Abstract
Somatic hypermutation (SHM) in immunoglobulin gene (Ig) variable (V) regions is critical for the maturation of the antibody response. It is dependent on the expression of activation-induced cytidine deaminase (AID) and translesion DNA polymerases in germinal center B cells as well as Ig V transcription, as regulated by the Ig heavy chain (H) intronic enhancer (iEµ) and the 3′ enhancer (3′Eα) region. We analyzed the role of these cis elements in SHM by stably transfecting Ramos human lymphoblastoid B cells with a rearranged human IgH chain VD (diversity) J (joining) DNA construct containing a VH promoter at the 5′ end and CH1 and CH2 exons of Cγ1 at the 3′ end. In this construct, mutations preferentially targeted dA/dT basepairs in the RGYW/WRCY hotspot. Most of the dA/dT mutations and accompanying dC/dG mutations were transitions. Deletion of iEµ resulted in decreased SHM which could be partially restored by insertion of the IgH hs1,2 enhancer. Other two 3′Eα enhancers, hs3-hs4, did not significantly increase the mutation frequency, but further strengthened the dA/dT bias. The frequency and spectrum of the mutations were independent of the genomic integration of the transgene or V gene transcription level. Thus, we have established a novel in vitro system to analyze SHM and identify the role of multiple cis-regulatory elements in regulating dA/dT biased SHM. This model system will be useful to further address the role of other cis-regulating elements and recruited trans-acting factors in expressing the modalities of SHM.
Keywords: 3′ Heavy chain enhancer, cis-Regulatory elements, iEµ, Intronic Ig µ, enhancer, SHM, Somatic hypermutation
1. Introduction
During B cell development in the bone marrow, recombination-activating genes 1 (RAG1) and RAG2-mediated DNA recombination of germline immunoglobulin V, D and J genes underlies the generation of the high degree of diversity (in excess of 109) of the B cells receptor (BCR) for antigen and the primary antibody repertoire. The impact of antigen in the specialized microenvironment of the germinal center leads to diversification of such a primary BCR repertoire and the generation of antibodies with higher affinities through somatic hypermutation (SHM) and different immunological effector functions through class switch DNA recombination (CSR). SHM targets the V(D)J sequence immediately downstream of the Ig V promoter and extends to the intronic DNA downstream the JH, Jκ and Jλ region. It inserts mutations at a rate of 10−4 to 10−3 change/base/cell division and displays a striking preference for the RGYW/WRCY mutational hotspot (Rada et al., 1994; Rogerson, 1994; Wagner et al., 1995; Foster et al., 1999; Diaz and Casali, 2002). CSR substitutes the IgH Cµ region with downstream Cγ, Cα and Cε regions, thereby generating antibodies with different effector functions.
SHM and CSR are dependent on activation-induced cytidine deaminase (AID) (Muramatsu et al., 2000; Revy et al., 2000), which is strictly expressed in differentiating germinal center B cells (Muramatsu et al., 1999). AID is a sequelog (Varshavsky, 2004) of the RNA-editing cytidine deaminase APOBEC1 (Muramatsu et al., 1999). As such, AID was thought to edit an unknown mRNA precursor to yield novel mRNAs, which would in turn encode endonucleases to cleave targeted DNA at hypermutating V(D)J regions or switch (S) regions (RNA editing hypothesis) (Honjo et al., 2002, 2004; Doi et al., 2003; Begum et al., 2004; Nagaoka et al., 2005). However, mounting evidence indicates that AID directly deaminates DNA dC residues (Petersen-Mahrt et al., 2002; Pham et al., 2003), yielding dU:dG mispairs, which are replicated over or dealt with either the base excision repair (BER) or the mismatch repair (MMR) pathways to introduce mutations through the intervention of translesion DNA polymerases, such as pol ζ, pol η and pol θ (Rada et al., 2004). In addition, accumulating evidence suggests that mutations can be introduced by the same translesion DNA polymerases (Zan et al., 2001; Diaz and Casali, 2002; Diaz and Lawrence, 2005) while repairing DNA breaks, including double stranded DNA breaks (DSBs) involving resected ends generated through AID-dependent DNA deamination (Bross et al., 2000; Papavasiliou and Schatz, 2000; Wu et al., 2003; Zan et al., 2003; Nagaoka et al., 2005; Xu et al., 2005).
SHM depends on V gene transcription (Peters and Storb, 1996; Fukita et al., 1998), as suggested by the greatly diminished frequency of mutations in the IgH locus when the V gene promoter is removed (Fukita et al., 1998), and conversely, by unchanged level of SHM in V regions if the endogenous promoter is replaced with a transcriptionally active heterologous promoter (Betz et al., 1994; Tumas-Brundage and Manser, 1997). B cell specific VH gene transcription is regulated by the IgH intronic enhancer (iEµ), which is located 5′ of Sµ (Banerji et al., 1983; Gillies et al., 1983) and recruits multiple transcription factors, including proteins of the E-box and POU families (Ernst and Smale, 1995). A second IgH transcription regulatory region is located 2 kb downstream of the Cα gene in the mouse and consists of four B cell-specific DNase I hypersensitivity (hs) sites, hs3a, hs1,2, hs3b, and hs4, each of which is referred to as an IgH 3′Eα enhancer. The hs1,2 enhancer lies at the center of a 25kb dyad symmetric region with two virtually identical hs3a and hs3b located at the two termini (Chauveau and Cogne, 1996; Saleque et al., 1997). In the human, a duplicated 3′Eα region separates two CH gene clusters, resulting in a germline CH arrangement of Cµ-Cδ–Cγ3-Cγ1-Cα1–3′Eα-Cγ2-Cγ4-Cε–Cα2–3′Eα. Each 3′Eα region consists of three hs sites in the 5′-hs3-hs1,2-hs4–3′ configuration, with two hs1,2 enhancers being inverted in respect to each other (Chen and Birshtein, 1997; Mills et al., 1997). Different transcription factors, such as E2A, Oct, Ets, and NF-κB family proteins, regulate the IgH transcription through their binding motifs in the 3′Eα region at different B-cell differentiation stages (Khamlichi et al., 2000; Linderson et al., 2001; Schaffer et al., 2003; Kim et al., 2004; Sepulveda et al., 2004).
