Abstract
The regulatory connections between the circadian clock and hormone signaling are essential to understand, as these two regulatory processes work together to time growth processes relative to predictable environmental events. Gibberellins (GAs) are phytohormones that control many growth processes throughout all stages of the plant life cycle, including germination and flowering. An increasing number of examples demonstrate that the circadian clock directly influences GA biosynthesis and signaling. EARLY FLOWERING 3 (ELF3) participates in a tripartite transcriptional complex known as the Evening Complex (EC). In this capacity, ELF3 is fundamental to core circadian clock activity, as well as time-of-day specific regulation of genes directly responsible for growth control, namely the PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5 genes. Here we show that the GA biosynthesis inhibitor paclobutrazol substantially reduces the long hypocotyl and petiole phenotypes of Arabidopsis elf3 mutants. In addition, loss of ELF3 activity causes upregulation of the key GA biosynthesis genes GA20ox1 and GA20ox2. Moreover, GA20ox1 and GA20ox2 expression depends strongly on the redundant activities of PIF4 and PIF5. These findings indicate that the defining growth phenotypes of elf3 mutants arise from altered GA biosynthesis due to misregulation of PIF4 and PIF5. These observations agree with recent work linking increased GA production with the elongated growth phenotypes of the barley elf3 mutant. Thus, the role of the EC in regulation of GA biosynthesis and signaling in eudicots is shared with monocots and, therefore, is a highly conserved mechanism for growth control.
Keywords: circadian clock, early flowering 3, gibberellin, phytohormone, phytochrome interacting factor, PIF4, plant growth
Abbreviations
- CO
CONSTANS
- Col-0
Columbia
- EC
Evening Complex
- ELF3
EARLY FLOWERING 3
- ELF4
EARLY FLOWERING 4
- EMS
ethyl methanesulfonate
- FT
FLOWERING LOCUS T
- GA
gibberellin
- GA20ox
gibberellin 20-oxidase
- GA3ox
gibberellin 3-oxidase
- LD
long day
- LUX
LUX ARRHYTHMO
- PAC
paclobutrazol
- PIF
PHYTOCHROME INTERACTING FACTOR
- qPCR
quantitative RT-PCR
- SD
short day
- WT
wild type
- ZT
Zeitgeiber Time
Introduction
Plants must coordinate growth processes with the predictable daily environmental changes accompanying daytime and nighttime.1,2 The plant circadian clock accomplishes this by imparting rhythmic patterns with approximately 24-hour periods to biological processes and their underlying genes, which confines these processes to defined times within the day.3-6 The plant circadian clock is set (or entrained) by light and temperature cues, while a core molecular oscillator comprised of transcription-translation feedback loops sustains rhythmic activity.6,7 The molecular interactions within the oscillator compose of a series of primarily repressive regulatory events, which involve numerous proteins that themselves have peak abundances and activities at specific times within the day.8,9 Progress in the last two decades has significantly advanced the understanding of how the core clock functions; however, many mechanistic connections between the core molecular oscillator and physiological outputs remain incompletely understood.
A fundamental role of the circadian clock is to communicate timing information to diverse “output” processes, including growth, development, and metabolism (for reviews, see refs. 10-12). An example of direct coupling between the core oscillator and growth outputs involves the activity of the Arabidopsis thaliana evening complex (EC).13,14 The tripartite EC protein complex directly represses daytime-phased genes and allows clock progression from day to night.13-16 At least 3 different dusk-expressed proteins compose the EC: LUX ARRHYTHMO/PHYTOCLOCK 1 (LUX/PHY1), EARLY FLOWERING 4 (ELF4), and EARLY FLOWERING 3 (ELF3).13,17-19 LUX is a MYB-like GARP transcription factor that targets the EC to promoters by sequence-specific binding of cis elements upstream of many day-expressed genes.14,18-20 Recruitment of the EC to promoters also requires NOX/BOA, a MYB transcription factor in the same family as LUX.14,20,21 The exact biochemical functions of both ELF3 and ELF4 are presently unclear, as neither protein has recognizable functional domains; however, both are required for EC activity since daytime-phased genes are highly expressed throughout the night in elf3 mutant Arabidopsis seedlings.15,16,22,23
In addition to control of core clock genes, the EC regulates expression of genes encoding important components of growth control networks, including the PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5 genes.13,14 PIF4 and PIF5, along with other members of the PIF transcription factor family, are central regulatory hubs where multiple internal and external cues converge and are integrated to control diverse plant growth processes, including germination, skotomorphogenesis, chloroplast differentiation, shade-avoidance, and flowering (for review, see ref. 24). PIF protein abundance and activity is defined by a combination of circadian clock-directed gene expression and post-translational mechanisms that detect light, temperature, and phytohormone presence.25-28 Together, these regulatory mechanisms tightly constrain the bulk of PIF4 and PIF5 activity in young seedlings to a short time period just prior to dawn.25,29 The circadian clock generates rhythmic PIF4 and PIF5 expression through EC-imposed repression at dusk that subsequently declines over the pre-dawn hours as EC activity feeds back to inhibit expression of LUX and ELF4.13,20 As a result, PIF4 and PIF5 gene expression increases in the hours before dawn and continues throughout the day so that PIF4 and PIF5 transcripts have peak expression at approximately midday.25,30 However, PIF4 and PIF5 protein accumulation is maximal only immediately before dawn because accumulated PIF proteins are rapidly degraded in the first light of day by a phytochrome B- and ubiquitination-dependent pathway.25,26,31,32
