The plasma membrane targeting of Lgl, a key polarity and tumor suppressor protein, is mediated by electrostatic interactions between a polybasic motif in Lgl and phospholipids on the plasma membrane, and this mechanism is regulated by hypoxia and aPKC-phosphorylation.
Abstract
Lethal giant larvae (Lgl) plays essential and conserved functions in regulating both cell polarity and tumorigenesis in Drosophila melanogaster and vertebrates. It is well recognized that plasma membrane (PM) or cell cortex localization is crucial for Lgl function in vivo, but its membrane-targeting mechanisms remain poorly understood. Here, we discovered that hypoxia acutely and reversibly inhibits Lgl PM targeting through a posttranslational mechanism that is independent of the well-characterized atypical protein kinase C (aPKC) or Aurora kinase–mediated phosphorylations. Instead, we identified an evolutionarily conserved polybasic (PB) domain that targets Lgl to the PM via electrostatic binding to membrane phosphatidylinositol phosphates. Such PB domain–mediated PM targeting is inhibited by hypoxia, which reduces inositol phospholipid levels on the PM through adenosine triphosphate depletion. Moreover, Lgl PB domain contains all the identified phosphorylation sites of aPKC and Aurora kinases, providing a molecular mechanism by which phosphorylations neutralize the positive charges on the PB domain to inhibit Lgl PM targeting.
Introduction
Lethal giant larvae (Lgl) was one of the first genetically identified tumor suppressors in Drosophila melanogaster, as tissues in lgl mutant larvae show neoplastic transformation with overproliferative and metastatic phenotypes. Lgl was later also identified as a key regulator of cell polarity and asymmetric cell divisions (Wirtz-Peitz and Knoblich, 2006; Froldi et al., 2008). Lgl is well conserved structurally and functionally between Drosophila and vertebrates (Froldi et al., 2008). Although the molecular mechanisms underlying the biological activity of Lgl remain to be fully characterized, plasma membrane (PM) or cortical localization of Lgl appears to be essential. First, a temperature-sensitive Lgl (Lglts) becomes cytosolic at the restrictive temperature (Manfruelli et al., 1996). Second, loss of PM targeting of hLgl has been seen in multiple human cancer cells (Schimanski et al., 2005; Lisovsky et al., 2009). Finally, phosphorylation by atypical PKC (aPKC) inhibits Lgl membrane targeting and is crucial for regulating Lgl functions in cell polarization, asymmetric cell division, and cell migration (Betschinger et al., 2003, 2005; Plant et al., 2003). However, molecular mechanisms mediating the PM targeting of Lgl remain to be elucidated. It has been proposed that unphosphorylated Lgl is in an open and active conformation capable of binding cortical actin network to target Lgl to PM, whereas phosphorylation on the conserved serine residues by aPKC induces a self-folded and inactive conformation that disassociates Lgl from PM (Betschinger et al., 2005). Nonetheless, direct genetic and biochemical evidence supporting this model have not been demonstrated to date.
We recently generated a functional lgl::GFP knock-in allele in Drosophila using an established genomic engineering approach (Huang et al., 2009). Live imaging experiments with Lgl::GFP revealed unexpectedly that hypoxia acutely and reversibly inhibits the PM targeting of Lgl in epithelial cells. Loss of Lgl PM targeting under hypoxia is controlled by a posttranslational mechanism that is independent of phosphorylation or the cortical actin network. Led by such observations, we identified that in both Drosophila and mammalian cells, the PM targeting of Lgl is mediated by electrostatic interactions between a positively charged polybasic (PB) domain in Lgl and the negatively charged inositol phospholipids in the PM. Such PB domain–mediated PM binding provides a direct and efficient molecular mechanism for regulating Lgl PM targeting and function by phosphorylations, which neutralize the positive charges of PB domain, and by hypoxia and intracellular ATP depletion, which reduce the levels of PM inositol phospholipids.
Results
Acute and reversible regulation of Lgl PM targeting by hypoxia in Drosophila epithelial cells
Homozygous lgl::GFP knock-in flies are viable, healthy, and fertile, indicating that the Lgl::GFP fusion protein expressed under its endogenous promoter fully substitutes the wild-type Lgl throughout development (Huang et al., 2009). Under normal conditions, Lgl::GFP in embryonic epithelia predominantly localizes to basolateral PM/cell cortex (Fig. 1 A). However, we fortuitously discovered that Lgl::GFP in embryos became cytosolic under hypoxic conditions. Using an environmentally controlled micro-imaging chamber, we confirmed that under hypoxic (0.5 or 1% O2) conditions, Lgl::GFP completely diffused from PM to the cytosol within 30–60 min (Fig. 1 A). Surprisingly, once normoxia was resumed, Lgl::GFP rapidly and completely relocalized back to PM within minutes (Fig. 1 A). Lgl diffusion under hypoxia is also seen in larval, pupal, and adult epithelial tissues, and we developed protocols for robust ex vivo hypoxia assays in follicular epithelia from dissected adult female ovaries (Fig. 1 B and Videos 1 and 2). We confirmed that loss of Lgl PM targeting is not caused by disruption of cell polarity or the cortical actin network under hypoxia, as subcellular localization of several key cell polarity proteins including DE-Cad::GFP (Huang et al., 2009), β-catenin::GFP (Clyne et al., 2003; Huang et al., 2011b), Bazooka(dPar-3)::GFP, and Dlg::GFP (Kelso et al., 2004; Buszczak et al., 2007) remain unchanged under hypoxia (Fig. S1, A–C). MoeAD::GFP (Bloor and Kiehart, 2002) and Sqh::GFP (Royou et al., 2004), two well-established markers for the actomyosin network, also remain unchanged under hypoxia (Fig. 1 C). Loss of PM-localized Lgl under hypoxia is not caused by the fusion of GFP moiety either, as endogenous Lgl showed similar diffusion from PM under hypoxia (Fig. 1 D). Our data revealed for the first time that hypoxia acutely and reversibly inhibits the potential PM targeting of Lgl protein in vivo.
Figure 1.
Acute and reversible subcellular relocalization of Lgl::GFP under hypoxia. (A) Lgl::GFP localization in late embryonic epithelial cells under normoxia (i.e., 21% O2), 60 min of 0.5% O2 hypoxia, and 10 min of post-hypoxia reoxygenation with air. (B) Lgl::GFP subcellular localization in adult female ovary follicular cells under normoxia, 35 min of 0.5% O2 hypoxia, and 2 min of post-hypoxia reoxygenation with air. Histone::RFP (i.e., His2Av::RFP; red) indicates nuclear DNA. (C) Actin cytoskeleton (labeled by MoeAD::GFP) and cortical myosin network (labeled by Sqh::GFP) do not show obvious disruptions under hypoxia. In hypoxia imaging assays, MoeAD::GFP or sqh::GFP embryos were mixed with lgl::GFP his2Av::RFP embryos, which were used to confirm the effective hypoxic exposures (n = 8). (D) Hypoxia-treated lgl::GFP or wild-type embryos were immunostained by anti-GFP or anti-Lgl antibody together with anti-Baz antibody, with normoxia-treated embryos as controls (n = 8). (E) A representative FRAP sample of Lgl::GFP in late-stage (stage 15) embryo going through hypoxia and reoxygenation. Image is in inverted grayscale for better presentation. (F) FRAP results of Lgl::GFP in embryos treated with hypoxia/reoxygenation (n = 6) and in embryos staying in normoxia (n = 8). *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant. Bars, 5 µm. Error bars represent means ± SEM.
Hypoxia inhibits Lgl PM targeting via a posttranslational mechanism that is modulated by HIF pathway but is independent of phosphorylations
Hypoxia regulates the nuclear relocalization of hypoxia-inducible factor 1α (HIF1α) through inhibiting its protein degradation induced by prolyl hydroxylase domain (PHD) proteins (Kaelin and Ratcliffe, 2008). To investigate whether loss of Lgl PM targeting under hypoxia also involves Lgl protein degradation and synthesis, we performed whole-cell FRAP assays (Huang et al., 2011b) by photobleaching the Lgl::GFP in a patch of epithelium so that Lgl::GFP can only be recovered through de novo protein synthesis. As shown in Fig. 1 (E and F), unlike unbleached cells, photobleached cells never recovered their Lgl::GFP under hypoxia and subsequent reoxygenation. Whole-cell FRAP assays in fact revealed that Lgl::GFP has an extremely slow turnover rate even under normoxia (Fig. 1 F). Thus, loss of PM-targeted Lgl::GFP under hypoxia involves no detectable Lgl::GFP synthesis and degradation, suggesting that hypoxia directly inhibits the PM targeting of Lgl. We then tested whether on the other hand HIF1α is required for inhibiting the PM targeting of Lgl::GFP using Drosophila HIF1α (namely, sima or dHIFα) or PHD (fatiga or hph) mutants (Lavista-Llanos et al., 2002; Centanin et al., 2005). Inhibition of PM Lgl::GFP by hypoxia is unchanged in sima or fatiga sima double mutant embryos (Fig. 2 A) but is delayed in Sima-overexpressing follicular cells (Fig. S2 A) or in fatiga mutants that have high Sima levels as a result of loss of Fatiga (Fig. 2 A; Lavista-Llanos et al., 2002). Our data suggest that high levels of Sima proteins promote rather than inhibit the PM targeting of Lgl under hypoxia (see below).