The iEµ has been implicated in SHM by the findings that it could replace the 3′ λ enhancer to induce SHM of a λ transgene in mice (Klotz and Storb, 1996), and by the finding that iEµ increased SHM in a human pre-B lymphoma cell line, 18–81, transfected with a rearranged Igµ gene under the regulation of a thimidine kinase (tk) promoter and a 3′ κ chain enhancer (Bachl et al., 1998). In mice, the addition of hs1,2 to transgenes, even to those containing iEµ, did not increase mutation frequency to level comparable to that of endogenous IgH (Tumas-Brundage et al., 1997; Terauchi et al., 2001). Further addition of hs3b-hs4 increased the mutation load by eight-folds, suggesting a role of hs3b-hs4 in SHM (Terauchi et al., 2001). However, in knockout mice lacking the hs3b-hs4 region, the mutation efficiency in the JH intronic region is similar in the wildtype and targeted allele, suggesting that the function of hs3b-hs4 can be compensated by other IgH locus cis-elements (Morvan et al., 2003).
These preliminary data from different models prompted us to undertake a systematic analysis of the contribution of the IgH cis-elements in SHM. We reasoned that a human IgH construct that can be transfected into Ramos B cells, a spontaneously hypermutating human Burkitt’s lymphoma cell line (Sale and Neuberger, 1998), would be a good vehicle to analyze the function of the iEµ, hs1,2 and hs3-hs4 enhancers in different combinations. We further reasoned that regulation of transcription of such IgH constructs by its own promoter would have higher physiological relevance. To this end, we cloned the VHDJH DNA from a hybridoma that secrets a human IgG mAb against rabies virus, mAb57 (Ueki et al., 1990; Ikematsu et al., 1993) in a “physiological” configuration, that is, flanked 5′ by its own VH promoter and 3′ by the iEµ enhancer, followed by a recombined Sµ-Sγ1 region preceding the first two exons of the Cγ1 cluster. This was immediately followed downstream by the 3′Eα hs1,2 or hs3-hs4 enhancers. Different iterations of this construct, containing or not containing the iEµ and/or 3′ enhancers, were used to stably transfect into Ramos B cells. This Burkitt’s lymphoma cell line displays a germinal center-like phenotype and spontaneously hypermutates the rearrange and expressed Ig V(D)J genes in vitro. The frequency and the nature of VH region mutations in stable Ramos B cell transfectants were analyzed to address the role of the iEµ and 3′Eα enhancers in SHM.
2. Materials and methods
2.1. Plasmid construction
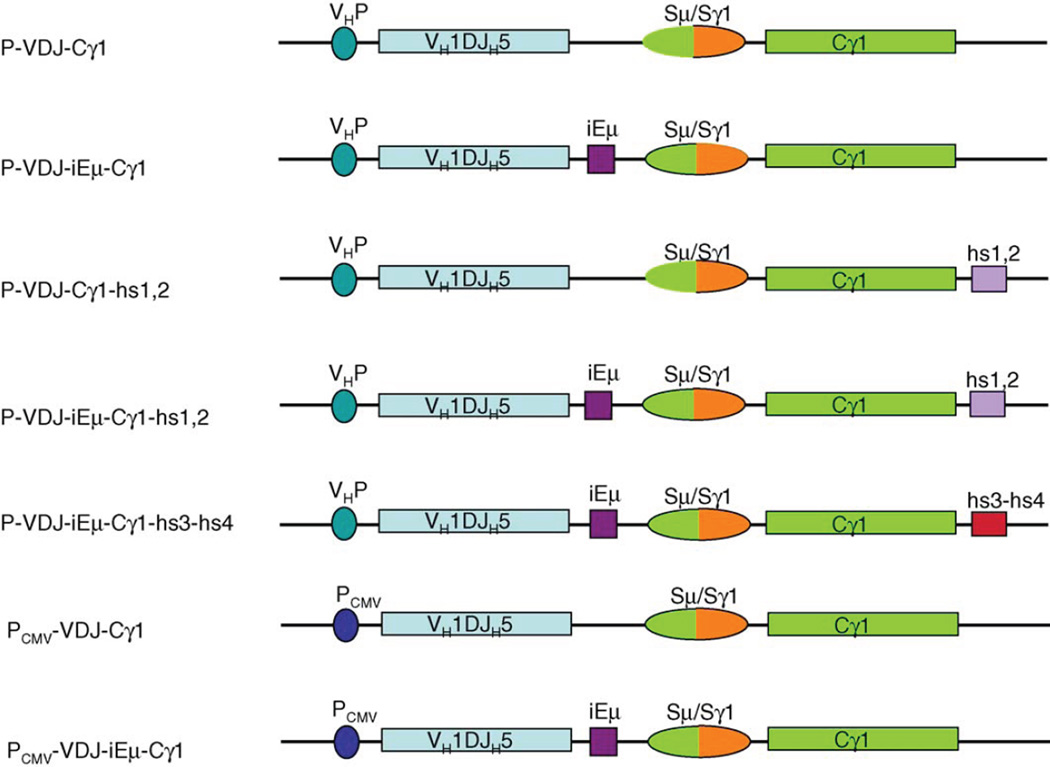
Genomic DNA was extracted using the QIAamp DNA mini kit (Qiagen Inc.). The 5′-flanking VH1 promoter, rearranged Vh1-DXP′1-Jh5, iEµ and first two exons of switched Cγ1 sequence DNA (P-VDJ-iEµ-Cγ1) were amplified en block by Elongase (Invitrogen Corp.) from the genomic DNA of the mAb57-secreting hybridoma, using the primer A (forward primer with KpnI site: 5′-AGAGCTGGGTACCGCAGGATT-TAGGGCTTGGTCTC-3′) and primer B (reverse primer with EcoRI site: 5′-AGAGCTGAATTCTTGGAGATGGTTTTCTC-GATG-3′). A modified pcDNA3.1 vector (Invitrogen Corp., Carlsbad, CA) with the 667 bp MluI-NheI fragment of CMV promoter–enhancer excised, was used to clone and express P-VDJ-iEµ-Cγ1. To construct P-VDJ-Cγ1 which lacks iEµ, the rearranged VH1-DXP′1-JH5 DNA and the Sµ/Sγ1-Cγ1 DNA were amplified separately, using the primers A and C (reverse primer with PacI site: 5′-AGAGCTTAATTAA-AAGGCATCGGAAATCCACAG-3′), D (forward primer with Pad site: 5′-AGAGCTTAATTAAACTACCCAGAGCT-GGGATGCG-3′) and B, respectively, and ligated. To construct P-VDJ-iEµ-Cγ1-hs1,2, P-VDJ-hs1,2 and P-VDJ-iEµ-Cγ1-hs3-hs4, human hs1,2 or hs3-hs4 DNA (kindly provided by Dr. Edward E. Max) were ligated to the 3′ end of P-VDJ-iEµ-Cγ1 or VDJ-Cγ1, correspondingly. To construct PCMV-VDJ-iEµ-Cγ1, PCMV-VDJ-Cγ1 and pEBVHis-VDJ-iEµ-Cγ1, unmodified pcDNA3.1 or pEBVHisB (Invitrogen Corp.) vectors were used.
