Abstract
Biological carbon dioxide fixation is an essential and crucial process catalyzed by both prokaryotic and eukaryotic organisms to allow ubiquitous atmospheric CO2 to be reduced to usable forms of organic carbon. This process, especially the Calvin-Bassham-Benson (CBB) pathway of CO2 fixation, provides the bulk of organic carbon found on earth. The enzyme ribulose 1,5-bisphosphate (RuBP) carboxylase/oxygenase (RubisCO) performs the key and rate-limiting step whereby CO2 is reduced and incorporated into a precursor organic metabolite. This is a highly regulated process in diverse organisms, with the expression of genes that comprise the CBB pathway (the cbb genes), including RubisCO, specifically controlled by the master transcriptional regulator protein CbbR. Many organisms have two or more cbb operons that either are regulated by a single CbbR or employ a specific CbbR for each cbb operon. CbbR family members are versatile and accommodate and bind many different effector metabolites that influence CbbR's ability to control cbb transcription. Moreover, two members of the CbbR family are further posttranslationally modified via interactions with other transcriptional regulator proteins from two-component regulatory systems, thus augmenting CbbR-dependent control and optimizing expression of specific cbb operons. In addition to interactions with small effector metabolites and other regulator proteins, CbbR proteins may be selected that are constitutively active and, in some instances, elevate the level of cbb expression relative to wild-type CbbR. Optimizing CbbR-dependent control is an important consideration for potentially using microbes to convert CO2 to useful bioproducts.
INTRODUCTION
The CbbR protein is a LysR-type transcriptional regulator (LTTR) that functions to control expression of genes of the CO2 fixation (cbb) operons that specify enzymes of the Calvin-Bassham-Benson (CBB) pathway. CbbR-dependent regulation occurs in diverse organisms, including nonsulfur and sulfur purple bacteria, marine and freshwater chemoautotrophic bacteria, cyanobacteria, methylotrophic bacteria, several varieties of hydrogen-oxidizing bacteria, and different Pseudomonas, Mycobacterium, and Clostridium strains (1–14). In addition, CbbR regulates carbon fixation gene expression in chloroplasts of eukaryotic red algae (15).
For many prokaryotic and eukaryotic organisms, CO2 is often the sole source of carbon, with the CBB pathway acting as the paramount metabolic pathway that enables such organisms to synthesize all the building blocks and macromolecules required for life. The net goal of the enzymes of the CBB cycle is to provide one triose phosphate molecule as the fundamental reduced form of useable carbon from an intake of three CO2 molecules. Although several enzymes are dedicated to the CBB pathway, ribulose 1,5-bisphosphate (RuBP) carboxylase/oxygenase (RubisCO) is the enzyme that catalyzes the actual “fixation” of CO2 onto the ene-diol form of RuBP, resulting in the production of two phosphorylated three-carbon molecules of 3-phosphoglyceric acid (3-PGA). Because this enzyme is a relatively poor catalyst and must contend with CO2 and O2, competing for the same active site under aerobic conditions, cells very often compensate by synthesizing huge amounts of RubisCO (e.g., up to 50% of the soluble protein under appropriate growth conditions [16]). Clearly, this is a highly regulated system, and under some physiological conditions, especially in bacteria, it is necessary for the organism to either upregulate or downregulate expression of genes of the CBB pathway. Transcriptional control of the cbb genes is thus vital because of the heavy energy demands and the burden of additional protein synthesis placed on the cell by CO2 assimilation. In bacteria, the master regulator protein in all cases is CbbR.
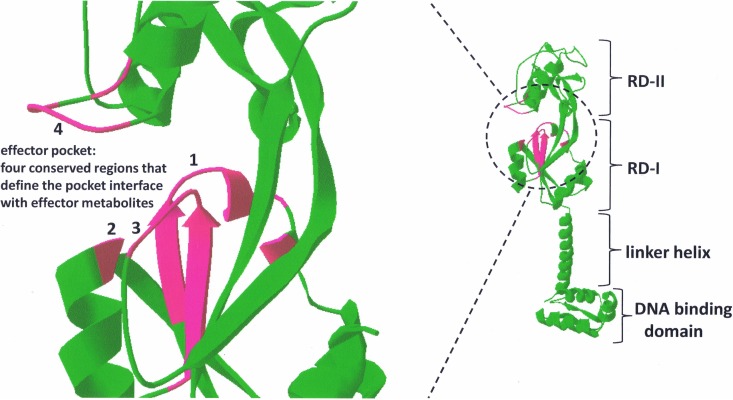
Like all LTTR family members, CbbR binds as a tetramer to the promoter region of the cbb operon. The generalized consensus DNA binding sequence for LTTRs is T-N11-A, and all CbbR proteins interact with this DNA binding motif (17). Typically, each of two DNA binding sites (within approximately 6 to 20 bp of each other) is bound by a CbbR dimer, creating a dimer of dimers (tetramer). Like all LTTRs, the CbbR protein structure is about 300 to 330 amino acids in length and is composed of three functional domains. There is a DNA binding domain (DBD) at the N terminus that binds to the cbb promoter region. The LTTR DBD is classified as a winged helix-turn-helix (HTH) motif (18, 19). All four HTH motifs within a CbbR tetramer interact with DNA when bound to the cbb promoter. A linker helix domain functions to connect the DBD and the recognition domains (RD) of the protein (RD-I and RD-II; also referred to as effector domains). The linker domain is a 30-amino-acid α-helix that operates as a rigid linker helix to prevent interaction between the DBD and the RD. The linker helix also contributes to dimer formation through coiled-coil interactions (20–26). Figure 1 illustrates a generalized structure for all CbbR proteins. Regions of conservation that distinguish CbbR proteins from other LTTRs are found in the recognition domains and are discussed below with respect to effector interactions.
FIG 1.
Proposed structure of CbbR. On the right is the generalized ribbon structure of the CbbR monomer based on the structure of the LTTR family member CbnR (20), illustrating the four major domains of the LTTR. On the left is the enlarged structure of the CbbR effector pocket. Four regions of the effector pocket, denoted 1 to 4, are highlighted in magenta, with residues of regions 1 to 4 conserved among CbbR family members. These four conserved regions define the effector pocket and are positioned at the interface between the effector metabolite(s) and CbbR. The conserved amino acid sequences (magenta) for each region are as follows: for region 1, GVVSTAKYFXP; for region 2, NR; for region 3, DLAIMGRPP; and for region 4, REXGSGTR (“X” represents a residue position that is not conserved). The effector pocket conservation also applies to CmpR, CcmR, and QscR, which are CbbR subfamily members. Conservation is high within the four regions of the effector pocket for all CbbRs examined: 81.5% for region 1, 95% for region 2, 86% for region 3, and 97% for region 4. DeepView/Swiss-Pdb Viewer software was used to generate the structural model.
cbbR AND cbb GENE ORGANIZATION
The cbbR gene, like most LTTR family members, is usually located directly upstream of the cbb operon that it regulates but in the opposite orientation. There are some notable exceptions to this general rule of gene organization. For example, cbbR of Rhodospirillum rubrum is in the same orientation as and adjacent to cbbM (encoding form II RubisCO); however, the orientation of cbbR is opposite that of the remainder of the cbb operon (4, 27). Another interesting exception is provided by Hydrogenophilus thermoluteolus; there, cbbR is located within a split cbb operon in an orientation opposite that of all the cbb genes. Rhodobacter capsulatus also presents an interesting situation where this organism contains two cbbR genes encoding two distinct CbbR proteins (CbbRI and CbbRII) that regulate two separate cbb operons, one of which, along with its cognate cbbR gene, was apparently derived from a chemoautotrophic ancestor (7, 28). In the chemoautotroph Hydrogenovibrio marinus, there also are two cbbR genes (cbbR1 and cbbRm) and three cbb operons (cbbLS-1, cbbLS-2, and cbbM), with cbbR1 located upstream and in an orientation opposite that of cbbLS-1, while the cbbRm gene is located upstream of but in the same orientation as cbbM (10, 29–31). The cbbLS-2 operon contains genes encoding carboxysomes and is expressed under conditions of low CO2 concentrations, independently of CbbR1 or CbbRm regulation (30, 31). On the other hand, CbbR1 and CbbRm of H. marinus may be involved in repressing expression of carboxysome genes contained within the CbbLS-2 operon at high levels of CO2 (31). Finally, CbbR has also been shown to regulate the expression of carboxysome genes in Acidithiobacillus ferrooxidans, a chemoautotrophic gammaproteobacterium that characteristically grows in acidic environments (3, 13). Notably, the single CbbR from A. ferrooxidans regulates four distinct cbb operons (3, 13).