Figure 2.
Inhibition of Lgl PM targeting by hypoxia is modulated by HIF pathway but is independent of phosphorylation events. (A) Lgl::GFP showed normal subcellular relocalization in sima mutant embryos under hypoxia and reoxygenation (air). In fatiga mutant embryos, Lgl::GFP showed no significant diffusion at 60 min of hypoxia treatment, and only become diffused at 120 min of hypoxia. Lgl::GFP showed complete diffusion in fatiga sima double mutant embryos at 60 min of hypoxia and recovered normally during reoxygenation (n = 6). (B) Lgl::GFP show normal subcellular redistribution under hypoxia in either par-6 or aPKC mutant follicular epithelial cell clones (n = 4). Mutant cells of par-6 are marked by the absence of nuclear GFP, whereas aPKC mutant cells are marked by the absence of nuclear His2Av::RFP. (C) PM targeting of LglS5A::GFP is acutely and reversibly inhibited under hypoxia in follicular epithelial cells. Loss of RFP marks lgS5A::GFP clones, whereas wild-type lgl::GFP twin clones are labeled by increased expression of RFP (indicated by white asterisks). **, P < 0.01; ***, P < 0.001. Bars, 5 µm. Error bars represent means ± SEM.
Surprisingly, hypoxia readily inhibits Lgl::GFP PM targeting in either par-6 or aPKC mutant follicular cell clones (Fig. 2 B and Fig. S2 B). We also generated a lglS5A::GFP knock-in allele in which all five conserved serines phosphorylatable by aPKC or Aurora kinases (see Fig. 5 A; Bell et al., 2015; Carvalho et al., 2015) have been mutated to alanines. PM localization of LglS5A::GFP showed acute and reversible inhibition by hypoxia identical to Lgl::GFP (Fig. 2 C). In addition, although aPKC also shows loss of PM localization under hypoxia (Fig. S2 C), overexpression of membrane-bound aPKCCAAX (Sotillos et al., 2004) did not rescue the loss of Lgl PM targeting under hypoxia (Fig. S2 D). Thus, we conclude that hypoxia directly inhibits Lgl PM targeting through a posttranslational mechanism that is modulated by Sima protein levels but is independent of phosphorylation events by aPKC/Aurora kinases or physical interactions between Lgl and aPKC.
Figure 5.
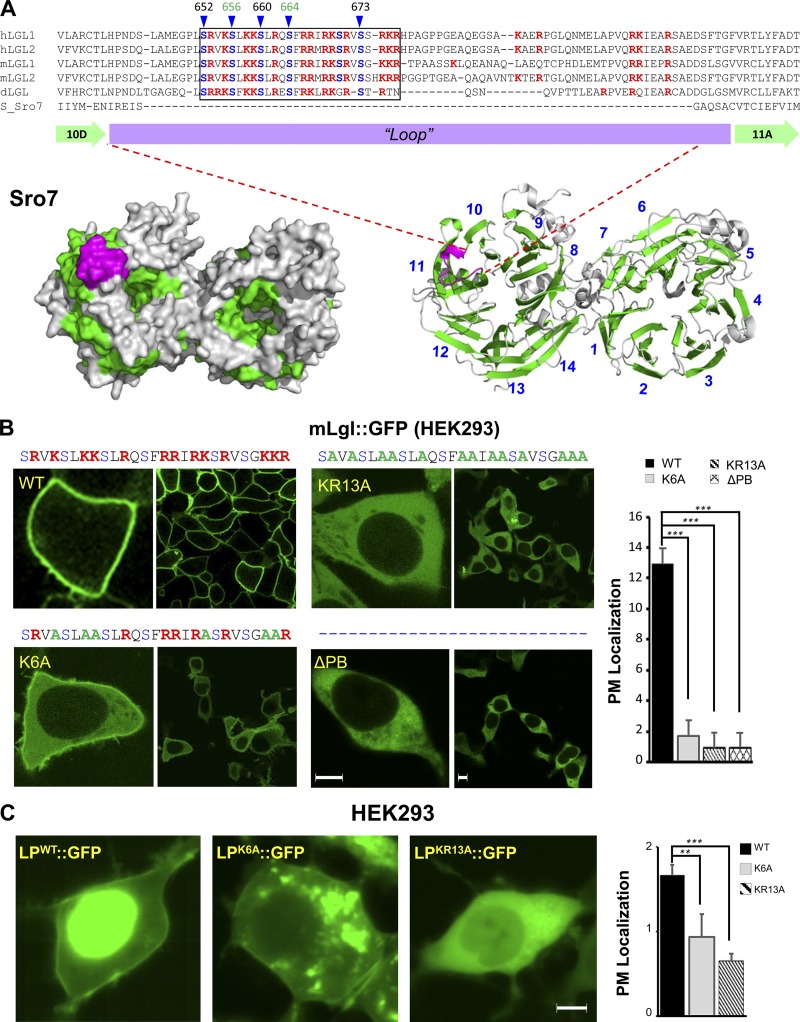
An evolutionarily conserved PB domain in Lgl mediates its PM targeting. (A) Lgl contains a conserved PB domain. (Bottom) Surface and cartoon models of Sro7 structure (Protein Data Bank accession no. 2OAJ; Hattendorf et al., 2007) rendered by PyMOL software. β sheets are in green to highlight the WD40 blades, of which each contains four β sheets. The “loop” in between WD40 blade 10 and 11 is in magenta. (Top) Alignment of the loop regions between 10D and 11A β sheets in Lgl or Sro7. Positively charged Lys and Arg residues within the loop sequences are in red, and PB domains are boxed. Blue arrowheads point to the conserved Ser residues (in blue) that can be phosphorylated by aPKC (Ser residues in green numbers are also Aurora kinase phosphorylation sites). Ser residues were numbered based on Drosophila Lgl (dLgl) isoform A (gi24464586). hLgl1/2, human Lgl1/2 (gi 62912476 and 40674459); mLgl1/2, mouse Lgl1/2 (gi 56800067 and 21703874); S_Sro7, Saccharomyces cerevisiae Sro7 (gi 6325289). Sequence alignments are modified from Hattendorf et al. (2007). Complete sequence alignments can be found in Fig. S2 by Hattendorf et al. (2007). (B) Subcellular localization of mLgl::GFP (WT), mLglΔPB::GFP (ΔPB), mLglK6A::GFP (K6A; all six lysines mutated to alanine), and mLglKR13A::GFP (KR13A; all 13 lysines and arginines mutated to alanine) in HEK293 cells (n = 6). (C) GFP fused with wild-type loop of mLgl (LoopWT::GFP), but not loops carrying K6A or KR13A mutations (LoopK6A::GFP and LoopKR13A::GFP, respectively), targets to the PM in HEK293 cells. LoopWT::GFP is also nuclear (n = 6 for each sample). **, P < 0.01; ***, P < 0.001. Bars, 5 µm. Error bars represent means ± SEM.
Reduction of intracellular ATP is sufficient to inhibit PM targeting of Lgl in both Drosophila and mammalian cells
Hypoxia acutely and reversibly inhibits ATP production in Drosophila embryos, and the time course of intracellular ATP level reduction and recovery (DiGregorio et al., 2001) is very similar to that of Lgl::GFP (Fig. 1 F). Indeed, both Lgl::GFP and endogenous Lgl diffused from PM to cytosol in embryos and female ovaries treated with cyanide and antimycin A (AM), which inhibit ATP production (Fig. 3, A and B; and Fig. S3 A; DiGregorio et al., 2001), and we confirmed that hypoxia and cyanide treatments all reduced intracellular ATP levels in embryos (Fig. S3 B). Thus, reduction of intracellular ATP levels appears to mediate the hypoxia-induced inhibition of Lgl PM targeting. In addition, live imaging using a custom micro-imaging chamber showed that mouse Lgl::GFP (mLgl::GFP) localized strongly to the PM under normoxia in both HEK293 and Madin-Darby canine kidney (MDCK) cells, diffused into cytosol after one to several hours of hypoxic exposure and quickly retargeted to PM within 30 min of reoxygenation (Fig. 3 C and Videos 3 and 4). Blocking ATP production in HEK293 cells by 2-deoxyglucose (2-DG) and AM caused loss of mLgl::RFP PM targeting that was reversed by drug removal, whereas a palmitoylation-based membrane marker GAP43::GFP (Okada et al., 1999) remained on the PM (Fig. 3 D). Finally, endogenous hLgl-2 was also lost from PM in hypoxia-treated MCF-7 cells (Fig. S3 C). Thus, acute and reversible inhibition of Lgl PM targeting by hypoxia and intracellular ATP reduction is conserved in both Drosophila and mammalian cells. We speculate that high Sima protein levels likely have made ATP production in fatiga(dPHD) mutant embryos or Sima-overexpression follicular cells more resistant to hypoxia (Aragonés et al., 2008), resulting in delayed loss of Lgl::GFP from PM.
Figure 3.