2.2. Ramos B cells and transfection conditions
The human Burkitt’s lymphoma B cell line Ramos was maintained in RPMI 1640 supplemented with 10% FBS, 2mM l-glutamine, 100 units/ml penicillin and 100 µg/ml streptomycin. Transfection was performed with Cellfectin reagent (Invitrogen Corp.). Forty-eight hours after transfection, G418 was added to a final concentration of 0.5 mg/ml. Cells were seeded into a 24-well plate at a density of 105 cells/ml. Twelve days later, G418-resistant cell cultures were grown in media with 0.2 mg/ml G418. Ramos B cells transfected with pEBVHisB-VDJ-iEµ-Cγ1 was selected in media containing 50 µg/ml hygromycin (Sigma, St. Louis, MO).
2.3. PCR amplification of transfected DNA
After a 30-day culture, genomic DNA was extracted from at least two independent cultures of Ramos B cells and transfected genes were amplified by PCR using Elongase. The forward primers were specific for multicloning sites at the vectors used: E (5′-GTTTAAACTTAAGCTTGGTACC-3′) for pcDNA3.1 with CMV promoter excised, F (5′-GCTGGCTAGCGTTTAAAC-TT-3′) for pcDNA3.1 vector and G (5′-GACGATAAGGAT-CCGAGCTCGAGATCT-3′) for pEBVHisB vector. The reverse primer was H (5′-ATGTAGGCTGTGCTCGTGGATTCG-3′), located within the VH1 region. The PCR products were purified and cloned into the pCR-Blunt II-TOPO vector (Invitrogen Corp.) and transformed into Top10 competent cells (Invitrogen Corp.). Bacterial colonies were screened by PCR using primers I (forward, 5′-AGGTTCCTCTTTGTGGTGGC-3′) and H, and VH1 gene positive colonies were subjected to single-strand conformational polymorphism (SSCP) analysis.
2.4. Detection of mutated DNA sequences by SSCP and DNA sequencing
For SSCP analysis, performed on Genephor Electrophoresis Unit (Amersham Biosciences), cloned DNA was amplified by PCR using the primers described above. Five microliter PCR products were mixed with 5 µl denaturing solution (95% formamide, 0.05% xylene cyanol and 0.04% bromophenol blue), denatured at 98 °C for 8 min, quickly chilled on ice, and loaded onto GeneGel Excel 12.5/24 kit (Amersham Biosciences). DNA with mutations displayed an altered electrophoretic mobility on the SSCP gel and the corresponding plasmid DNA was subjected to sequence analysis using the Taq DiDeoxy Terminator Cycle Sequencing Kit and 373 Automatic Sequencer (Applied Biosystems).
2.5. Semiquantitative RT-PCR detecting transcription of transfected genes and AID
RNA was extracted from Ramos B cells using RNAeasy Mini Kit (Qiagen Inc.). Total RNA (5 µg) was used as the template for synthesis of first-strand cDNA using the Superscript Preamplification System (Invitrogen Corp.). Serial diluted cDNA was used as templates for PCR using primers I (forward) and J (reverse: 5’-ACGGTCCCCCCAGGAGTTCAGGTAG-3′, within CH2 of Cγ1) for the transfected genes. Primers to detect AID and β-actin transcripts by RT-PCR were reported (Zan et al., 2003).
2.6. Statistical analysis
Statistical t-test was performed using the Prism software (GraphPad Software) and returned p-value was used to detect statistically significant differences.
3. Results
3.1. SHM in an assembled VHDJH-Cγ1 construct
To identify the role of different IgH locus cis-elements in SHM, we established an in vitro system that would allow us to test different DNA elements for their ability to support SHM. To this end, we cloned the complete 5′ sequence of the rearranged IgH of mAb57 into a modified pcDNA3.1 vector whose cytomegalovirus immediate early promoter (CMV promoter) was excised. The cloned DNA fragment contains the endogenous VH1 promoter (VHP), rearranged VH1-DXP′1-JH5 gene segment, iEµ and its flanking matrix attachment regions (MARs), post-CSR junction Sµ/Sγ1 sequence, and the first and the second exons of Cγ1 (P-VDJ-iEµ-Cγ1) (Fig. 1). Thirty days after transfection of the plasmid containing P-VDJ-iEµ-Cγ1 into Ramos B cells, the VH1-DXP′1-JH5 region was amplified from selected cells, submitted to SSCP analysis and sequenced. The endogenous rearranged and expressed VH4-DXP′1-JH6 region of Ramos B cells was subjected to the similar sequencing analysis.
Fig. 1.
Schematic illustration of different constructs containing the rearranged VH1-DXP′1-JH5, post-switched Sµ/Sγ1 region, the first two exons of the Cγ1 region and different combinations of cis-regulating elements. cis-Regulating elements, such as the endogenous VH promoter, iEµ, hs1,2, hs3-hs4 and CMV promoter, were indicated.
By SSCP analysis, 14.1% of Ramos VH4-DXP′1-JH6 sequences showed one or more mutations and the mutational frequency was about 5.6 × 10−4 mutation/bp (Table 1). If only unique mutations were counted, this yielded a mutation rate of 2.8 × 10−5 mutation/bp/cell generation, which is comparable to the SHM rate previously reported in Ramos B cells (Sale and Neuberger, 1998; Zhang et al., 2001) and in the hypermutating pre-B cell line 18–81 (Bachl and Wabl, 1996; Bachl et al., 2001). Likewise, among 192 sequences of VH1-DXP′1-JH5 region in the P-VDJ-iEµ-Cγ1, 24 (12.5%) had one or more mutation and the mutational frequency was 4.4 × 10−4 mutation/bp (Table 1), which yielded a mutation rate of 2.2 × 10−5 mutation/bp/cell generation. These mutation rates were almost 10-folds greater than those that could have been generated by the Elongase, whose error rate is 2.3 × 10−6 change/base/cycle. Thus, the assembled VHDJH-Cγ1 construct underwent SHM at a frequency comparable to that of the endogenous Ramos VH4-DXP′1-JH6 segment.
Table 1.