PIF4 and PIF5, along with PIF1 and PIF3, contribute to elongation growth in dark-grown seedlings.25,29,33 However, PIF4 and PIF5 mainly drive elongation growth in green seedlings. An important activity of these transcription factors is to positively regulate hypocotyl elongation in pre-dawn hours under diurnal conditions.25,34,35 The typical elongated hypocotyl phenotype of arrhythmic Arabidopsis clock mutants, like elf3, elf4, and lux, is largely due to high, nearly constitutive PIF4 and PIF5 expression.13,25 Continuous gene expression causes greater PIF protein accumulation throughout the entire night and consequent growth enhancement, rather than the normal restriction of growth to pre-dawn hours.13,25 As a result, elf3-2 pif4-101 and elf3-2 pif5-1 double mutants have shorter hypocotyls relative to elf3-2 seedlings, and the elf3-2 pif4-101 pif5-1 triple mutant has a hypocotyl length indistinguishable from wild type (WT) seedlings.13
PIF transcription factors promote growth by binding to G-box and E-box variant cis-elements in the promoters of target genes, including genes involved in phytohormone biosynthesis and signaling.35,36 PIF1, PIF3, PIF4, and PIF5 together regulate thousands of genes, including many genes involved in hormone biosynthesis and signaling.36-38 PIF4, PIF5, and PIF7 activate expression of TAA1, CYP79B2, YUC8, and YUC9 genes, which encode enzymes required for the rate-limiting steps of distinct auxin biosynthetic pathways.35,39,40
GA biosynthesis is highly regulated, primarily though transcriptional control of gibberellin 20-oxidase (GA20ox) and gibberellin 3-oxidase (GA3ox) genes, which encode enzymes for catalysis of the final reactions in the production of bioactive GA1 and GA4.41 In a series of oxidation steps, GA20ox enzymes convert GA12 and GA53 to GA9 and GA20, respectively. GA3ox enzymes then produce GA1 and GA4 from these substrates. Of the 5 paralogous GA20ox genes in the Arabidopsis genome, GA20ox1 and GA20ox2 are primarily required for GA biosynthesis, while GA20ox3 contributes to a lesser degree.42,43 GA20ox1 is most important, as loss of it alone reduces internode elongation, while single mutants of the others do not affect growth.42 The contribution of GA20ox2 is revealed only when combined with a ga20ox1 mutant. Similarly, loss of GA20ox3 exacerbates the phenotypes of a ga20ox1 mutant.43 At present, the transcriptional networks that regulate GA20ox and GA3ox expression in Arabidopsis are incompletely understood. PIF1/PIL5 represses GA3ox1 and GA3ox2.37,44 PIF1/PIL5 also binds to GAI and RGA promoters, which encode repressors of GA signaling, and promotes expression of these genes, but apparently does not bind to the promoters of other GA biosynthetic genes, including GA3ox1, GA3ox2, GA2ox2, and any of the GA20ox paralogs.44
While the circadian clock influences PIF gene expression, and PIF proteins impact hormone signaling, how these components work together as a module is unclear. For example, mutants of Arabidopsis elf3, which by definition are mutants for EC activity, are well-studied and characterized with respect to their effect on core circadian clock function, but the impact of losing ELF3 activity (i.e., EC function) on growth regulatory pathways is less defined, aside from a clear role in direct regulation of PIF4 and PIF5 expression.13,22 The Arabidopsis elf3-1 allele, like all elf3 null mutants, has pale green leaves and cotyledons, in addition to elongated hypocotyl and petiole growth.13,45-47 This constellation of phenotypes resembles plants with an increased GA response.48,49 The putative GA hyper-response phenotype in elf3 is especially pronounced when plants are grown in short day (SD; 8 hours light/16 hours dark) photoperiods.45 GA biosynthesis and signaling is deeply entwined with the circadian clock. The daily expression pattern of many GA metabolism and signaling genes is rhythmic.5 GA sensitivity and responses also are limited to discrete times of day by the action of the circadian clock.5,50-53 Additionally, the growth phenotypes of a barley elf3 mutant, which include elongated coleoptiles and extended pale-green leaves, result from greater GA abundance.54 Here, we show that elongated growth phenotypes in Arabidopsis elf3 mutants are GA-dependent. In addition, we demonstrate an important regulatory connection between the EC and GA-dependent growth that involves the growth-promotion activity of PIF4 and PIF5.
Results
GA biosynthesis contributes to the elf3-1 phenotype
The Arabidopsis null elf3-1 mutant was tested for GA biosynthesis-dependent phenotypes by assessing the consequences of paclobutrazol (PAC) treatment on hypocotyl elongation in WT and elf3-1 seedlings. PAC inhibits an initial enzymatic step in GA biosynthesis, which is upstream of the action of GA20ox and GA3ox enzymes.55 As expected, untreated elf3-1 seedlings have severely elongated hypocotyls that are nearly 3 times longer than WT under SD conditions (Fig. 1A and B). Five days of PAC treatment significantly diminishes the hyper-elongation phenotype in elf3-1 seedlings and the strength of the effect is dependent on the PAC concentration. Increasing the PAC concentrations from 0.1 to 10 μM progressively suppresses hypocotyl elongation in elf3-1 seedlings to the point where the difference between WT and elf3-1 is only 30% at 1 and 10 μM PAC (Fig. 1A and B). The strong inhibitory effect of PAC on elf3-1 hypocotyl elongation indicates that the majority of this growth is due to enhanced GA biosynthesis. Notably, the difference between WT and elf3-1 seedlings at both 1 and 10 μM PAC remains significant (unpaired t-test, p < 0.05), which shows that additional factors beyond GA make a small contribution to hypocotyl growth in the elf3 mutant.
Figure 1.
