Abstract
Study of the diarrhoea-causing pathogen Cryptosporidium has been hindered by a lack of genetic-modification and culture tools. A description of genome editing and propagation methods for the parasite changes this picture.
A common saying to stymied travellers in New England is, “You can’t get there from here.” Until recently, this was also true for scientific travellers wishing to study the widespread diarrhoeal agent Cryptosporidium using modern molecular genetics. But on page 477 of this issue, Vinayak et al.1 show that indeed ‘one can get there’. Their report of genetic modification of these unicellular organisms using CRISPR/Cas9 technology opens up a bold new era in the study of this pathogen.
The genus Cryptosporidium includes several species that infect humans and other mammals. These protozoan parasites are recognized as being among the most important diarrhoeal pathogens2,3, accounting for more than 10% of global child mortality and often infecting people who have compromised immune systems. Infections occur worldwide in association with contaminated water. One notable example in the United States was the ‘bug that made Milwaukee famous’ — an outbreak that affected the entire city in 1993 (ref. 4).
Cryptosporidial infections arise from the ingestion of parasites at the thick-walled cyst stage (oocyst) of their life cycle. After surviving the harsh conditions of the stomach, an oocyst ‘excysts’ and releases the infective and replicative form, the sporozoites, which divide in the intestinal lining, in turn generating cysts that are shed in the faeces. Cryptosporidia are members of the Apicomplexa group of protozoan parasites, and diverged early from their better-studied apicomplexan relatives Toxoplasma and the malaria parasite Plasmodium. They thus present numerous evolutionary novelties, including differences in fundamental cell biology (their lack of an organelle called the apicoplast is one example), in their infectious cycle, and in their genome, which at around 3,950 genes is much smaller than that of other apicomplexans5–7. The Cryptosporidium genome contains several essential genes acquired by lateral transfer from other microorganisms5–7, which perhaps reflects the parasite’s intimacy with intestinal bacteria. Collectively, these features provide exciting opportunities for basic research as well as for identifying cellular pathways relevant to therapy — but both these tasks have been made difficult by a lack of genetic tools.
The true challenge, however, was not the molecular technology but the limitations of working with Cryptosporidium, which cannot be cultured long term in vitro. Instead, oocysts must be isolated from infected calves or purchased commercially. Cysts can be stored for months, but excysted parasites that are inoculated onto mammalian-cell monolayers for growth undergo one or two rounds of replication at most. This narrow time window has profoundly hindered experimental manipulation2.
Vinayak et al.1 have dramatically improved this state of affairs. They made a series of optimizations to existing genetic-modification techniques that establish the basic parameters for successful transient transfection of Cryptosporidium sporozoites. This procedure introduces a segment of DNA (in this case, a plasmid) encoding a gene of interest that is then expressed by the cell for a short time. The authors verified successful transfections using a marker gene that encodes the protein luciferase, which produces bioluminescence in the presence of the appropriate substrate. This marker is fused to a gene conferring resistance to neomycin-class antibiotics, which provides a means of selecting transfected cells.
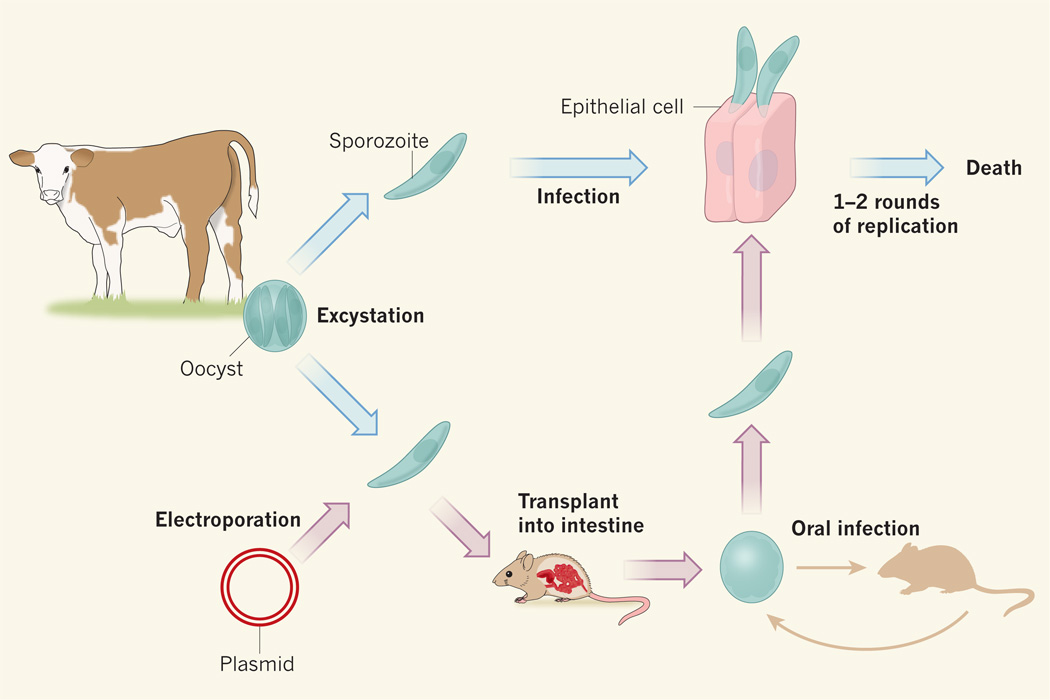
Not content with achieving reproducible transient transfection, Vinayak et al. proceeded to overcome the narrow experimental window. During in vitro culture, Cryptosporidium does not generate the thick-walled cyst forms that survive in the faeces and the stomach, but the researchers bypassed this biological block by inoculating the manipulated sporozoites directly back into the intestines of immunodeficient mice, in which the parasites propagated and produced oocysts (Fig. 1).
