Impaired turnover of newly synthesized mitochondrial proteins of the oxidative phosphorylation complexes leads to protein over-accumulation in the inner mitochondrial membrane, thereby generating a stress that dissipates the mitochondrial membrane potential and therefore compromises organelle and cellular fitness.
Abstract
Mitochondrial ribosomes synthesize a subset of hydrophobic proteins required for assembly of the oxidative phosphorylation complexes. This process requires temporal and spatial coordination and regulation, so quality control of mitochondrial protein synthesis is paramount to maintain proteostasis. We show how impaired turnover of de novo mitochondrial proteins leads to aberrant protein accumulation in the mitochondrial inner membrane. This creates a stress in the inner membrane that progressively dissipates the mitochondrial membrane potential, which in turn stalls mitochondrial protein synthesis and fragments the mitochondrial network. The mitochondrial m-AAA protease subunit AFG3L2 is critical to this surveillance mechanism that we propose acts as a sensor to couple the synthesis of mitochondrial proteins with organelle fitness, thus ensuring coordinated assembly of the oxidative phosphorylation complexes from two sets of ribosomes.
Introduction
Synthesis of the mitochondrial respiratory chain complexes requires coordinated gene expression from two genomes and two sets of ribosomes. Approximately99% of the mitochondrial proteome is nuclear encoded (Pagliarini et al., 2008), but a compact mitochondrial genome has been maintained within the organelle of metazoans to ensure transcription and translation of a small number of proteins required for assembly into multisubunit respiratory chain complexes (Christian and Spremulli, 2012; Fox, 2012). The extreme hydrophobicity of these mitochondrially encoded polypeptides and the cotranslational insertion of metal moieties have been hypothesized to account for the retention of a mitochondrial genome (Woodson and Chory, 2008). Therefore, to prevent disruptions in proteostasis, such a system requires temporal and spatial regulation to coordinate folding, membrane insertion, and quality control of de novo proteins after they exit mitochondrial ribosomes (mito-ribosomes).
Mitochondrial protein synthesis occurs on a dedicated set of ribosomes of dual genetic origin. The ribosomal RNA component is encoded by the mitochondrial genome, whereas in mammals the ∼80 mito-ribosomal proteins are entirely nuclear encoded (Cavdar Koc et al., 2001; Koc et al., 2001, 2010; Sharma et al., 2003; Brown et al., 2014; Greber et al., 2014a,b; Amunts et al., 2015). This mitochondrial translation machinery is of proteobacterial descent, but has some unique features not observed in any other protein-synthesizing system (Koc et al., 2010; Christian and Spremulli, 2012). There is mounting evidence that pharmacological or genetic disruptions to mitochondrial protein synthesis have a direct effect on mammalian cell proliferation and fitness, which suggests the presence of an intracellular circuit coupling mito-ribosomal function to cell proliferation (Battersby and Richter, 2013).
We have been investigating the molecular basis linking mitochondrial protein synthesis to mammalian cell proliferation using the antibiotic actinonin as a model system. In mammalian cells, actinonin impairs mitochondrial translation and arrests cell proliferation (Lee et al., 2004; Escobar-Alvarez et al., 2010; Richter et al., 2013). The connection between these two phenotypes is caused by a specific dysfunction arising from mito-ribosomes during translation elongation and not from the absence of translation (Lee et al., 2004; Escobar-Alvarez et al., 2010; Richter et al., 2013). Within 6 h of actinonin treatment, there is stalling of mitochondrial protein synthesis in tandem with fragmentation of the mitochondrial reticulum. The growth defect arises after these mitochondrial disturbances. All of the phenotypes are suppressed by coincubating with chloramphenicol, the well-known mitochondrial translation inhibitor that binds in the ribosomal A site, preventing translation elongation (Richter et al., 2013). This suggests that the stress originates only during mitochondrial protein synthesis but is downstream of the peptidyl transferase center of mito-ribosomes.
Actinonin is a peptide mimetic that resembles small peptides with formylated methionine at the N termini, as is found for proteins in bacteria and those encoded in mitochondrial and chloroplast genomes. This drug is a well-known inhibitor of peptide deformylase (Pdf), the key enzyme required for removal of the formyl group from the starter methionine as part of the N-terminal methionine excision pathway (Fieulaine et al., 2011). The factors for this pathway are found in prokaryotes, chloroplasts, and mitochondria (Giglione and Meinnel, 2001) and affect protein half-life in bacteria and chloroplasts (Meinnel et al., 2006; Adam et al., 2011).
Here, we identify the basis by which actinonin impairs mitochondrial protein synthesis and compromises cell fitness. We show that the deleterious effects of actinonin are not a result of the inhibition of mitochondrial Pdf and the loss of deformylation activity on mitochondrial proteins. Instead, our data demonstrate that actinonin impairs the turnover of de novo mitochondrial proteins inserted into the inner membrane of the organelle, in particular two subunits of the ATP synthase Mt-Atp6 and Mt-Atp8. This disbalance of polypeptides creates stress to the inner membrane that leads to dissipation of the potential across it, thereby generating a severe impingement to organelle homeostasis and cellular fitness. We demonstrate that Afg3l2 (m-AAA) and Oma1 are critical proteases for this pathway. By combining pharmacological and genetic approaches, we have identified a novel mechanism in mitochondria to maintain proteostasis of the inner membrane that we propose acts as a sensor to effectively couple the synthesis of mitochondrial proteins with organelle fitness to ensure coordinated assembly of the respiratory chain complexes from two sets of ribosomes.
Results
Previously, we established a threshold effect to the actinonin impairment of mitochondrial translation that occurs within 6 h of treatment, leading to mito-ribosome stalling and Opa1-dependent fragmentation of the mitochondrial reticulum (Richter et al., 2013). Importantly, this stress does not lead to enhanced transcription (Richter et al., 2013) or synthesis of factors (Fig. S1) associated with the mitochondrial unfolded protein response (Houtkooper et al., 2013), but it is the trigger for inhibiting cell proliferation and mito-ribosomal decay (Battersby and Richter, 2013; Richter et al., 2013). The mechanism by which actinonin induces a stress downstream of the peptidyl transferase center of mito-ribosomes only during mitochondrial protein synthesis was unknown, so we set out to establish the molecular basis for this drug’s effect on mitochondrial gene expression.
Actinonin induces a time-dependent aberrant accumulation of de novo mitochondrial proteins inserted into the membrane
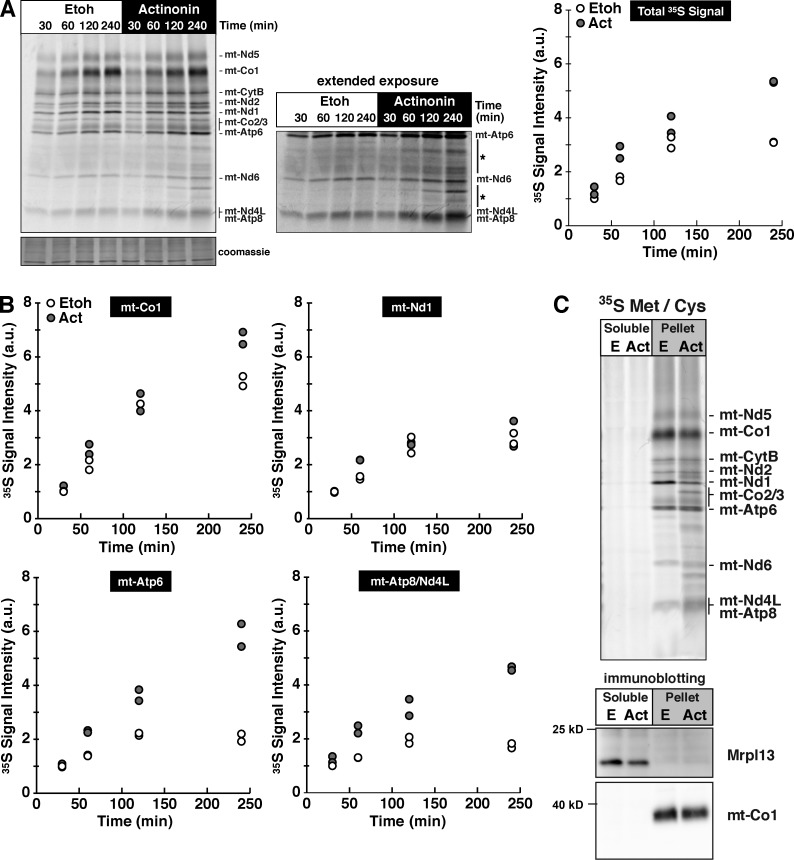
Actinonin induces mito-ribosome stalling that results in a progressive loss of radiolabel incorporation into mitochondrial polypeptides (Richter et al., 2013), but the mechanism accounting for the effect was unknown. To investigate this in more detail, we metabolically labeled mouse embryonic fibroblasts (MEFs) with a time-dependent pulse of [35S]methionine/cysteine from 30 to 240 min in the presence or absence of actinonin (Fig. 1 A). Extended pulse labeling of mitochondrial proteins reflects both protein synthesis and turnover. Surprisingly, during this extended labeling period with actinonin, we observed an increase in the total radiolabel incorporation (Fig. 1 A) that appears to be a result of selected proteins, in particular the two subunits of the ATP synthase mt-Atp6 and mt-Atp8 (Fig. 1 B). We also noted the appearance of translation products of unusual size lower down in the gel that could result from either proteolytic processing or stalled synthesis (Fig. 1 A).
Figure 1.
Actinonin induces a time-dependent aberrant accumulation of mitochondrial polypeptides in the inner membrane. (A, left) Pulse 35S metabolic labeling with ethanol (Etoh) or actinonin (Act) in MEFs for the indicated time points. The asterisks indicate the accumulation of novel translation products. (right) Signal quantification determined for all translation products relative to the signal intensity at 30 min in the control from two independent experiments. (B) Quantification of 35S signal intensity with time of selected mitochondrial polypeptides. (C, top) Sodium carbonate extraction of 35S-labeled mitochondrial translation products. (bottom) Immunoblotting for membrane-anchored mt-Co1 and ribosomal protein Mrpl13. a.u., arbitrary units.
All 13 of the mitochondrial proteins are highly hydrophobic and require cotranslational insertion into the inner mitochondrial membrane (Christian and Spremulli, 2012). To test whether the radiolabeled mitochondrial polypeptides were inserted into the inner mitochondrial membrane, we metabolically labeled MEFs with 35S for a 3-h extended pulse and then used sodium carbonate extraction to distinguish between proteins associated with the membrane and those integrated within the membrane. All of the mitochondrial translation products generated in the presence of actinonin were inserted into the membrane, including the aberrant-sized polypeptides (Fig. 1 C). Mito-ribosomes were only found in the soluble fraction, consistent with the understanding that this protein-synthesizing machine only associates with the membrane and does not possess any structural components anchored into the lipid bilayer (Liu and Spremulli, 2000; Sharma et al., 2003; Brown et al., 2014; Greber et al., 2014a,b; Amunts et al., 2015).
A reduced rate of mitochondrial translation elongation suppresses de novo protein accumulation and stress on the inner membrane
If the aberrant accumulation of mitochondrial proteins in the inner mitochondrial membrane is a key response to the actinonin effects on mitochondrial translation, decreasing the rate of translation elongation should delay or suppress the effects downstream of mito-ribosomes. To test this hypothesis, we took advantage of cultured human cells from patients with defects in mitochondrial protein synthesis where the rate of translation elongation is decreased but not mito-ribosome assembly.