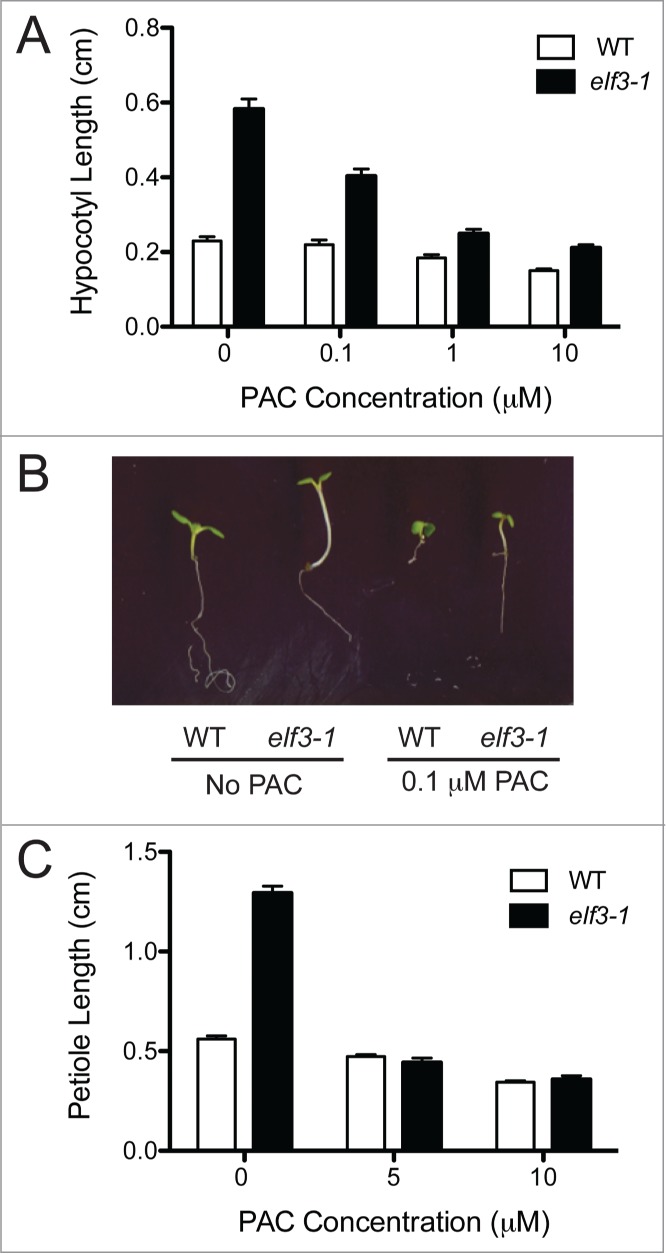
Inhibition of GA biosynthesis reduces elongated growth in the elf3-1 mutant. (A) Hypocotyl lengths of 7 day-old WT (open bars) and elf3-1 (black bars) seedlings grown under SD on the indicated concentrations of GA biosynthesis inhibitor PAC for the last 5 d before measurement. Shown is the mean (±SEM) from 3 independent experimental replicates of n = 20 seedlings each. (B) Seven day-old WT and elf3-1 seedlings grown in SD without PAC or with 0.1 μM PAC grown as indicated. (C) Mean (±SEM) petiole lengths of 28 day-old WT and elf3-1 plants exposed to the indicated PAC concentration for 3 weeks. Measurements are from a single experimental replicate (n = 15) grown in SD.
PAC treatment of older plants confirms that other growth phenotypes of elf3-1 are also GA biosynthesis-dependent. The petiole length of 28 day-old elf3-1 plants is more than twice that of age-matched WT plants (Fig. 1C). On the other hand, petiole length of the two genotypes is indistinguishable in the presence of either 5 or 10 μM PAC (Fig. 1C). Taken together, these observations show that GA biosynthesis is a major contributing factor to the Arabidopsis elf3-1 elongation growth phenotype.
GA biosynthesis genes are highly expressed in elf3 mutants
The sensitivity of hypocotyl and petiole elongation to PAC in elf3-1 indicates that the pronounced growth of these two organs in the mutant arises from higher than normal GA production. Since the elf3-1 mutant causes large-scale changes in transcription for circadian clock-regulated genes, a potential mechanism for enhanced GA production in the elf3-1 background is through misregulation of genes required for GA biosynthesis.
Expression of GA20ox1 and GA20ox2 exhibits a rhythmic pattern in WT with a time of peak expression that depends on the photoperiod conditions. In SD conditions, the transcript of each rises throughout the night and peaks at dawn (Fig. S1A and B). Interestingly, the pattern for each gene is changed in long day (LD; 16 hours light/8 hours darkness) photoperiods: expression begins to rise at dawn and the time of maximal expression is shifted 8 hours into the day (Fig. S1A and B). Thus, GA20ox1 and GA20ox2 expression is adjusted according to photoperiod, which is consistent with the circadian clock contributing to the expression behavior of these genes.
Loss of ELF3 activity causes strong upregulation of both GA20ox1 and GA20ox2 expression in SD conditions, but has little effect on the expression of the other GA20ox paralogs. Quantitative RT-PCR (qPCR) analysis of GA20ox1 expression at dawn (Zeitgeiber Time 0 (ZT0) hours) in two different elf3 mutant alleles, elf3-1 and elf3-2, reveals significantly higher transcript levels in each mutant background compared to WT seedlings (unpaired t-test, p < 0.05 and p < 0.0005, respectively) (Fig. 2A). GA20ox2 is also significantly induced in elf3-1 and elf3-2 to nearly the same degree as GA20ox1 (unpaired t-test, p < 0.01 and p < 0.05) (Fig. 2B). On the other hand, neither mutant allele changes the expression behavior of GA20ox3, GA20ox4, or GA20ox5 to any significant degree (Fig. 2C–E). GA3ox1 encodes the enzyme acting immediately downstream of GA20ox that is responsible for producing bioactive GAs. Expression of this gene is also not changed by either mutant (Fig. 2F). Therefore, ELF3 is required to repress expression of both GA20ox1 and GA20ox2, which indicates that EC activity normally suppresses these genes.
Figure 2.