Hypoxia and reduction of intracellular ATP levels inhibit the PM targeting of Lgl in both Drosophila and mammalian cells. (A) lgl::GFP embryos were treated with PBS (control) or 0.5% cyanide (CN). Embryos were double labeled by anti-Baz (red) and anti-GFP or anti-HA (green) antibodies (n = 6). (B) Dissected ovaries from lgl::GFP females that were treated with 1 µM AM or 0.5% cyanide (n = 6). PBS and 1% DMSO served as controls. (C) Inhibition of mLgl::GFP PM targeting in hypoxia-treated MDCK or HEK293 cells (n = 4). (D) Inhibition of ATP production by 2-DG and AM in HEK293 cells induced loss of mLgl::RFP from PM that can be reversed when drugs were washed out. PM localization of GAP::GFP is unaffected (n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant. Bars, 5 µm. Error bars represent means ± SEM.
An evolutionarily conserved PB domain is essential for Lgl PM targeting
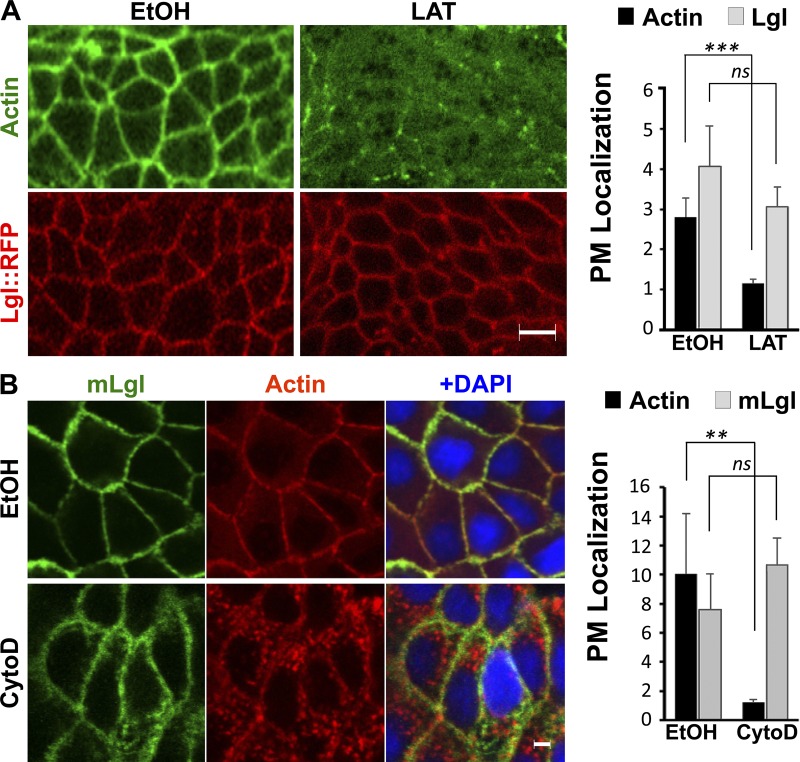
Given that hypoxia inhibits Lgl PM targeting without disrupting the cortical actin network (Fig. 1 C), we decided to determine whether the cortical actin network is indeed required for Lgl PM targeting as reported previously (Betschinger et al., 2005). We repeated the latrunculin treatment used by Betschinger et al. (2005) to disrupt F-actin in Drosophila embryos. Despite severe disruption of cortical F-actin indicated by the loss of MoeAD::GFP (Fig. 4 A) or phalloidin staining (Fig. S3 D), Lgl (shown by immunostaining; Fig. S3 D), Lgl::GFP (Fig. S3 D), and Lgl::mCherry (shown by live imaging; Fig. 4 A) remain localized on the PM in treated embryos. We also depolymerized F-actin in MDCK cells by cytochalasin D, and mLgl::GFP still remained on PM (Fig. 4 B). Our data strongly suggest that PM targeting of Lgl does not depend on an intact cortical actin network.
Figure 4.
Lgl membrane targeting does not require intact actin cytoskeleton. (A) Drosophila embryos of lgl::RFP MoeAD::GFP were treated with 10 µM latrunculin-A (LAT) with 0.5% ethanol (EtOH) as control. Each pair of control and treated samples was captured under identical imaging parameters and processed identically. Actin was visualized by MoeAD::GFP (n = 4). (B) MDCK cells were treated with 25 µg/ml cytochalasin D (CytoD), with 0.5% ethanol as control. Actin was visualized by TRITC-phalloidin staining (n = 4). Each pair of control and treated samples was captured under identical imaging parameters and processed identically. **, P < 0.01; ***, P < 0.001. ns, not significant. Bars, 5 µm. Error bars represent means ± SEM.
What is the cortical actin–independent mechanism that targets Lgl to the PM? We noticed that the sequence containing the conserved aPKC-phosphorylatable serines is in fact a typical PB domain in which half of the amino acids are either Arg or Lys residues (Fig. 5 A). It has been well demonstrated that PB domains can mediate PM-specific targeting through electrostatic interactions between their positively charged Arg or Lys residues and the negatively charged phosphatidylinositol phosphates (PIPs) such as phosphatidylinositol 4-phosphate (PI4P) and phosphatidylinositol 4,5-bisphosphate (PIP2), which are uniquely enriched in PM (Heo et al., 2006; Yeung et al., 2008; Hammond et al., 2012). The PB domain in Lgl is absent in the yeast Lgl homologue Sro7 but is highly conserved between mammalian and Drosophila Lgl proteins (Fig. 5 A). In addition, according to Hattendorf et al. (2007), both Sro7 and Lgl proteins form two β propellers that each contains seven WD40 repeats, and the PB domain in Lgl was predicted as part of a loop between the 10th and 11th WD40 blades of the second β propeller. The Sro7 structure also suggests that the Lgl loop is likely accessible for binding PM or proteins like aPKC (Fig. 5 A). Moreover, PB domain in Lgl contains hydrophobic residues such as leucine and phenylalanine (Fig. 5 A), a key feature that has been shown to enhance PB domain’s binding to PM and to prevent the PB domain from acting as a nuclear localization signal (Heo et al., 2006).
Consistently, we found that the mLglΔPB::GFP mutant that deletes the PB domain is completely cytosolic in HEK293 and MDCK cells (Fig. 5 B and Fig. S4 A). We then generated mLglK6A::GFP and mLglKR13A::GFP by replacing either all the Lys or all the Lys plus Arg residues in the PB domain with alanines, respectively. mLglK6A::GFP showed dramatically reduced PM targeting, whereas mLglKR13A::GFP is completely cytosolic (Fig. 5 B and Fig. S4 A). GFP fused with the wild-type loop of mLgl, but not loops carrying K6A or KR13A mutations, also showed PM targeting in HEK293 cells (Fig. 5 C). To confirm that this PB domain–targeting mechanism is also conserved in Drosophila, we made similar knock-in mutants lglΔPB::GFP and lglKR12A::GFP and, as expected, both mutant proteins showed drastically reduced PM targeting in mutant follicular cell clones (Fig. 6 A). The residual PM localization of LglΔPB::GFP or LglKR12A::GFP is sensitive to hypoxia (Fig. 6 B), suggesting an additional electrostatic interaction–based targeting mechanism likely acting in partial redundancy to the main PB domain in Lgl. Nonetheless, both lglKR12A::GFP and lglΔPB::GFP are homozygous lethal with lgl phenotype, and their follicular cell mutant clones show overproliferation and loss of polarity phenotypes (Fig. 6 C and Fig. S4 C) similar to lglnull clones. Previous studies suggest that Lgl may function as an inhibitive substrate of aPKC (Rolls et al., 2003; Wirtz-Peitz and Knoblich, 2006; Grzeschik et al., 2010); therefore, phenotypes by lglΔPB::GFP and lglKR12A::GFP could be caused by elevated aPKC activity, as LglKR12A and LglΔPB are unlikely able to bind and inhibit aPKC. Nonetheless, phenotypes in lglKR12Amutant cells are not rescued by knocking down aPKC (Fig. S4 D). Collectively, our data indicate that PB domain–mediated PM targeting is specifically required for Lgl functions in vivo.
Figure 6.
Lgl PB domain is essential for its PM targeting and functions in Drosophila. (A) Subcellular localization of LglΔPB::GFP (ΔPB) and LglKR12A::GFP (KR12A; all lysines and arginines mutated to alanine) in Drosophila follicular cells (n = 8 each). Cells homozygous of lglΔPB::GFP or lglKR12A::GFP were labeled by the loss of RFP. (B) Residual PM targeting of LglKR12A::GFP is also reversibly inhibited under hypoxia in follicular epithelial cells. Loss of RFP marks lglKR12A::GFP clones, whereas wild-type lgl::GFP twin clones are labeled by increased expression of RFP (indicated by white asterisks). (C) Follicular cell clones of LglΔPB::GFP (ΔPB) or LglKR12A::GFP (KR12A) show overproliferation and loss of polarity. Polarity protein dPatj (red) localizes to the apical side in wild-type follicular cells but is mislocalized in lglΔPB::GFP or lglKR12A::GFP mutant cells (labeled by the loss of GFP; blue), which also become multilayered. White boxes in the right of most panels highlight the area enlarged in the left panels. ***, P < 0.001. Bars, 5 µm. Error bars represent means ± SEM.