While the foregoing represent some very interesting situations where CbbR plays an important physiological role, in addition to regulating cbb gene expression, the usual situation is that a single cbbR gene is used to exclusively regulate the two major cbb operons that are found in most prokaryotic organisms. Many phototrophic and chemoautotrophic organisms contain multiple RubisCO genes, usually encoding distinct form I (CbbLS) and form II (CbbM) enzymes that function to fix CO2 at low and high CO2 levels, respectively (32–35). The most thoroughly studied examples where a single CbbR regulates cbb operons containing distinct form I and form II RubisCO genes are Rhodobacter sphaeroides (5, 36, 37) and Rhodopseudomonas palustris (11). Several additional autotrophic bacterial species, including Ralstonia eutropha strain H16, contain two cbb operons regulated by the product of a single cbbR gene. R. eutropha has a well-characterized cbb regulon where a single cbbR gene controls both chromosomal and megaplasmid-borne cbb genes (2); however, the RubisCO enzymes encoded by these separate operons are virtually identical (38). Mycobacterium sp. strain JC1 DSM 3803 also has two cbb operons regulated by one CbbR, but the cbbR gene is directly downstream of and in the same orientation as cbbLS-1 (12, 39).
PHYLOGENETIC RELATEDNESS OF CbbRS AND CLOSELY RELATED LTTRS
Amino acid identities accurately reflect the general relatedness of CbbR proteins from different organisms. Yet there is a striking drift of amino acid homologies among the CbbR family similar to the general lack of amino acid identity within the LTTR family as a whole. Despite these differences in primary sequence, there is a high degree of structural and conformational similarity of the monomeric, dimeric, and tetrameric states of all LTTR proteins. Regions of residue similarity and identity within the LTTR and CbbR families include the DBD (HTH motif), regions defining the effector pocket, and areas of the protein important for the formation of dimers and tetramers. As determined on the basis of amino acid identities, the CbbR subfamily also includes QscR, CmpR, and CcmR, three closely related LTTRs that are more similar to some CbbRs than some CbbRs are to each other. QscR regulates the expression of two operons involved in the one-carbon serine assimilatory pathway of some methylotrophic bacteria (40, 41). CmpR regulates transcription of operons involved with bicarbonate transport in cyanobacteria (42) and specifically regulates expression of certain genes involved in the CO2-concentrating mechanism (CCM) (43–46). The CCM allows cyanobacteria to actively transport HCO3− into the cytoplasm and then into the carboxysome. Subsequently, carboxysomal carbonic anhydrase catalyzes the conversion of HCO3− to CO2 such that CO2 becomes highly concentrated in this microcompartment and is readily made available and saturates the active site of RubisCO (47). In Synechocystis sp. strain PCC 6803, CmpR activates transcription of the cmpABCD operon (high-affinity bicarbonate transporter), but another CbbR-like protein, CcmR, represses expression of the ndhD3, ndhF3, and chpY genes which are required for expression of the inducible high-affinity CO2 transporter, NDH-13 (48, 49).
Total amino acid identities for individual CbbRs range from 22% to 56%, with the majority of identities falling between 35% and 45%. The CbbRs with the highest identities include CbbR of Bradyrhizobium japonicum and CbbR of R. palustris at 56%, CbbR of Allochromatium vinosum and CbbR of Methylococcus capsulatus at 55%, CbbR of R. sphaeroides and CbbRII of R. capsulatus at 54%, and CmpR of Synechococcus elongatus PCC 7942 and CcmR of Synechocystis PCC 6803 at 54%. The phylogenetic analysis of CbbR proteins is in good agreement with the overall phylogenetic relationship of microorganisms that possess cbbR genes.
INTERACTIONS WITH CbbR: THE CASE OF DUELING TRANSCRIPTION FACTORS
There are several studies that show LTTR interactions with RNA polymerase or sigma factors (50–55), but there are few examples of interactions of LTTR proteins (not including CbbRs) with other transcriptional regulators (56, 57). By and large, this is a testament to the ability of LTTRs to independently and adequately regulate their operons in the prokaryotic kingdom. However, in the case of some phototrophic bacteria, regulation of cbb expression is much more complex, imposing additional layers of regulation on the energetically costly process of CO2 assimilation. There are two well-studied systems that illustrate this regulatory complexity: interaction of CbbR with additional (and different) transcription regulators in R. sphaeroides and R. palustris (58–61).
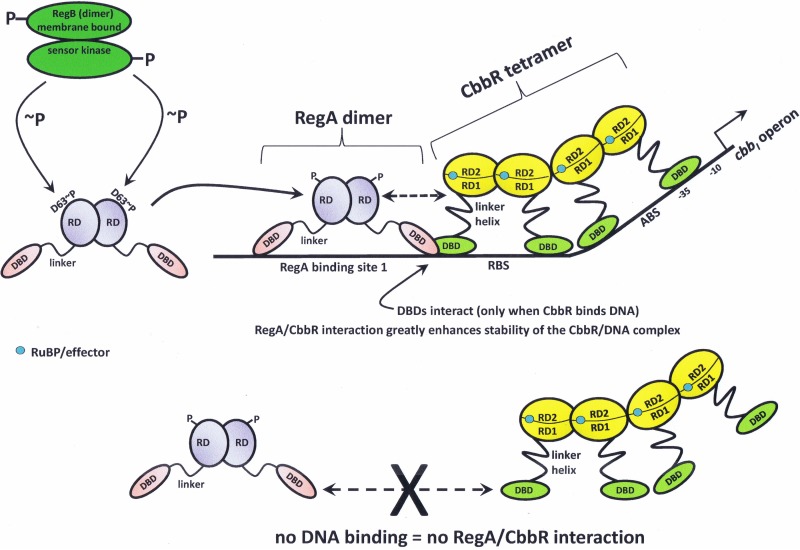
In R. sphaeroides, CbbR interacts with the RegA response regulator, which is part of a global two-component system (RegA/RegB) that controls expression of both the cbbI and cbbII operons of this organism (58, 62). In addition to the cbb regulon, RegA and its cognate sensor kinase, RegB, maintain control over several operons involved with energy-related (redox) metabolism in phototrophic nonsulfur purple bacteria (63, 64). Thus, the response regulator of this two-component system, RegA, binds to multiple sites within the promoter regions of both cbb operons of R. sphaeroides (37, 65). This scenario is similar to that seen with the hemA gene in R. sphaeroides, where RegA and FnrL bind the hemA promoter at positions where FnrL takes the place of CbbR in the hemA promoter and phosphorylation of RegA changes the affinity of RegA for the hemA promoter (66). For the cbbI operon of R. sphaeroides, it has been demonstrated that RegA has a higher affinity for the promoter when RegA binding site 3 is present, and site 3 is also necessary for optimal expression of the cbbI operon in vivo (58, 65). It was further shown that RegA greatly enhances the stability of the CbbR/promoter DNA complex and that CbbR must be bound to the cbb promoter in order to interact with RegA but that it is not necessary for RegA to be bound to DNA to interact with CbbR (58). This scenario presumably prevents interactions between the two regulator proteins unless CbbR is bound to the cbb promoter, pointing to a finely tuned attenuation strategy limiting potential nonspecific interactions between the two proteins. It may be surmised that a deleterious situation would be avoided if the proteins did not interact unless bound to the cbb operon promoter. Adding to the complexity, some studies have shown that phosphorylation of RegA greatly increases its DNA binding stability, while other studies have illustrated that phosphorylation of RegA enhances activation of transcription (64). Cross-linking experiments, using a bifunctional binding compound, indicated that RegA and CbbR form a stoichiometric complex, results that were buttressed by gel mobility shift assays that also showed specific interactions between RegA and CbbR-bound DNA (58). In addition, extensive mutational analyses provided evidence that CbbR and RegA interact with each other through their DBDs (59). A model for this rather complex regulatory scenario, which is based on several lines of evidence and provides a likely explanation for how CbbR and RegA interact to regulate cbb gene expression and, subsequently, CO2 fixation in R. sphaeroides, is presented in Fig. 2. Also considered in this model is the involvement of various small-molecule effectors that influence CbbR function (Fig. 2) (discussed below).