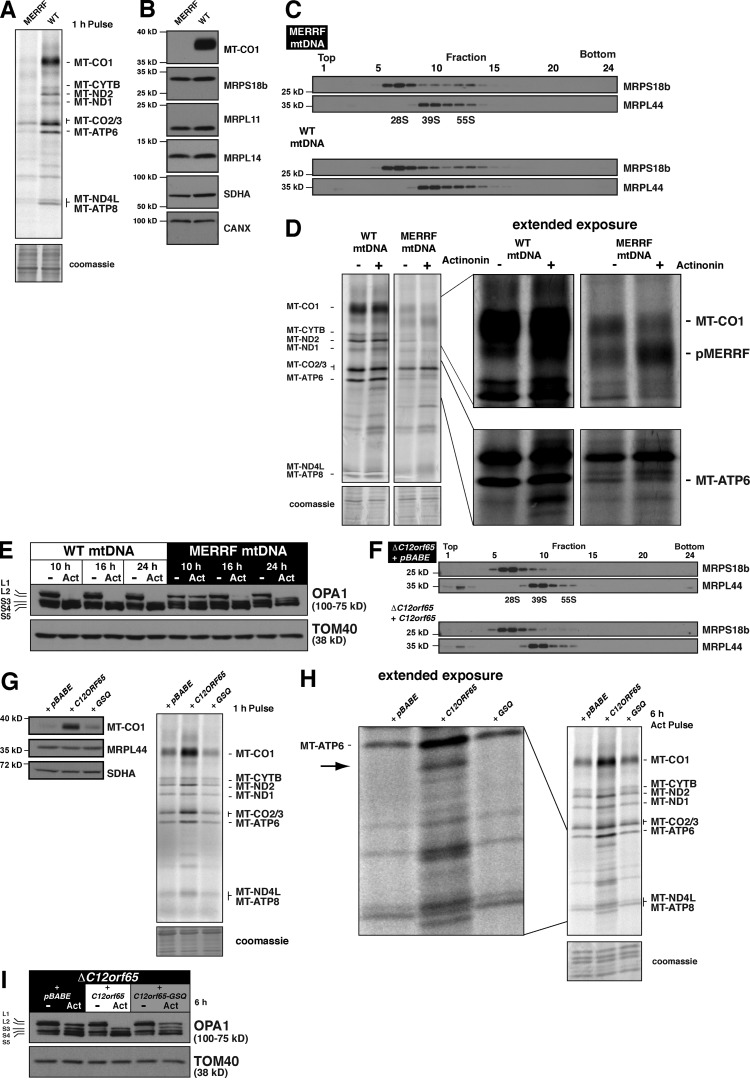
We first used cultured human myoblasts homoplasmic for the pathogenic 8344 A>G mitochondrial (mt) DNA mutation and a matched control with the same patient nuclear background, only homoplasmic for wild-type mtDNA (Sasarman et al., 2008). The 8344 A>G mutation in the MT-TK gene leads to a defect in the aminoacylation of mitochondrial tRNALys and is associated with the MERRF (myoclonus epilepsy associated with ragged-red fibers) syndrome (Boulet et al., 1992; Enriquez et al., 1995). Insufficient aminoacylated tRNALys promotes mito-ribosome stalling at lysine codons during translation elongation with a profound defect in mitochondrial protein synthesis (Fig. 2 A; Boulet et al., 1992; Enriquez et al., 1995). A consequence of this translation defect is undetectable steady-state levels of mitochondrially synthesized MT-CO1 (Fig. 2 B) when the mtDNA mutation is homoplasmic in cells. However, this mutation does not affect the stability of mito-ribosomal proteins (Fig. 2 B) or assembly of the small or large mito-ribosomal subunits (Fig. 2 C). Extended metabolic labeling with 35S in cells homoplasmic for the MERFF mtDNA showed a reduction in the accumulation of radiolabeled proteins with actinonin (Fig. 2 D), in particular MT-ATP6. We also noticed that actinonin leads to an enhanced accumulation of pMERRF (Fig. 2 D), a hallmark of mitochondrial protein synthesis with the 8344 A>G mutation that is truncated MT-CO1 stalled in synthesis (Boulet et al., 1992; Enriquez et al., 1995).
Figure 2.
A reduced rate of mitochondrial elongation delays the effects of actinonin on mitochondrial membrane stress. (A) Translation gel of human myoblasts homoplasmic for the 8344 A>G MERRF mutation or wild-type mtDNA metabolically labeled with 35S for 1 h. (B) Immunoblot of human myoblasts homoplasmic for the MERRF mutation or wild-type mtDNA with the indicated antibodies. MT-CO1 is used to represent the steady-state abundance of the mitochondrially synthesized proteins. (C) Immunoblotting of sucrose density gradient fractions of human myoblasts homoplasmic for the MERRF mtDNA mutation or wild-type mtDNA. (D) Translation gel of human myoblasts homoplasmic for the 8344 A>G MERRF mutation or wild-type mtDNA metabolically labeled with 35S for 3 h in the presence or absence of actinonin. pMERRF is stalled synthesis of MT-CO1, characteristic of the 8344 A>G MERRF mutation. (E) Immunoblot of human myoblasts homoplasmic for the MERRF mtDNA mutation or wild-type mtDNA treated with ethanol or actinonin for 10–24 h. (F) Immunoblotting of sucrose density gradient fractions of human fibroblasts with the C12orf65 R84X mutation retrovirally transduced with empty vector (pBABE) or wild-type C12orf65 cDNA. (G, left) Immunoblot of whole cell lysates from human fibroblasts with the C12orf65 R84X mutation retrovirally transduced with empty vector, wild-type C12orf65 cDNA, or GSQ-C12orf65 (GSQ) cDNA. (right) The same human fibroblasts metabolically labeled with 35S for 1 h with the standard protocol. (H) Translation gel of human fibroblasts from G metabolically labeled with 35S for 6 h in the presence of actinonin. The arrow indicates aberrant translation product generated with actinonin treatment. (I) Immunoblotting of whole cell lysates from human fibroblasts in G treated with or without actinonin for 6 h. CANX, calnexin; WT, wild type.
In tandem with the loss of mitochondrial protein synthesis, actinonin also induces a time-dependent processing of the long isoforms of OPA1, the dynamin-related GTPase important for orchestrating fusion (Wong et al., 2000; Olichon et al., 2003) and fission of the mitochondrial inner membrane (Anand et al., 2014), so that by 6 h of treatment long isoforms are not immunodetectable (Richter et al., 2013). This processing event is completely dependent on protein synthesis on mito-ribosomes because it was lost after genetic ablation of mito-ribosomes or by coincubating with chloramphenicol (Richter et al., 2013); the latter blocks translation elongation in the ribosomal A site. In myoblasts with the 100% wild-type mtDNA, the L1 and L2 OPA1 isoforms are completely lost after 6 h of actinonin, whereas the myoblasts homoplasmic with the MERRF mtDNA have delayed OPA1 processing (Fig. 2 E).
Next, we used human patient fibroblasts homozygous for the R84X mutation in C12orf65 (Antonicka et al., 2010), one of four mammalian mitochondrial class I release factors (Duarte et al., 2012). The absence of C12orf65 was reported to affect the rate of mitochondrial translation elongation, but with no adverse effect on the steady-state abundance of mito-ribosomal proteins (Antonicka et al., 2010). We confirmed this observation and also demonstrate that the mutation has no effect on the assembly of mito-ribosomal subunits (Fig. 2 F). The conserved GGQ domain in class I release factors is required for hydrolyzing the ester bond of a peptidyl tRNA (Handa et al., 2010; Kogure et al., 2012). Site-directed mutagenesis of the catalytic GGQ motif to GSQ showed that this activity is required to maintain the rate of mitochondrial translation elongation, which also can be assessed by the steady-state abundance of MT-CO1 (Fig. 2 G). Metabolic labeling of these cells with an extended pulse of 35S in the presence of actinonin delays the accumulation of aberrant translation products (Fig. 2 H), including MT-ATP6, and suppressed the OPA1 processing (Fig. 2 I). Collectively, these data suggest that modulating the rate of translation elongation of mito-ribosomes has a direct effect on the aberrant accumulation of de novo mitochondrial proteins and the response time to inner membrane stress (OPA1 processing) when treating cells with actinonin.
Aberrant accumulation of de novo mitochondrial proteins dissipates the mitochondrial membrane potential and activates the Oma1 protease
In mammals, Opa1 is posttranslationally processed by the Oma1 and Yme1L proteases under steady-state conditions into membrane-anchored and soluble isoforms (Song et al., 2007; Ehses et al., 2009; Head et al., 2009; Quirós et al., 2012; Anand et al., 2014). The long membrane-anchored Opa1 isoforms are essential for maintaining fusion of the inner mitochondrial membrane and keeping a tubular morphology of the organelle (Cipolat et al., 2004; Meeusen et al., 2006; Westermann, 2010). Stress can also trigger cleavage of the long Opa1 isoforms by the metalloprotease Oma1 in response to loss of mitochondrial membrane potential with the uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or from heat stress (Ehses et al., 2009; Head et al., 2009; Quirós et al., 2012; Baker et al., 2014).
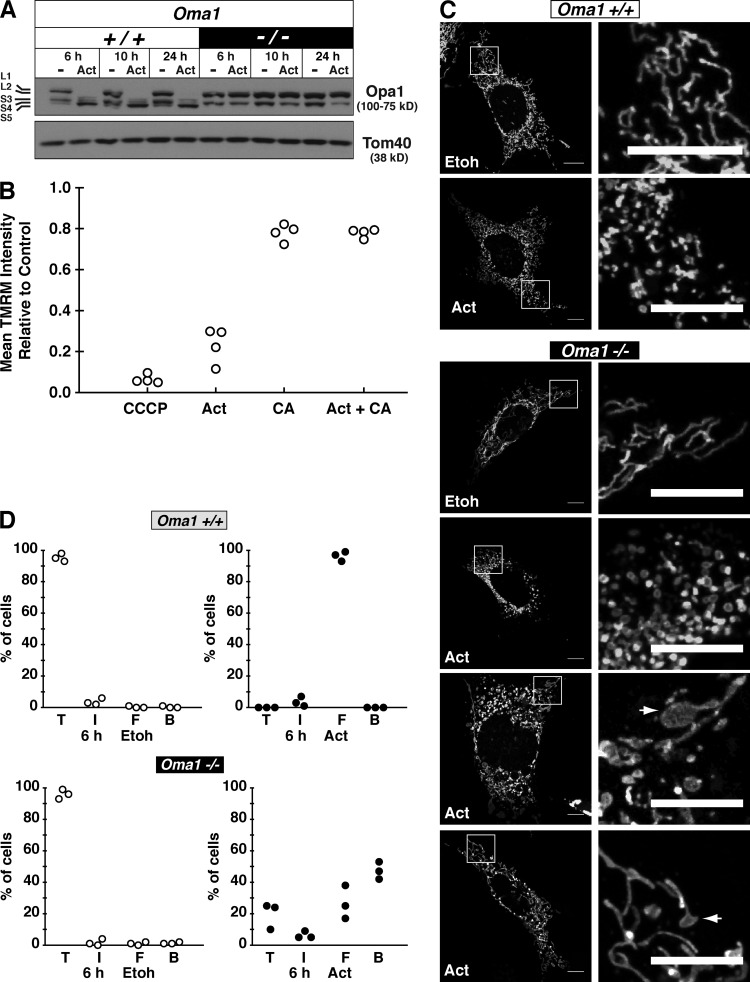
To test whether the Opa1 processing we observed was the result of Oma1 activation, we treated Oma1 knockout (KO) MEFs (Quirós et al., 2012) with actinonin. In the absence of Oma1, there was no Opa1 processing even after extended treatment with the drug (Fig. 3 A). To determine how actinonin induced the Oma1 metalloprotease, we first assessed the mitochondrial membrane potential by staining cells with the potentiometric dye tetramethylrhodamine methyl ester (TMRM) combined with flow cytometry analysis. After 6 h of actinonin treatment, we observed a profound decrease in mitochondrial membrane potential (Fig. 3 B). In contrast, the complete inhibition of mitochondrial protein synthesis with chloramphenicol did not induce such an effect (Fig. 3 B). Moreover, cotreating cells with chloramphenicol and actinonin suppressed the loss of mitochondrial membrane potential (Fig. 3 B). This finding is consistent with our previous results showing that cotreating cells with actinonin and chloramphenicol blocked the Opa1 processing and growth inhibition (Richter et al., 2013).