Targeting of Lgl requires PIPs on PM
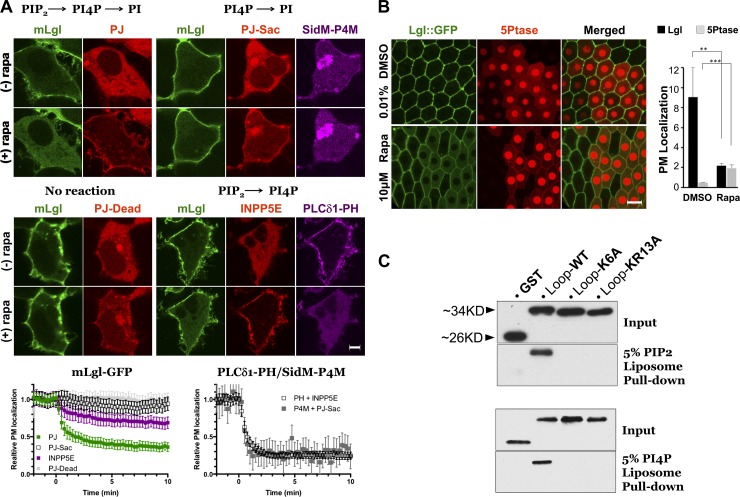
We then investigated whether PIPs that are uniquely enriched on PM such as PI4P and PIP2 are required for targeting Lgl to PM. We first treated cells with ionomycin and phenylarsine oxide (PAO), which have been well characterized for inhibiting the electrostatic binding of polycationic proteins to PM by disrupting the PM phospholipids (Yeung et al., 2008; Hammond et al., 2012). Both treatments diffused PM-localized mLgl::RFP in HEK293 cells and Lgl::GFP in Drosophila follicular cells, as well as PLCδ-PH::GFP marker, which specifically binds to PIP2 (Fig. S5, A and B). Second, we used an inducible heterodimerization system in which the addition of rapamycin recruits to the PM a hybrid phosphatase Pseudojanin (PJ) that acutely depletes both PIP2 and PI4P (Hammond et al., 2012). As shown in Fig. 7 A, rapamycin-triggered PM recruitment of PJ, but not enzymatically dead PJ, acutely and completely diffuses mLgl::GFP from PM in HEK293 cells (Video 5). Depleting PIP2 alone by 5-phosphatases (INPP5E) also significantly diffuses mLgl::GFP from PM, but this is substantially less complete than when combined with PI4P depletion, despite complete PIP2 elimination as indicated by the dissociation of a coexpressed iRFP::PLCδ-PH. Depleting PI4P alone by PJ-Sac phosphatase shows complete dissociation of a specific PI4P biosensor, iRFP::SidM-P4M (Hammond et al., 2014), yet a barely detectable reduction of PM mLgl::GFP (Fig. 4 A). We also performed similar dimerization assays ex vivo in Drosophila follicular cells coexpressing PM-bound anchor protein Lck-FRB::CFP and cytosolic mRFP::FKBP-5Ptase (i.e., INPP5E; Reversi et al., 2014). The addition of rapamycin to the medium also triggered PM recruitment of 5Ptase and concomitant reduction of Lgl::GFP from PM in follicular cells in dissected Drosophila ovaries (Fig. 7 B). Finally, we performed co-sedimentation assays using PIP2- and PI4P-containing liposomes and purified GST fusion proteins with mLgl loop containing either wild-type PB domain or PB domains carrying K6A or KR13A mutations. Wild-type PB domain is capable of binding both liposomes, whereas K6A or KR12A mutants are not (Fig. 7 C). In summary, our data indicate that PM targeting of Lgl is mediated by direct binding between its PB domain and inositol phospholipids PIP2 and PI4P, which are uniquely enriched at the PM. Our data also suggest that, although PIP2 appears to be the primary lipid for targeting Lgl to PM, depletion of PI4P is required for complete inhibition of Lgl PM targeting.
Figure 7.
PM targeting of Lgl requires inositol phospholipids on the PM. (A) HEK293 cells expressing mLgl::GFP or Lyn11-FRB::CFP and RFP::FKBP12-PJ (PJ) to degrade PI4P and PIP2, RFP::FKBP12-PJ-Sac (PJ-Sac) to degrade PI4P, mRFP::FKBP12-INPP5E (INPP5E) to degrade PIP2, or FKBP12-PJ-dead::RFP (PJ-dead) as a control. The addition of rapamycin induced membrane recruitment of the FKBP12-tagged enzymes. Only combined depletion of PIP2 and PI4P causes complete diffusion of mLgl::GFP, whereas complete dissociation of iRFP::PLCδ-PH and iRFP::SidM-P4M indicated complete depletion of PIP2 or PI4P by INPP5E and PJ-Sac, respectively. Graphs are means ± SEM of 11 ≤ n ≤ 19 cells from three independent experiments. (B) In Drosophila follicular cells expressing nuclear RFP, mRFP::FKBP-5Ptase (5Ptase) and Lck-FRB::CFP (not depicted), rapamycin addition induced PM recruitment of 5Ptase and dislocalization of Lgl::GFP from PM. Images were captured at 40 min after rapamycin or DMSO addition (n = 4 for each sample). (C) Liposome pull-down assays of GST and GST fusions of Lgl loop (LoopWT), LoopK6A, or LoopK12A. **, P < 0.01; ***, P < 0.001. Bars, 5 µm. Error bars represent means ± SEM.
Hypoxia and reduction of intracellular ATP also inhibit the PM targeting of phospholipid-binding proteins
PIP2 levels on PM acutely depend on intracellular ATP levels (Poggioli et al., 1983), suggesting that hypoxia inhibits Lgl PM targeting by reducing phospholipid levels on the PM through ATP depletion. Thus, hypoxia or inhibition of ATP production should also disrupt the PM targeting of other proteins that require electrostatic binding to PM inositol phospholipids. Indeed, PIP2 and PIP3 markers PLCδ-PH::GFP and GRP1-PH::GFP (i.e., tGPH; Britton et al., 2002) showed acute and reversible loss of PM targeting under hypoxia (Fig. 8 A) in Drosophila follicular cells. Inhibiting ATP production in HEK293 cells also dislocalizes PLCδ-PH::GFP from PM (Fig. 8 B). Numb, a key protein regulated by asymmetric cell division, also contains a basic domain that has been shown to bind PIP2 in vitro (Dho et al., 1999), and Drosophila Numb::GFP showed acute and reversible loss of PM targeting under hypoxia (Fig. 8 A) similar to Lgl::GFP. In addition, our result that PM localization of aPKC is also sensitive to hypoxia (Fig. S2 C) is consistent with observations that aPKC becomes dislocalized in PIP2-deficient cells (Claret et al., 2014). Thus, hypoxia or reduction of intracellular ATP levels can generally inhibit the PM targeting of proteins that require binding to PM inositol phospholipids.
Figure 8.
Hypoxia and reduction of intracellular ATP levels acutely deplete inositol phospholipids on PM. (A) Hypoxia reversibly inhibits PM targeting of PLCδ-PH::GFP, GRP1-PH::GFP, and Numb::GFP in Drosophila follicular cells (n = 4). (B) Inhibition of ATP production by 2-DG and AM in HEK293 cells induced loss of PLCδ::PH::GFP from PM. ***, P < 0.001. Bars, 5 µm. Error bars represent means ± SEM.
Reducing hLgl levels increases cell survival under hypoxia
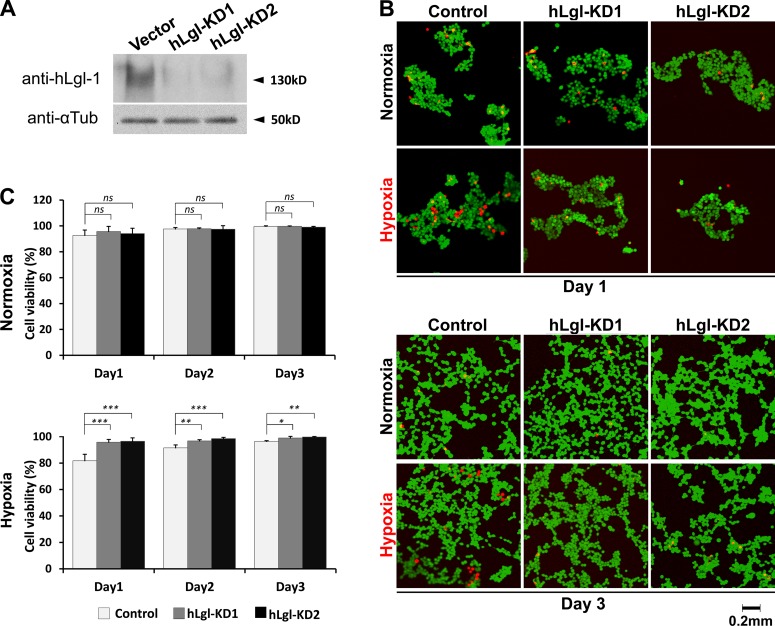
Loss of PM targeting under hypoxia likely inhibits the biological activity of Lgl, suggesting a possibility that reducing Lgl activity may in fact be beneficial for cells to survive hypoxia. We used an established hypoxia-induced cell death assay in HEK293 cells (Chen et al., 2003) to investigate whether reducing Lgl activity by knocking down its protein levels promotes cell survival under hypoxia. We generated two stable HEK293 cell lines, hLgl-KD1 and hLgl-KD2, in which the endogenous human hLgl-1 protein levels were significantly reduced by the expression of shRNAs against hLgl-1 (Fig. 9 A). We cultured these cell lines in 0.5% O2 hypoxic incubator for 3 d, and cell viability was determined daily by calcein AM and ethidium homodimer-1 staining. As shown in Fig. 9 (B and C), compared with the control, shRNA line hLgl-KD1 and hLgl-KD2 showed significantly reduced cell death especially in the first day of hypoxic incubation, suggesting that reducing Lgl activity benefits cell survival under hypoxia.