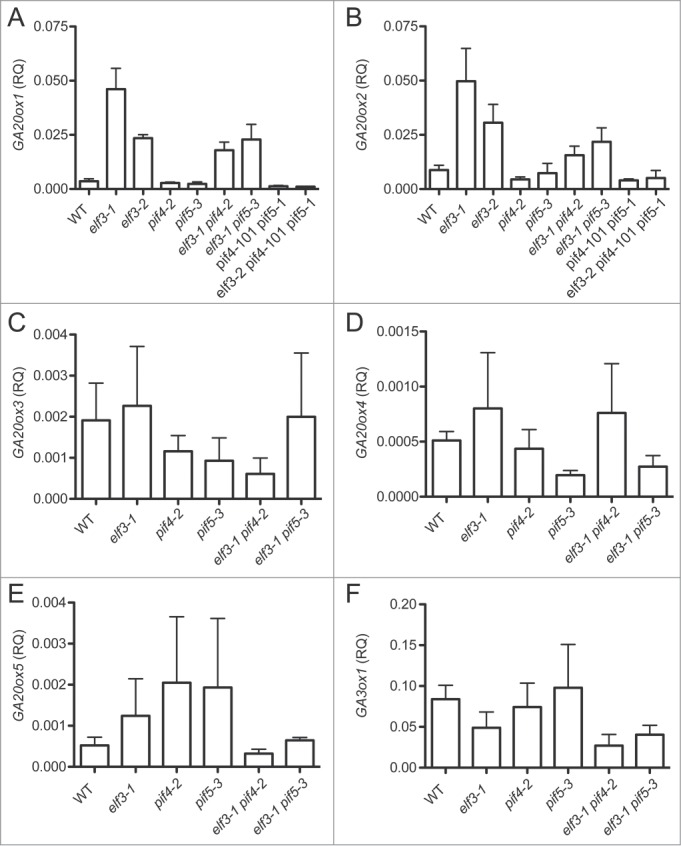
Elevated pre-dawn expression of GA biosynthetic genes in elf3 mutants requires PIF4 and PIF5. Transcript abundance for (A) GA20ox1, (B) GA20ox2, (C) GA20ox3, (D) GA20ox4, (E) GA20ox5 and (F) GA3ox1 in the indicated genotypes at 15 minutes before ZT0 (lights-on) under SD conditions. Transcript levels were measured by qPCR analysis. Genotypes are depicted on x-axis and y-axis displays relative quantitation (RQ) of transcript normalized to PDF2 according to Materials and Methods. Shown is the mean (±SEM) of 3 independent experimental replicates.
GA20ox expression requires the transcription factors PIF4 and PIF5
To identify factors regulating growth downstream of elf3-1 and the EC, a suppressor screen was carried out where ethyl methanesulfonate (EMS) mutagenized elf3-1 plants grown in SD conditions were scored for restoration of WT-like growth habit. One suppressor line (s41) exhibited hypocotyl, petiole, and leaf shape that resembled WT plants (Fig. S2A and B). Mapping of the s41 suppressor mutation identified a C to T transition in the PIF4 gene that converts an arginine at position 182 to a premature stop codon (Fig. S2C and D). To confirm mutation of pif4 was responsible for the suppressor phenotype and to further study the genetic interaction between the ELF3 and PIF4 genes, elf3-1 was crossed to the pif4-2 reference allele. Like the previously described elf3-2 pif4-101 mutant of Nusinow et al.,13 the elf3-1 pif4-2 double mutant and the s41 suppressor have comparable phenotypes, including reduced hypocotyl elongation, shorter petioles, and WT-like leaf shape.
Since the characteristic growth phenotypes of elf3-1 and elf3-2 both coincide with elevated GA20ox expression and depend on PIF4 and PIF5 activity, we tested whether either PIF4 or PIF5 influence GA20ox gene expression in WT and elf3 mutant backgrounds. Combining either the pif4-2 or pif5-3 mutant with the elf3-1 allele reduces the high GA20ox1 and GA20ox2 transcript levels of the single elf3-1 mutant by 30% and 50% for elf3-1 pif4-2 and elf3-1 pif5-3, respectively (Fig. 2A and B). Although the difference is apparent by eye, statistical testing shows only the changes for GA20ox2 are statistically significant between the two populations (unpaired t-test, p < 0.05). In contrast, the single pif4-2 and pif5-3 alleles alone cause little change in GA20ox1 and GA20ox2 expression. Importantly, GA20ox1 and GA20ox2 transcript accumulation in elf3-2 pif4-101 pif5-1 triple mutant seedlings is strongly reduced relative to the elf3-2 parent (unpaired t-test, p < 0.0005 and p < 0.05, respectively) and the levels of each are near that of WT seedlings (Fig. 2A and B). Similar analysis for GA20ox3, GA200ox4, GA20ox5, and GA3ox1 reveals no significant effect on expression caused by pif4 and pif5 mutants (Fig. 2C–F).
Like GA20ox1 and GA20ox2, PIF4 and PIF5 are expressed in a rhythmic fashion.25,56 A comparison of the expression profiles for PIF4, PIF5, GA20ox1 and GA20ox2 reveals striking similarities between them (Fig. S2). For all, the timing of peak expression is determined by photoperiod. In SD conditions, expression of all 4 genes begins after dusk and continues to rise until dawn, at which point expression drops for each, aside from PIF4. Compared to the pattern in SD conditions, LD photoperiods cause a pronounced shift in peak expression into the day. The tight correlation in expression between the PIF transcription factor genes and the GA20ox genes is consistent with PIF4 and PIF5 playing a role in regulation of these GA biosynthesis genes. All together, the findings here indicate that EC influences GA biosynthesis through control of GA20ox1 and GA20ox2 expression in a pathway that requires the redundant action of PIF4 and PIF5.
Auxin biosynthesis and signaling genes are highly expressed in elf3, which requires PIF4 and PIF5 activity
The phytohormone auxin is an established regulator of GA biosynthesis for several plant processes, including hypocotyl elongation.57 It is possible that enhanced GA biosynthesis in elf3 arises from misregulation of genes for auxin biosynthesis and signaling that then leads to elevated expression of GA20ox1 and GA20ox2. YUCCA8 (YUC8) and TAA1 are genes encoding enzymes for rate-limiting steps in auxin biosynthesis and each is subject to PIF4 regulation.35,39,40 YUC8 expression is significantly elevated in elf3-2 compared to WT (Fig. 3A; unpaired t-test, p < 0.05); on the other hand, TAA1 expression is unchanged in the elf3 mutant background (Fig. 3B). The high YUC8 expression in elf3-2 is dependent on PIF4 and PIF5, as YUC8 transcript levels in the elf3-2 pif4-101 pif5-1 triple mutant are below those in WT (Fig. 3A). Thus, PIF4 and PIF5 are responsible for elevated YUC8 expression in the elf3 mutant.