Figure 3.
Actinonin induces a loss of mitochondrial membrane potential and activation of Oma1. (A) Immunoblot of wild-type and KO Oma1 MEFs treated with actinonin or ethanol (−) for the indicated time. (B) Quantification of mitochondrial membrane potential with TMRM staining in HEK293 cells treated for 6 h with chloramphenicol (CA) and actinonin (Act). Ethanol-treated cells were used as the control, and CCCP was used as a positive control to dissipate the membrane potential. The data represent four independent experiments. (C) Representative confocal images of mitochondrial morphology from Oma1 wild-type or KO MEFs treated with ethanol or actinonin for 6 h. The arrows indicate bulbous mitochondrial morphology. Bars, 10 µm. (D) Quantification of mitochondrial morphology from Oma1 wild-type or KO MEFs treated with ethanol (Etoh) or actinonin for 6 h. Data from three independent experiments with 100 cells counted per experiment. B, bulbous; F, fragmented; I, intermediate (mix of fragmented and tubular); T, tubular.
The inability to proteolytically process the long Opa1 isoforms in the Oma1 KO cells altered the morphological response to mitochondrial shape after actinonin treatment (Fig. 3, C and D). Most Oma1 KO cells did not maintain tubular mitochondria, but were also not completely fragmented compared with wild-type cells (Fig. 3 D). A striking observation was the appearance of bulbous protrusions in the mitochondrial reticulum only in the Oma1 KO cells that accounted for >40% of mitochondrial morphologies (Fig. 3, C and D). We have never observed this response before in wild-type human or mouse cells treated with actinonin (Richter et al., 2013). Thus, actinonin induces an aberrant accumulation of de novo mitochondrial proteins that occurs in tandem with dissipation of the mitochondrial membrane potential and Oma1 activation.
Dissipation of the mitochondrial membrane potential blocks translation elongation
How does the loss of membrane potential affect mitochondrial translation elongation? A previous study demonstrated that mitochondrial membrane potential is important for in organello mitochondrial protein synthesis and turnover, with differential effects depending on the uncoupler (Côté et al., 1990). Because actinonin induces a loss of membrane potential within 6 h (Fig. 3 B) that also occurs in tandem with a progressive loss of radiolabel incorporation into mitochondrial polypeptides (Richter et al., 2013), we investigated the connection between these two parameters in more detail.
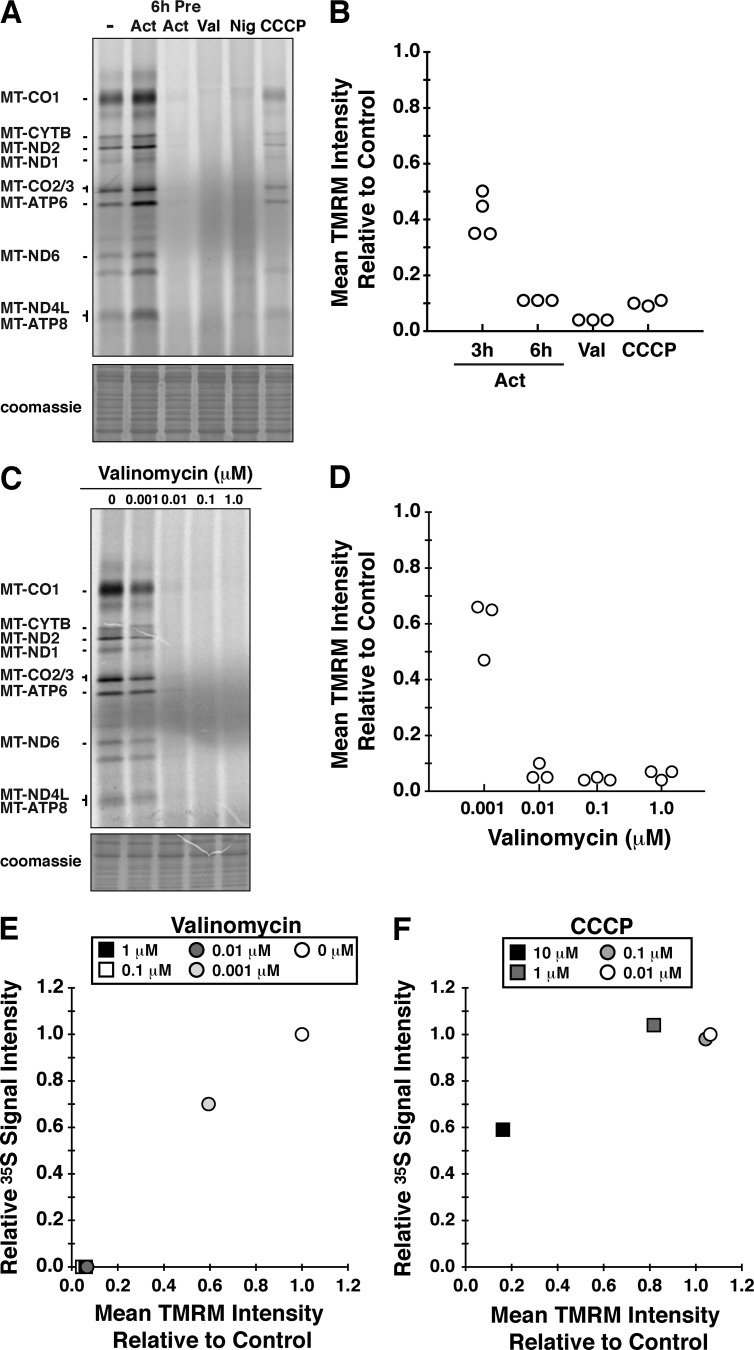
We treated cells with several uncouplers while simultaneously labeling mitochondrial proteins with [35S]methionine/cysteine for 45 min in the presence of anisomycin to inhibit cytoplasmic protein synthesis (Fig. 4 A). The effect of these mitochondrial uncouplers on mitochondrial translation was compared with cells either pretreated for 6 h with actinonin before the 35S pulse or cotreated with actinonin during the 45-min pulse. Several observations stand out from this experiment. The coincubation of actinonin in the pulse enhanced the radiolabeling of the polypeptides (Fig. 4 A), whereas the 6-h pretreatment with actinonin robustly blocked mitochondrial translation elongation similar to dissipating the mitochondrial membrane potential with valinomycin (Fig. 4, A and C). Not all ionophores appear to disrupt mitochondrial translation elongation equally, as the effect with CCCP was more modest than with valinomycin (Fig. 4 A) and consistent with a previous study for isolated mitochondria (Cote et al., 1990).
Figure 4.
Mitochondrial translation is dependent on membrane potential. (A) HEK293 cells were metabolically labeled with 35S for a 45-min pulse and separated by SDS-PAGE. Cells were treated under a variety of conditions: DMSO (−); actinonin (Act) only during the pulse; pretreating with actinonin for 6 h before the pulse (6h Pre Act); 1-µM valinomycin (Val); 2.5-µM nigericin (Nig); and 10-µM CCCP. (B) Scatter plot of flow cytometry analysis with TMRM staining of HEK293 cells treated for 45 min with CCCP and valinomycin, and for 3 and 6 h with actinonin relative to the signal in untreated cells (control). All data originate from independent experiments. (C) Metabolic labeling with 35S for 45 min in HEK293 cells with a titration of valinomycin from 0 to 1 µM. (D) Corresponding flow cytometry analysis with TMRM staining of HEK293 cells treated for 45 min with valinomycin from 0 to 1 µM. Quantification from three independent experiments. (E and F) The relationship between translation performance (relative signal intensity from metabolic labeling) and membrane potential (mean TMRM intensity) for valinomycin (E) and CCCP (F). The data are representative of three independent experiments.
To further explore how membrane potential affects mitochondrial translation elongation, we performed a titration of valinomycin and CCCP. The effects of valinomycin on membrane potential resulted in a corresponding loss of mitochondrial translation elongation (Fig. 4, C–E). In contrast, the response on mitochondrial translation differs with a CCCP titration (Fig. 4 F). The data suggest the progressive stalling of mito-ribosomes by actinonin (Richter et al., 2013) is a result of mitochondrial uncoupling, and the severity of the inhibitory effect may be dependent on the specific cation.
Actinonin impairs turnover of de novo mitochondrial proteins
Next, we turned our attention to the accumulation of the two ATP synthase subunits (Fig. 1 B). In mammals, these two proteins are translated from a bicistronic mRNA with overlapping start and stop codons by a process directly coupled to the assembly of the F1 component of the complex. In the absence of the F1 complex, MT-ATP6 and MT-ATP8 are not stable, so they are rapidly degraded (Rak et al., 2011). This response was proposed as a quality control mechanism to prevent a proton leak across the inner membrane arising from the abnormal accumulation of these subunits within the membrane (Rak et al., 2011).
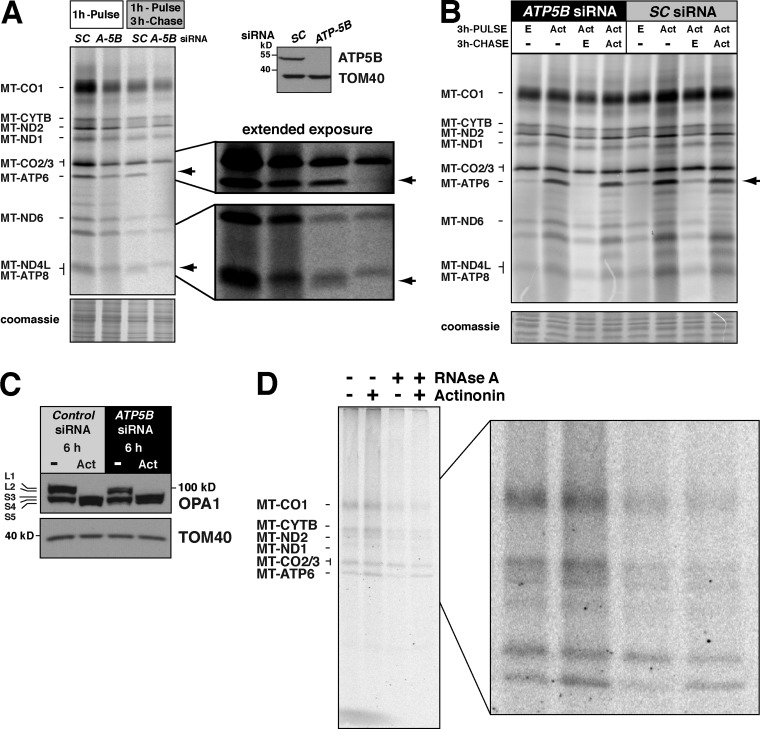
To test whether actinonin treatment could affect the turnover of MT-ATP6 and MT-ATP8, cells were transfected with an siRNA against the F1 subunit ATP5B and assessed 5 d after knockdown. Consistent with the previous study, the stability of MT-ATP6 and MT-ATP8 was dependent on the F1 component (Fig. 5 A; Rak et al., 2011). After a 3-h chase, no radiolabeled MT-ATP6 or MT-ATP8 could be detected (Fig. 5 A). Next, we treated these cells with actinonin to test whether the turnover of MT-ATP6 and MT-ATP8 was affected. As we hypothesized, the rapid turnover of these proteins was blocked when treated with actinonin (Fig. 5 B), which also led to OPA1 processing (Fig. 5 C). Thus, the abnormal accumulation of MT-ATP6 and MT-ATP8 appears to arise from impaired turnover.