Figure 9.
Reducing hLgl levels increases cell survival under hypoxia. (A) Protein levels of hLgl-1 are severely reduced in hLgl-KD1 and hLgl-KD2 stable lines expressing hLgl-1 shRNA, but not in the control cells that express scrambled shRNA. αTubulin (αTub) is the loading control. (B) Cell death in control, hLgl-KD1, and hLgl-KD2 cells incubated under normoxia or hypoxia (1% O2/5%CO2), respectively. Live cells are labeled by calcein AM (green), and dead cells are labeled by ethidium homodimer-1 (red). Cell death decreases after day 1 likely because old dead cells were lost by medium washes, which are required before dye staining. (C) Quantification of the cell viability after normoxia or hypoxia exposures in stable HEK293 cell lines expressing hLgl-1 shRNA (hLgl-KD1 and hLgl-KD2) or scrambled shRNA (Control) as shown in B. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant. Bars, 5 µm. Error bars represent means ± SEM.
Discussion
In this report, we identified an evolutionarily conserved phosphorylatable PB domain that directly targets Lgl to the PM (Fig. 10). The PB domain in Lgl contains all the conserved phosphorylation sites of aPKC and Aurora kinases, providing a straightforward molecular mechanism by which phosphorylations on serine residues directly inhibit Lgl PM targeting by neutralizing the positive charges of Arg and Lys in the PB domain. Such a regulatory mechanism makes Lgl remarkably similar to another tumor suppressor, myristoylated alanine-rich C kinase substrate protein (MARCKS), whose PM targeting requires its PB effector domain, which is also a PKC substrate (Arbuzova et al., 2002; Bickeböller et al., 2015). In fact, adding the effector domain of MARCKS rescues the loss of PM targeting of mLglΔPB::GFP (Fig. S4 B). The lack of such PB domain in yeast Sro7 suggests that this is an evolutionarily acquired feature in metazoan Lgl, and our results highlight that phosphorylatable PB domain serves a conserved regulatory mechanism to control the PM targeting of Lgl and MARCKS. Based on our data, the cortical actin network is not required for Lgl PM targeting, and the strong PM targeting of nonphosphorylatable LglS5A suggests that phosphorylation-dependent binding of Lgl to Dlg (Zhu et al., 2014) is unlikely essential for Lgl PM localization either. However, our results emphasize the critical roles of PM PIP2 in regulating cell polarity, as reported recently in Drosophila (Claret et al., 2014).
Figure 10.
Hypoxia and phosphorylations regulate the PB domain–mediated PM targeting of Lgl.
Hypoxia and reduction of intracellular ATP levels reduce the PM inositol phospholipid levels, therefore inhibiting PM targeting of proteins such as Lgl that require binding to PIPs. To our knowledge, Lgl is the first polarity protein identified for such a PB domain-based PM targeting mechanism. Nonetheless, many proteins localize to the PM through electrostatic binding to phospholipids (Heo et al., 2006; Yeung et al., 2008; Hammond et al., 2012), and our data suggest that potential disruption of their PM localization under hypoxia may have profound consequences for cell survival and function. Cancer cells in solid tumors often need to overcome and proliferate in hypoxic environments caused by poor angiogenesis (Keith and Simon, 2007), and inhibiting the electrostatic interaction–mediated PM targeting may act as a potential mechanism for cancer cells to repress the activities of tumor suppressors such as Lgl under hypoxia.
Materials and methods
Fly stocks and genetics
Drosophila stocks: sqh-GFP::MoeAD (“MoeAD::GFP,” which is a fusion between Moesin actin–binding domain and GFP ubiquitously expressed under sqh promoter; provided by D. Kiehart, Duke University, Durham, NC), y w sqhAX; sqh::GFP (ubiquitously expressing Sqh::GFP fusion under sqh promoter in sqhAX background), w par-6Δ226 FRT9-2/FM6 (provided by J. Knoblich, Institute of Molecular Pathology, Vienna, Austria), y; simaKG07607/TM3, Sb Ser (Bloomington Stock Center [BL] no. 14640), fatiga(Hph)02255ry/TM3, Sb Ser (BL no. 11561), y w; aPKCk06403/CyO (BL no. 10622), w; His2Av::mRFP (BL no. 23651), y w Ubi-GFPNLS FRT9-2 (BL no. 5154), w; Act5C(FRT.CD2)-Gal4, UAS-RFP/TM3, Sb (BL no. 30558), w; UAS-Sima (provided by P. Wappner, FCEyN-Universidad de Buenos Aires, Buenos Aires, Argentina), UAS-PLCδ-PH::GFP (BL no. 39693), UAS-GRP1-PH::GFP(tGPH) (BL no. 8163), UAS-Numb::GFP (provided by F. Roegiers, Fox Chase Cancer Center, Philadelphia, PA) and UAS-aPKC-RNAi (BL no. 25946). w UASp>mRFP::FKBP-5Ptase and w; UASp>Lck-FRB::CFP were provided by S. De Renzis (European Molecular Biology Laboratory, Heidelberg, Germany; Reversi et al., 2014). Cy2-Gal4 (driving ubiquitous expression in follicular cells in late-stage ovaries; provided by G. Schupbach, Princeton University, Princeton, NJ). Generation of lgl::GFP knock-in alleles was described previously (Huang et al., 2009). Generation of lgl::mCherry, lglS5A::GFP, lglΔPB::GFP, and lglKR12A::GFP knock-in alleles was performed according to a modified genomic engineering approach (Huang et al., 2011a). Each lgl knock-in allele contains an ∼7-kb genomic DNA of lgl that was modified in pGE-attB vector and inserted back into lgl locus through øC31-mediated DNA integration (Huang et al., 2009). Drosophila cultures and genetic crosses were performed at 25°C. Clonal expression of Numb::GFP was achieved by heat-shocking w hs-FLP/+; UAS-Numb::GFP/Act5C(FRT.CD2)-Gal4, UAS-RFP females to activate Act5C-Gal4. Detailed information about the strains from Bloomington Stock Center can be found in FlyBase.
Live imaging of Lgl::GFP under hypoxia in Drosophila and mammalian cells
Embryos were collected overnight at 25°C and dechorinated before imaging. Ovaries were dissected in halocarbon oil (no. 95) from adult females at the desired age. Embryos or ovaries were mounted in halocarbon oil on a gas-permeable slide that ensures efficient gas exchange between the mounted samples and the environment (Huang et al., 2011b). The slide was then mounted in an air-tight custom-made micro chamber that allows live imaging of samples under confocal microscope. Oxygen levels inside the chamber were controlled by air or custom O2/N2 flowing at ∼1–5 cc/s. Gas flow rates were constantly monitored throughout the imaging session. All custom hypoxic gas mixtures are balanced with N2. Imaging assays were performed at room temperature (25°C) on a confocal microscope (PL APO 40× oil objective; NA = 1.25; TCS-NT; Leica) with Leica software (TCS-NT), a confocal microscope (40× oil objective; NA = 1.3; LSM 510; Carl Zeiss) with Carl Zeiss software, or a confocal microscope (40× Uplan FL N oil objective; NA = 1.3; FV1000; Olympus) with Olympus software (FV10-ASW). HEK293 cells were cultured in glass-bottom dishes (In Vitro Scientific) with a gas-permeable membrane mounted above the cell. An air-tight custom-made micro chamber cap was used to seal the dish for the live confocal imaging assays under hypoxia. Polarized MDCK cells were cultured on 12-mm diameter Transwell filters (Corning). After cells reached confluence, the filter was cut and then mounted with a gas-permeable membrane in a special imaging micro chamber for live imaging under hypoxia at 37°C on a confocal microscope (40× oil objective; NA = 1.3; LSM 510; Carl Zeiss) with Carl Zeiss software or a confocal microscope (Plan Fluo 40× oil objective; NA = 1.3; A1; Nikon) with NIS-Elements AR software. All hypoxic gas mixtures for cultured cell live imaging were balanced with N2 plus 5% CO2. All confocal images were processed in Adobe Photoshop software.
Quantification of Lgl::GFP PM localization and subcellular redistribution
PM localizations were measured in ImageJ (National Institutes of Health [NIH]). For each cell, mean intensities of PM, cytosol, and background (outside the cell) were traced and measured using line and area measurement tools in ImageJ. The PM localization was then calculated as the ratio of (PM − background)/(cytosol − background). To measure Lgl::GFP subcellular redistribution over the hypoxia treatment in Fig. S1 (B and C), we developed a custom ImageJ macro (see supplemental material) that quantifies the PM localization of Lgl::GFP as PM localization index (PI), which is calculated as GFPpx0/GFPpx2, where GFPpx0 and GFPpx2 are the mean GFP intensities at the cell cortex and at two pixels away, respectively. The degree of Lgl::GFP cytosolic diffusion at 60 min of hypoxia treatment is quantified as diffusion index (DI) = (PI0minhypoxia − 1)/(PI60minhypoxia − 1). Conversely, recovery index (RI) of Lgl::GFP at 10 min of post-hypoxia reoxygenation is quantified as RI = (PI10minnormoxia − 1)/(PI60minhypoxia − 1).