Mutation frequency and mutation rate in VHDJH regions
| Mutated genes | Mutated/total sequences | Total mutations/total bps | Mutation frequency (mutation/bp) |
Mutation rate (mutations/bp/ce 11 division)a |
|---|---|---|---|---|
| Endogenous VH4-DXP′1-JH6 | ||||
| Ramosb | 34/240 | 40/71,280 | 5.6 × 10−4 | 2.8 × 10−5 |
| Ramosc | 2.7 × 10−5 | |||
| Exogenous VH1-DXP′1-JH5 | ||||
| P-VDJ-iEµ-Cγ1 | 24/192 | 31/70,848 | 4.4 × 10−4 | 2.2 × 10−5 |
| P-VDJ-Cγ1 | 26/386 | 29/141,696 | 2.0 × 10−4 | 1.0 × 10−5 |
| P-VDJ-iEµ-Cγ1-hs1,2 | 36/336 | 44/123,984 | 3.5 × 10−4 | 1.8 × 10−5 |
| P-VDJ-Cγ1-hs1,2 | 47/432 | 50/159,408 | 3.1 × 10−4 | 1.6 × 10−5 |
| P-VDJ-iEµ-Cγ1-hs3-hs4 | 26/192 | 27/70,848 | 3.8 × 10−4 | 1.9 × 10−5 |
| PCMV-VDJ-iEµ-Cγ1Cγ 1 | 12/96 | 29/35,424 | 8.2 × 10−4 | 8.2 × 10−5d |
| PCMV-VDJ-Cγ1 | 11/96 | 16/35,424 | 4.4 × 10−4 | 4.4 × 10−5d |
| pEBVHis-VDJ-iEµ-Cγ1 | 11/96 | 12/35,424 | 3.4 × 10−4 | 3.4 × 10−5d |
Mutation rate calculated based on 20 cell generations with a doubling time of 36 h.
Data from this study.
Data from Sale and Neuberger (1998).
Mutation rate calculated based on 10 cell generations.
3.2. Preferentially targeting of dA/dT by mutations in the exogenous VH1-DXP′ 1-JH5 DNA in Ramos B cells
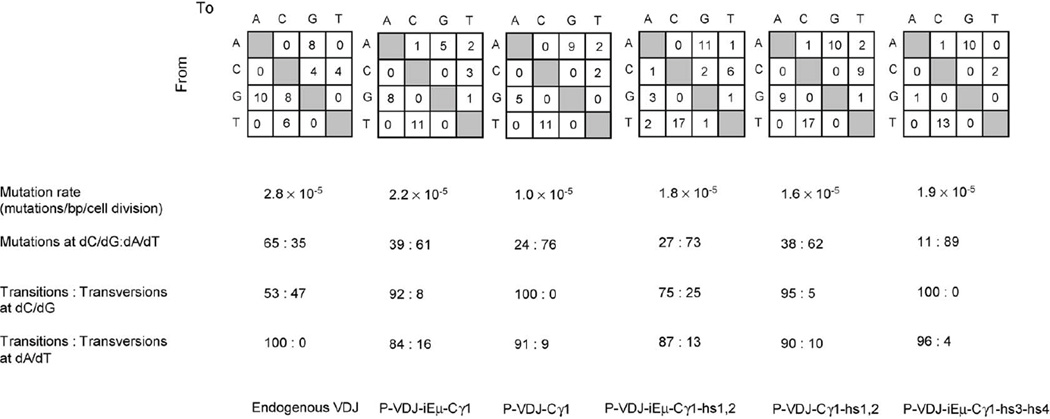
The sequence of VH1-DXP′1-JH5 was analyzed to determine whether the mutation pattern of the exogenous DNA was different from that of the endogenous VH gene. In the endogenous Ramos VH4-DXP′1-JH6, 65% of mutations were dC/dG mutations without a preference for transitions or transversions, while all dA/dT mutations (35%) were dA → dG or dT → dC transitions (Fig. 2). In contrast, mutations in the transfected VH1-DXP′1-JH5 DNA (P-VDJ-iEµ-Cγ1) (Fig. 1) showed a strong preference for dA/dT residues (61%). Eighty-four percent of these dA/dT mutations were transitions, and so were 92% of the accompanying dC/dG mutations.
Fig. 2.
Mutation rate and spectrum in endogenous Ramos B cells VH4-DXP′1-JH6 and transfected VH1-DXP′1-JH5 gene segments. The number and nature of independent mutations scored in the endogenous VH4-DXP’1-JH6 or VH 1-DXP′1-JH5 from transfected contructs, as indicated at the bottom, are depicted in the upper squares. Tabulated are mutation rate, the ratio of mutations at dC/dG vs. at dA/dT, and the ratio of transitions vs. transversions at both dA/dT and dC/dG mutations.
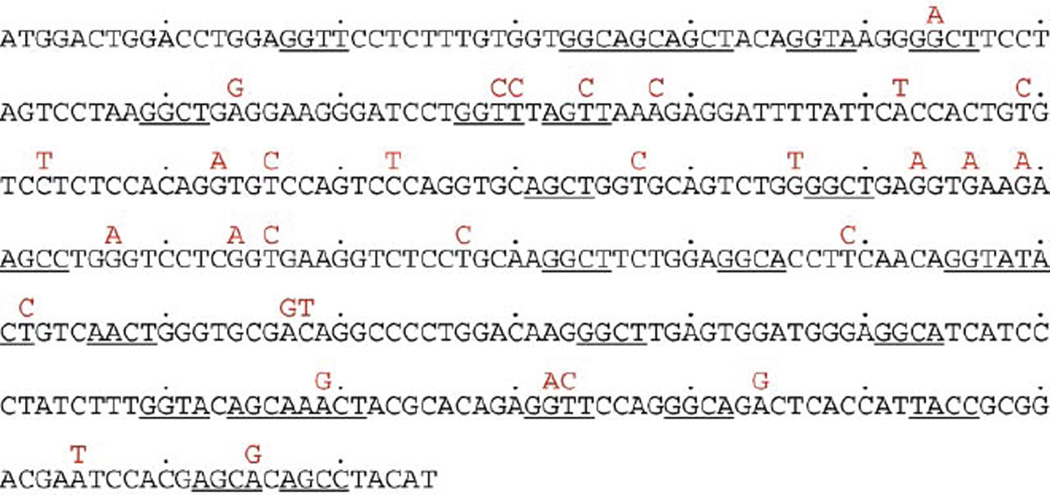
SHM preferentially targets the RGYW/WRCY mutational hotspot. In VH 1-DXP′ 1-JH5, there are 27 RGYW/WRCY motifs which account for 27.5% of the unmutated sequence (Fig. 3). Sequence analysis showed that 47% of the dA/dT mutations segregated within RGYW/WRCY, in excess of 25% of dA/dT that are within RGYW/WRCY in the germline sequence (p < 0.05). In contrast, only 16.7% of the dC/dG mutations segregated within RGYW/WRCY, while 28.1% of the overall dC/dG are within RGYW/WRCY in the germline sequence. Thus, in the transfected VH1-DXP′1-JH5 DNA, preferential targeting of the RGYW/WRCY hotspot was a feature of dA/dT mutations.
Fig. 3.
