Abstract
Now it is well recognized that the microRNA (miRNA) expression is altered in response to internal (developmental or hormonal) or external stimuli such as biotic and abiotic stresses in plants. It is also known that several miRNAs are induced in response to deficiency of specific nutrients within the plant or in the external sources, i.e., soil/nutrient media. For instance, P-deprivation induces miR399, S-deprivation induces miR395 and Cu-deprivation induces miR398, miR397 and miR408 in several plant species. Although the transcription factors (PHR1, SLIM1 and SPB7) that regulate these nutrient-deprivation inducible miRNAs have been identified but the upstream biochemical pathway that activates them is relatively unknown. In a recent study, we demonstrated for the first time that redox signaling plays a critical role in S-deprivation-inducible miR395 expression in Arabidopsis. In this short review, we draw additional hypotheses for the involvement of redox signaling and/or reactive oxygen species (ROS) signaling in inducing other nutrient or stress-responsive miRNAs in plants.
Keywords: acclimation, arabidopsis, micrornas, oxidative stress, redox signaling
Being sessile organisms, plants must adjust their metabolism and growth to changing environment fairly quickly, as such, stress sensing and signaling are most critical steps in this process.1 One of the early responses to stress also include changes in redox (reduction-oxidation) homeostasis (redox imbalance). Stress-induced redox imbalances initiate compensatory responses via redox signaling network either to readjust redox homeostasis or to repair oxidative damage as well as in modulating gene expression associated with acclimation to stress.2 One important mechanism of redox-dependent signaling is based on the reversible oxidation and reduction of crucial Cys residues, which is a well-conserved feature among living organisms.3 In this thiol-based signaling network, redox cues convert the cysteine thiols to disulfide, sulfenic, sulfinic and sulfonic acid, nitrosothiol or glutathionylated forms.4
In a stricter sense thiol redox regulation and ROS signaling differ despite their intimate linkage through thiol peroxidases and the Halliwell-Asada-cycle.5 Distinguishing these 2 is challenging and, therefore, these terminologies have been used rather vaguely and interchangeably in the literature. Intracellular thiol redox status regulates a variety of cellular and molecular events such as the activity of proteins, signal transduction, transcription and several other cellular functions. On the other hand, besides inducing a shift in redox balance, oxidative stress can directly affects enzyme activities, signaling, gene expression, lipid peroxidation, and even cell death. Thus, often the effects of redox changes overlap with those of oxidative stress, therefore, it is hard to distinguish whether the observed changes in cellular responses are due to ROS signaling (oxidative stress) or due to stress-induced redox imbalance or both6.
In a cell, energy supply and consumption maintains the redox equilibrium and any imbalance between these processes could lead to redox imbalance. Under normal conditions, feedback regulation between upstream and downstream activities in a biochemical pathway adjusts this redox equilibrium, but such maintenance will often fail under stress conditions resulting in more reduced or oxidized states of the NADP/NADPH.7 The altered NADPH redox milieu will feed into the thiol protein redox network via NADPH-dependent thioredoxin reductases (NTRs) and NADPH-dependent glutathione reductases, which subsequently modify the redox state of the target proteins.4 Thus far, more than 400 thiol proteins are known in plants that can interact with thioredoxin or glutaredoxin or undergo major conformational changes upon shifting from a reduced to oxidized state.4 The target proteins vary widely in their functions ranging from metabolism to signaling to translation and transcription7.
Plant growth, development and adaptation to biotic and abiotic stresses depend on an accurate regulation of gene expression both in time and space. Recently the genome-encoded miRNAs emerged as important regulators of gene expression associated with acclimation to stress including nutrient deprivation as the abundances of various miRNAs are altered under stress.8,9 In a few cases, even the transcription factors (PHB, SPB, and SLIM1) that bind to the promoter regions of nutrient-responsive miRNAs have been characterized. However the upstream signaling pathway that regulates/activates these transcription factors is completely unknown. In our earlier report using mutants partially defective in redox pathways, redox signaling was shown to be responsible for the induction of miR395 under S deprivation.10 This was the first characterization of upstream biochemical pathway(s) involved in regulation of a miRNA expression in plants.
Changes in cellular redox properties have been observed under a variety of biotic, abiotic stresses including nutrient deprivation. It has been reported previously that the reduction potential was increased in spinach leaves subjected to S- and P-deprivation11 and N-deprivation.12 The readjustment of redox homeostasis fails when excessively available electrons are transferred to oxygen producing reactive oxygen species (ROS), establishing strong electron sinks and thus inducing oxidative stress. The thiol redox homeostasis is maintained by NADPH-dependent thioredoxin reductases (NTRa, -b and -c) - thioredoxin (TRX) and glutathione reductase - GSH-dependent glutaredoxin (GRX) systems that modify target proteins facilitating stress responses in plants. Compared to wild-type miR395 levels, in single (cad2: γ-glutamyl cysteine synthetase involved in GSH biosynthesis), double (ntra/ntrb) and triple (ntra/ntrb/cad2) mutants grown under S- deprived conditions approximately 20, 40 and 45% decrease in induction of miR395 levels, respectively, was noted suggesting the involvement of redox signaling in miR395 expression under S-deprivation.10 Only partial decrease even in double and triple mutants suggested potential redundancy between the TRX and GRX systems in plants. Thus, Jagadeeswaran et al.10 established a link between redox signaling and the SLIM1 transcription factor that regulates miR395 expression during S-deprivation in Arabidopsis. It could also be shown that miR395 is induced by arsenic and copper, both of which impose oxidative stress.10 Expression analysis of miR395 under excess arsenic and copper exposure in single (cad2), double (ntra/ntrb) and triple (ntra/ntrb/cad2) mutants should tentatively reveal whether or not miR395 induction under oxidative stress is due to redox or ROS signaling.