Figure 5.
Actinonin impairs the turnover of de novo mitochondrial protein synthesis. (A, left) Standard protocol for metabolic labeling of HEK293 cells after ATP5B (A-5B) siRNA knockdown or scrambled control (SC) with a chase. Arrows indicate the position of MT-ATP6 and MT-ATP8. (right) SDS-PAGE immunoblot of whole cell lysates with representative antibodies. (B) Modified protocol for extended metabolic labeling of HEK293 cells after ATP5B siRNA knockdown for a 3-h pulse and chase in the indicated conditions (Act, actinonin; E, ethanol). (C) Immunoblot of whole cell lysates of ATP5B siRNA knockdown treated with actinonin or ethanol for 6 h. (D) HEK293 cells metabolically labeled for 45 min with [35S]methionine/cysteine with and without actinonin and then precipitated with CTAB. Each sample was split equally in two with one half treated with RNase A before addition of CTAB. A representative image of three independent experiments is shown.
We have previously shown that actinonin induces stalling of mitochondrial ribosomes and the accumulation of mitochondrial polypeptidyl-tRNA (Richter et al., 2013). To test whether any of the abnormal accumulation of MT-ATP6 was attributable to nascent chains, we adopted the method of Siegel and Walter (1988) for precipitation of nascent chains generated with in vitro translation systems. The method uses the positively charged detergent hexadecyltrimethylammonium bromide (CTAB) to precipitate RNA (Hobden and Cundliffe, 1978). Thus, polypeptidyl chains covalently linked to a tRNA in the mito-ribosomal P site will precipitate with CTAB. We metabolically labeled cells for 45 min with 35S in the presence or absence of actinonin during the pulse. Each sample was split equally into two, with one half treated with RNase A as a control to distinguish precipitation based upon the presence of a tRNA. Both halves of each sample were treated with CTAB, and the precipitated material was separated on 12–20% SDS-PAGE (Fig. 5 D). In this experiment, we observed a 2.1-fold (±0.5 [SD]; n = 3) increase in RNase A–sensitive radiolabeled MT-ATP6 precipitated with CTAB when actinonin was included during pulse labeling of mitochondrial proteins (Fig. 5 D). This indicates that actinonin leads to the accumulation of mitochondrially synthesized nascent chains in the inner membrane.
The deleterious effects of actinonin on mitochondrial translation are not mediated via Pdf inhibition
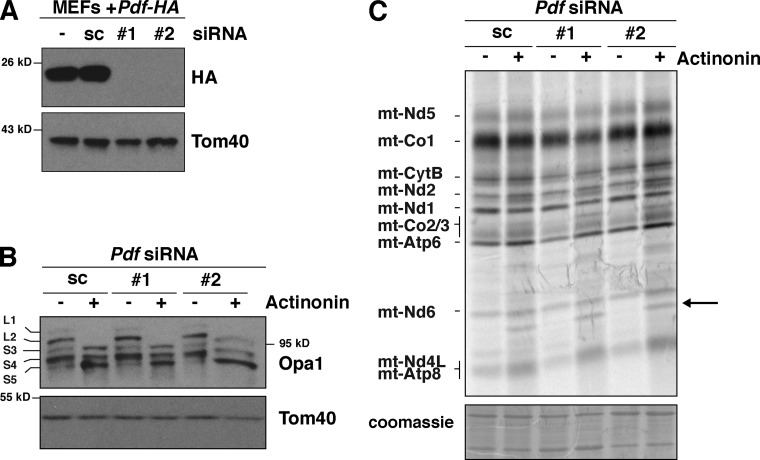
Mitochondrial Pdf has been considered the in vivo target of actinonin in mammalian cells because of the established inhibitory role of this antibiotic on the enzyme. This enzyme is not a core component of mito-ribosomes, and previously we found that actinonin led to the accumulation of Pdf on mito-ribosomes in tandem with translation elongation stalling (Richter et al., 2013). Nonetheless, support for a direct role of Pdf mediating the deleterious effects of actinonin in mitochondria was lacking. To resolve this issue, we silenced Pdf expression by siRNA. Currently, we do not have a reliable antibody against Pdf, so we validated the knockdown efficacy of two siRNA constructs in MEFs stably transduced with a retrovirus expressing Pdf with a C-terminal HA tag (Fig. 6 A). Both siRNA constructs produced a robust knockdown of Pdf. Loss of Pdf had no effect on the actinonin-induced Opa1 processing (Fig. 6 B) or the impaired quality control of newly synthesized mitochondrial proteins (Fig. 6 C). These data indicate that the inhibitory effects of actinonin on de novo mitochondrial protein turnover are not mediated by the lack of deformylation or through the trapping of Pdf on mito-ribosomes, but instead must be a result of the inhibition of other enzymes involved in coordinating the folding and turnover of mitochondrial proteins inserted into the inner membrane.
Figure 6.
Deleterious effects of actinonin on mitochondrial protein synthesis do not arise from inhibiting Pdf. (A) Immunoblot of whole cell lysates of Pdf knockdown with independent siRNAs compared with scrambled control (sc) or without siRNA (−) in MEFs ectopically expressing an HA-tagged Pdf. (B) Immunoblotting of Pdf knockdown MEFs treated with or without actinonin for 6 h. (C) Extended 35S metabolic labeling for 3 h in MEFs with Pdf knockdown treated with and without actinonin. The arrow indicates accumulation of aberrant translation product.
Oma1 proteolytically processes selected de novo mitochondrial proteins
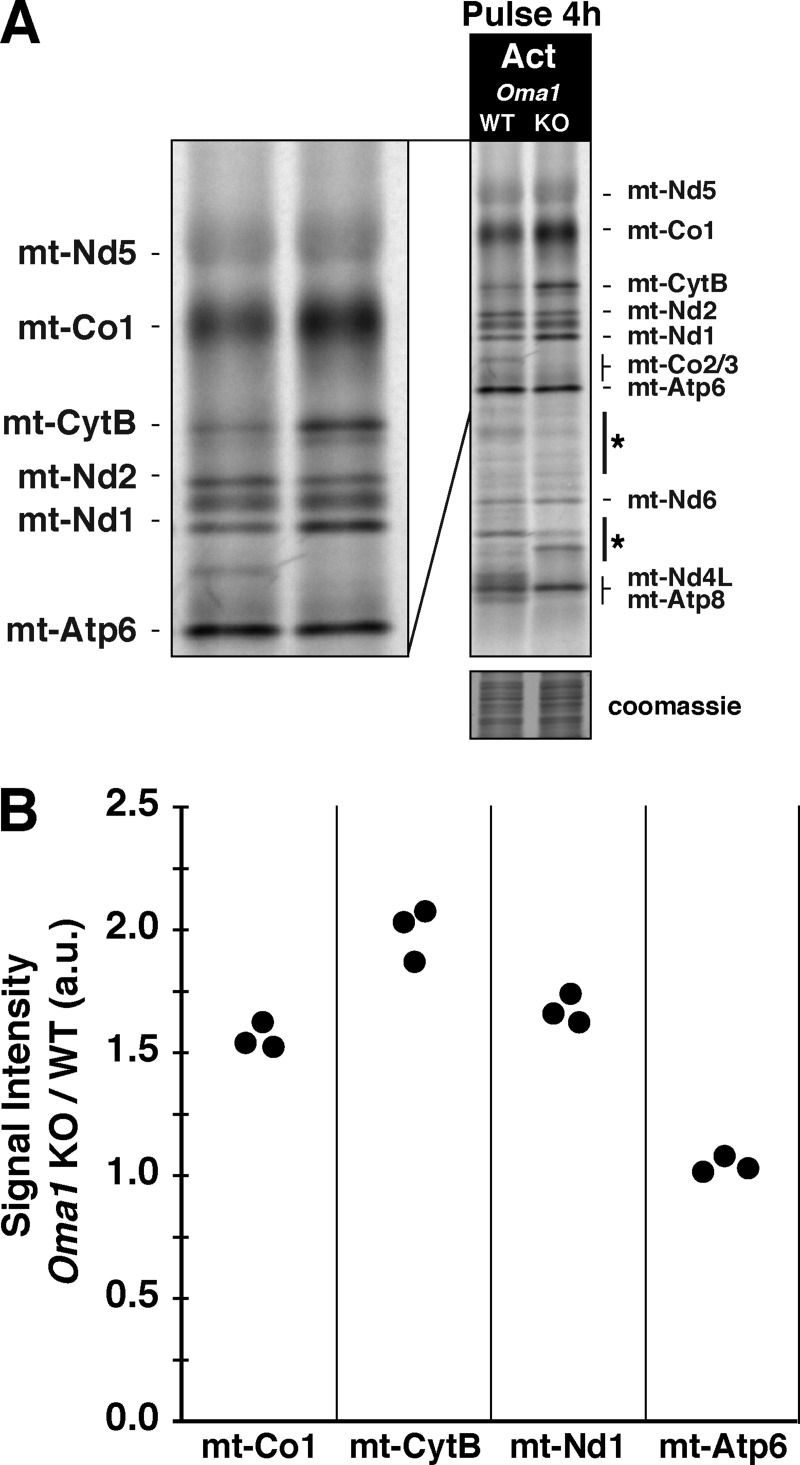
Abnormal protein accumulation can be toxic, so mitochondria possess an abundant and diverse set of compartmentalized proteases for protein quality control (Baker et al., 2011). We revisited the role of the Oma1 metalloprotease in response to aberrant protein accumulation generated by actinonin because the protease has been shown to play a role in the selective degradation of mt-Co1 in yeast cytochrome c oxidase assembly mutants (Khalimonchuk et al., 2012). Oma1 KO MEFs metabolically labeled with 35S for an extended pulse in the presence of actinonin displayed a greater accumulation of mt-Co1, mt-CytB, and mt-Nd1 compared with wild-type MEFs, whereas there was no effect on the abundance of mt-Nd2 or mt-Atp6 (Fig. 7 A). Quantification of these experiments shows that the absence of Oma1 activity leads to a 1.5–2-fold increase in selected mitochondrial proteins when cells were treated with actinonin (Fig. 7 B). These data point to a role for Oma1 in modulating the accumulation of selected de novo mitochondrial proteins in the inner membrane.
Figure 7.
Oma1 proteolytically processes selective de novo mitochondrial proteins. (A) Mitochondrial translation gel of Oma1 wild-type or KO MEFs labeled with 35S for 4 h with actinonin. The asterisks indicate aberrant radiolabeled mitochondrial proteins. (B) Quantification of A from three independent experiments. The data represent the ratio of the signal intensity of Oma1 KO to the wild type of the listed individual polypeptides set to the level of mt-Nd2, whose abundance does not differ. a.u., arbitrary units.
AFG3L2 modulates the abundance of de novo MT-ATP6 and maintains inner membrane integrity
What other proteases might be critical to the regulation of mitochondrial protein abundance in the inner membrane? The membrane-anchored i-AAA and m-AAA proteases flank opposite sides of the mitochondrial inner membrane and provide compartmentalized quality control via their chaperone-like and proteolytic activity to maintain proteostasis (Baker et al., 2011). Both proteases have been implicated in the turnover of the mitochondrially synthesized proteins, although the m-AAA appears to play a greater role affecting the turnover of many unassembled proteins in yeast (Käser and Langer, 2000).