Figure 3.
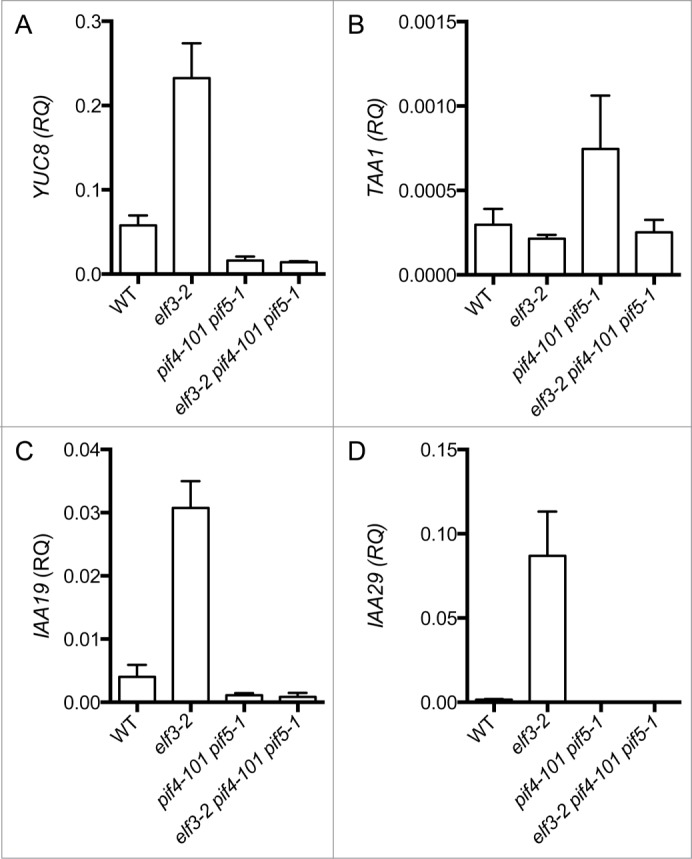
High expression of YUC8, IAA19, IAA29 indicate potential elevated auxin biosynthesis and signaling in elf3 mutants that requires PIF4 and PIF5. Transcript abundance for (A) TAA1, (B) YUC8, (C) IAA19 and (D) IAA28 in the indicated genotypes at 15 minutes before ZT0 (lights-on) under SD conditions. Transcript levels were measured by qPCR analysis. Genotypes are depicted on x-axis and y-axis displays relative quantitation (RQ) of transcript normalized to PDF2 according to Materials and Methods. Shown is the mean (±SEM) of 3 independent experimental replicates.
PIF4 and PIF5 also modify auxin sensitivity in hypocotyls to regulate growth.57 To evaluate whether elevated auxin biosynthesis or signaling is apparent in the elf3 mutant, the expression the Aux/IAA genes IAA19 and IAA29 was evaluated in WT and elf3-2. These Aux/IAA genes are regulated by PIF4 and PIF5.58 Indeed, both IAA19 and IAA29 are highly upregulated in elf3-2 (Fig. 3C and D; unpaired t-test, p < 0.05). Addition of the pif4-101 and pif5-1 alleles suppresses this phenotype, which indicates that PIF4 and PIF5 are needed for elevated IAA19 and IAA29 expression in the elf3-2 background (Fig. 3C and D). These observations demonstrate that modified expression of auxin biosynthesis and signaling genes due to upregulation of PIF4 and PIF5 may underlie the GA-dependent growth phenotypes in elf3.
Discussion
Delineating connections between the circadian clock and hormone signaling is essential to understanding how plants time growth processes relative to predictable environmental events. GA is a fundamental regulator of many growth processes throughout all stages of the plant life cycle. An increasing number of examples show that the clock influences GA biosynthesis and signaling.51-54 The findings here show that inactivating the EC alters expression of GA biosynthesis genes and that the defining growth phenotypes of Arabidopsis elf3 mutants depend on GA biosynthesis. elf3 mutants have high transcript levels of the GA biosynthesis genes GA20ox1 and GA20ox2, which are normally tightly controlled to maintain GA homeostasis. Moreover, the timed presence of GA20ox1 and GA20ox2 transcripts in seedlings is strongly influenced by the transcription factors PIF4 and PIF5. In this role, PIF4 and PIF5 act redundantly. The similarities between the observations in Arabidopsis here and the demonstration that increased GA production in the barley elf3 mutant drives elongated growth (see ref. 54), indicates that the EC has similar, if not identical, regulatory roles over GA biosynthesis and signaling in eudicots and monocots.
The findings here indicate that GA rhythms are conferred by EC-dependent nighttime repression of key GA biosynthesis genes and this is a fundamental means by which the clock regulates GA biosynthesis and GA-dependent growth. Defects in nighttime repression of barley GA metabolism genes occur when ELF3 function is lost.54 In SD photoperiods, the GA20ox2 transcript level in WT barley increases just after dawn and continues to rise into the early part of the day, a time when EC activity is not present.13,54 In contrast, GA20ox2 expression in a barley elf3 mutant was significantly higher throughout the night. The time of maximal difference between mutant and WT was in the pre-dawn to dawn time window (ZT21-0), at which time GA2ox3 expression in elf3 was about 4 times higher than in WT at pre-dawn (ZT21) but not near midday (ZT5). Corresponding to these expression changes, dawn levels of GA19, GA20, and GA1, which are substrates and products of GA20ox, as well as the immediate degradation product GA8, are all significantly higher in the barley elf3 mutant.54 Similar to GA20ox2 in WT barley, Arabidopsis GA20ox1 and GA20ox2 expression rises after dawn and continues to increase for 4–8 hours when seedlings are grown under 12-hour day or LD conditions, while expression peaks at dawn in SD (Fig. S1A and B). Both elf3-1 and elf3-2 Arabidopsis mutants grown in short days have significantly higher GA20ox1 and GA20ox2 transcript abundance immediately before dawn compared to WT (Fig. 2A and B). In these conditions, the growth phenotypes of elf3-1 are also particularly severe and GA biosynthesis is important for manifestation of these phenotypes, since the GA biosynthesis inhibitor PAC attenuates these (Fig. 1). Therefore, ELF3 regulates plant growth by affecting GA biosynthesis, presumably together with ELF4 and LUX in the EC.