Somatic point-mutations in the VH1-DXP′1-JH5 region of the P-VDJ-iEµ-Cγ 1 construct. The mutations are above the germline VH1-DXP′1-JH5 sequence; the RGYW/WRCY motifs in the germline VH1-DXP′1-JH5 are underlined.
3.3. Differential regulation of SHM by iEµ, hs1,2 and hs3-hs4 enhancers
Since the hypermutation machinery was active in Ramos B cells and could target the exogenous P-VDJ-iEµ-Cγ1 DNA, we used different iterations of the P-VDJ-iEµ-Cγ1 construct containing different combination of cis-regulatory elements (Fig. 1) to dissect the effects of such elements on SHM. Substitution of the endogenous VH1 promoter with the CMV promoter (PCMV-VDJ-iEµ-Cγ1) did not alter the proportion of mutated sequences (12.5%), but increased the overall mutational frequency from 4.4 × 10−4 to 8.2 × 10−4 mutation/bp (p < 0.005) (Table 1). The PCMV-VDJ-iEµ-Cγ1-transfected cells were cultured for 10 generations before sequence analysis, yielding a mutation rate of 8.2 × 10−5 mutation/bp/cell generation, which was about fourfold greater than that of P-VDJ-iEµ-Cγ1, indicating that the Ig VH promoter was not essential and could be functionally replaced by a heterologous promoter in SHM.
Deletion of iEµ (P-VDJ-Cγ1) significantly dampened SHM by decreasing both the proportion of mutated sequences, from 12.5 to 6.7%, and the overall mutation frequency, from 4.4 × 10−4 to 2.0 × 10−4 mutation/bp (p<0.05) (Table 1). A comparable reduction in SHM was also evident when comparing the mutational frequencies of PCMV-VDJ-iEµ-Cγ1 and its counterpart PCMV-VDJ-Cγ1 (8.2 × 10−4 and 4.4 × 10−4 mutation/bp, respectively) (p < 0.05), suggesting that iEµ positively regulated SHM in the IgH locus.
We next examined the contribution of the hsl,2 and hs3-hs4 enhancers to SHM. Addition of the hsl,2 (P-VDJ-iEµ-Cγ1-hs1,2) (Fig. 1) did not significantly affect the rate or the spectrum of mutations in VH1-DXP′1-Jh5 (Fig. 2). In the presence of hs1,2, deletion of iEµ from P-VDJ-iEµ-Cγ1-hs1,2 (P-VDJ-Cγ1-hs1,2) (Fig. 1) resulted in only a 11% decrease of mutation rate (Fig. 2; Table 1). This contrasted with the 50% reduction of mutations in P-VDJ-Cγ1 compared to P-VDJ-iEµ-Cγ1, suggesting that hs1,2 could at least partially compensate for the lack of iEµ function. Addition of the hs3-hs4 region (P-VDJ-iEµ-Cγ1-hs3-hs4) (Fig. 1) did not change the mutation rate, but further swayed the mutational spectrum towards dA/dT mutations (Fig. 2; Table 1). Indeed, transition mutations at dA/dT accounted for 85% of total mutations in P-VDJ-iEµ-Cγ1-hs3-hs4, as compared to 59% in P-VDJ-iEµ-Cγ1 (p<0.05). Thus, different 3′Eα enhancer elements had different role in SHM by affecting either mutation efficiency or spectrum.
3.4. SHM is independent of chromosomal integration or transcription level of transfected genes
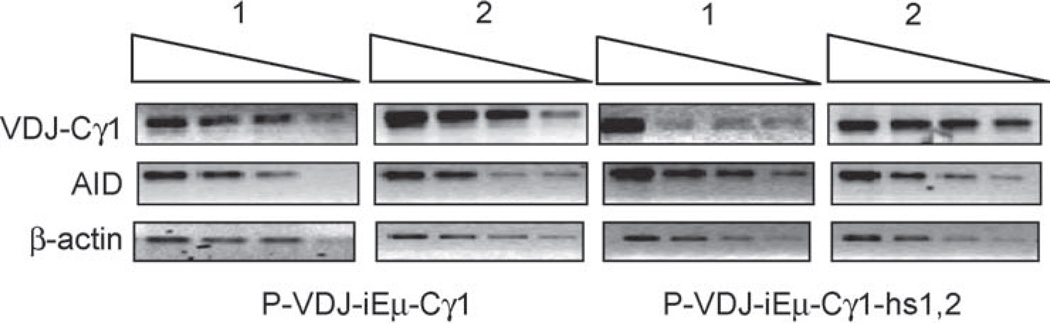
To rule out the possibility that the mutation rate of transfected genes was dependent on chromosomal integration sites, we cloned the P-VDJ-iEµ DNA into the pEBVHis vector which can replicate and persist as an episome in mammalian cells. The mutation rate of the VH1-DXP′1-JH5 segment in the episomal pEBVHis-P-VDJ-iEµ plasmid was 3.4 × 10−5 mutation/bp/cell generation (Table 1), a rate comparable to that of the VH1-DXP′1-JH5 DNA cloned into the pcDNA3.1 vector and integrated into chromosomal DNA. Also, there was no significant difference in the nature of mutations, including the preferential dA/dT mutations and bias of transitions over transversions (data not shown). To determine whether the mutation rate was correlated with the level of transcription, we analyzed the level of VHDJH-Cγ1 transcript encoded by P-VDJ-iEµ-Cγ1 or P-VDJ-iEµ-Cγ1-hs1,2 in two independent transfectants of each construct. Although all transfectants had similar mutation frequencies at the VH1-DXP′1-JH5 region (data not shown), the VHDJH-Cγ1 cDNA level was within an eight-fold range and the AID cDNA level did not vary significantly (Fig. 4). Thus, SHM in the assembled P-VDJ-iEµ-Cγ1 construct was not significantly affected by the site of its chromosomal integration or transcription level.
Fig. 4.
Expression of the VH1-DXP′1-JH5, AID and β-actin in two independent transfectants of each of P-VDJ-iEµ-Cγ1 and P-VDJ-iEµ-Cγ1-hs1,2, as analyzed by semi-quantitative RT-PCR using serially two-fold diluted cDNA as templates.
4. Discussion
We show here that SHM occurs in the V region of the rear-ranged human IgH locus that was stably transfected into Ramos B cells. Surprisingly, SHM in the transfected VH1-DXP′1-JH5 DNA showed a strong dA/dT bias. Such a dA/dT bias was not restricted to the integrated transgene because mutations in the same VH1-DXP′1-JH5 DNA maintained episomally also targeted dA/dT. The dA/dT bias was characteristic to the mutations in the VH 1-DXP′ 1-JH5 construct, whether integrated or in a episomal form, as mutations in the endogenous VH4-DXP′1-JH6 DNA displays a characteristic dC/dG bias (Sale and Neuberger, 1998).