FIG 2.
Transcriptional regulation of the cbb operons of Rhodobacter sphaeroides. CbbR and RegA interact on the cbbI and cbbII promoters. There are four RegA binding sites upstream of the cbbI operon and six RegA binding sites upstream of the cbbII operon (37, 65). RegA DNA binding site 1 and the RegA binding site (RBS) overlap upstream of the cbbI transcriptional start site. The DBDs of CbbR and RegA interact and generate a CbbR/RegA/DNA complex at the cbbI promoter (59). The interaction of RegA with CbbR greatly increases the stability of the CbbR/DNA complex (58). CbbR does not interact with RegA if CbbR is not bound to the cbb promoter (58). RuBP (positive effector) is shown within the effector pocket of a CbbR monomer; the pocket is a small cleft formed between RD-1 and RD-2. Dashed arrows represent interactions with CbbR. ABS, activation binding site. RD-1 and RD-2, recognition domain 1 and recognition domain 2, respectively. DBD, DNA binding domain. RD (RegA), receiver domain. ∼P, phosphorylation at residue D63 of RegA. The −10 and −35 regions of the cbbI promoter are indicated.
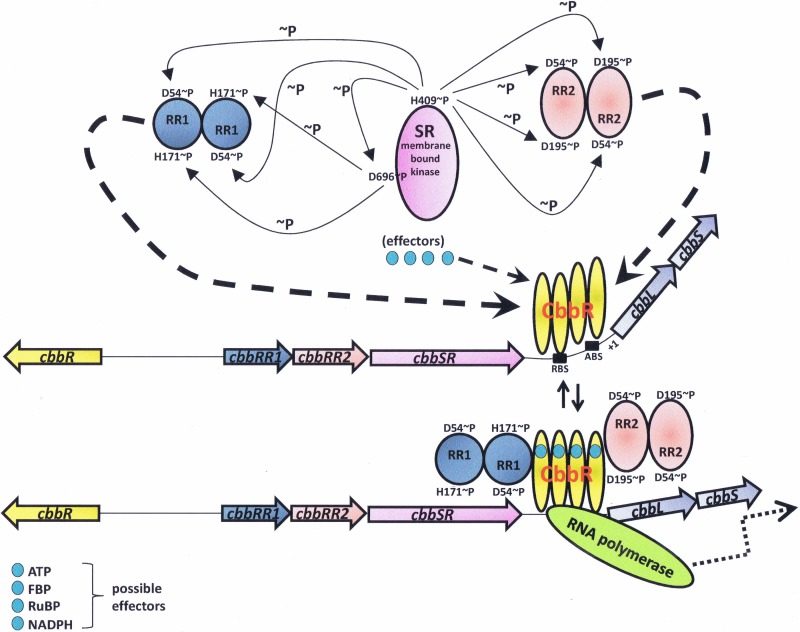
R. palustris represents an interesting and even more complex regulatory system involving CbbR and several additional protein regulators. While R. palustris also contains a Reg-type two-component system (67), it is not clear what this system regulates in this organism, as it apparently does not control photosystem biosynthesis as in R. sphaeroides and R. capsulatus (J. T. Beatty and F. R. Tabita, unpublished observations), nor is there any evidence to date to indicate that the Reg system controls cbb gene expression in R. palustris (J. L. Gibson and F. R. Tabita, unpublished observations). However, the regulation of the cbbI operon in R. palustris has proven to be extremely complex, involving a novel three-protein two-component system (11, 68). The regulatory proteins involved, CbbR, CbbRR1, CbbRR2, and CbbSR, are all encoded by genes that are closely juxtaposed within the cbbI operon region, with cbbR divergently transcribed from cbbRR1, cbbRR2, and cbbSR, which are immediately upstream from the cbbL and cbbS genes encoding the large and small subunits, respectively, of form I RubisCO (11). The CbbSR protein is a large transmembrane sensor kinase which, like many sensor kinases, is capable of autophosphorylation. In addition, CbbSR contains a consensus phosphate acceptor site, with a conserved aspartate-containing motif typical of many response regulators. Studies have shown that, upon autophosphorylation at a specific histidine residue, the phosphate may be transferred to the acceptor site of CbbSR (11, 68). Thus, this large protein basically acts as its own two-component system. However, CbbSR also catalyzes phosphorylation of both response regulators, CbbRR1 and CbbRR2 (11), the specificity for which is influenced by specific PAS domains on CbbSR (68). Both physiological/genetic and in vitro studies indicate that CbbRR1 and CbbRR2 bind to CbbR and influence cbbI gene transcription (60). In addition, various effector molecules influence these interactions in a concentration-dependent fashion and stabilize CbbR binding to the cbbLS promoter. CbbR/CbbRR1 interactions enhance the binding affinity of CbbR to the promoter, and CbbRR1, in concert with effectors ATP, RuBP, and fructose-1,6-bisphosphate, stabilizes the CbbR/promoter complex (69). A model for CbbR/CbbRR1/CbbRR2 interaction proposes that CbbRR2 acts as an antiactivator in the absence of effectors and that CbbRR1, by binding to CbbR and altering the conformation of CbbR, thus prevents CbbR from binding the cbbLS promoter (61). The presence of CbbRR1 and effectors negates the effect that CbbRR2 has on CbbR and allows binding of CbbR to the promoter and subsequent expression of the cbbI operon (61). A model summarizing the information relative to CbbR involvement with all these additional factors in R. palustris is presented (Fig. 3). Surface plasmon resonance (SPR) experiments were crucial to providing quantitative results concerning the effects of effectors on protein interactions as well as forming the basis for interpreting the interplay between CbbR, CbbRR1, CbbRR2, and effectors and how these factors all influence the regulation of the cbbLS promoter in R. palustris (60, 61, 69). Indeed, it is interesting that such a very complex system has been adopted by R. palustris to ensure control of the expression of form I RubisCO (cbbL and cbbS) and the genes of the cbbI operon. Of note, the cbbII operon, including the gene (cbbM) encoding form II RubisCO, does not appear to be controlled by CbbR or by CbbRR1, CbbRR2, or CbbSR (11).
FIG 3.
Transcriptional regulation of the cbbI operon of Rhodopseudomonas palustris and the role of the four regulatory factors in the expression of cbbI under autotrophic conditions. SR is the membrane-bound sensor kinase (CbbSR) that autophosphorylates and catalyzes phosphorylation of the two response regulators, RR1 (CbbRR1) and RR2 (CbbRR2). RR1 and RR2 subsequently interact with CbbR. CbbR binds the cbbI promoter at the recognition binding site (RBS) and the activation binding site (ABS). Potential positive effectors ATP, FBP, RuBP, and NADPH (68) are shown. Dashed arrows represent interactions with CbbR (60, 61, 69). Oppositely pointing solid arrows represent reversible interactions with CbbR. Dotted arrows represent transcriptional activation. Relaxation of the bend angle that CbbR imposes on the cbbI promoter is brought about by effector binding and precedes transcription. ∼P, phosphorylation at specific residues of CbbSR, CbbRR1, and CbbRR2 (68). The transcriptional start site is denoted +1.