Oxidative stress resulting from S-deficiency was confirmed by analyzing miR398 and the target Cu/Zn superoxide dismutase (CSD1) expression, which can serve as markers for oxidative stress.10 miR398 expression is down-regulated by the oxidative stress (ROS accumulation) in Arabidopsis13,14 but it was unknown whether and to which extent redox signaling is involved in this process. miR398 levels were almost unaffected in mutants defective in redox signaling when compared with wild type miR398 levels under S-deprivation.10 Therefore, only miR395 expression is compromised in mutants defective in redox signaling but not miR398 levels, which suggested that redox signaling controls the expression of miR395 but not miR398 under S-deprivation. These findings argue in favor that the ROS signaling (oxidative stress) and redox signaling are distinct under S-deprivation.
Salt stress, drought, ozone, hydrogen peroxide, excess levels of copper, iron, among others that generate ROS production have been shown to alter the expression of miRNAs in plants.9 Seven novel miRNA families that showed differential regulation under hydrogen peroxide stress in rice have been identified.15 An in depth comparative analysis in wild-type and mutants defective in redox signaling will reveal the involvement of redox in regulating the expression of nutrient and stress-responsive miRNAs (Fig. 1). Also such studies will help to distinguish between thiol redox and ROS-mediated regulation or even the extent of overlap between these 2 signaling pathways.
Figure 1.
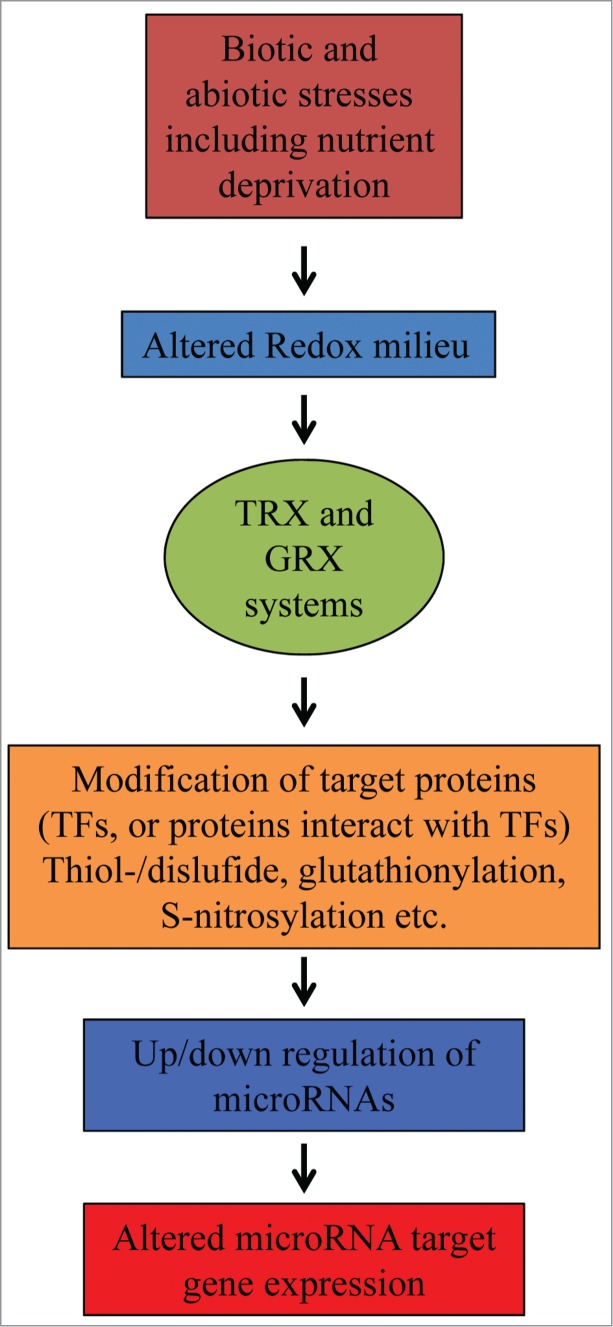
Generalized scheme for the involvement of redox signaling mediated regulation of miRNA expression and subsequent effect on their target gene expression in plants.