The i-AAA is a homooligomeric complex composed of Yme1l facing the intermembrane space (Leonhard et al., 2000). We treated Yme1l KO MEFs (Anand et al., 2014) with actinonin during an extended metabolic labeling with 35S and found that the overall response to the drug was identical to wild-type MEFs (Fig. S2 A). The data indicate that Yme1l function is likely not impaired by actinonin.
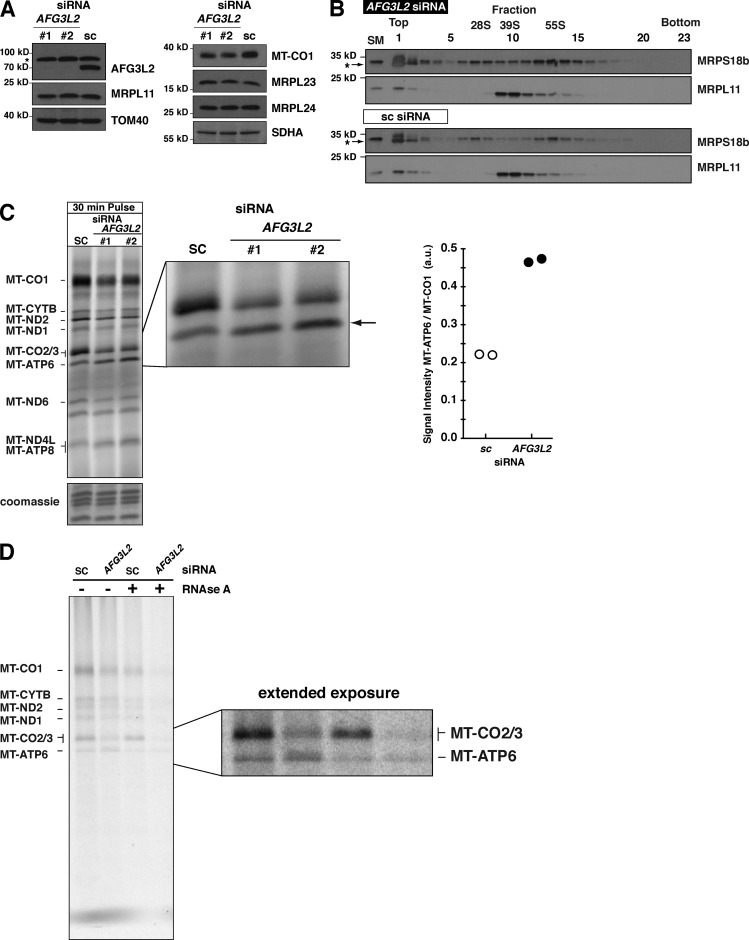
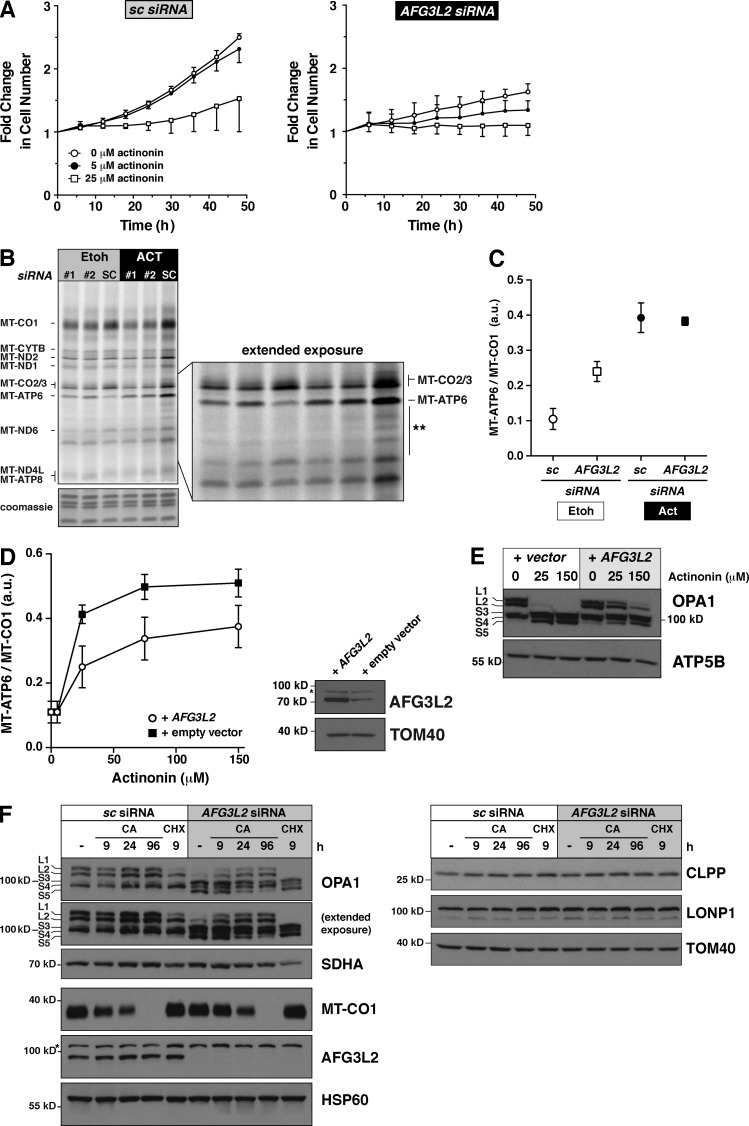
Next, we assessed the role of the m-AAA protease, which in humans is composed of homooligomers of AFG3L2 and heterooligomers of AFG3L2 with paraplegin (Koppen et al., 2007). In yeast, loss of the m-AAA protease function affects the maturation of Mrpl32 and generates a defect in mitochondrial protein synthesis (Nolden et al., 2005). After a 5-d siRNA knockdown of AFG3L2 in human HEK293 cells, we detected a decrease in the steady-state abundance of MT-CO1 as expected, but no decrease in the levels of large mito-ribosomal subunit proteins (Fig. 8 A), assembly of the large subunit, or monosome formation (Fig. 8 B). Short metabolic labeling pulses with 35S in these cells demonstrate a defect in translation elongation, but surprisingly, an increase in the abundance of MT-ATP6 (Fig. 8 C). Quantifying the increase in MT-ATP6 relative to MT-CO1 synthesis showed more than a twofold accumulation of the ATP synthase subunit in the absence of the m-AAA (Fig. 8 C).
Figure 8.
Loss of AFG3L2 induces aberrant accumulation of MT-ATP6. (A) Immunoblot of HEK293 whole cell lysates after AFG3L2 knockdown with two independent siRNAs compared with scrambled control (sc). The asterisk indicates unspecific signal detected with the AFG3L2 antibody. (B) Immunoblotting of sucrose density gradient fractions of AFG3L2 siRNA. The asterisks indicate unspecific signal in fractions 1 and 2 detected only with MRPS18b antibody with TCA precipitated samples. (C, left) A 30-min 35S metabolic labeling of cells in A. The arrow indicates MT-ATP6. (right) quantification of the signal intensity of MT-ATP6 relative to MT-CO1 in the same sample for two independent biological experiments of #2 AFG3L2 siRNA and scrambled control (sc). (D) HEK293 cells treated with the indicated siRNA metabolically labeled for 45 min with [35S] methionine/cysteine in the presence of anisomysin and then precipitated with CTAB. Each sample was split equally in two with one half treated with RNase A before addition of CTAB. A representative image of independent biological experiments is shown. a.u., arbitrary units.
To test whether any of the MT-ATP6 accumulation was attributable to nascent chains, as we observed with actinonin treatment (Fig. 5 D), AFG3L2 siRNA–treated cells were pulse-labeled for 45 min with 35S, and the lysates were precipitated with CTAB. In the absence of AFG3L2 function, there was an increase in RNase A–sensitive radiolabeled MT-ATP6 precipitated with CTAB (Fig. 8 D). These findings indicate that the m-AAA protease is important for preventing an overaccumulation of de novo MT-ATP6.
AFG3L2 abundance modulates the cellular effects of actinonin
The absence of AFG3L2 or loss-of-function mutations also triggers OPA1 processing by OMA1 (Ehses et al., 2009) and thus, surprisingly, impaired m-AAA protease function phenocopies many of the described actinonin effects on mitochondrial protein turnover and membrane integrity. This indicates that perhaps this protease may be within the pathway inhibited by the drug.
To investigate this hypothesis in more detail, we assessed cell growth in the absence of AFG3L2 and the response to actinonin. After a 5-d siRNA knockdown of AFG3L2, cell growth was measured for the next 50 h with a titration dose of actinonin (Fig. 9 A). The loss of AFG3L2 alone induces a profound defect in cell proliferation even in the absence of actinonin (Fig. 9 A). Moreover, these cells were now sensitive to low doses (5 µM) of actinonin compared with the control (Fig. 9 A).
Figure 9.
Overexpression of AFG3L2 suppresses actinonin effects on translation and OPA1 processing. (A) Growth curve of U2OS cells in actinonin after treatment with the indicated siRNA for 5 d. The data represent mean ± SD; n = 3. (B) A representative, extended 3-h 35S metabolic labeling in the presence or absence of actinonin after a 5-d siRNA knockdown of AFG3L2 in HEK293 cells. The asterisks represent aberrant translation products. (C) Quantification of the signal intensity of MT-ATP6 relative to MT-CO1 in B from independent biological experiments. Data represent mean ± SD; n = 4. (D, left) Quantification of the signal intensity of MT-ATP6 relative to MT-CO1 from 35S metabolic labeling from four independent biological experiments with AFG3L2 transient overexpression in HEK293 cells treated with different doses of actinonin for 3 h. Data represent mean ± SD; n = 4. (right) An immunoblot of cells. The asterisk is a nonspecific band detected with the AFG3L2 antibody. (E) An immunoblot of HEK293 cells treated with the indicated siRNA and actinonin doses. (F, left) A representative immunoblot of HEK293 cells treated with the indicated siRNA for 5 d with and without chloramphenicol (CA) or cycloheximide (CHX) for the indicated times. (right) Three panels of data generated from the same samples, only separated on different SDS-PAGE and transferred to independent membranes. a.u., arbitrary units.
Next, we performed an extended 35S metabolic labeling of AFG3L2 siRNA–treated cells that showed a persistent elevated level of MT-ATP6 even in the absence of the drug (Fig. 9, B and C). The addition of actinonin to these cells led to a further increase in MT-ATP6 abundance, although the effect does not appear to be additive (Fig. 9, B and C).
Because the absence of AFG3L2 function appears to sensitize cells to actinonin, we reasoned that, in contrast, increasing the abundance of AFG3L2 should suppress the drug effects on mitochondrial protein accumulation and the downstream effects on mitochondrial membranes. To test this hypothesis, we transiently overexpressed the AFG3L2 cDNA and then treated cells with different doses of actinonin during extended 35S metabolic labeling. The increase in AFG3L2 suppressed the accumulation of MT-ATP6 with actinonin treatment (Fig. 9 D) and also delayed the OPA1 processing (Fig. 9 E).