POSTRANSLATIONAL REGULATION OF CbbR FUNCTION: THE ROLE OF EFFECTOR METABOLITES
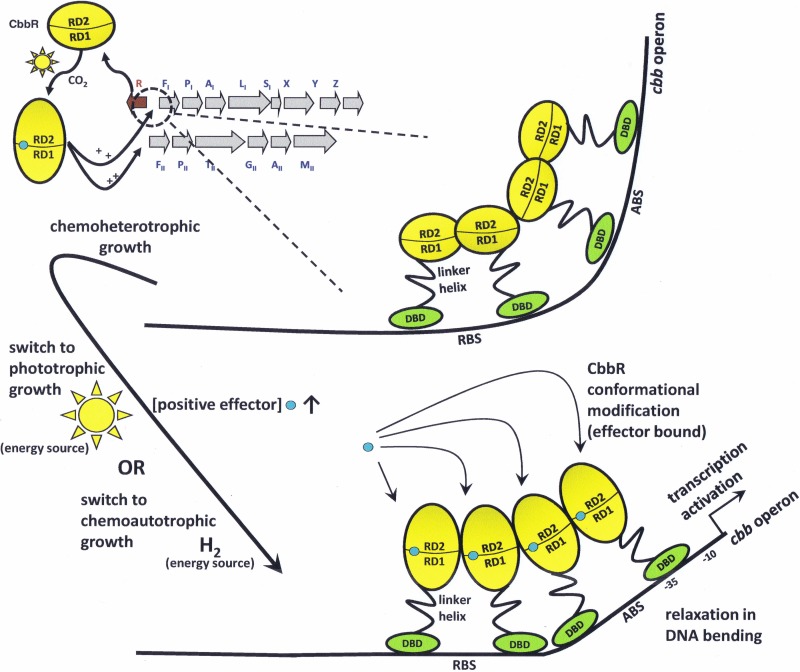
As prototypical LTTRs, CbbRs require a bound coinducer molecule or effector to activate transcription from the cbb promoter (17, 19). Common to most of the members of the LTTR family, the effector usually is an intermediate metabolite of the pathway that is regulated (17, 19). Effector binding occurs in a small cleft formed between RD-I and RD-II (Fig. 1) and is a hallmark of the LTTR family (22, 70–80). A recent study using nondenaturing mass spectrometry has illustrated for the first time that an LTTR tetramer binds four molecules of its effector in a stepwise pattern while bound to DNA (81). Binding of the effector produces a change in the angle at which an LTTR bends the promoter DNA to which it is bound (17, 19). For most scenarios, this change in DNA bend angle initiates contact or alters contact with the RNA polymerase, activating transcription. Effector binding to the LTTR can also inhibit transcription (19, 82). Studies illustrating DNA bend angle modification or initiation of transcription brought about by effector binding have been reported for some CbbRs (8, 76, 82, 83). The effector molecules may be different for each CbbR, depending on the organism. Figure 4 illustrates the relaxation of the cbb promoter bend angle imposed by CbbR subsequent to conformational changes elicited by effector (RuBP) binding after the switch to autotrophic growth for R. sphaeroides, R. capsulatus, Xanthobacter flavus, and H. thermoluteolus (8, 76, 82, 83).
FIG 4.
Consequences of relaxation of the promoter DNA bend angle upon effector binding (blue circle) to CbbR. For four CbbR-containing bacteria, a change in the promoter DNA bend angle has been shown to occur upon effector binding to the effector pocket of the protein. In R. sphaeroides, as illustrated here, CbbR undergoes a conformational change upon binding of a positive effector such as RuBP as a result of switching from heterotrophic to photo- or chemoautotrophic CO2-dependent growth conditions, thus allowing a relaxation of DNA bending and subsequent transcription of the cbb genes. Under appropriate physiological conditions, RuBP concentrations greatly increase and RuBP binds to CbbR of R. sphaeroides (76) and CbbRI of R. capsulatus (84), while phosphoenolpyruvate (PEP), FBP, and 3-PGA concentrations greatly increase and each binds to CbbRII of R. capsulatus during photoautotrophic growth (85). NADPH concentrations rapidly increase in X. flavus (83, 87) and H. thermoluteolus (8) during chemoautotrophic growth. This relaxation of the DNA bend angle leads to appropriate contact with RNA polymerase and activates transcription of the cbb operon. RBS, recognition binding site; ABS, activation binding site. RD-1 and RD-2, recognition domains 1 and 2, respectively.
In R. sphaeroides, RuBP, which of course is a unique metabolite of the CBB pathway, was suggested to be the effector for CbbR. The suggestion was based on a study employing a strain deleted for form I and form II RubisCO, leading to an accumulation of RuBP and a subsequent increase in transcription for both the cbbI and cbbII operons (84). Additionally, in vitro, the CbbR from R. sphaeroides was shown to alter the angle at which it bends the cbbI promoter DNA in the presence of RuBP, as illustrated by a change in the mobility of the CbbR/DNA complex in gel mobility shift assays (76). DNase I footprint analysis also demonstrates that RuBP improves protection of the cbbI promoter by CbbR (76).
For R. capsulatus, the metabolites that may influence CbbR-mediated expression present a more complex situation. Since R. capsulatus has two CbbRs (CbbRI and CbbRII) regulating two cbb operons (cbbI and cbbII, respectively) (7), each CbbR has its own set of effector molecules. In gel mobility shift assays, expression of CbbRI was shown to result in a significant increase in binding to the cbbI promoter DNA in the presence of 3-phosphoglycerate, 2-phosphoglycolate, ATP, KH2PO4, and RuBP and a small increase in binding to the cbbI promoter in the presence of NADPH, fructose-6-phosphate, fructose-1,6-bisphosphate (FBP), and ribose-5-phosphate (85). DNase I footprint analyses illustrated modified protection of the cbbI promoter DNA by CbbRI in the presence of RuBP, indicating a change in conformation of CbbRI and suggesting an altered bend angle of the cbbI promoter DNA (85). For CbbRII, gel mobility shifts demonstrated enhanced binding to the cbbII promoter in the presence of RuBP, 2-phosphoglycolate, 3-phosphoglycerate, phosphoenolpyruvate, and FBP (85). DNase I footprint analyses illustrated modified protection of the cbbII promoter DNA by CbbRII in the presence of FBP, phosphoenolpyruvate, and 3-phosphoglycerate, indicating a change in conformation of CbbRII and suggesting an altered bend angle of the cbbII promoter DNA (85).
Several compounds act as effectors of CbbR from R. palustris. Gel mobility shift assays demonstrated that ATP, FBP, RuBP, and NADPH all enhance binding of CbbR to the cbbLS promoter and that phosphoenolpyruvate inhibits binding of CbbR to the promoter (69). Quantitative SPR studies provided rate constant information and verified that R. palustris CbbR exhibits greater affinity for the cbbLS promoter in the presence of RuBP, FBP, ATP, and NADPH (61, 69).
Based on in vitro transcription experiments, the presence of phosphoenolpyruvate was shown to severely inhibit transcription of the cbb promoter by CbbR in R. eutropha strain H16 (82). This makes the effector metabolite, phosphoenolpyruvate, a corepressor for CbbR in R. eutropha, binding CbbR in the effector pocket but with the opposite effect on transcription. Gel mobility shift studies indicated that phosphoenolpyruvate enhances binding of CbbR to the cbb promoter (82, 86). Recent results also indicate that RuBP, ATP, and NADPH increase binding of wild-type CbbR to the cbb promoter of R. eutropha (86). Several mutant CbbRs with single-amino-acid substitutions near the effector pocket have reduced binding affinities in the presence of phophoenolpyruvate, RuBP, and ATP (86). Two other organisms, Xanthobacter flavus and Hydrogenophilus thermoluteolus, have CbbRs that show altered promoter DNA bending or increased promoter affinity in the presence of NADPH (8, 83, 87).
The CbbR from Cyanidioschyzon merolae (referred to as plastid-encoded transcription factor Ycf30) displays increased binding affinity for its promoter in the presence of NADPH and RuBP, as reported using gel mobility assays (15). In vivo experiments in permeabilized chloroplasts also indicated that RubisCO gene transcription is activated by 3-phosphoglyceric acid, RuBP, and NADPH (15). Ycf30 controls expression of the nucleus-independent RubisCO operon in chloroplasts in this red alga (15).
Studies have demonstrated that CmpR and CcmR can use the same effector molecules that are utilized by many cyanobacterial CbbRs. CmpR from S. elongatus PCC 7942 has high affinity for 2-phosphoglycolate and low affinity for RuBP, as illustrated in gel mobility shift studies that demonstrated enhanced binding of CmpR to the cmp operon regulatory region (45). CcmR from Synechocystis PCC 6803 regulates the promoter regions of a variety of genes involved in the CCM through the use of NADP+ and α-ketoglutarate as effectors (49). SPR studies illustrated that both NADP+ and α-ketoglutarate enhanced binding of CcmR to the ndhF3 promoter, the regulatory region for several genes involved in high-affinity CO2 uptake (49). Similarly to the case with R. eutropha, where CbbR utilizes phosphoenolpyruvate as a corepressor (82), CcmR binds NADP+ and α-ketoglutarate as corepressors to repress expression of the genes involved in the inducible high-affinity CCM of Synechocystis sp. strain PCC 6803 (49).