The link between oxidative stress signaling and induction of miRNAs has also been known in animals,16–18 although the involvement of redox singling per se has not been studied. Thus, this short review may induce more studies to address the involvement of thiol-based-redox signaling in regulating miRNA expression in plants and animals. Such studies should also consider other redox- and ROS-related cell parameters such as the redox pair ascorbate and dehydroascorbate or oxylipins that likely interfere with the described mechanisms.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We sincerely thank Prof. Dr. Karl-Josef Dietz, University of Bielefeld, Germany for critical reading of the manuscript and constructive suggestions.
Funding
Oklahoma Agricultural Experiment Station supported the research in Sunkar laboratory. SKP acknowledges the Indo-US Science and Technology Forum (IUSSTF) for the award of postdoctoral fellowship.
References
- 1.Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci 2005; 24:23-58; http://dx.doi.org/ 10.1080/07352680590910410 [DOI] [Google Scholar]
- 2.Dietz K-J, Pfannschmidt T. Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol 2011; 155:1477-85; PMID:21205617; http://dx.doi.org/ 10.1104/pp.110.170043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmstorm KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol 2014; 15:411-21; PMID:24854789; http://dx.doi.org/ 10.1038/nrm3801 [DOI] [PubMed] [Google Scholar]
- 4.Dietz K-J. Redox regulation of transcription factors in plant stress acclimation and development. Antiox Redox Signal 2014; 21:1356-72; PMID:24182193; http://dx.doi.org/ 10.1089/ars.2013.5672 [DOI] [PubMed] [Google Scholar]
- 5.Asada K. 1999. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50, 601-639; PMID:15012221; http://dx.doi.org/ 10.1146/annurev.arplant.50.1.601 [DOI] [PubMed] [Google Scholar]
- 6.Noda T, Iwakiri R, Fujimoto K, Aw TY. Induction of mild intracellular redox imbalance inhibits proliferation of CaCo-2 cells FASEB J 2001; 15:2131-9; PMID:11641239; http://dx.doi.org/ 10.1096/fj.01-0131com [DOI] [PubMed] [Google Scholar]
- 7.Dietz K-J. Redox-dependent regulation, Redox control and oxidative damage in plant cells subjected to abiotic stress. Meth Mol Biol 2010; 639:57-70; PMID:20387040; http://dx.doi.org/ 10.1007/978-1-60761-702-0_4 [DOI] [PubMed] [Google Scholar]
- 8.Sunkar R, Zhu JK. Micro RNAs and short-interfering RNAs in plants. J Integr Plant Biol 2007; 49:817-26; http://dx.doi.org/ 10.1111/j.1744-7909.2007.00499.x [DOI] [Google Scholar]
- 9.Sunkar R, Li Y, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci 2012; 17:196-203; PMID:22365280; http://dx.doi.org/ 10.1016/j.tplants.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 10.Jagadeeswaran G, Li Y-F, Sunkar R. Redox signaling mediates the expression of a sulfate-deprivation-inducible miR395 in Arabidopsis. Plant J 2014; 77:85-96; PMID:24164591; http://dx.doi.org/ 10.1111/tpj.12364 [DOI] [PubMed] [Google Scholar]
- 11.Dietz K-J, Heilos L. Carbon metabolism in spinach leaves as affected by leafage and phosphorus and sulfur metabolism. Plant Physiol 1990; 93:1219-25; PMID:16667581; http://dx.doi.org/ 10.1104/pp.93.3.1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wingler A, Brwonhill E, Pourtau N. Mechanisms of the light-dependent induction of cell death in tobacco plants with delayed senescence. J Exp Bot 2005; 56:2897-905; PMID:16157651; http://dx.doi.org/ 10.1093/jxb/eri284 [DOI] [PubMed] [Google Scholar]
- 13.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down-regulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006; 18:2051-65; PMID:16861386; http://dx.doi.org/ 10.1105/tpc.106.041673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagadeeswaran G., Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 2009; 229:1009-14; PMID:19148671; http://dx.doi.org/ 10.1007/s00425-009-0889-3 [DOI] [PubMed] [Google Scholar]
- 15.Li T., Li H., Zhang Y-X. and Liu J-Y. (2001). Identification and analysis of seven H2O2-responsive miRNAs and 32 new miRNAs in the seedlings of rice ( Oryza sativa L. ssp. Indica). Nucleic Acids Res 39: 2821-33; PMID:21113019; http://dx.doi.org/ 10.1093/nar/gkq1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorescu D, Griendling KK. Reactive oxygen species, mitochondria, and NAD(P)H oxidases in the development and progression of heart failure. Congest Heart Fail 2002; 3:132-40; PMID:12045381; http://dx.doi.org/ 10.1111/j.1527-5299.2002.00717.x [DOI] [PubMed] [Google Scholar]
- 17.Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvements of microRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem 2009; 284:7903-13; PMID:19158092; http://dx.doi.org/ 10.1074/jbc.M806920200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radic Biol Med 2013; 64:20-30; PMID:23712003; http://dx.doi.org/ 10.1016/j.freeradbiomed.2013.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]