Figure 1. Modification and culture of Cryptosporidium.
The strong-walled oocyst form of Cryptosporidium parasites can be isolated from the faeces of infected calves. Oocysts can be induced to excyst to release the sporozoite form, which will infect cultured mammalian epithelial cells, but the sporozoites undergo only one or two rounds of replication before they die. Vinayak et al.1 have improved on this limited in vitro system in two ways. They have developed techniques for genetically modifying the sporozoite form — using electroporation to introduce foreign DNA in the form of a plasmid bearing the sequences required for CRISPR-based genome editing. And they show that these modified sporozoites will replicate when directly transplanted into the intestines of mice, and can be recovered as modified oocysts, which can be collected from mouse faeces for analysis in culture, or used to inoculate new mice to maintain the line indefinitely.
For stable genomic modifications, in which the introduced DNA is incorporated into the genome, rather than relying on the parasite’s own mechanisms for doing this, the authors turned to the genetic ‘tool de jour’ — the CRISPR/Cas9 system, a genome-editing approach that has proved effective in almost all organisms tested, including protozoan parasites. Another series of clever optimizations established the functionality and utility of this system in Cryptosporidium. Eventually, transfection of sporozoites with both the luciferase–neomycin-resistance fusion gene and DNA encoding the CRISPR/Cas9 machinery, followed by infection of mice with the sporozoites and treatment with the neomycin analogue paromomycin, led to the recovery from mouse faeces of antibiotic-resistant parasites stably expressing an integrated luciferase gene.
This first demonstration of genetically engineered Cryptosporidium introduces a method that is primed for real-world applications, already enabling in vitro or in vivo assays for monitoring parasite survival after drug or other treatments. The authors further demonstrated the utility of CRISPR/Cas9 by using it in the sporozoites to ablate expression of thymidine kinase, one of the few enzymes used by Cryptosporidium to generate nucleotides8. These experiments showed that this enzyme’s activity provides a bypass for the activity of another enzyme, dihydrofolate reductase, which accounts for the relative ineffectiveness of antifolate drugs against Cryptosporidium compared with other apicomplexan parasites.
The success of Vinayak and colleagues’ study lies not so much in the novelty or insight of particular steps, but rather in the systematic and incisive integration of them all towards what had been considered an impossible goal. As such, this is a textbook study on how to tackle a previously intractable pathogen, and it will serve as a model for future attempts with other disease-causing organisms.
The approach is by no means perfect — it is cumbersome and time-consuming to generate genetically modified cell lines by passaging them through mice, and the parasites can be studied only following recovery of cysts from faeces. But one can imagine many advances and future directions, such as using CRISPR-based systems to generate and probe panels of mutated parasites simultaneously. Perhaps high on the list of priorities will be the generation of modified parasites that can replicate and differentiate indefinitely in vitro. A second challenge is that genes required for parasite survival inside host cells cannot be ablated in order to study their mechanism; however, the importation of RNA- or protein-based regulatory strategies from other apicomplexans should overcome this.
So, having found how to ‘get there’, the application of Cryptosporidium genetic modification will greatly increase our understanding of the pathogen’s basic biology and virulence, and provide key information and validation for the development of improved vaccines and therapeutics.
References
- 1.Vinayak S, et al. Nature. 2015;523:447–480. [Google Scholar]
- 2.Checkley W, et al. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Striepen B. Nature. 2013;503:189–191. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- 4.MacKenzie WR, et al. N. Engl. J. Med. 1994;331:161–167. [Google Scholar]
- 5.Abrahamsen MS, et al. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 6.Bouzid M, Hunter PR, Chalmers RM, Tyler KM. Clin. Microbiol. Rev. 2013;26:115–134. doi: 10.1128/CMR.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu P, et al. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 8.Sun XE, et al. J. Biol. Chem. 2010;285:15916–15922. doi: 10.1074/jbc.M110.101543. [DOI] [PMC free article] [PubMed] [Google Scholar]