In addition to being GA-dependent, enhanced hypocotyl and petiole growth in elf3-1 requires the activity of PIF4 and PIF5. Together pif4 and pif5 mutant alleles restore normal hypocotyl length to elf3 mutants (see ref. 13), as well as nearly WT expression for GA20ox1 and GA20ox2 (Fig. 2). Elevated GA20ox1 and GA20ox2 transcript levels in elf3-1 likely arise from the enhanced PIF protein accumulation common to this mutant, especially in SD photoperiods.25 Therefore, PIF4 and PIF5 positively affect GA20ox1 and GA20ox2 expression and this is revealed in the elf3 mutant. Collectively, these results indicate that the clock regulates PIF gene expression to temporally control GA-dependent growth.
Considering how PIF protein activity is controlled in a GA-dependent manner, a feed-forward mechanism may enhance the growth phenotypes caused by EC inactivation. DELLA proteins repress GA signaling (for review see ref. 59), in part by direct binding of PIF proteins to block their activity.28,60 This inhibition is attenuated in the presence of GA, by ubiquitin-26S proteasome mediated degradation of DELLA proteins.61-64 In the elf3 background, it is possible that the greater PIF protein abundance at an unusual time of day could overcome normal DELLA protein inhibition due a lack of sufficient DELLA to bind all PIF proteins; consequently, more PIF proteins could be available to activate GA biosynthesis. Furthermore, increased GA levels may maintain low levels of DELLA proteins, which can result in even more PIF activity.
Despite the fact that PIFs directly regulate many growth-related genes, PIF4 and PIF5 may not bind directly to the promoters of GA20ox1 and GA20ox2. Maximum PIF4 and PIF5 protein accumulation occurs just before dawn, because the proteins are destabilized by light.25,26 In contrast, GA20ox expression levels remain elevated into the daylight hours under 12-hour day and LD conditions, a time when PIF4 and PIF5 are at their lowest ebb (Supplementary Fig. 1A and B; see ref. 25). Furthermore, G-box or modified E-box sequences are not present in the promoter regions of GA20ox1 and GA20ox2 (B.C.T, unpublished observations).
Although a direct regulatory link for PIF4 and PIF5 is not established for GA20ox1 and GA20ox2, loss of PIF4 and PIF5 activity impacts the expression of genes for auxin biosynthesis and signaling. TAA1 and YUC8 encode enzymes for the rate-limiting steps in auxin biosynthesis and each gene is subject to PIF4 and PIF5 control.35,39,40 High YUC8 expression in the elf3 background is dependent on PIF4 and PIF5 (Fig. 3A). PIF4 and PIF5 also modify auxin sensitivity in hypocotyls to regulate growth.57 Indeed, auxin-regulated genes are over-represented among genes differentially expressed between pif4 pif5 double mutants and WT.65,66 Auxin is an established regulator of GA biosynthesis for some plant processes, including hypocotyl elongation. Exogenous application of auxin increases transcript abundance for GA20ox1, GA20ox2, GA3ox1, and 3 of 7 GA2ox paralogs in Arabidopsis seedlings.67,68 Furthermore, overexpression of YUCCA1 produces elongated hypocotyls and this phenotype is suppressed by PAC treatment.67 Thus, auxin-dependent hypocotyl growth requires GA biosynthesis and PAC inhibits the effects of exogenous auxin. Circadian-regulated genes are over-represented in auxin responsive genes, with peak phases at pre-dawn to dawn (ZT22-2), which are times when hypocotyl growth is most active.57,69 In elf3-2, the Aux/IAA genes IAA19 and IAA29 are highly expressed only when PIF4 and PIF5 are active (Fig. 3C and D). Therefore, the EC potentially acts through PIF4 and PIF5 to control auxin biosynthesis and this module ultimately regulates GA biosynthesis.
The work presented here establishes an EC-dependent role for GA biosynthesis in control of elongation growth in Arabidopsis. Photoperiod-insensitive early flowering, however, is another defining aspect of elf3 mutants in Arabidopsis, barley, and pea.45,54,65 An established cause of this early flowering phenotype is constitutively high CONSTANS (CO) expression that promotes high expression of FLOWERING LOCUS T (FT)70. Since recent findings show that PIF4, and possibly PIF5, directly bind to the FT promoter to accelerate flowering (see refs. 27, 71), the high PIF levels throughout the night in elf3 mutant backgrounds may also result in elevated FT expression under some environmental conditions, especially those that include elevated temperature.71 Alternatively, PIF-dependent accumulation of GA may also induce flowering through a GA-dependent flowering pathway.72 The early flowering phenotype in barley elf3 is GA-dependent and some flowering time genes respond to GA.54 While in the past, flowering in elf3 has been largely attributed to mis-regulation of the photoperiod pathway and CO, this classic early flowering mutant can now be re-examined for its reproductive defects in the context of PIF transcription factors and GA.
This work demonstrates that many factors work together in a temporally regulated module to control plant growth. These circadian clock-hormone modules are an importation aspect of plant biology, as plants must time growth and developmental processes relative to daily environmental cycles. While the mechanistic role that ELF3 plays in the circadian clock is increasingly better understood in recent years, the connections between the clock and physiological outputs remain at the forefront of many investigations in chronobiology.