Hypoxia and drug treatment of embryos and dissected ovaries
Drug treatments of embryos were performed as described previously (Huang et al., 2011b). In brief, embryos were collected overnight, dechorionated by bleach, and washed. Then embryos were placed into a 1:1 mixture of octane and Schneider’s medium containing drugs. DMSO was used as control. Embryos were shaken at 400 rpm for 30 min and rinsed with octane briefly before imaging. Ovaries from three to five females were dissected in 1× PBS, transferred to a drop of 20 µl Schneider’s medium containing 0.5% cyanide or 1 µM AM. After 30 min, ovaries were washed with Schneider’s medium and imaged live in air-permeable conditions, as described previously (Huang et al., 2011b). In brief, embryos or ovaries were placed on an air-permeable membrane (YSI Membrane Model 5793; YSI Inc.) that is sealed by vacuum grease over a 10 × 10–mm cut-through window on a custom-made plastic slide. After placing on the coverslip, the open membrane ensures the sufficient air exchange to samples during the imaging session.
Generation of mutant clones in follicular epithelia
Mutant follicular cell clones of lglΔPB::GFP, lglKR12A::GFP, aPKCk06403, and par-6Δ226 were generated by the routine FLP/FRT technique. Clones were induced by heat-shocking young females at 37°C for 1 h, and ovaries were dissected 3 d after heat shock.
Immunostaining and confocal imaging
Immunostaining of embryos and adult ovaries were performed as described previously (Huang et al., 2009). Primary antibodies: rabbit anti-GFP (Huang et al., 2009), 1:1,500; chicken anti-GFP (Aves Lab), 1:1,000; rabbit anti-Lgl (d-300; Santa Cruz Biotechnology, Inc.), 1:200; guinea pig against N-terminal 80 aa of Baz (Huang et al., 2009), 1:500; rabbit and rat anti-Par6 (Huang et al., 2009), 1:500; rabbit and mouse anti-dPatj, 1:1,000 (Zhou and Hong, 2012); mouse anti-Dlg (4F3, DSHB), 1:50; rabbit anti-aPKC (Santa Cruz Biotechnology, Inc.), 1:1,000; mouse anti-hLgl-2 (H00003993-M06; Abnova), 1:500; and rabbit anti–E-cadherin (3195S; Cell Signaling Technology), 1:200. Secondary antibodies: Cy2-, Cy3-, or Cy5-conjugated goat anti–rabbit IgG, anti–chicken IgG, goat anti–rat IgG, goat anti–mouse IgG, and goat anti–guinea pig IgG (all at 1:400; Jackson ImmunoResearch Laboratories, Inc.). Images were collected on confocal microscopes (FV1000; Olympus; Center for Biological Imaging, University of Pittsburgh Medical School) and processed in Adobe Photoshop for compositions.
Quantifying intracellular ATP levels in hypoxia- and cyanide-treated embryos
Dechorinated embryos were weighted before hypoxia or cyanide treatments and were snap-frozen immediately in liquid nitrogen at the end of the treatments. After adding 250 µl of 6% perchloric acid per 20–40 mg embryos, embryos were homogenized and incubated on the ice for 10 min. After 10 min of spin-down at 12,000 g at 4°C, supernatant was neutralized to pH 7 by adding 44 µl 2M K2CO3, incubated for 30 min on ice, and centrifuged at 12,000 g for 10 min at 4°C. Supernatant was filtered using a 0.45-µm spin filter by 12,000 g for 5 min at 4°C and stored at −80°C until HPLC injection. Concentrations of ATP, ADP, and AMP were measured by HPLC according to established methods (Celotto et al., 2011). HPLC separation of adenylate nucleotides was performed on 20 µl of extract using a gradient-mobile phase on a Waters XBridge Shield RP18 column and the following conditions: flow rate of 0.8 µl/min, detection wavelength of 257 nm, and column temperature of 30°C. The gradient consisted of 100% Buffer A from 0 to 6.5 min, 100% Buffer B from 6.5 to 12.5 min, and 100% Buffer A from 12.5 to 25 min. Buffer A was 50 mM NH4H2PO4, pH 5.7; Buffer B was 60:40 acetonitrile/H2O. Adenylate standards were linear through the range of 100–250 µM. Retention times were ∼4.2, 4.7, and 6.2 min for ATP, ADP, and AMP, respectively.
Liposome pull-down assays
The GST and GST fusion proteins were purified from bacteria BL21. After a 1-h induction of 0.1 mM IPTG under 25°C, bacteria were harvested and sonicated on ice. The GST proteins were purified by glutathione agarose (Thermo Fisher Scientific). Liposomal binding assays were performed as described in Kim et al. (2008). Liposomes were prepared via mixing lipids PC, PS, PE, cholesterol, and PIP2 or PI4P (all purchased from Avanti Polar Lipids, Inc.) at the proportions of 37.5, 10, 37.5, 10, and 5.0%. The mixture was dried and resuspended to a final concentration of 1 mg/ml of total phospholipids in Hepes buffer (50 mM Hepes, 100 mM NaCl, and 0.5 mM EDTA). After 30 min of sonication, liposomes were centrifuged at 16,000 g for 10 min and resuspended in binding buffer (20 mM Hepes, 120 mM KCl, 20 mM NaCl, 1 mM EGTA, 1 mM MgCl, and 1 mg/ml BSA). 50 µl of liposome suspension was mixed with 0.1 µg of purified protein and incubated at room temperature for 15 min. Liposomes were pelleted at 16,000 g for 10 min, and bound proteins were analyzed by Western blot.
Drug treatments in mammalian cells
Ionomycin and PAO treatments: HEK293 cells were seeded in a glass-bottom dish and cotransfected with R-pre::GFP (21179; Addgene) or PLCδ::GFP (17274; Addgene) and mLgl::RFP. 24 h after transfection, cells were treated with 10 µM ionomycin or 10 µM PAO. For 2-DG and AM treatments, HEK293 cells were seeded in a glass-bottom dish and cotransfected with GAP-43::GFP and mLgl::RFP. 24 h after transfection, the cells were serum and glucose starved for another 12 h, and then treated with 1 µM 2-DG and 10 mM AM. For cytochalasin D treatment, stable line MDCK-mLgl-GFP cells were treated with either 25 µg/ml cytochalasin D or 0.5% EtOH (solvent control) for 30 min. Cells were then fixed and stained with TRITC-phalloidin to label the F-actin. Images were taken by a confocal microscope (Plan Fluo 40 × oil objective; NA = 1.3; A1; Nikon) by NIS-Elements AR software under room temperature.
Generation of hLgl shRNA knockdown cell lines
hLgl1-specific shRNA with a 19-nucleotide targeting sequence (5′-GCGCGAAGACCAAGTTCAA-3′) was synthesized and cloned into pLKO1-TRC vector according to the instructions from Addgene. A scrambled control shRNA (5′-CCTAAGGTTAAGTCGCCCTCG-3′) in the same expression vector was purchased from Addgene. HEK293 cells were transfected using X-treme Gene 9 DNA transfection reagent (Roche). After 24 h, cultures were switched to complete medium containing 2 µg/ml puromycin. 1 wk after transfection, isolated cell clones were picked up and cultured in a 24-well plate to expand into clones. Cell lines were screened by Western blot using anti–hLgl-1 antibody (Cell Signaling Technology) at a 1:1,000 dilution. Details of molecular cloning of the plasmids used in this paper are available upon request.
Hypoxia-induced cell death assays in HEK293 cells
HEK293-hLgl-KD1, HEK293-hLgl-KD2, and HEK293 control cells were cultured in DMEM supplemented with 10% FBS and 200 µg/ml G418. For the hypoxia experiments, cells were seeded in 6-well plates at 106 cells/well and cultured under normoxic conditions for 1 d. After they reached ∼50–60% confluence, the medium was replaced by fresh DMEM plus 10% FBS, and plates were then placed into an incubator with 1% O2, 5% CO2, and 94% N2 for up to 4 d. Cell viability was determined after 1, 2, and 3 d using a live/dead viability/cytotoxicity kit (L-3224; Molecular Probes). In accordance with the manufacturer’s protocol, cells were washed with PBS once before being subjected to calcein AM (3 µM) and ethidium homodimer-1 (4 µM) staining for 40 min under room temperature. Culture plates were examined under a confocal microscope (LSM 510; Carl Zeiss) using a 40× oil objective (NA = 1.3). For quantification, three individual experiments were performed with live and dead cells counted using ImageJ.