Finally, in the methylotrophic bacterium Methylobacterium extorquens AM1, QscR regulates two serine-cycle pathway operons, qsc1 and qsc2, and also regulates the expression of a third gene, glyA (40, 41). Intermediate metabolites of the serine-cycle and traditional-energy metabolites (effectors of CbbRs) were found not to be effectors of QscR (40). Formyl-tetrahydrofolate, an intermediate for formaldehyde assimilation which is linked to the serine cycle, was shown to be a candidate effector for QscR (41). Gel mobility shift assays demonstrated that formyl-tetrahydrofolate enhances binding of QscR to the promoters of both the qsc1 and qsc2 serine-cycle operons (41).
To better understand how effector molecules interact with CbbR, it is instructive to consider an enlargement of the effector pocket structure of the CbbR subfamily as a distinguishing feature to separate CbbRs from other LTTRs (Fig. 1). The four conserved regions that define the effector pocket contain positively charged residues, usually arginine (sometimes lysine), and polar residues that attract and accommodate negatively charged effectors. The conserved amino acid sequences (highlighted in Fig. 1 in magenta) for each region are as follows: for region 1, GVVSTAKYFXP; for region 2, NR; for region 3, DLAIMGRPP; and for region 4, REXGSGTR (“X” represents a residue position that is not conserved) (Fig. 1). All analyzed bacterial CbbRs utilize similar effector metabolites that have negatively charged phosphate moieties, usually two phosphate moieties, or that may be organic acids that contain two negatively charged acid groups (Table 1). Many of the CbbR effectors, such as RuBP, 3-PGA, FBP, 2-phosphoglycolate, and 2-phosphoglycerate, are metabolites of the CBB pathway and would be expected to be present at higher concentrations in the cell during active biosynthetic CO2 assimilation.
TABLE 1.
CbbRs and subfamily members from various organisms and their effectors
| Source and protein | Effector metabolite(s) (reference[s]) |
|---|---|
| R. sphaeroides CbbR | RuBPa,b (76) |
| R. capsulatus | |
| CbbRI | RuBP,c,b PEP,b 3-PGA,b 2-phosphoglycolate,b 2-phosphoglycerate,b ATP,b KH2PO4b (85) |
| CbbRII | FBP,c 3-PGA,c,b PEP,c,b RuBP,b 2-phosphoglycolateb (85) |
| R. eutropha CbbR | PEPb,d (corepressor), RuBP,b ATP,b NADPHb (82, 86) |
| R. palustris CbbR | RuBP,b,e ATP,b,e FBP,b,e NADPH,b PEP (inhibits DNA binding) (61, 69) |
| X. flavus CbbR | NADPHa,b (83, 87) |
| H. thermoluteolus CbbR | NADPHa (8) |
| C. merolae CbbR | RuBP,b,d NADPH,b,d 3-PGAd (15) |
| S. elongatus CmpR | RuBP,b 2-phosphoglycolateb (45) |
| Synechocystis PCC 6803 CcmR | NADP+,e α-ketoglutaratee (49) |
| M. extorquens QscR | Formyl-tetrahydrofolateb (41) |
Metabolite that allows a change in the bend angle CbbR imposes on promoter DNA via gel mobility shift assay.
Metabolite that increases CbbR binding affinity for promoter DNA (i.e., enhances stability of CbbR on promoter DNA) via gel mobility shift assay.
Metabolite that alters the region of promoter DNA protected by CbbR via DNase I footprinting/protection assay.
Metabolite that allows CbbR to activate (or inhibit) transcription from the cbb promoter via in vitro transcription assay.
Metabolite that increases CbbR binding affinity for promoter DNA (i.e., enhances stability of CbbR on promoter DNA) via SPR.
CONSTITUTIVELY ACTIVE CbbR PROTEINS
LTTR constitutive activity may be defined as activation of gene expression under conditions that normally repress gene transcription, typically in the absence of the LTTR's effector. When certain residues were altered, various LTTR proteins were found to constitutively activate gene expression; each LTTR appears to be unique with respect to which amino acid substitutions confer constitutive activity. This is probably a logical adaptation, as one might assume that the residues that are important for effector binding or for specific interactions with target DNA might be specific for each LTTR. (“LTTR*” denotes an LTTR variant with constitutive activity.) Many of these amino acid substitutions are centered at the effector pocket, but substitutions in other areas of the LTTR, such as at residues within the linker helix or hinge region or throughout RD-I and RD-II, can generate constitutive activity (70, 72, 76, 86, 88–94). Typically, single-amino-acid substitutions encompass the vast majority of the reported changes identified for constitutive proteins, and most constitutive proteins bind their effectors but may behave differently from the wild-type LTTR in gel mobility shift assays, DNase I footprinting assays, or in vitro transcription assays (76, 88, 89, 92–94). Amino acid substitutions that confer constitutive activity are thought to change the conformation of the LTTR tetramer to mimic the conformation seen when it is bound with the effector or to change the conformation of the LTTR/promoter complex to produce a favorable interaction with RNA polymerase to activate transcription. Studies of LTTR* proteins from several LTTR family members, including NodD, AmpR, OccR, CysB, OxyR, NahR, GtlR, XapR, and Cbl, have been previously published (70–72, 88–96).
A large set of CbbR* variants from both R. sphaeroides and R. eutropha have been isolated (76, 86). Constitutive proteins were generated by specific biological selection strategies involving the use of a reporter construct containing the cbb promoter fused to the lacZ open reading frame (ORF) integrated into the genome of a cbbR deletion strain for both organisms. The mutated cbbR proteins contain mutations that encode CbbR* proteins. For R. sphaeroides, several of the amino acid substitutions that confer constitutive activity clustered around the effector pocket which proved to be critical in defining the effector pocket for LTTRs (76). Several of the CbbR* proteins interact differently with promoter DNA in the presence of RuBP (effector) compared to wild-type CbbR. Interestingly, several of the CbbR* proteins activate expression of the cbbI promoter to a much greater extent than wild-type CbbR under conditions of cbb activation (76). Under conditions repressive for cbb activation (chemoheterotrophic growth), the amounts of cbb expression produced by the CbbR* proteins differed greatly; that is to say, some CbbR* proteins were better than others at activating gene expression.
CbbR* proteins from R. eutropha with amino acid substitutions located in all regions of the protein except the DBD were isolated. Substitutions were localized in the effector pocket, throughout RD-I and RD-II, and in the C terminus, and several residue changes were located in the linker helix. All of these were previously characterized (86). One particular CbbR* of interest is a truncation, leaving only the DBD and the linker helix to act as a transcriptional regulator. Nonetheless, this truncated CbbR* was able to support growth under chemoautotrophic conditions and to activate the cbb operons under repressive conditions in R. eutropha (86). This truncated protein illustrates that the DBD/linker helix region of CbbR is sufficient to activate expression from the cbb promoter, demonstrating that either the DBD or the linker helix or both make contact with the RNA polymerase (86). Similarly to some CbbR* proteins from R. sphaeroides, some of the CbbR* proteins from R. eutropha activated expression at levels severalfold greater than those seen with wild-type CbbR under autotrophic growth conditions (76, 86). The CbbRs of R. sphaeroides and R. eutropha exhibit only 35.6% identity, and each species appears to have a specific suite of residue changes that lead to CbbR* activity. Indeed, conserved residues whose presence is known to result in constitutive activity in R. sphaeroides CbbR did not confer constitutive activity when similar residues were changed in R. eutropha CbbR (86).
CONCLUDING REMARKS
CbbR controls the assimilation of carbon in autotrophic bacteria. It is the master regulator of the CBB CO2 assimilation pathway, playing an essential role to ensure that the cbb genes are actively transcribed. It is clear that CbbR must be posttranslationally modified, and there are various ways in which this is accomplished, including the binding of small-molecule effectors as well as interactions with other transcription factors. Studies that investigate the interaction of other proteins with CbbR will advance understanding of how CO2 fixation is regulated and of how LTTRs regulate transcription in general. CbbR also plays an important role in ensuring that CO2-assimilatory organisms generate essential carbon metabolic intermediates that can be subsequently diverted into the synthesis of economically and globally meaningful biological molecules, such as biofuels. Constitutive CbbR variants have been proven to greatly increase the level of expression from the cbb promoter; thus, additional modification of the CbbR protein will further enhance the power of the cbb promoter as a tool for the production of biological compounds (76, 86). Other LTTRs are also amenable to constitutive modification, which may be important for the enhanced expression of other pathways in bacteria, since LTTRs are the most common transcriptional regulators in prokaryotes.
