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York, U.S.A: 2001. [Google Scholar]
- Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, Sauer DG, Smith KM, Baker SE, Freitag M, Kubicek CP. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol. 2012;84:1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Kothe GO, Matsen CB, Khlafallah TK, Adhvaryu KK, Hemphill M, Freitag M, Motamedi MR, Selker EU. The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenetics Chromatin. 2008;1:5. doi: 10.1186/1756-8935-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Sancar G, Dekhang R, Sullivan CM, Li S, Tag AG, Sancar C, Bredeweg EL, Priest HD, McCormick RF, Thomas TL, Carrington JC, Stajich JE, Bell-Pedersen D, Brunner M, Freitag M. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for neurospora white collar complex. Eukaryot Cell. 2010;9:1549–1556. doi: 10.1128/EC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol. 2011;31:2528–2542. doi: 10.1128/MCB.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Smith AD. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics. 2011;27:870–871. doi: 10.1093/bioinformatics/btr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer JL, El Ghalid M, Glaser N, Ollivier B, Linglin J, Grandaubert J, Balesdent MH, Connolly LR, Freitag M, Rouxel T, Fudal I. Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans. PLoS Genet. 2014;10:e1004227. doi: 10.1371/journal.pgen.1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Studt L, Schmidt FJ, Jahn L, Sieber CM, Connolly LR, Niehaus EM, Freitag M, Humpf HU, Tudzynski B. Two histone deacetylases, FfHda1 and FfHda2, are important for secondary metabolism and virulence in Fusarium fujikuroi. Appl Environ Microbiol. 2013;79:7719–7734. doi: 10.1128/AEM.01557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, Zala M, McDonald BA, Wang J, Schierup MH. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 2011;21:2157–2166. doi: 10.1101/gr.118851.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Teytelman L, Ozaydin B, Zill O, Lefrancois P, Snyder M, Rine J, Eisen MB. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. Mapping Protein/DNA Interactions by Cross-Linking [Internet] Paris: Institut national de la santé et de la recherche médicale; 2001. ChIP with Native Chromatin: Advantages and Problems Relative to Methods Using Cross-Linked Material. [PubMed] [Google Scholar]
- Van Holde KE. Chromatin, Springer series in molecular biology. New York: 1989. [Google Scholar]
- Wittenberg AH, van der Lee TA, Ben M’barek S, Ware SB, Goodwin SB, Kilian A, Visser RG, Kema GH, Schouten HJ. Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS One. 2009;4:e5863. doi: 10.1371/journal.pone.0005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P, Sieber CM, von Bargen KW, Studt L, Niehaus EM, Espino JJ, Huß K, Michielse CB, Albermann S, Wagner D, Bergner SV, Connolly LR, Fischer A, Reuter G, Kleigrewe K, Bald T, Wingfield BD, Ophir R, Freeman S, Hippler M, Smith KM, Brown DW, Proctor RH, Münsterkötter M, Freitag M, Humpf HU, Güldener U, Tudzynski B. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- Yang F, Li W, Jørgensen HJ. Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PloS ONE. 2013;8:e81606. doi: 10.1371/journal.pone.0081606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.