Induction of FKBP12-phosphatase and Lyn11-FRB::CFP dimerization in HEK293 cells
HEK293 cells cultured in 35-mm glass-bottom dishes (In Vitro Scientific) as described in the preceding section were transiently transfected with 1 µg of total DNA with 3 µg lipofectamine 2000 in Opti-MEM according to the manufacturer’s instructions. cDNAs included the Lyn11-FRB::CFP recruiter, FKBP12-phosphatases (Hammond et al., 2012), iRFP::PLCδ-PH (Idevall-Hagren et al., 2012), and iRFP::SidM-P4M (Hammond et al., 2014) as indicated. After 22–26 h, they were imaged in Fluoro-Brite medium (Life Technologies) using a confocal laser-scanning microscope (A1R; Nikon) though a 100×, NA/1.45 plan apochromatic objective lens. Time-lapse imaging began 2 min before bath addition of 1 µM rapamycin. PM localization was calculated from the ratio of fluorescence within the PM (defined by a binary mask derived from the CFP::Lyn11-FRB image) to the whole cell, and is expressed relative to the mean before rapamycin addition. The procedure has been described in detail previously (Hammond et al., 2014).
Ex vivo induction of mRFP::FKBP-5Ptase and Lck-FRB::CFP dimerization in Drosophila follicular cells
Young females of w UASp>mRFP::FKBP-5Ptase/+; lgl::GFP hs-FLP/+; Act5C(FRT.CD2)-Gal4 UAS-RFP/UASp>Lck-FRB::CFP were heat-shocked at 37°C for 1 h, and 4 d later, their ovaries were dissected in 1× PBS and transferred to a drop of 20 µl Schneider’s medium containing either 0.01% DMSO or 10 µM rapamycin. Ovaries were then imaged live in air-permeable conditions, as described in the preceding section.
Genotypes of Drosophila samples in figures
Fig. 1: (A) w; lgl::GFP. (B) w; lgl::GFP FRTG13 His2Av::mRFP. (C) w; sqh-GFP::MoeAD (MoeAD::GFP) and sqhAX; sqh::GFP. (D) w; lgl::GFP and w1118. (E) w; lgl::GFP. Fig. 2: (A) w; lgl::GFP/+; simaKG07607; w; lgl::GFP/+; fatiga02255 and w; lgl::GFP/+; fatiga1 simaKG07607. (B) w par-6Δ226 FRT9-2/w ubi-GFPNLS FRT9-2; lgl::GFP hs-FLP38/lgl::GFP and w hs-FLP/+; lgl::GFP FRTG13 aPKCK06403/lgl::GFP FRTG13 His2Av::mRFP; (C) w hs-FLP/+; lgl::GFP ubi-RFPNLS FRT40A/lglS5A::GFP FRT40A. Fig. 3: (A and B) w; lgl::GFP. Fig. 4: (A) w; lgl::mCherry sqh-GFP::MoeAD (“MoeAD::GFP”). Fig. 6: (A) w hs-FLP/+; LglΔPB::GFP FRT40A/ubi-RFPNLS FRT40A; w hs-FLP/+; lglKR12A::GFP FRT40A/ubi-RFPNLS FRT40A and w; lgl::GFP. (B) w hs-FLP/+; lgl::GFP ubi-RFPNLS FRT40A/LglKR12A::GFP FRT40A. (C) w hs-FLP/+; LglΔPB::GFP FRT40A/ubi-RFPNLS FRT40A and w hs-FLP/+; lglKR12A::GFP FRT40A/ubi-RFPNLS FRT40A. Fig. 7: (B) y w UASp-mRFP::FKbP-5Ptas/+; lgl::GFP hs-FLP/+; Act5C(FRT.CD2)-Gal4 UAS-RFP/UASp-Lck-FRB::CFP. Fig. 8: (A) Cy2-Gal4/UAS-PLCδ-PH::GFP; Cy2-Gal4/UAS-GRP1-PH::GFP and hs-FLP/+; UAS-Numb::GFP/Act5C(FRT.CD2)-Gal4, UAS-RFP. Fig. S1: (A) w; shg::GFP; w arm::GFP; w baz::GFP and w dlg::GFP. Fig. S2: (A) w; lgl::GFP hs-FLP38/lgl::GFP UAS-Sima; Act5C(FRT.CD2)-GAL4 UAS-RFP/+. (B) w par-6Δ226 FRT9-2/w Ubi-GFPNLS FRT9-2; lgl::GFP hs-FLP38 and w hs-FLP; lgl::GFP FRTG13 aPKCK06403/lgl::GFP FRTG13 His2Av::RFP. (C) w; lgl::GFP. (D) w; lgl::GFP hs-FLP38/lgl::GFP UAS-aPKCCAAX; Act5C(FRT.CD2)-GAL4, UAS-RFP/+. Fig. S3: (A) w1118. (D) w1118 and w; lgl::GFP. Fig. S4: (C) w hs-FLP/+; lgl::GFP ubi-RFPNLS FRT40A/lglΔPB::GFP FRT40A and w hs-FLP/+; lgl::GFP ubi-RFPNLS FRT40A/lglKR12A::GFP FRT40A. (D) w; Gal80 FRT40A/lglnull FRT40A; UAS-aPKC-RNAi/hs-FLP Act5C(FRT.CD2)-Gal4 UAS-RFP and w; Gal80 FRT40A/lglKR12A FRT40A; UAS-aPKC-RNAi/hs-FLP Act5C(FRT.CD2)-Gal4 UAS-RFP. Fig. S5: (A and B) w; lgl::GFP.
Online supplemental material
Fig. S1 shows that hypoxia does not disrupt cell polarity in Drosophila embryonic cells. Fig. S2 shows that inhibition of Lgl PM targeting by hypoxia is modulated by Sima activity but is independent of aPKC/Par-6 complex. Fig. S3 shows that hypoxia reduces intracellular ATP levels to inhibit Lgl PM targeting in Drosophila embryonic epithelial cells and in human MCF-7 breast cancer cells. Fig. S4 shows that the PB domain is both essential for Lgl PM targeting and its functions in vivo. Fig. S5 shows that PM phospholipids are required for targeting Lgl. Video 1 shows the reversible inhibition of Lgl::GFP and His2Av::RFP in follicular cells under hypoxia. Video 2 shows PM relocalization of Lgl::GFP during post-hypoxia reoxygenation. Video 3 shows the loss of mLgl::GFP PM targeting in MDCK cells under hypoxia. Video 4 shows the recovery of PM targeting of mLgl::GFP in hypoxia-treated HEK293 cells. Video 5 shows the loss of mLgl::GFP PM targeting by rapamycin-induced PM recruitment of PJ::RFP in HEK293 cells. “ImageJ_Macro-1.txt” is the ImageJ macro used to measure cortical Lgl::GFP localization. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503067/DC1.
Supplementary Material
Acknowledgments
We are grateful to Patty Chiu and Dr. Kenneth Hallows for technical assistance; Drs. Tamas Balla, Sergio Grinstein, Pablo Wappner, Jurgern Knoblich, Stefano De Renzis, Fabrice Roegiers, and Daniel Kiehart for reagents and fly stocks; Dr. Simon Watkins and University of Pittsburgh Medical School Center for Biological Imaging (supported by NIH grant 1S10OD019973-01) for generous imaging and microscopy support; Drs. Patrick Thibodaux and Marijn Ford for PyMOL visualization; Bloomington Stock Center for fly stocks; and Developmental Studies Hybridoma Bank for antibodies.