The spectrum of mutations is determined by two stages of SHM process: (i) the initiating stage that generates DNA lesions; (ii) the repair stage that involves MMR and error-prone DNA polymerases (Xu et al., 2005). A prevailing model of SHM entails a first stage in which AID directly deaminates dC by AID (initiating stage) to yield dU:dG mispairs. Such dU:dG lesions can be “replicated over” yielding dC/dG transitions (Phase 1a), or, upon dU deglycosylation by a uracil deglycosylase (UDG), give rise to abasic sites that are bypassed by translesion DNA polymerases, yielding both transition and transversions (Phase 1b) (Rada et al., 2004; Diaz and Lawrence, 2005; Neuberger et al., 2005; Zan et al., 2005). Alternatively, dU:dG mismatches can trigger the MMR pathway, which entails the excision of a stretch of nucleotides by a yet to be identified endonuclease and exonuclease ExoI and subsequent “patch repair” by the translesion DNA polymerases, such as pol θ and pol η, to generate mutations at dA/dT (Phase 2). If MMR in Phase 2 cannot cope with the rate of dU:dG mismatch emergence, error-prone repair or “replicating over” of the excessive dU:dG lesions would then lead to dC/dG biased mutations (Bachl and Wabl, 1996; Martin et al., 2002). While in Ramos B cells the Msh2 activity is seemingly normal (Sale and Neuberger, 1998), recruitment of Msh2 to the endogenous VH4-DXP′1-JH6 DNA might be inefficient, leading to ineffective MMR and lack of dA/dT mutations. The dA/dT-biased mutations in the transfected VH1-DXP′ 1-JH5 DNA suggest that Msh2, recruited by specific but yet identified features of VH1-DXP′1-JH5, triggers MMR in Phase 2. Translesion DNA polymerases, pol η in particular, would be recruited by Msh2 to insert mismatches mostly at dA/dT Indeed, pol η physically interacts with Msh2 (Wilson et al., 2005) and displays a preference in inserting transition at dA/dT (Rogozin et al., 2001; Zeng et al., 2001; Delbos et al., 2005; Martomo et al., 2005), consistent with the predominant dA/dT transitions in the VH 1-DXP′ 1-JH5 DNA. In turn, the newly generated mismatches at dA/dT could trigger additional rounds of MMR, thereby introducing more mutations at dA/dT residues. The excess transition mutations at dC/dG in the VH 1-DXP′1-JH5 DNA contrast with the comparable frequency of transitions and transversions at dC/dG in the endogenous VH4-DXP′1-JH6 DNA. As the MMR in Phase 2 is the preferred pathway to repair the dU:dG lesions in the VH 1-DXP′ 1-JH5 DNA, access of Phase 1b to the dU:dG lesions would be limited, leaving the “replicating over” as the only way to introduce mutations at dC/dG. Thus, the relative engagement of different pathways to repair AID-mediated dU:dG lesions would be determined by cis DNA sequences and would dictate the spectrum of mutations.
Another possible cause of the lack of dC/dG bias in the VH1-DXP′1-JH5 DNA is the significantly decreased targeting at dC/dG during the initiating stage. Indeed, the dA/dT mutations in VH 1-DXP′ 1-JH5 segregate within RGYW/WRCY “hot-spots”, while the dC/dG mutations do not show such a “hotspot” focusing. A dA/dT-biased targeting may involve DNA cleavage by a yet-to-be-defined endonuclease, perhaps encoded by an AID-edited mRNA (“RNA editing” hypothesis) (Honjo et al., 2002, 2004; Doi et al., 2003; Begum et al., 2004; Nagaoka et al., 2005), and would entail the generation of resected DSBs. Such resected ends have been shown to preferentially target RGYW/WRCY motifs and are repaired by translesion DNA polymerases (Zan et al., 2001, 2003; Xu et al., 2005).
Mutation rate of PCMV-VDJ-iEµ-Cγ1 is four-fold greater than that of P-VDJ-iEµ-Cγ1, which is consistent with previous findings that the Ig VH promoter is not essential for SHM and can be functionally replaced by a heterologous promoter (Betz et al., 1994; Tumas-Brundage and Manser, 1997). SHM of VH1-DXP′1-JH5 DNA was differentially regulated by the iEµ, hs1,2 and hs3-hs4 enhancers. Compared to that of P-VDJ-Cγ1, the increased mutation frequencies in P-VDJ-iEµ-Cγ1 and P-VDJ-iEµ-Cγ1-hs1,2 was unlikely due to possible transcription enhancing effects of iEµ or hs1,2 since the transcription level of VH1-DXP′1-JH5 could vary by as much as eight-fold among Ramos B cells with comparable mutation frequencies. This finding is in apparent contrast with a previous report suggesting that increased transcription levels induces higher mutation rates in 18–81 cells using an inducible GFP-transgene expression system (Bachl et al., 2001). This difference could be explained by the different cell lines used and/or promoters and genes analyzed. A κ transgene with a short and defined insert containing two E-box motifs “CAGGTG” in the V region showed a mutation load four-fold greater than that of the transgene in which “CAGGTG” was replaced with “AAGGTG” (Michael et al., 2002, 2003). However, the static state mRNA level of two transgenes inversely correlated to their mutability, consistent with the notion that enhancer motifs modulate SHM efficiency independent of there transcription enhancing function. Instead, it is more likely that trans-factors recruited by iEµ or hs1,2 participate in either the initial targeting or the lesion repairing stages by forming protein complexes with critical factors such as AID or Msh2. The complex formation between RNA polymerase II and AID (Nambu et al., 2003) may be bridged by such a fraras-factor as well. Alternatively, the recruitment of DNA-binding proteins would modify the DNA architecture, thereby making the IgH region more accessible to the SHM machinery, as suggested by iEµ mediated-induction of chromatin conformational alternation (Nikolajczyk et al., 1996, 1999). Indeed, the binding of the heterodimeric nucleolin/hnRNP D to iEµ bends DNA through contacts with the DNA minor groove (Hanakahi and Maizels, 2000), a mechanism similar to that employed by HMG1 and HMG2 to bend DNA and modulate the RAG1- and RAG2-mediated cleavage of DNA during the V(D)J recombination (Sawchuk et al., 1997; van Gent et al., 1997).