The link between the loss of m-AAA function and OPA1 processing is well established, but the mechanism triggering OMA1 activation in this case has been elusive (Baker et al., 2011). Because of the phenotype similarities between the actinonin response and the loss of AFG3L2, we posited that perhaps the loss of quality control during mitochondrial protein synthesis by the m-AAA was the key stress that leads to OMA1 activation and mitochondrial fragmentation. If this interpretation is correct, inhibiting mitochondrial protein synthesis should suppress OPA1 processing. To test this hypothesis, we treated AFG3L2 knockdown cells with chloramphenicol and found that eliminating mitochondrial protein synthesis for as little as 9 h restored the long OPA1 isoforms (Fig. 9 F). This was specific for mitochondrial protein synthesis because inhibiting cytosolic ribosomes with cycloheximide had no effect (Fig. 9 F). Adding chloramphenicol to Yme1l KO MEFs had no effect on Opa1 processing, suggesting specificity of the stress from mitochondrial protein synthesis was toward the m-AAA (Fig. S2 B). Moreover, we found that knockdown of AFG3L2 or the inhibition of mitochondrial protein synthesis did not affect the steady-state abundance of several factors implicated in the mitochondrial unfolded protein response (Fig. 9 F). Collectively, these findings demonstrate a critical role for the m-AAA in the quality control of mitochondrial protein synthesis to ensure that de novo proteins generated by mito-ribosomes do not overaccumulate in the inner membrane of the organelle and thereby compromise cell fitness.
Discussion
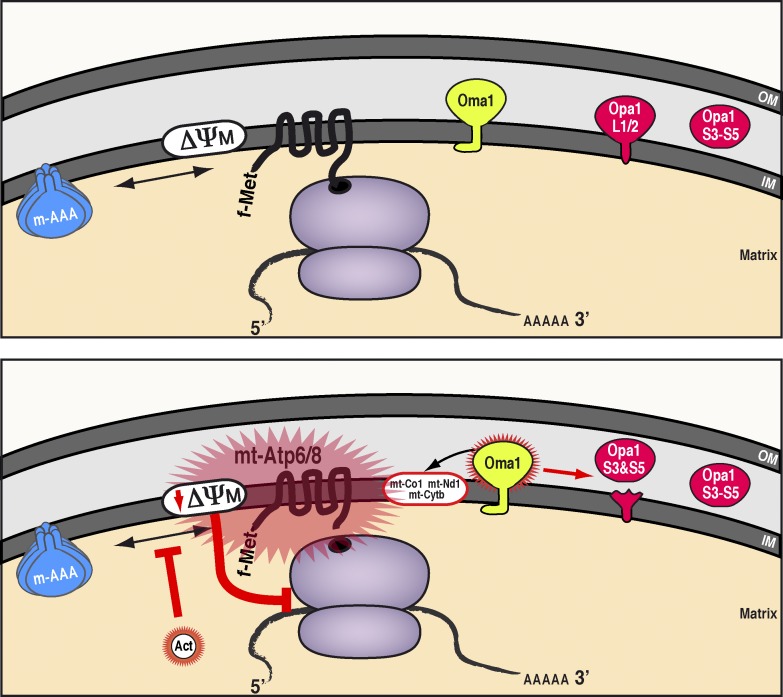
This study demonstrates how dysfunctional quality control of protein synthesis by mito-ribosomes can fundamentally compromise mitochondrial and cellular fitness. Here, we identify a regulatory mechanism for safeguarding the integrity of the mitochondrial inner membrane from mitochondrial protein synthesis by preventing the overaccumulation of de novo mitochondrial proteins (Fig. 10).
Figure 10.
Model for quality control of mitochondrial protein synthesis and the mechanism for actinonin inhibition. (top) Wild-type scenario in mitochondria: the m-AAA protease ensures de novo proteins synthesized on mito-ribosomes do not overaccumulate in the inner membrane. (bottom) Impairing the turnover of de novo mitochondrial proteins either by actinonin treatment or the loss of m-AAA leads to polypeptide overaccumulation in the inner membrane, in particular the two subunits of the ATP synthase, mt-Atp6 and mt-Atp8. The membrane stress can dissipate the mitochondrial membrane potential, which in turn activates the metalloprotease Oma1 that processes the long Opa1 isoforms required for inner membrane fusion and modulates the abundance of selected de novo mitochondrial proteins. Prolonged disruptions to the mitochondrial inner membrane feed back onto mitochondrial translation elongation by stalling mito-ribosomes and preventing further protein synthesis. IM, inner membrane; ΔΨM, mitochondrial membrane potential; OM, outer membrane.
The m-AAA protease plays a central role in this translational quality control pathway to limit the accumulation of de novo mitochondrial proteins. This function appears coupled to the stress-activated metalloprotease OMA1. Disturbances to the inner membrane activate OMA1, leading to two effects: remodeling of mitochondrial membranes via regulatory processing of OPA1 and selective turnover of de novo mitochondrial proteins. Persistent disruptions to inner membrane integrity dissipate the mitochondrial membrane potential, leading to stalling of translation elongation on mito-ribosomes. We propose that this mechanism acts as a circuit breaker within mitochondria to prevent further insults to the inner membrane from mitochondrial protein synthesis and, in the process, ensures balanced protein synthesis required for assembly of the mitochondrial respiratory chain and ATP synthase.
The m-AAA protease is a hexamer consisting of AFG3L2 alone or partnered with paraplegin. In mice, there is a third subunit, Afg3l1, that forms both homo- and heterooligomers (Koppen et al., 2007; Baker et al., 2011). The AAA domain is thought to have chaperone-like activity required for extracting and unfolding substrates destined for proteolysis in the cavity of this complex (Leonhard et al., 2000). Genetic studies in both yeast and mammalian cells point to the AAA domain as being critical to the function of the protease complex. Yeast deletion mutants of the m-AAA generate an ATP synthase assembly defect, which is not caused by impaired synthesis of MT-ATP6 (Paul and Tzagoloff, 1995). This phenotype can be rescued with an m-AAA that is proteolytically dead but functionally wild type in the AAA domain (Arlt et al., 1996), suggesting the role as a chaperone is critical to ATP synthase assembly. Ectopic expression of AFG3L2 with a mutation in the Walker B motif (E408Q) of the AAA domain in a wild-type background induces mitochondrial fragmentation and growth arrest in proliferating cells, whereas expression of a proteolytic mutant (E575Q) has no effect on either phenotype (Ehses et al., 2009). A recent study with another AAA protease complex (ClpXP) has shown that the chaperone function of the mitochondrial ClpX subunit alone was required for protein maturation of aminolevulinic acid synthase, which is integral to heme biosynthesis (Kardon et al., 2015). This evidence points to the m-AAA to potentially function as a chaperone to selected mitochondrial proteins exclusive to its proteolytic activity role. Clearly, future experiments will be needed to establish such a possibility during mitochondrial protein synthesis.
The only difference between the actinonin responses we describe and the genetic disruption to m-AAA function appears to be the loss of mitochondrial membrane potential (Ehses et al., 2009). However, work in mammalian cells has shown that in the absence of m-AAA activity, the ATP synthase can function in reverse to maintain the membrane potential (Maltecca et al., 2012).
At this stage, we do not know whether the actinonin effect is direct or indirect on the m-AAA. Target substrates of AAA complexes can bind directly to the unfoldase domain of these proteases or be delivered via adapters (Truscott et al., 2011). Because actinonin is a peptidomimetic of the formylated N termini of mitochondrial polypeptides, any protein handling these termini is a potential target of the drug. Based on our data, we favor a model whereby actinonin impairs a factor acting as an adapter and thus prevents delivery of novel mitochondrial proteins to the m-AAA complex. At this point, this interpretation is purely speculative, and future work is required to clarify our hypothesis.
Ultimately, the integrity of the inner membrane and the potential across it are the most important requirements for mitochondrial and cellular fitness (Harbauer et al., 2014). In metazoans, all of the mitochondrially synthesized proteins are cotranslationally inserted into the inner membrane, so it is paramount for coordinated regulation of protein synthesis on mito-ribosomes. How a cell responds to inner membrane stress will depend on the intrinsic ability to regulate mitochondrial protein synthesis and the turnover of these hydrophobic proteins. Because both parameters may vary qualitatively and quantitatively across different cell types (Battersby and Richter, 2013), the ability to respond to proteotoxic and membrane stress may account for some of the tissue specificity observed in human mitochondrial diseases with dysfunctional mitochondrial protein synthesis (Martinelli et al., 2009).
In plants, damage to the integrity of the inner mitochondrial membrane has been posited as a mechanism for cytoplasmic male sterility (CMS), which results from the expression of toxic chimeric fusion of open reading frames of mitochondrially encoded genes often involving one of the ATP synthase subunits (Hanson and Bentolila, 2004). Restorers of CMS are nuclear-encoded factors that can rescue the mitochondrial dysfunction in a dominant manner by preventing the synthesis of the toxic fusion proteins or by modulating their deleterious effects post- or cotranslationally (Hanson and Bentolila, 2004). In sugar beets, a CMS open reading frame encoding a 39-kD protein containing mt-ATP6 leads to the assembly of a 200-kD oligomeric complex in the inner membrane and mitochondrial dysfunction (Yamamoto et al., 2005). The restorer for this CMS was recently cloned and identified as a plant homologue of Oma1 (Matsuhira et al., 2012). Thus, the mechanism of impaired quality control of de novo mitochondrial proteins in the inner membrane we show here may account for the organelle dysfunction observed in some CMS varieties.
Our findings show how inner membrane stress is coupled to cell proliferation, but the relevance of proteotoxic mitochondrial membrane stress to organelle dysfunction may be a common theme in biology, so the regulatory mechanisms we identified could also be extended to other cell types and eukaryotic systems.
Materials and methods
Cell culture
Wild-type MEFs were generated from BALB/c mice (Richter et al., 2013). The Oma1 KO MEFs and matching wild-type control were on a C57BL/6 background (Quirós et al., 2012). The Yme1L KO MEFs and matching wild-type control were on a C57BL/6 background (Anand et al., 2014). The MEF cell lines, human fibroblasts (R84X C12orf65; a gift from E. Shoubridge, McGill University, Montreal, Quebec, Canada), and HEK293 were all cultured at 37°C in DMEM (Lonza) with high glucose supplemented with 10% FBS, 1× glutamax, and 50 mg/ml uridine. Human myoblast cultures homoplasmic for the MERRF mutation and respective controls (a gift from E. Shoubridge) were grown in myoblast medium (Lonza). All cells were tested for mycoplasma infection and were clean for the experiments. Cells were treated with 150-µM actinonin unless indicated otherwise, 200 µg/ml chloramphenicol, or carrier (100% ethanol or DMSO). The mitochondrial uncoupler CCCP (Sigma-Aldrich) was used at a dose of 10 µM, and the ionophores were used at 2.5-µM nigericin and 1-µM valinomycin (Sigma-Aldrich). Stealth siRNA against ATP5B, AFG3L2, Pdf, and scrambled sequences were obtained from Life Technologies. siRNAs were transfected on days 1 and 3 with Lipofectamine RNAiMAX (Thermo Fisher Scientific), and cells were collected on day 5 for analysis unless indicated otherwise. The wild-type AFG3L2 cDNA was obtained from the Genome Biology Unit of the University of Helsinki and transiently transfected into HEK293 cells.
Growth curves of U2OS cells were generated using the Cell-IQ monitoring culture platform and the Cell-IQ Analyser Pro-Write software (Chip-Man Technologies Ltd.). In brief, cells were grown on a 24-well plate and continuously imaged over 48 h. A total of six nonoverlapping images were collected per well, calculating the total number of cells at each time point. In each independent experiment, there were a total of four replicates per condition. Growth curves were performed in three independent experiments.