ACKNOWLEDGMENTS
This work was supported by grant DE-FG02-08ER15976 from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, and ARPA-E grant DE-AR000009 from the U.S. Department of Energy.
REFERENCES
- 1.Viale AM, Kobayashi H, Akazawa T, Henikoff S. 1991. rbcR, a gene coding for a member of the LysR family of transcriptional regulators, is located upstream of the expressed set of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in the photosynthetic bacterium Chromatium vinosum. J Bacteriol 173:5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Windhövel U, Bowien B. 1991. Identification of cfxR, an activator gene of autotrophic CO2 fixation in Alcaligenes eutrophus. Mol Microbiol 5:2695–2705. doi: 10.1111/j.1365-2958.1991.tb01978.x. [DOI] [PubMed] [Google Scholar]
- 3.Kusano T, Sugawara K. 1993. Specific binding of Thiobacillus ferrooxidans RbcR to the intergenic sequence between the rbc operon and the rbcR gene. J Bacteriol 175:1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falcone DL, Tabita FR. 1993. Complementation analysis and regulation of CO2 fixation gene expression in a ribulose 1,5-bisphosphate carboxylase-oxygenase deletion strain of Rhodospirillum rubrum. J Bacteriol 175:5066–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson JL, Tabita FR. 1993. Nucleotide sequence and functional analysis of CbbR, a positive regulator of the Calvin cycle operons of Rhodobacter sphaeroides. J Bacteriol 175:5778–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Bergh ERE, Dijkhuizen L, Meijer WG. 1993. CbbR, a LysR-type transcriptional activator, is required for expression of the autotrophic CO2 fixation enzymes of Xanthobacter flavus. J Bacteriol 175:6097–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paoli GC, Vichivanives P, Tabita FR. 1998. Physiological control and regulation of the Rhodobacter capsulatus cbb operons. J Bacteriol 180:4258–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terazono K, Hayashi NR, Igarashi Y. 2001. CbbR, a LysR-type transcriptional regulator from Hydrogenophilus thermoluteolus, binds two cbb promoter regions. FEMS Microbiol Lett 198:151–157. doi: 10.1111/j.1574-6968.2001.tb10635.x. [DOI] [PubMed] [Google Scholar]
- 9.Wei X, Sayavedra-Soto LA, Arp DJ. 2004. The transcription of the cbb operon in Nitrosomonas europaea. Microbiology 150:1869–1879. doi: 10.1099/mic.0.26785-0. [DOI] [PubMed] [Google Scholar]
- 10.Toyoda K, Yoshizawa Y, Arai H, Ishii M, Igarashi Y. 2005. The role of two CbbRs in the transcriptional regulation of three ribulose-1,5-bisphosphate carboxylase/oxygenase genes in Hydrogenovibrio marinus strain MH-110. Microbiology 151:3615–3625. doi: 10.1099/mic.0.28056-0. [DOI] [PubMed] [Google Scholar]
- 11.Romagnoli S, Tabita FR. 2006. A novel three-protein two-component system provides a regulatory twist on an established circuit to modulate expression of the cbbI region of Rhodopseudomonas palustris CGA010. J Bacteriol 188:2780–2791. doi: 10.1128/JB.188.8.2780-2791.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Park DO, Park SW, Hwang EH, Oh JI, Kim YM. 2009. Expression and regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in Mycobacterium sp. strain JC1 DSM 3803. J Microbiol 47:297–307. doi: 10.1007/s12275-008-0210-3. [DOI] [PubMed] [Google Scholar]
- 13.Esparza M, Cárdenas JP, Bowien B, Jedlicki E, Holmes DS. 2010. Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans, carbon fixation in A. ferrooxidans. BMC Microbiol 10:229. doi: 10.1186/1471-2180-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Zhang Y, Pohlmann EL, Li J, Roberts GP. 2011. The poor growth of Rhodospirillum rubrum mutants lacking RubisCO is due to the accumulation of ribulose-1,5-bisphophate. J Bacteriol 193:3293–3303. doi: 10.1128/JB.00265-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minoda A, Weber APM, Tanaka K, Miyagishima SY. 2010. Nucleus-independent control of the rubisco operon by the plastid-encoded transcription factor Ycf30 in the red alga Cyanidioschyzon merolae. Plant Physiol 154:1532–1540. doi: 10.1104/pp.110.163188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis RJ. 1979. The most abundant protein in the world. Trends Biochem Sci 4:241–244. doi: 10.1016/0968-0004(79)90212-3. [DOI] [Google Scholar]
- 17.Schell MA. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol 47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 18.Harrison SC, Aggarwal AK. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem 59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 19.Maddocks SE, Oyston PCF. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 20.Muraoka S, Okumura R, Ogawa N, Nonaka T, Miyashita K, Senda T. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J Mol Biol 328:555–566. doi: 10.1016/S0022-2836(03)00312-7. [DOI] [PubMed] [Google Scholar]
- 21.Monferrer D, Tralau T, Kertesz MA, Dix I, Sola M, Uson I. 2010. Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol Microbiol 75:1199–1214. doi: 10.1111/j.1365-2958.2010.07043.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Lou Z, Fu S, Yang A, Shen H, Li Z, Feng Y, Bartlam M, Wang H, Rao Z. 2010. Crystal structure of ArgP from Mycobacterium tuberculosis confirms two distinct conformations of full-length LysR transcriptional regulators and reveals its function in DNA binding and transcriptional regulation. J Mol Biol 396:1012–1024. doi: 10.1016/j.jmb.2009.12.033. [DOI] [PubMed] [Google Scholar]
- 23.Taylor JL, De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. 2012. The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH. Mol Microbiol 83:457–470. doi: 10.1111/j.1365-2958.2011.07919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruangprasert A, Craven SH, Neidle EL, Momany C. 2010. Full-length structures of BenM and two variants reveal different oligomerization schemes for LysR-type transcriptional regulators. J Mol Biol 404:568–586. doi: 10.1016/j.jmb.2010.09.053. [DOI] [PubMed] [Google Scholar]
- 25.Vinson C, Acharya A, Taparowsky EJ. 2006. Deciphering B-zip transcription factor interactions in vitro and in vivo. Biochim Biophys Acta 1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Batchelor JD, Lee PS, Wang AC, Doucleff M, Wemmer DE. 2013. Structural mechanism of GAF-regulated σ54 activators from Aquifex aeolicus. J Mol Biol 425:156–170. doi: 10.1016/j.jmb.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nargang F, McIntosh L, Somerville C. 1984. Nucleotide sequence of the ribulose bisphosphate carboxylase gene from Rhodospirillum rubrum. Mol Gen Genet 193:220–224. doi: 10.1007/BF00330671. [DOI] [Google Scholar]
- 28.Paoli GC, Morgan NS, Tabita FR, Shively JM. 1995. Expression of the cbbLcbbS and cbbM genes and distinct organization of the cbb Calvin cycle structural genes of Rhodobacter capsulatus. Arch Microbiol 164:396–405. [PubMed] [Google Scholar]
- 29.Chung SY, Yaguchi T, Nishihara H, Igarashi Y, Kodama T. 1993. Purification of form L2 rubisCO from a marine obligately autotrophic hydrogen-oxidizing bacterium, Hydrogenovibrio marinus strain MH-110. FEMS Microbiol Lett 109:49–54. [DOI] [PubMed] [Google Scholar]
- 30.Yaguchi T, Chung SY, Igarashi Y, Kodama T. 1994. Cloning and sequence of the L2 form of RubisCO from a marine obligately autotrophic hydrogen-oxidizing bacterium, Hydrogenovibrio marinus strain MH-110. Biosci Biotech Biochem 58:1733–1737. doi: 10.1271/bbb.58.1733. [DOI] [PubMed] [Google Scholar]
- 31.Yoshizawa Y, Toyoda K, Arai H, Ishii M, Igarashi Y. 2004. CO2-responsive expression and gene organization of three ribulose-1,5-bisphosphate carboxylase/oxygenase enzymes and carboxysomes in Hydrogenovibrio marinus strain MH-110. J Bacteriol 186:5685–5691. doi: 10.1128/JB.186.17.5685-5691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson GMF, Tabita FR. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microbiol Lett 146:13–22. doi: 10.1111/j.1574-6968.1997.tb10165.x. [DOI] [PubMed] [Google Scholar]