Although deletion of either iEµ or hs1,2 decreased mutation rates in the VH 1-DXP′ 1-JH5 DNA, a combination of the two elements did not synergize to increase the frequency of mutations or alter the spectrum of mutations. The comparable mutation frequency and pattern of P-VDJ-iEµ-Cγ1 and P-VDJ-Cγ1-hs1,2 are consistent with a previous study in transgenic mice showing comparable mutation frequency and distribution in a rearranged transgene containing the mouse hs1,2 enhancer (Terauchi et al., 2001). Our findings suggest that iEµ and hs1,2 act in pathways that are at least overlapping, possibly by recruiting the same protein factor(s). A sequence survey for trans-factor binding sites (http://www.cbil.upenn.edu/tess/) reveals several motifs shared by iEµ and hs1, including the µE3 motif “CATGTG”, which recruits the E-box factor TFE3 (Ernst and Smale, 1995). Interestingly, this motif is also present in some VH promoters and κ light chain enhancer (Peterson and Calame, 1989), suggesting a possible role of TFE3 in the V region transcription and SHM.
As we show in this study, in addition to their role in effecting long-distance regulation of CSR in mice (Pinaud et al., 2001), hs3-hs4 also regulates SHM in human B cells. The hs3-hs4 enhancer contributed a stronger dA/dT bias in the VH1-DXP′1-JH5 DNA without increasing the mutation load, an overall effect that is different from that of iEµ or hs1,2. It is likely that a different set of protein factors are recruited to modulate the activity of either initial lesion-generating factors or error-prone DNA polymerases during repair. Our findings are apparently at odd with a previous study showing an hs3b-hs4-mediated increase, by as much as 10-folds, of the mutation rate in the absence of alternations in mutation spectrum in transgenic mice (Terauchi et al., 2001). However, they are in agreement with another study showing no appreciable change of SHM after deletion of hs3b-hs4 enhancers in a knockout mice strain (Morvan et al., 2003). An explanation for these conflicting results would be that in addition to difference in genes analyzed, human hs3 or hs4 and mouse hs3a, 3b or hs4 region are differentially regulated and exert distinct functions in SHM by virtue of different proteins recruited. It is notable that transgenes carrying mutations at the PU.1 or NF-EM5 binding motifs in the IgK 3′ enhancer also displayed biased dA/dT mutations (Kodama et al., 2001), further supporting the notion that discrete binding motifs for trans-acting factors play an important role in defining mutation patterns. In addition to outlining elements and mechanisms that underlie SHM targeting of dA/dT, our system can be used to precisely define the roles of additional cis-elements and trans-factors mediating the expressing characteristic modalities of SHM.
Acknowledgements
We thank Junli Feng for excellent technical assistance. This work was supported by Grants NIH AR 40908, AI 45011 and AI 60573 to P. Casali.
Abbreviations
- AID
activation-induced cytidine deaminase
- BCR
B cell receptor
- BER
base excision repair
- DSB
double strand break
- iEµ
intronic Ig µ enhancer
- Ig
immunoglobulin
- MMR
mismatch repair
- SHM
somatic hypermutation
- SSCP
single-strand conformational polymorphism
References
- Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- Bachl J, Olsson C, Chitkara N, Wabl M. The Ig mutator is dependent on the presence, position, and orientation of the large intron enhancer. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2396–2399. doi: 10.1073/pnas.95.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachl J, Wabl M. An immunoglobulin mutator that targets GC base pairs. Proc. Nat. Acad. Sci. U.S.A. 1996;93:851–855. doi: 10.1073/pnas.93.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffher W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Begum NA, Kinoshita K, Kakazu N, Muramatsu M, Nagaoka H, Shinkura R, Biniszkiewicz D, Boyer LA, Jaenisch R, Honjo T. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 2004;305:1160–1163. doi: 10.1126/science.1098444. [DOI] [PubMed] [Google Scholar]
- Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene; critical role for the intronic enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Brass L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- Chauveau C, Cogne M. Palindromic structure of the IgH 3’ locus control region. Nat. Genet. 1996;14:15–16. doi: 10.1038/ng0996-15. [DOI] [PubMed] [Google Scholar]
- Chen C, Birshtein BK. Virtually identical enhancers containing a segment of homology to murine 3’IgH-E(hs1,2) lie downstream of human Ig Cα1 and Cα2 genes. J. Immunol. 1997;159:1310–1318. [PubMed] [Google Scholar]
- Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Casali P. Somatic immunoglobulin hypermutation. Curr. Opin. Immunol. 2002;14:235–240. doi: 10.1016/s0952-7915(02)00327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26:215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Doi T, Kinoshita K, Ikegawa M, Muramatsu M, Honjo T. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2634–2638. doi: 10.1073/pnas.0437710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Smale ST. Combinatorial regulation of transcription II: the immunoglobulin mu heavy chain gene. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Foster SJ, Dorner T, Lipsky PE. Somatic hypermutation of VκJκ rearrangements: targeting of RGYW motifs on both DNA strands and preferential selection of mutated codons within RGYW motifs. Eur. J. Immunol. 1999;29:4011–4021. doi: 10.1002/(SICI)1521-4141(199912)29:12<4011::AID-IMMU4011>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Hanakahi LA, Maizels N. Transcriptional activation by LR1 at the Emu enhancer and switch region sites. Nucleic Acids Res. 2000;28:2651–2657. doi: 10.1093/nar/28.14.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Honjo T, Muramatsu M, Fagarasan S. AID: how does it aid antibody diversity? Immunity. 2004;20:659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Ikematsu H, Harindranath N, Ueki Y, Notkins AL, Casali P. Clonal analysis of a human antibody response. J. Immunol. 1993;150:1325–1337. [PMC free article] [PubMed] [Google Scholar]
- Khamlichi AA, Pinaud E, Decourt C, Chauveau C, Cogne M. The 3’ IgH regulatory region: a complex structure in a search for a function. Adv. Immunol. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- Kim EC, Edmonston CR, Wu X, Schaffer A, Casali P. The HoxC4 homeodomain protein mediates activation of the immunoglobulin heavy chain 3’ hs1,2 enhancer in human B cells. Relevance to class switch DNA recombination. J. Biol. Chem. 2004;279:42258–42269. doi: 10.1074/jbc.M407496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz EL, Storb U. Somatic hypermutation of a lambda 2 trans-gene under the control of the lambda enhancer or the heavy chain intron enhancer. J. Immunol. 1996;157:4458–4463. [PubMed] [Google Scholar]
- Kodama M, Hayashi R, Nishizumi H, Nagawa F, Takemori T, Sakano H. The PU.l and NF-EM5 binding motifs in the Igκ 3’ enhancer are responsible for directing somatic hypermutations to the intrinsic hotspots in the transgenic Vκ gene. Int. Immunol. 2001;13:1415–1422. doi: 10.1093/intimm/13.11.1415. [DOI] [PubMed] [Google Scholar]