Radioisotope labeling of mitochondrial translation
Mitochondrial protein synthesis can be analyzed in cultured cells by metabolic labeling with [35S]methionine/cysteine in the presence of emetine or anisomycin to inhibit cytoplasmic ribosomes (Leary and Sasarman, 2009; Richter et al., 2013). Typically, at least 8 of the 13 mammalian proteins can be robustly identified with this approach, but there are differences between mouse and human in the migration of individual polypeptides through SDS-PAGE (Leary and Sasarman, 2009). Pulse labeling of mitochondrial translation products in cultured cells was performed as described previously (Richter et al., 2013) with the following modifications. In the first approach, cells were pretreated with 100 µg/ml anisomycin to inhibit cytoplasmic translation and then pulsed with [35S]methionine/cysteine (EasyTag; PerkinElmer) for 15–240 min in the presence or absence of actinonin, chloramphenicol, or ethanol added directly in the pulse. In the second approach used for ATP5B siRNA, HEK293 cells were first incubated in labeling medium lacking methionine and cysteine for 25 min, and then anisomycin was added along with 50-µM puromycin for 15 min. The medium was replaced with fresh labeling medium containing anisomycin and [35S]methionine/cysteine in the presence or absence of actinonin. All samples were treated with benzonase according to the manufacturer’s instructions and then mixed with gel loading buffer (186-mM Tris-HCl, pH 6.7, 15% glycerol, 2% SDS, 0.5 mg/ml bromophenol blue, and 6% β-mercaptoethanol). A 12–20% gradient SDS-PAGE was used to separate samples, which were then dried for exposure with a phosphor screen and scanned with a Typhoon 9400 or Typhoon FLA 7000 (GE Healthcare) for quantification. Gels were rehydrated in water and Coomassie stained to confirm loading.
Identification of mitochondrial nascent chains
The approach of Siegel and Walter (1988) to identify nascent chains with in vitro translation systems was applied to mitochondrial protein synthesis with the following modifications. A 10-cm plate of cells was pretreated with 100 µg/ml anisomycin to inhibit cytoplasmic translation and then pulsed with [35S]methionine-cysteine for 45 min, and 400 µg/ml chloramphenicol was added for 5 min before collecting cells. Cell pellets were snap frozen in liquid nitrogen and stored at −80°C until processing. Cells were lysed in PBS, 1% dodecyl-maltoside, complete protease inhibitor (Roche), and 400 µg/ml chloramphenicol and then centrifuged at 4°C for 30 min at 18,000 g. TCA was added to the supernatant for precipitation. A sample was centrifuged at 4°C for 30 min at 18,000 g, and the pellet was then washed twice with acetone/HCl (19:1). The pellet was solubilized first in 6.4-M urea, 0.6% SDS, and 5-mM sodium acetate, pH 5.0, and then diluted 2.2-fold with water. At this point, each sample was split into two tubes (A and B), only one of which contained RNase A (Life Technologies). Both tubes were incubated for 10 min at 30°C and then placed on ice, and 100 µl of 10% CTAB and 500 µl of 500-mM sodium acetate, pH 5.0, were added. Samples were incubated for 10 min at 30°C and then centrifuged for 30 min at 18,000 g at RT. Each tube was washed first with 1% CTAB and sodium acetate, pH 5.0, and then twice with 19:1 acetone/HCl. Pellets were air dried and solubilized in 15 µl of 8-M urea and 45 µl of translation loading buffer (186-mM Tris-Cl, pH 6.7, 15% glycerol, 2% SDS, 0.5 mg/ml bromophenol blue, and 6% β-mercaptoethanol). A 12–20% gradient SDS-PAGE was used to separate samples, which were then dried for exposure with a phosphor screen and scanned with a Typhoon 9400 or Typhoon FLA 7000 (GE Healthcare) for quantification.
Retroviral expression
The full-length cDNA of human C12orf65 was obtained from E.A. Shoubridge in the retroviral expression vector pBABE-puro. Site-directed mutagenesis was used to mutate the GGQ domain to GSQ (Agilent Technologies). Retrovirus was generated by transient transfection of retroviral plasmids into the Phoenix amphotropic packaging line (Swift et al., 2001). Virus was collected and then used to infect recipient cells (immortalized human patient fibroblasts homozygous for R84X mutation in C12orf65). Cells were used directly in experiments after selection with puromycin.
Immunoblotting and antibodies
Cells were solubilized in PBS, 1% dodecyl maltoside, 1-mM PMSF, and complete protease inhibitor (Roche). Protein concentrations were measured by the Bradford assay (Bio-Rad Laboratories). Equal amounts of proteins were separated by Tris-glycine SDS-PAGE and transferred to nitrocellulose by semidry transfer. Primary antibodies were incubated overnight at 4°C and detected the following day with secondary HRP conjugates using ECL with film. The following primary antibodies were used. Rabbit anti-Mrpl13 was a gift from N. Göran Larsson (Metodiev et al., 2009); from the Proteintech Group, all antibodies were raised in rabbits: anti-AFG3L2 (14631-1-AP), anti-Atp5B (17247-1-AP), anti-ClpP (15698-1AP), anti-Mrpl11 (15543-1-AP), anti-Mrpl23 (11706-1-AP), anti-Mrpl24 (16224-1-AP), anti-Mrpl44 (16394-1-AP), anti-Mrps18b (16139-1-AP), anti-Mrps27 (17280-1-AP), and anti-Yme1l (11510-1-AP); from Sigma-Aldrich, rabbit anti-LonP1 (HPA002192), Mrpl14 (SAB4502786), and anti-HA (H6908); from MitoSciences/Abcam, mouse anti–mt-Co1 (1D6E1A8) and Sdha (C2061); from BD, mouse anti-Opa1 (612606); from Stressgen, rabbit anti-Canx (SPA-860); and from Santa Cruz Biotechnology, Inc., rabbit anti-Tom40 (sc-11414) and goat anti-Hsp60 (sc-1052). The secondary antibodies from Jackson ImmunoResearch Laboratories, Inc. were donkey anti–goat IgG (705-035-003), goat anti–mouse IgG (115-035-146), and goat anti–rabbit IgG (111-035-144). Representative data were cropped in Photoshop (Adobe) with only linear corrections applied.
TMRM staining
HEK293 cells were treated with actinonin, chloramphenicol, and ethanol (as a control) and then stained with 200-nM TMRM (Life Technologies). Cells treated with 10-µM CCCP were used a positive control. After TMRM staining, cells were analyzed by flow cytometry (Accuri C6; BD). At least three independent experiments were performed and quantified.
Isokinetic sucrose gradient assays
Cells were lysed (50-mM Tris, pH 7.2, 10-mM Mg(Ac)2, 40-mM NH4Cl, 100-mM KCl, 1% DDM, and 1-mM PMSF) for 20 min on ice followed by centrifugation for 20 min at 20,000 g at 4°C. The supernatant was loaded on top of a 16-ml linear 10–30% sucrose gradient (50-mM Tris, pH 7.2, 10-mM Mg(Ac)2, 40-mM NH4Cl, 100-mM KCl, and 1-mM PMSF) and centrifuged for 15 h at 4°C and 74,400 g (SW 32.1 Ti; Beckman Coulter). 24 equal volume fractions were collected from the top and TCA precipitated. Samples were separated by SDS-PAGE for immunoblotting.
Microscopy
MEFs were transiently transfected with GFP-omp25 (Nemoto and De Camilli, 1999) and then treated on the following day with antibiotics or ethanol. Cells were fixed in 4% paraformaldehyde and mounted with DABCO/MOWIOL on glass slides for confocal imaging with an inverted microscope (TCS CARS SP8 motorized DMI 6000; Leica) at RT using a 63× HC PL APO CS2 (1.2 NA) water objective with the Argon (488 nm) laser. Images were acquired with a Hybrid GaAsP detector (HyD1; Leica) with LAS AF software (Leica) and then exported into ImageJ (National Institutes of Health) using the Fiji plugin to apply brightness and contrast adjustments.
Sodium carbonate extraction
Sodium carbonate extraction was performed as described previously (Fujiki et al., 1982) with the following modification. MEFs were metabolically labeled as described for the mitochondrial translation assays using the second approach. In brief, 400 µg of protein was resuspended in 10-mM Hepes, pH 7.4, to which an equal volume of 200-mM Na2CO3 was added, and then mixed and incubated on ice for 30 min. Samples were centrifuged at 230,000 g (50.4Ti; Beckman Coulter) for 60 min at 4°C. The supernatant was collected, precipitated with TCA, and resuspended in 2× translation loading buffer (186-mM Tris-Cl, pH 6.7, 15% glycerol, 2% SDS, 0.5 mg/ml bromophenol blue, and 6% β-mercaptoethanol). Pellets were rinsed with water and then resuspended in 2× translation loading buffer with sonication.
Online supplemental material
Fig. S1 shows an Hsp60 immunoblot of MEFs treated with different antibiotic inhibitors of mitochondrial protein synthesis. Fig. S2 shows metabolic labeling and Opa1 processing in wild-type or Yme1l KO MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201504062/DC1.
Supplementary Material
Acknowledgments
We thank the following people for generously sharing reagents: E.A. Shoubridge, C. Lopez-Otin (Universidad de Oviedo, Oviedo, Asturias, Spain), and T. Langer (University of Cologne, Cologne, Germany). We thank R. Jokinen, A. Wartiovaara, H.T. Jacobs, and V. Paavilainen for discussion, the Biomedicum Imaging Unit for the confocal facilities, and the Genome Biology Unit for the cDNA clones.