Methods
Plant growth
All plants were in the Columbia-0 (Col-0) background. The elf3-2 mutant is described by Nusinow et al.42 pif4-2 and pif5-3 were a gift from Dr. Peter Quail (Plant Gene Expression Center, Albany, CA, USA; see refs. 73, 74), and elf3-2, pif4-101 pif5-1, and elf3-2 pif4-101 pif5-1 seeds were a gift from Dr. Dmitri Nusinow (Donald Danforth Plant Science Center, St. Louis, MO, USA). Seeds for all experiments were surface-sterilized in a 50% bleach solution and stratified for 2–5 d in the dark at 4°C before use. All seedlings were grown in Percival growth chambers (Percival Scientific, www.percival-scientific.com), set to constant 22°C and SD conditions (8 h light:16 h darkness) with cool white light at 100 μmol/m2/s.
PAC treatments
Seeds were sown at ZT0 on 100 mm × 15 mm square plates (BD Biosciences, www.bdbiosciences.com) containing 50 ml of MS medium at pH 5.8 with 0.8% type I micropropagation agar (Caisson Laboratories, www.caissonlabs.com). For hypocotyl measurements, seedlings were grown for two full days on MS plates and then were transferred at ZT0 on day 3 under sterile conditions to either MS plates or MS plates containing paclobutrazol (Chem Service, www.chemservice.com) at concentrations ranging from 0.1 to 10 μM. Seedlings were allowed to grow for 5 additional days, then images of seedlings were made by scanning the plates with a flatbed scanner and hypocotyl length calculated from the images using ImageJ software (http://imagej.nih.gov/ij/). For petiole measurements, seeds were sown into Phytatray boxes (Sigma Aldrich, www.sigmaaldrich.com) containing 250 mL MS media as above. After one week, seedlings were transferred to boxes containing paclobutrazol concentrations ranging from 0 to 10 μM. After an additional 3 weeks at these conditions, images were taken of the plants and measurements made from the images using ImageJ software.
Quantitative real-time PCR
Seeds for qPCR experiments were sown onto sterile filter paper (Whatman, www.whatman.com) and grown on MS plates as above. Fifteen minutes before ZT0, while seedlings were in the dark, entire seedlings were harvested under a green LED light, placed into 1.5 ml microcentrifuge tubes, and immediately frozen in liquid N2. Frozen samples were pulverized in 1.5 mL tubes with micropestles. RNA was isolated with the Qiagen RNeasy Kit and contaminating genomic DNA digested with the Qiagen RNase-free DNase Set according to the manufacturer's protocol (Qiagen, www.qiagen.com/us/). First-strand cDNA was prepared from 1 μg of total RNA with the Maxima Universal First Strand cDNA Synthesis kit (Thermo Scientific, www.thermofisher.com/) and diluted 1:5 in RNase-free water prior to use. Transcript levels were determined with qPCR using Bio-Rad SsoAdvanced Universal Supermix and a CFX96 Real-Time PCR Detection System according to manufacturer's protocols (Bio-Rad, www.bio-rad.com). Transcript levels of target genes were calculated using the equation RQ = 2[Ct (Control) − Ct (Experimental)], where Ct is the mean threshold cycle for each sample. The transcript from PDF2 served as the normalization control (AT1G13320).75 Primer sequences are in Table S1.
Acknowledgements
We thank Claire Bendix and Elana Skeers for assistance with experiments.
Funding
This work was supported by the United States Department of Agriculture [5335-21000-026-00D to F.G.H] and the National Institutes of Health [F32GM083536- 01 to B.T.]. J.F. was supported in part by a grant to the Keck Science Department from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program.
Supplementaral Materials
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005; 309:630-3; PMID:16040710; http://dx.doi.org/ 10.1126/science.1115581 [DOI] [PubMed] [Google Scholar]
- 2. de Montaigu A, Toth R, Coupland G. Plant development goes like clockwork. Trends Genet 2010; 26:296-306; PMID:20483501; http://dx.doi.org/ 10.1016/j.tig.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 3. Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. . Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 2008; 4:e14; PMID:18248097; http://dx.doi.org/ 10.1371/journal.pgen.0040014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000; 290:2110-3; PMID:11118138; http://dx.doi.org/ 10.1126/science.290.5499.2110 [DOI] [PubMed] [Google Scholar]
- 5. Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 2008; 9:R130; PMID:18710561; http://dx.doi.org/ 10.1186/gb-2008-9-8-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AA. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 2007; 30:333-49; PMID:17263778; http://dx.doi.org/ 10.1111/j.1365-3040.2006.01627.x [DOI] [PubMed] [Google Scholar]
- 7. McClung CR. Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep 2014; 6:2; PMID:24592314; http://dx.doi.org/ 10.12703/P6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pokhilko A, Fernandez AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 2012; 8:574; PMID:22395476; http://dx.doi.org/ 10.1038/msb.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fogelmark K, Troein C. Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Computational Biology 2014; 10:e1003705; PMID:25033214; http://dx.doi.org/ 10.1371/journal.pcbi.1003705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farre EM, Weise SE. The interactions between the circadian clock and primary metabolism. Curr Opin Plant Biol 2012; 15:293-300; PMID:22305520; http://dx.doi.org/ 10.1016/j.pbi.2012.01.013 [DOI] [PubMed] [Google Scholar]
- 11. Nozue K, Maloof JN. Diurnal regulation of plant growth. Plant Cell Environ 2006; 29:396-408; PMID:17080594; http://dx.doi.org/ 10.1111/j.1365-3040.2005.01489.x [DOI] [PubMed] [Google Scholar]
- 12. Stitt M, Zeeman SC. Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol 2012; 15:282-92; PMID:22541711; http://dx.doi.org/ 10.1016/j.pbi.2012.03.016 [DOI] [PubMed] [Google Scholar]
- 13. Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011; 475:398-402; PMID:21753751; http://dx.doi.org/ 10.1038/nature10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signal Behav 2012; 7:170-3; PMID:22307044; http://dx.doi.org/ 10.4161/psb.18766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thines B, Harmon FG. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci U S A 2010; 107:3257-62; PMID:20133619; http://dx.doi.org/ 10.1073/pnas.0911006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 2011; 21:120-5; PMID:21236675; http://dx.doi.org/ 10.1016/j.cub.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002; 419:74-7; PMID:12214234; http://dx.doi.org/ 10.1038/nature00954 [DOI] [PubMed] [Google Scholar]