- Linderson Y, French NS, Neurath MF, Pettersson S. Context-dependent Pax-5 repression of a PU.1/NF-kB regulated reporter gene in B lineage cells. Gene. 2001;262:107–114. doi: 10.1016/s0378-1119(00)00546-1. [DOI] [PubMed] [Google Scholar]
- Martin A, Bardwell PD, Woo C, Fan M, Schulman MJ, Scharff MD. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 2002;14:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase η- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael N, Martin TE, Nicolae D, Kim N, Padjen K, Zhan P, Nguyen H, Pinkert C, Storb U. Effects of sequence and structure on the hypermutability of immunogloblin genes. Immunity. 2002;16:123–134. doi: 10.1016/s1074-7613(02)00261-3. [DOI] [PubMed] [Google Scholar]
- Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- Mills FC, Harindranath N, Mitchell M, Max EE. Enhancer complexes located downstream of both human immunoglobulin Cα genes. J. Exp. Med. 1997;186:854–858. doi: 10.1084/jem.186.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan CL, Pinaud E, Decourt C, Cuvillier A, Cogne M. The immunoglobulin heavy-chain locus hs3b and hs4 3’ enhancers are dispensable for VDJ assembly and somatic hypermutation. Blood. 2003;102:1421–1427. doi: 10.1182/blood-2002-12-3827. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand V, Anant S, Sugai M, Kinoshita K, Davidson N, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Nagaoka H, Ito S, Muramatsu M, Nakata M, Honjo T. DNA cleavage in immunoglobulin somatic hypermutation depends on de novo protein synthesis but not on uracil DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2022–2027. doi: 10.1073/pnas.0409491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Di Noia JM, Beale RC, Williams GT, Yang Z, Rada C. Somatic hypermutation at A·T pairs: polymerase error versus dUTP incorporation. Nat. Rev. Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- Nikolajczyk BS, Nelsen B, Sen R. Precise alignment of sites required for mu enhancer activation in B cells. Mol. Cell. Biol. 1996;16:4544–4554. doi: 10.1128/mcb.16.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolajczyk BS, Sanchez JA, Sen R. ETS protein-dependent accessibility changes at the immunoglobulin µ heavy chain enhancer. Immunity. 1999;11:11–20. doi: 10.1016/s1074-7613(00)80077-1. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Calame K. Proteins binding to site C2 (µE3) in the immunoglobulin heavy-chain enhancer exist in multiple oligomeric forms. Mol. Cell. Biol. 1989;9:776–786. doi: 10.1128/mcb.9.2.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogne M. Localization of the 3’ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the a/t-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Rada C, Gonzalez-Fernandez A, Jarvis JM, Milstein C. The 5’ boundary of somatic hypermutation in a Vκ gene is in the leader intron. Eur. J. Immunol. 1994;24:1453–1457. doi: 10.1002/eji.1830240632. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio A, Brousse N, Muramatsu M, Notarangelo L, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rogerson BJ. Mapping the upstream boundary of somatic mutations in rearranged immunoglobulin transgenes and endogenous genes. Mol. Immunol. 1994;31:83–98. doi: 10.1016/0161-5890(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat. Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- Sale JE, Neuberger MS. TdT-accessible breaks are scattered over the immunoglobulin V domain in constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- Saleque S, Singh M, Little RD, Giannini SL, Michaelson JS, Birshtein BK. Dyad symmetry within the mouse 3′ IgH regulatory region includes two virtually identical enhancers (Cα3′E and hs3) J. Immunol. 1997;158:4780–4787. [PubMed] [Google Scholar]
- Sawchuk DJ, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig MC, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J. Exp. Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer A, Kim EC, Wu X, Zan H, Testoni L, Salamon S, Cerutti A, Casali P. Selective inhibition of class switching to IgG and IgE by recruitment of the HoxC4 and Oct-1 homeodomain proteins and Ku70/Ku86 to newly identified ATTT cis-elements. J. Biol. Chem. 2003;278:23141–23150. doi: 10.1074/jbc.M212952200. [DOI] [PubMed] [Google Scholar]
- Sepulveda MA, Emelyanov AV, Birshtein BK. NF-κB and Oct-2 synergize to activate the human 3’ Igh hs4 enhancer in B cells. J. Immunol. 2003;172:1054–1064. doi: 10.4049/jimmunol.172.2.1054. [DOI] [PubMed] [Google Scholar]
- Terauchi A, Hayashi K, Kitamura D, Kozono Y, Motoyama N, Azuma T. A pivotal role fo DNase I-sensitive regions 3b and/or 4 in the induction of somatic hypermutation of IgH genes. J. Immunol. 2001;167:811–820. doi: 10.4049/jimmunol.167.2.811. [DOI] [PubMed] [Google Scholar]
- Tumas-Brundage K, Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J. Exp. Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumas-Brundage KM, Vora KA, Manser T. Evaluation of the role of the 3’α heavy chain enhancer in VH gene somatic hypermutation. Mol. Immunol. 1997;34:367–378. doi: 10.1016/s0161-5890(97)00065-5. [DOI] [PubMed] [Google Scholar]
- Ueki Y, Goldfarb IS, Harindranath N, Gore M, Koprowski H, Notkins AL, Casali P. Clonal analysis of a human antibody response. Quantitation of precursors of antibody-producing cells and generation and characterization of monoclonal IgM, IgG, and IgA to rabies virus. J. Exp. Med. 1990;171:19–34. doi: 10.1084/jem.171.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. ‘Spalog’ and ‘sequelog’: neutral terms for spatial and sequence similarity. Curr. Biol. 2004;14:R181–R183. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Wagner SD, Milstein C, Neuberger MS. Codon bias targets mutation. Nature. 1995;376:732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, Gearhart PJ. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Feng J, Komori A, Kim E, Zan H, Casali P. Immunoglobulin somatic hypermutation: double-strand DNA breaks. AID and error-prone DNA repair. J. Clin. Immunol. 2003;23:235–246. doi: 10.1023/a:1024571714867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Fulop Z, Zhong Y, Evinger AJ, III, Zan H, Casali P. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann. N. Y Acad. Sci. 2005;1050:146–162. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik M, Diaz M, Casali P. The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Shima N, Xu Z, Al-Qhatani A, Evinger AJ, III, Zhong Y, Schimenti JC, Casali P. The translesion polymerase θ plays a dominant role in Ig gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Winter DB, Kasmer KH, Lehmann AR, Gearhart PJ. DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bardwell PD, Woo C, Poltoratsky V, Scharff MD, Martin A. Clonal instability of V region hypermutation in the Ramos Burkitt’s lymphoma cell line. Int. Immunol. 2001;13:1175–1184. doi: 10.1093/intimm/13.9.1175. [DOI] [PubMed] [Google Scholar]