B.J. Battersby received funding from the Jane and Aatos Erkko Foundation, the Academy of Finland, and the University of Helsinki. U. Richter is an Academy of Finland postdoctoral fellow. F. Suomi was supported by the Ella and Georg Ehrnrooth Foundation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- CMS
- cytoplasmic male sterility
- CTAB
- hexadecyltrimethylammonium bromide
- KO
- knockout
- MEF
- mouse embryonic fibroblast
- MERRF
- myoclonus epilepsy associated with ragged-red fibers
- peptide deformylase
- TMRM
- tetramethylrhodamine methyl ester
References
- Adam Z., Frottin F., Espagne C., Meinnel T., and Giglione C.. 2011. Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell. 23:3745–3760. 10.1105/tpc.111.087239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A., Brown A., Toots J., Scheres S.H., and Ramakrishnan V.. 2015. The structure of the human mitochondrial ribosome. Science. 348:95–98. 10.1126/science.aaa1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R., Wai T., Baker M.J., Kladt N., Schauss A.C., Rugarli E., and Langer T.. 2014. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204:919–929. 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka H., Ostergaard E., Sasarman F., Weraarpachai W., Wibrand F., Pedersen A.M., Rodenburg R.J., van der Knaap M.S., Smeitink J.A., Chrzanowska-Lightowlers Z.M., and Shoubridge E.A.. 2010. Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am. J. Hum. Genet. 87:115–122. 10.1016/j.ajhg.2010.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H., Tauer R., Feldmann H., Neupert W., and Langer T.. 1996. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 85:875–885. 10.1016/S0092-8674(00)81271-4 [DOI] [PubMed] [Google Scholar]
- Baker M.J., Tatsuta T., and Langer T.. 2011. Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3:a007559 10.1101/cshperspect.a007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.J., Lampe P.A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., and Langer T.. 2014. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33:578–593. 10.1002/embj.201386474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby B.J., and Richter U.. 2013. Why translation counts for mitochondria – retrograde signalling links mitochondrial protein synthesis to mitochondrial biogenesis and cell proliferation. J. Cell Sci. 126:4331–4338. 10.1242/jcs.131888 [DOI] [PubMed] [Google Scholar]
- Boulet L., Karpati G., and Shoubridge E.A.. 1992. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet. 51:1187–1200. [PMC free article] [PubMed] [Google Scholar]
- Brown A., Amunts A., Bai X.C., Sugimoto Y., Edwards P.C., Murshudov G., Scheres S.H., and Ramakrishnan V.. 2014. Structure of the large ribosomal subunit from human mitochondria. Science. 346:718–722. 10.1126/science.1258026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavdar Koc E., Burkhart W., Blackburn K., Moseley A., and Spremulli L.L.. 2001. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 276:19363–19374. 10.1074/jbc.M100727200 [DOI] [PubMed] [Google Scholar]
- Christian B.E., and Spremulli L.L.. 2012. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim. Biophys. Acta. 1819:1035–1054. 10.1016/j.bbagrm.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S., Martins de Brito O., Dal Zilio B., and Scorrano L.. 2004. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 101:15927–15932. 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté C., Boulet D., and Poirier J.. 1990. Expression of the mammalian mitochondrial genome. Role for membrane potential in the production of mature translation products. J. Biol. Chem. 265:7532–7538. [PubMed] [Google Scholar]
- Duarte I., Nabuurs S.B., Magno R., and Huynen M.. 2012. Evolution and diversification of the organellar release factor family. Mol. Biol. Evol. 29:3497–3512. 10.1093/molbev/mss157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J.C., Westermann B., Rugarli E.I., and Langer T.. 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187:1023–1036. 10.1083/jcb.200906084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez J.A., Chomyn A., and Attardi G.. 1995. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNALys and premature translation termination. Nat. Genet. 10:47–55. 10.1038/ng0595-47 [DOI] [PubMed] [Google Scholar]
- Escobar-Alvarez S., Gardner J., Sheth A., Manfredi G., Yang G., Ouerfelli O., Heaney M.L., and Scheinberg D.A.. 2010. Inhibition of human peptide deformylase disrupts mitochondrial function. Mol. Cell. Biol. 30:5099–5109. 10.1128/MCB.00469-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieulaine S., Boularot A., Artaud I., Desmadril M., Dardel F., Meinnel T., and Giglione C.. 2011. Trapping conformational states along ligand-binding dynamics of peptide deformylase: The impact of induced fit on enzyme catalysis. PLoS Biol. 9:e1001066 10.1371/journal.pbio.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T.D. 2012. Mitochondrial protein synthesis, import, and assembly. Genetics. 192:1203–1234. 10.1534/genetics.112.141267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Fowler S., Shio H., Hubbard A.L., and Lazarow P.B.. 1982. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: Comparison with endoplasmic reticulum and mitochondrial membranes. J. Cell Biol. 93:103–110. 10.1083/jcb.93.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C., and Meinnel T.. 2001. Organellar peptide deformylases:Universality of the N-terminal methionine cleavage mechanism. Trends Plant Sci. 6:566–572. 10.1016/S1360-1385(01)02151-3 [DOI] [PubMed] [Google Scholar]
- Greber B.J., Boehringer D., Leibundgut M., Bieri P., Leitner A., Schmitz N., Aebersold R., and Ban N.. 2014a The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 515:283–286. [DOI] [PubMed] [Google Scholar]
- Greber B.J., Boehringer D., Leitner A., Bieri P., Voigts-Hoffmann F., Erzberger J.P., Leibundgut M., Aebersold R., and Ban N.. 2014b Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 505:515–519. 10.1038/nature12890 [DOI] [PubMed] [Google Scholar]
- Handa Y., Hikawa Y., Tochio N., Kogure H., Inoue M., Koshiba S., Güntert P., Inoue Y., Kigawa T., Yokoyama S., and Nameki N.. 2010. Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J. Mol. Biol. 404:260–273. 10.1016/j.jmb.2010.09.033 [DOI] [PubMed] [Google Scholar]
- Hanson M.R., and Bentolila S.. 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 16:S154–S169. 10.1105/tpc.015966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer A.B., Zahedi R.P., Sickmann A., Pfanner N., and Meisinger C.. 2014. The protein import machinery of mitochondria—a regulatory hub in metabolism, stress, and disease. Cell Metab. 19:357–372. 10.1016/j.cmet.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Head B., Griparic L., Amiri M., Gandre-Babbe S., and van der Bliek A.M.. 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187:959–966. 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobden A.N., and Cundliffe E.. 1978. The mode of action of alpha sarcin and a novel assay of the puromycin reaction. Biochem. J. 170:57–61. 10.1042/bj1700057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., and Auwerx J.. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497:451–457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon J.R., Yien Y.Y., Huston N.C., Branco D.S., Hildick-Smith G.J., Rhee K.Y., Paw B.H., and Baker T.A.. 2015. Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell. 161:858–867. 10.1016/j.cell.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käser M., and Langer T.. 2000. Protein degradation in mitochondria. Semin. Cell Dev. Biol. 11:181–190. 10.1006/scdb.2000.0166 [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O., Kim H., Watts T., Perez-Martinez X., and Winge D.R.. 2012. Oligomerization of heme o synthase in cytochrome oxidase biogenesis is mediated by cytochrome oxidase assembly factor Coa2. J. Biol. Chem. 287:26715–26726. 10.1074/jbc.M112.377200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc E.C., Burkhart W., Blackburn K., Moyer M.B., Schlatzer D.M., Moseley A., and Spremulli L.L.. 2001. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 276:43958–43969. 10.1074/jbc.M106510200 [DOI] [PubMed] [Google Scholar]
- Koc E.C., Haque M.E., and Spremulli L.L.. 2010. Current views of the structure of the mammalian mitochondrial ribosome. Isr. J. Chem. 50:45–59. 10.1002/ijch.201000002 [DOI] [Google Scholar]
- Kogure H., Hikawa Y., Hagihara M., Tochio N., Koshiba S., Inoue Y., Güntert P., Kigawa T., Yokoyama S., and Nameki N.. 2012. Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65. Proteins. 80:2629–2642. 10.1002/prot.24152 [DOI] [PubMed] [Google Scholar]
- Koppen M., Metodiev M.D., Casari G., Rugarli E.I., and Langer T.. 2007. Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia. Mol. Cell. Biol. 27:758–767. 10.1128/MCB.01470-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary S.C., and Sasarman F.. 2009. Oxidative phosphorylation: Synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol. Biol. 554:143–162. 10.1007/978-1-59745-521-3_10 [DOI] [PubMed] [Google Scholar]
- Lee M.D., She Y., Soskis M.J., Borella C.P., Gardner J.R., Hayes P.A., Dy B.M., Heaney M.L., Philips M.R., Bornmann W.G., et al. 2004. Human mitochondrial peptide deformylase, a new anticancer target of actinonin-based antibiotics. J. Clin. Invest. 114:1107–1116. 10.1172/JCI200422269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., and Langer T.. 2000. Membrane protein degradation by AAA proteases in mitochondria: Extraction of substrates from either membrane surface. Mol. Cell. 5:629–638. 10.1016/S1097-2765(00)80242-7 [DOI] [PubMed] [Google Scholar]
- Liu M., and Spremulli L.. 2000. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J. Biol. Chem. 275:29400–29406. 10.1074/jbc.M002173200 [DOI] [PubMed] [Google Scholar]
- Maltecca F., De Stefani D., Cassina L., Consolato F., Wasilewski M., Scorrano L., Rizzuto R., and Casari G.. 2012. Respiratory dysfunction by AFG3L2 deficiency causes decreased mitochondrial calcium uptake via organellar network fragmentation. Hum. Mol. Genet. 21:3858–3870. 10.1093/hmg/dds214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli P., La Mattina V., Bernacchia A., Magnoni R., Cerri F., Cox G., Quattrini A., Casari G., and Rugarli E.I.. 2009. Genetic interaction between the m-AAA protease isoenzymes reveals novel roles in cerebellar degeneration. Hum. Mol. Genet. 18:2001–2013. 10.1093/hmg/ddp124 [DOI] [PubMed] [Google Scholar]
- Matsuhira H., Kagami H., Kurata M., Kitazaki K., Matsunaga M., Hamaguchi Y., Hagihara E., Ueda M., Harada M., Muramatsu A., et al. 2012. Unusual and typical features of a novel restorer-of-fertility gene of sugar beet (Beta vulgaris L.). Genetics. 192:1347–1358. 10.1534/genetics.112.145409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., and Nunnari J.. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 127:383–395. 10.1016/j.cell.2006.09.021 [DOI] [PubMed] [Google Scholar]
- Meinnel T., Serero A., and Giglione C.. 2006. Impact of the N-terminal amino acid on targeted protein degradation. Biol. Chem. 387:839–851. 10.1515/BC.2006.107 [DOI] [PubMed] [Google Scholar]
- Metodiev M.D., Lesko N., Park C.B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., and Larsson N.G.. 2009. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9:386–397. 10.1016/j.cmet.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Nemoto Y., and De Camilli P.. 1999. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 18:2991–3006. 10.1093/emboj/18.11.2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E.I., and Langer T.. 2005. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 123:277–289. 10.1016/j.cell.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., and Lenaers G.. 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278:7743–7746. 10.1074/jbc.C200677200 [DOI] [PubMed] [Google Scholar]
- Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 134:112–123. 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.F., and Tzagoloff A.. 1995. Mutations in RCA1 and AFG3 inhibit F1-ATPase assembly in Saccharomyces cerevisiae. FEBS Lett. 373:66–70. 10.1016/0014-5793(95)00979-J [DOI] [PubMed] [Google Scholar]
- Quirós P.M., Ramsay A.J., Sala D., Fernández-Vizarra E., Rodríguez F., Peinado J.R., Fernández-García M.S., Vega J.A., Enríquez J.A., Zorzano A., and López-Otín C.. 2012. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 31:2117–2133. 10.1038/emboj.2012.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M., McStay G.P., Fujikawa M., Yoshida M., Manfredi G., and Tzagoloff A.. 2011. Turnover of ATP synthase subunits in F1-depleted HeLa and yeast cells. FEBS Lett. 585:2582–2586. 10.1016/j.febslet.2011.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U., Lahtinen T., Marttinen P., Myöhänen M., Greco D., Cannino G., Jacobs H.T., Lietzén N., Nyman T.A., and Battersby B.J.. 2013. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr. Biol. 23:535–541. 10.1016/j.cub.2013.02.019 [DOI] [PubMed] [Google Scholar]
- Sasarman F., Antonicka H., and Shoubridge E.A.. 2008. The A3243G tRNALeu(UUR) MELAS mutation causes amino acid misincorporation and a combined respiratory chain assembly defect partially suppressed by overexpression of EFTu and EFG2. Hum. Mol. Genet. 17:3697–3707. 10.1093/hmg/ddn265 [DOI] [PubMed] [Google Scholar]
- Sharma M.R., Koc E.C., Datta P.P., Booth T.M., Spremulli L.L., and Agrawal R.K.. 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 115:97–108. 10.1016/S0092-8674(03)00762-1 [DOI] [PubMed] [Google Scholar]
- Siegel V., and Walter P.. 1988. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 7:1769–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., and Chan D.C.. 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178:749–755. 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S., Lorens J., Achacoso P., and Nolan G.P.. 2001. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell–based systems. In Current Protocols in Immunology J.E. Coligan, editor. John Wiley & Sons Inc., New York. [DOI] [PubMed] [Google Scholar]
- Truscott K.N., Bezawork-Geleta A., and Dougan D.A.. 2011. Unfolded protein responses in bacteria and mitochondria: A central role for the ClpXP machine. IUBMB Life. 63:955–963. 10.1002/iub.526 [DOI] [PubMed] [Google Scholar]
- Westermann B. 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11:872–884. 10.1038/nrm3013 [DOI] [PubMed] [Google Scholar]
- Wong E.D., Wagner J.A., Gorsich S.W., McCaffery J.M., Shaw J.M., and Nunnari J.. 2000. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151:341–352. 10.1083/jcb.151.2.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., and Chory J.. 2008. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9:383–395. 10.1038/nrg2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M.P., Kubo T., and Mikami T.. 2005. The 5′-leader sequence of sugar beet mitochondrial atp6 encodes a novel polypeptide that is characteristic of Owen cytoplasmic male sterility. Mol. Genet. Genomics. 273:342–349. 10.1007/s00438-005-1140-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.