- 33.Kusian B, Bowien B. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol Rev 21:135–155. doi: 10.1111/j.1574-6976.1997.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 34.Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71:576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. 2008. Distinct form I, II, III, and IV RubisCO proteins from the three kingdoms of life provide clues about RubisCO evolution and structure/function relationships. J Exp Bot 59:1515–1514. [DOI] [PubMed] [Google Scholar]
- 36.Dubbs JM, Tabita FR. 1998. Two functionally distinct regions upstream of the cbbI operon of Rhodobacter sphaeroides regulate gene expression. J Bacteriol 180:4903–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubbs JM, Tabita FR. 2003. Interactions of the cbbII promoter-operator region with CbbR and RegA (PrrA) regulators indicate distinct mechanisms to control expression of the two cbb operons of Rhodobacter sphaeroides. J Biol Chem 278:16443–16450. doi: 10.1074/jbc.M211267200. [DOI] [PubMed] [Google Scholar]
- 38.Kusian B, Bednarski R, Husemann M, Bowien B. 1995. Characterization of the duplicate ribulose-1,5-bisphosphate carboxylase genes and cbb promoters of Alcaligenes eutrophus. J Bacteriol 177:4442–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SW, Hwang EH, Jang HS, Lee JH, Kang BS, Oh JI, Kim YM. 2009. Presence of duplicate genes encoding a phylogenetically new subgroup of form I ribulose 1,5-bisphosphate carboxylase/oxygenase in Mycobacterium sp. strain JC1 DSM 3803. Res Microbiol 160:159–165. doi: 10.1016/j.resmic.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Kalyuzhnaya MG, Lidstrom ME. 2003. QscR, a LysR-type transcriptional regulator and CbbR homolog, is involved in regulation of the serine cycle genes in Methylobacterium extorquens AM1. J Bacteriol 185:1229–1235. doi: 10.1128/JB.185.4.1229-1235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalyuzhnaya MG, Lidstrom ME. 2005. QscR-mediated transcriptional activation of serine cycle genes in Methylobacterium extorquens AM1. J Bacteriol 187:7511–7517. doi: 10.1128/JB.187.21.7511-7517.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S-I. 2001. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol 183:1891–1898. doi: 10.1128/JB.183.6.1891-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodger FJ, Badger MR, Price GD. 2003. Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol 133:2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi Y, Yamaguchi O, Omata T. 2004. Roles of CmpR, a LysR family transcriptional regulator, in acclimation of the cyanobacterium Synechococcus sp. strain PCC 7942 to low-CO2 and high-light conditions. Mol Microbiol 52:837–845. doi: 10.1111/j.1365-2958.2004.04021.x. [DOI] [PubMed] [Google Scholar]
- 45.Nishimura T, Takahashi Y, Yamaguchi O, Suzuki H, Maeda S-I, Omata T. 2008. Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC7942. Mol Microbiol 68:98–109. doi: 10.1111/j.1365-2958.2008.06137.x. [DOI] [PubMed] [Google Scholar]
- 46.López-Igual R, Picossi S, Lopéz-Garrido J, Flores E, Herrero A. 2012. N and C control of ABC-type bicarbonate transporter Cmp and its LysR-type transcriptional regulator CmpR in a heterocyst-forming cyanobacterium, Anabaena sp. Environ Microbiol 14:1035–1048. doi: 10.1111/j.1462-2920.2011.02683.x. [DOI] [PubMed] [Google Scholar]
- 47.Rae BD, Long BM, Badger MR, Price GD. 2013. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev 77:357–379. doi: 10.1128/MMBR.00061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodger FJ, Bryant DA, Price GD. 2007. Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp. strain PCC 7002: role of NdhR/CcmR. J Bacteriol 189:3335–3347. doi: 10.1128/JB.01745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daley SME, Kappell AD, Carrick MJ, Burnap RL. 2012. Regulation of the cyanobacterial CO2-concentrating mechanism involves internal sensing of NADP+ and α-ketoglutarate levels by transcription factor CcmR. PLoS One 7:e41286. doi: 10.1371/journal.pone.0041286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tao K, Fujita N, Ishihama A. 1993. Involvement of the RNA polymerase alpha subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol Microbiol 7:859–864. doi: 10.1111/j.1365-2958.1993.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 51.McFall SM, Chugani SA, Chakrabarty AM. 1998. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene 223:257–267. doi: 10.1016/S0378-1119(98)00366-7. [DOI] [PubMed] [Google Scholar]
- 52.Park W, Jeon CO, Madsen EL. 2002. Interaction of NahR, a LysR-type transcriptional regulator, with the alpha subunit of RNA polymerase in the naphthalene degrading bacterium, Pseudomonas putida NCIB 9816-4. FEMS Microbiol Lett 213:159–165. doi: 10.1111/j.1574-6968.2002.tb11300.x. [DOI] [PubMed] [Google Scholar]
- 53.Deghmane AE, Giorgini D, Maigre L, Taha MK. 2004. Analysis in vitro and in vivo of the transcriptional regulator CrgA of Neisseria meningitidis upon contact with target cells. Mol Microbiol 53:917–927. doi: 10.1111/j.1365-2958.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 54.Lochowska A, Iwanicka-Nowicka R, Zaim J, Witkowska-Zimny M, Bolewska K, Hryniewicz MM. 2004. Identification of activating region (AR) of Escherichia coli LysR-type transcription factor CysB and CysB contact site on RNA polymerase alpha subunit at the cysP promoter. Mol Microbiol 53:791–806. doi: 10.1111/j.1365-2958.2004.04161.x. [DOI] [PubMed] [Google Scholar]
- 55.Stauffer LT, Stauffer GV. 2005. GcvA interacts with both the α and σ subunits of RNA polymerase to activate the Escherichia coli gcvB gene and the gcvTHP operon. FEMS Microbiol Lett 242:333–338. doi: 10.1016/j.femsle.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 56.Ghrist A, Heil G, Stauffer GV. 2001. GcvR interacts with GcvA to inhibit activation of the Escherichia coli glycine cleavage operon. Microbiology 147:2215–2221. doi: 10.1099/00221287-147-8-2215. [DOI] [PubMed] [Google Scholar]
- 57.Kovacikova G, Lin W, Skorupski K. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol Microbiol 53:129–142. doi: 10.1111/j.1365-2958.2004.04121.x. [DOI] [PubMed] [Google Scholar]
- 58.Dangel AW, Tabita FR. 2009. Protein-protein interactions between CbbR and RegA (PrrA), transcriptional regulators of the cbb operons of Rhodobacter sphaeroides. Mol Microbiol 71:717–729. doi: 10.1111/j.1365-2958.2008.06558.x. [DOI] [PubMed] [Google Scholar]
- 59.Dangel AW, Luther A, Tabita FR. 2014. Amino acid residues of RegA important for interactions with the CbbR-DNA complex of Rhodobacter sphaeroides. J Bacteriol 196:3179–3190. doi: 10.1128/JB.01842-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joshi GS, Bobst CE, Tabita FR. 2011. Unravelling the regulatory twist—regulation of CO2 fixation in Rhodopseudomonas palustris CGA010 mediated by atypical response regulators. Mol Microbiol 80:756–771. doi: 10.1111/j.1365-2958.2011.07606.x. [DOI] [PubMed] [Google Scholar]
- 61.Joshi GS, Zianni M, Bobst CE, Tabita FR. 2013. Regulatory twist and synergistic role of metabolic coinducer- and response regulator-mediated CbbR-cbbI interactions in Rhodopseudomonas palustris CGA010. J Bacteriol 195:1381–1388. doi: 10.1128/JB.02060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibson JL, Dubbs JM, Tabita FR. 2002. Differential expression of the CO2 fixation operons of Rhodobacter sphaeroides by the Prr/RegA two-component system during chemoautotrophic growth. J Bacteriol 184:6654–6664. doi: 10.1128/JB.184.23.6654-6664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joshi HM, Tabita FR. 1996. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci U S A 93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elsen S, Swem LR, Swem DL, Bauer CE. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev 68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dubbs JM, Bird TH, Bauer CE, Tabita FR. 2000. Interaction of CbbR and RegA* transcription regulators with the Rhodobacter sphaeroides cbbI promoter-operator region. J Biol Chem 275:19224–19230. doi: 10.1074/jbc.M002125200. [DOI] [PubMed] [Google Scholar]