This work was supported by grants NIH-NCRR R21RR024869 (to Y. Hong), NIH-NIGMS R01GM086423 (to Y. Hong), R01GM103369 (to M.J. Palladino), and RSG-11-120-01-CCG (to Y. Hong) from the American Cancer Society.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- 2-DG
- 2-deoxyglucose
- AM
- antimycin A
- aPKC
- atypical PKC
- HIF1α
- hypoxia-inducible factor 1α
- Lgl
- lethal giant larvae
- MDCK
- Madin-Darby canine kidney
- PAO
- phenylarsine oxide
- PB
- polybasic
- PHD
- prolyl hydroxylase domain
- PI4P
- phosphatidylinositol 4-phosphate
- PIP
- phosphatidylinositol phosphate
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PJ
- Pseudojanin
- PM
- plasma membrane
References
- Aragonés J., Schneider M., Van Geyte K., Fraisl P., Dresselaers T., Mazzone M., Dirkx R., Zacchigna S., Lemieux H., Jeoung N.H., et al. . 2008. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat. Genet. 40:170–180. 10.1038/ng.2007.62 [DOI] [PubMed] [Google Scholar]
- Arbuzova A., Schmitz A.A., and Vergères G.. 2002. Cross-talk unfolded: MARCKS proteins. Biochem. J. 362:1–12. 10.1042/bj3620001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G.P., Fletcher G.C., Brain R., and Thompson B.J.. 2015. Aurora kinases phosphorylate Lgl to induce mitotic spindle orientation in Drosophila epithelia. Curr. Biol. 25:61–68. 10.1016/j.cub.2014.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., and Knoblich J.A.. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 422:326–330. 10.1038/nature01486 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Eisenhaber F., and Knoblich J.A.. 2005. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr. Biol. 15:276–282. 10.1016/j.cub.2005.01.012 [DOI] [PubMed] [Google Scholar]
- Bickeböller M., Tagscherer K.E., Kloor M., Jansen L., Chang-Claude J., Brenner H., Hoffmeister M., Toth C., Schirmacher P., Roth W., and Bläker H.. 2015. Functional characterization of the tumor-suppressor MARCKS in colorectal cancer and its association with survival. Oncogene. 34:1150–1159. 10.1038/onc.2014.40 [DOI] [PubMed] [Google Scholar]
- Bloor J.W., and Kiehart D.P.. 2002. Drosophila RhoA regulates the cytoskeleton and cell-cell adhesion in the developing epidermis. Development. 129:3173–3183. [DOI] [PubMed] [Google Scholar]
- Britton J.S., Lockwood W.K., Li L., Cohen S.M., and Edgar B.A.. 2002. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell. 2:239–249. 10.1016/S1534-5807(02)00117-X [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A.D., Nystul T.G., Ohlstein B., Allen A., et al. . 2007. The carnegie protein trap library: A versatile tool for Drosophila developmental studies. Genetics. 175:1505–1531. 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho C.A., Moreira S., Ventura G., Sunkel C.E., and Morais-de-Sá E.. 2015. Aurora A triggers Lgl cortical release during symmetric division to control planar spindle orientation. Curr. Biol. 25:53–60. 10.1016/j.cub.2014.10.053 [DOI] [PubMed] [Google Scholar]
- Celotto A.M., Chiu W.K., Van Voorhies W., and Palladino M.J.. 2011. Modes of metabolic compensation during mitochondrial disease using the Drosophila model of ATP6 dysfunction. PLoS One. 6:e25823 10.1371/journal.pone.0025823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanin L., Ratcliffe P.J., and Wappner P.. 2005. Reversion of lethality and growth defects in Fatiga oxygen-sensor mutant flies by loss of hypoxia-inducible factor-alpha/Sima. EMBO Rep. 6:1070–1075. 10.1038/sj.embor.7400528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Behar K.L., Xu T., Fan C., and Haddad G.G.. 2003. Expression of Drosophila trehalose-phosphate synthase in HEK-293 cells increases hypoxia tolerance. J. Biol. Chem. 278:49113–49118. 10.1074/jbc.M308652200 [DOI] [PubMed] [Google Scholar]
- Claret S., Jouette J., Benoit B., Legent K., and Guichet A.. 2014. PI(4,5)P2 produced by the PI4P5K SKTL controls apical size by tethering PAR-3 in Drosophila epithelial cells. Curr. Biol. 24:1071–1079. 10.1016/j.cub.2014.03.056 [DOI] [PubMed] [Google Scholar]
- Clyne P.J., Brotman J.S., Sweeney S.T., and Davis G.. 2003. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 165:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dho S.E., French M.B., Woods S.A., and McGlade C.J.. 1999. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 274:33097–33104. 10.1074/jbc.274.46.33097 [DOI] [PubMed] [Google Scholar]
- DiGregorio P.J., Ubersax J.A., and O’Farrell P.H.. 2001. Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 276:1930–1937. 10.1074/jbc.M003911200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froldi F., Ziosi M., Tomba G., Parisi F., Garoia F., Pession A., and Grifoni D.. 2008. Drosophila lethal giant larvae neoplastic mutant as a genetic tool for cancer modeling. Curr. Genomics. 9:147–154. 10.2174/138920208784340786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N.A., Parsons L.M., Allott M.L., Harvey K.F., and Richardson H.E.. 2010. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20:573–581. 10.1016/j.cub.2010.01.055 [DOI] [PubMed] [Google Scholar]
- Hammond G.R.V., Fischer M.J., Anderson K.E., Holdich J., Koteci A., Balla T., and Irvine R.F.. 2012. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 337:727–730. 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond G.R.V., Machner M.P., and Balla T.. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205:113–126. 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattendorf D.A., Andreeva A., Gangar A., Brennwald P.J., and Weis W.I.. 2007. Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature. 446:567–571. 10.1038/nature05635 [DOI] [PubMed] [Google Scholar]
- Heo W.D., Inoue T., Park W.S., Kim M.L., Park B.O., Wandless T.J., and Meyer T.. 2006. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 314:1458–1461. 10.1126/science.1134389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A.M., and Hong Y.. 2009. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA. 106:8284–8289. 10.1073/pnas.0900641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ghosh P., Hatfull G.F., and Hong Y.. 2011a Successive and targeted DNA integrations in the Drosophila genome by Bxb1 and ϕC31 integrases. Genetics. 189:391–395. 10.1534/genetics.111.129247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Huang L., Chen Y.-J., Austin E., Devor C.E., Roegiers F., and Hong Y.. 2011b Differential regulation of adherens junction dynamics during apical-basal polarization. J. Cell Sci. 124:4001–4013. 10.1242/jcs.086694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall-Hagren O., Dickson E.J., Hille B., Toomre D.K., and De Camilli P.. 2012. Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA. 109:E2316–E2323. 10.1073/pnas.1211305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G. Jr., and Ratcliffe P.J.. 2008. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell. 30:393–402. 10.1016/j.molcel.2008.04.009 [DOI] [PubMed] [Google Scholar]
- Keith B., and Simon M.C.. 2007. Hypoxia-inducible factors, stem cells, and cancer. Cell. 129:465–472. 10.1016/j.cell.2007.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso R.J., Buszczak M., Quiñones A.T., Castiblanco C., Mazzalupo S., and Cooley L.. 2004. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32:D418–D420. 10.1093/nar/gkh014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.Y., Tang Z., Liu Q., Patel K.N., Maag D., Geng Y., and Dong X.. 2008. Pirt, a phosphoinositide-binding protein, functions as a regulatory subunit of TRPV1. Cell. 133:475–485. 10.1016/j.cell.2008.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavista-Llanos S., Centanin L., Irisarri M., Russo D.M., Gleadle J.M., Bocca S.N., Muzzopappa M., Ratcliffe P.J., and Wappner P.. 2002. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol. Cell. Biol. 22:6842–6853. 10.1128/MCB.22.19.6842-6853.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisovsky M., Dresser K., Baker S., Fisher A., Woda B., Banner B., and Lauwers G.Y.. 2009. Cell polarity protein Lgl2 is lost or aberrantly localized in gastric dysplasia and adenocarcinoma: an immunohistochemical study. Mod. Pathol. 22:977–984. 10.1038/modpathol.2009.68 [DOI] [PubMed] [Google Scholar]
- Manfruelli P., Arquier N., Hanratty W.P., and Sémériva M.. 1996. The tumor suppressor gene, lethal(2)giant larvae (1(2)g1), is required for cell shape change of epithelial cells during Drosophila development. Development. 122:2283–2294. [DOI] [PubMed] [Google Scholar]
- Okada A., Lansford R., Weimann J.M., Fraser S.E., and McConnell S.K.. 1999. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp. Neurol. 156:394–406. 10.1006/exnr.1999.7033 [DOI] [PubMed] [Google Scholar]
- Plant P.J., Fawcett J.P., Lin D.C., Holdorf A.D., Binns K., Kulkarni S., and Pawson T.. 2003. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5:301–308. 10.1038/ncb948 [DOI] [PubMed] [Google Scholar]
- Poggioli J., Weiss S.J., McKinney J.S., and Putney J.W. Jr. 1983. Effects of antimycin A on receptor-activated calcium mobilization and phosphoinositide metabolism in rat parotid gland. Mol. Pharmacol. 23:71–77. [PubMed] [Google Scholar]
- Reversi A., Loeser E., Subramanian D., Schultz C., and De Renzis S.. 2014. Plasma membrane phosphoinositide balance regulates cell shape during Drosophila embryo morphogenesis. J. Cell Biol. 205:395–408. 10.1083/jcb.201309079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M.M., Albertson R., Shih H.P., Lee C.Y., and Doe C.Q.. 2003. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163:1089–1098. 10.1083/jcb.200306079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J.C., Sullivan W., and Karess R.. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850. 10.1091/mbc.E03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski C.C., Schmitz G., Kashyap A., Bosserhoff A.K., Bataille F., Schäfer S.C., Lehr H.A., Berger M.R., Galle P.R., Strand S., and Strand D.. 2005. Reduced expression of Hugl-1, the human homologue of Drosophila tumour suppressor gene lgl, contributes to progression of colorectal cancer. Oncogene. 24:3100–3109. 10.1038/sj.onc.1208520 [DOI] [PubMed] [Google Scholar]
- Sotillos S., Díaz-Meco M.T., Caminero E., Moscat J., and Campuzano S.. 2004. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J. Cell Biol. 166:549–557. 10.1083/jcb.200311031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., and Knoblich J.A.. 2006. Lethal giant larvae take on a life of their own. Trends Cell Biol. 16:234–241. 10.1016/j.tcb.2006.03.006 [DOI] [PubMed] [Google Scholar]
- Yeung T., Gilbert G.E., Shi J., Silvius J., Kapus A., and Grinstein S.. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 319:210–213. 10.1126/science.1152066 [DOI] [PubMed] [Google Scholar]
- Zhou W., and Hong Y.. 2012. Drosophila Patj plays a supporting role in apical-basal polarity but is essential for viability. Development. 139:2891–2896. 10.1242/dev.083162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Shang Y., Wan Q., Xia Y., Chen J., Du Q., and Zhang M.. 2014. Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl. Cell Res. 24:451–463. 10.1038/cr.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.