- 66.Ranson-Olson B, Zeilstra-Ryalls JH. 2008. Regulation of the Rhodobacter sphaeroides 2.4.1 hemA gene by PrrA and FnrL. J Bacteriol 190:6769–6778. doi: 10.1128/JB.00828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres y Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 68.Romagnoli S, Tabita FR. 2007. Phosphotransfer reactions of the CbbRRS three-protein two-component system from Rhodopseudomonas palustris CGA010 appear to be controlled by an internal molecular switch on the sensor kinase. J Bacteriol 189:325–335. doi: 10.1128/JB.01326-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joshi GS, Zianni M, Bobst CE, Tabita FR. 2012. Further unravelling the regulatory twist by elucidating metabolic coinducer-mediated CbbR-cbbI promoter interactions in Rhodopseudomonas palustris CGA010. J Bacteriol 194:1350–1360. doi: 10.1128/JB.06418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burn JE, Hamilton WD, Wooten JC, Johnston AW. 1989. Single and multiple mutations affecting properties of the regulatory gene nodD of Rhizobium. Mol Microbiol 3:1567–1577. doi: 10.1111/j.1365-2958.1989.tb00142.x. [DOI] [PubMed] [Google Scholar]
- 71.Cebolla A, Sousa C, de Lorenzo V. 1997. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem 272:3986–3992. doi: 10.1074/jbc.272.7.3986. [DOI] [PubMed] [Google Scholar]
- 72.Jørgensen C, Dandanell G. 1999. Isolation and characterization of mutations in the Escherichia coli regulatory protein XapR. J Bacteriol 181:4397–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tyrrell R, Verschueren KH, Dodson EJ, Murshudov GN, Addy C, Wilkinson AJ. 1997. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure 5:1017–1032. doi: 10.1016/S0969-2126(97)00254-2. [DOI] [PubMed] [Google Scholar]
- 74.Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu S. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105:103–113. doi: 10.1016/S0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 75.Smirnova IA, Dian C, Leonard GA, McSweeney S, Birse D, Brzezinski P. 2004. Development of a bacterial biosensor for nitrotoluenes: the crystal structure of the transcriptional regulator DntR. J Mol Biol 340:405–418. doi: 10.1016/j.jmb.2004.04.071. [DOI] [PubMed] [Google Scholar]
- 76.Dangel AW, Gibson JL, Janssen AP, Tabita FR. 2005. Residues that influence in vivo and in vitro CbbR function in Rhodobacter sphaeroides and identification of a specific region critical for co-inducer recognition. Mol Microbiol 57:1397–1414. doi: 10.1111/j.1365-2958.2005.04783.x. [DOI] [PubMed] [Google Scholar]
- 77.Stec E, Witkowska-Zimny M, Hryniewicz MM, Neumann P, Wilkinson AJ, Brzozowski AM, Verma CS, Zaim J, Wysocki S, Bujacz GD. 2006. Structural basis of the sulphate starvation response in E. coli: crystal structure and mutational analysis of the cofactor-binding domain of the Cbl transcriptional regulator. J Mol Biol 364:309–322. doi: 10.1016/j.jmb.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 78.Ezezika OC, Haddad S, Clark TJ, Neidle EL, Momany C. 2007. Distinct effector-binding sites enable synergistic transcriptional activation by BenM, a LysR-type regulator. J Mol Biol 367:616–629. doi: 10.1016/j.jmb.2006.09.090. [DOI] [PubMed] [Google Scholar]
- 79.Balcewich MD, Reeve TM, Orlikow EA, Donald LJ, Vocadlo DJ, Mark BL. 2010. Crystal structure of the AmpR effector binding domain provides insight into the molecular regulation of inducible AmpC β-lactamase. J Mol Biol 400:998–1010. doi: 10.1016/j.jmb.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 80.Hayes RP, Moural TW, Lewis KM, Onofrei D, Xun L, Kang C. 2014. Structures of the inducer-binding domain of pentachlorophenol-degrading gene regulator PcpR from Sphingobium chlorophenolicum. Int J Mol Sci 15:20736–20752. doi: 10.3390/ijms151120736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vadlamani G, Thomas MD, Patel TR, Donald LJ, Reeve TM, Stetefeld J, Standing KG, Vocadlo DJ, Mark BL. 2015. The β-lactamase gene regulator AmpR is a tetramer that recognizes and binds the d-Ala-d-Ala motif of its repressor UDP-N-acetylmuramic acid (MurNAc)-pentapeptide. J Biol Chem 290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grzeszik C, Jeffke T, Schaferjohann J, Kusian B, Bowien B. 2000. Phosphoenolpyruvate is a signal metabolite in transcriptional control of the cbb CO2 fixation operons in Ralstonia eutropha. J Mol Microbiol Biotechnol 2:311–320. [PubMed] [Google Scholar]
- 83.van Keulen G, Girbal L, van den Bergh ERE, Dijkhuizen L, Meijer WG. 1998. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J Bacteriol 180:1411–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith SA, Tabita FR. 2002. Up-regulated expression of the cbbI and cbbII operons during photoheterotrophic growth of a ribulose 1,5-bisphosphate carboxylase-oxygenase deletion mutant of Rhodobacter sphaeroides. J Bacteriol 184:6721–6724. doi: 10.1128/JB.184.23.6721-6724.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dubbs P, Dubbs JM, Tabita FR. 2004. Effector-mediated interaction of CbbRI and CbbRII regulators with target sequences in Rhodobacter capsulatus. J Bacteriol 186:8026–8035. doi: 10.1128/JB.186.23.8026-8035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dangel AW, Tabita FR. 9 July 2015, posting date Amino acid substitutions in the transcriptional regulator CbbR lead to constitutively active CbbR proteins that elevate expression of the cbb CO2 fixation operons in Ralstonia eutropha (Cupriavidus necator) and identify regions of CbbR necessary for gene activation. Microbiology doi: 10.1099/mic.0.000131. [DOI] [PubMed] [Google Scholar]
- 87.van Keulen G, Ridder ANJA, Dijkhuizen L, Meijer WG. 2003. Analysis of DNA binding and transcriptional activation by the LysR-type transcriptional regulator CbbR of Xanthobacter flavus. J Bacteriol 185:1245–1252. doi: 10.1128/JB.185.4.1245-1252.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lochowska A, Iwanicka-Nowicka R, Plochocka D, Hryniewicz MM. 2001. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J Biol Chem 276:2098–2107. [DOI] [PubMed] [Google Scholar]
- 89.Akakura R, Winans SC. 2002. Constitutive mutations of the OccR regulatory protein affect DNA bending in response to metabolites released from plant tumors. J Biol Chem 277:5866–5874. doi: 10.1074/jbc.M110555200. [DOI] [PubMed] [Google Scholar]
- 90.Bartowsky E, Normark S. 1991. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC β-lactamase. Mol Microbiol 5:1715–1725. doi: 10.1111/j.1365-2958.1991.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 91.Cho K, Winans SC. 1993. Altered-function mutations in the Agrobacterium tumefaciens OccR protein and in an OccR-regulated promoter. J Bacteriol 175:7715–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kullik I, Toledano MB, Tartaglia LA, Storz G. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol 177:1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Belitsky BR, Sonenshein AL. 1997. Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J Bacteriol 179:1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bykowski T, van der Ploeg JR, Iwanicka-Nowicka R, Hryniewicz MM. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol Microbiol 43:1347–1358. doi: 10.1046/j.1365-2958.2002.02846.x. [DOI] [PubMed] [Google Scholar]
- 95.Hou B, Li F, Yang X, Hong G. 2009. A small functional intramolecular region of NodD was identified by mutation. Acta Biochim Biophys Sin 41:822–830. doi: 10.1093/abbs/gmp073. [DOI] [PubMed] [Google Scholar]
- 96.Colyer TE, Kredich NM. 1994. Residue threonine-149 of the Salmonella typhimurium CysB transcription activator: mutations causing constitutive expression of positively regulated genes of the cysteine regulon. Mol Microbiol 13:797–805. doi: 10.1111/j.1365-2958.1994.tb00472.x. [DOI] [PubMed] [Google Scholar]