- 18. Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci U S A 2005; 102:10387-92; PMID:16006522; http://dx.doi.org/ 10.1073/pnas.0503029102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 2005; 10:963-72; PMID:16164597; http://dx.doi.org/ 10.1111/j.1365-2443.2005.00892.x [DOI] [PubMed] [Google Scholar]
- 20. Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 2011; 21:126-33; PMID:21236673; http://dx.doi.org/ 10.1016/j.cub.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, et al. . BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 2011; 23:961-72; PMID:21447790; http://dx.doi.org/ 10.1105/tpc.111.084293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 2000; 408:716-20; PMID:11130072; http://dx.doi.org/ 10.1038/35047079 [DOI] [PubMed] [Google Scholar]
- 23. Hicks KA, Albertson TM, Wagner DR. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 2001; 13:1281-92; PMID:11402160; http://dx.doi.org/ 10.1105/tpc.13.6.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 2011; 16:19-28; PMID:20833098; http://dx.doi.org/ 10.1016/j.tplants.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nozue K, Covington M, Duek P, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007; 448:358-61; PMID:17589502; http://dx.doi.org/ 10.1038/nature05946 [DOI] [PubMed] [Google Scholar]
- 26. Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 1999; 400:781-4; PMID:10466729; http://dx.doi.org/ 10.1038/23500 [DOI] [PubMed] [Google Scholar]
- 27. Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, Wigge PA. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012; 484:242-5; PMID:22437497; http://dx.doi.org/ 10.1038/nature10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008; 451:480-4; PMID:18216857; http://dx.doi.org/ 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- 29. Soy J, Leivar P, Gonzalez-Schain N, Sentandreu M, Prat S, Quail PH, Monte E. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J 2012; 71:390-401; PMID:22409654, http://dx.doi.org/ 10.1111/j.1365-313X.2012.04992.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nomoto Y, Kubozono S, Miyachi M, Yamashino T, Nakamichi N, Mizuno T. A circadian clock- and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol 2012; 53:1965-73; PMID:23037004; http://dx.doi.org/ 10.1093/pcp/pcs141 [DOI] [PubMed] [Google Scholar]
- 31. Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 2006; 23:439-46; PMID:16885032; http://dx.doi.org/ 10.1016/j.molcel.2006.06.011 [DOI] [PubMed] [Google Scholar]
- 32. Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014; 344:1160-4; PMID:24904166; http://dx.doi.org/ 10.1126/science.1250778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 2008; 18:1815-23; PMID:19062289; http://dx.doi.org/ 10.1016/j.cub.2008.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 2008; 53:312-23; PMID:18047474; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- 35. Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, Lopez-Vidriero I, et al. . Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 2012; 71:699-711; PMID:22536829; http://dx.doi.org/ 10.1111/j.1365-313X.2012.05033.x [DOI] [PubMed] [Google Scholar]
- 36. Zhang YM, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH. A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub that Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in Arabidopsis. PLoS Genet 2013; 9:e1003244; PMID:23382695; http://dx.doi.org/ 10.1371/journal.pgen.1003244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 2009; 21:3535-53; PMID:19920208; http://dx.doi.org/ 10.1105/tpc.109.070672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A 2009; 106:7660-5; PMID:19380720; http://dx.doi.org/ 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun JQ, Qi L, Li Y, Chu J, Li C. PIF4-Mediated Activation of YUCCA8 Expression Integrates Temperature into the Auxin Pathway in Regulating Arabidopsis Hypcotyl Growth. PLoS Genet 2012; 8:e1002594; PMID:22479194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. . Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A 2011; 108:20231-5; PMID:22123947; http://dx.doi.org/ 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J 2012; 444:11-25; PMID:22533671; http://dx.doi.org/ 10.1042/BJ20120245 [DOI] [PubMed] [Google Scholar]
- 42. Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. . The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 2008; 53:488-504; PMID:18069939; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03356.x [DOI] [PubMed] [Google Scholar]
- 43. Plackett AR, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, et al. . Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 2012; 24:941-60; PMID:22427334; http://dx.doi.org/ 10.1105/tpc.111.095109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 2007; 19:1192-208; PMID:17449805; http://dx.doi.org/ 10.1105/tpc.107.050153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 1996; 10:691-702; PMID:8893545; http://dx.doi.org/ 10.1046/j.1365-313X.1996.10040691.x [DOI] [PubMed] [Google Scholar]
- 46. Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 1999; 17:63-71; PMID:10069068; http://dx.doi.org/ 10.1046/j.1365-313X.1999.00353.x [DOI] [PubMed] [Google Scholar]
- 47. Nefissi R, Natsui Y, Miyata K, Oda A, Hase Y, Nakagawa M, Ghorbel A, Mizoguchi T. Double loss-of-function mutation in EARLY FLOWERING 3 and CRYPTOCHROME 2 genes delays flowering under continuous light but accelerates it under long days and short days: an important role for Arabidopsis CRY2 to accelerate flowering time in continuous light. J Exp Bot 2011; 62:2731-44; PMID:21296763; http://dx.doi.org/ 10.1093/jxb/erq450 [DOI] [PubMed] [Google Scholar]
- 48. Wolf FT, Haber AH. Chlorophyll content of gibberellintreated wheat seedlings. Nature 1960; 186:217-8; PMID:13845637; http://dx.doi.org/ 10.1038/186217a0 [DOI] [PubMed] [Google Scholar]
- 49. Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol 1998; 118:773-81; PMID:9808721; http://dx.doi.org/ 10.1104/pp.118.3.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hisamatsu T, King RW, Helliwell CA, Koshioka M. The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol 2005; 138:1106-16; PMID:15923331; http://dx.doi.org/ 10.1104/pp.104.059055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blazquez MA, Trenor M, Weigel D. Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol 2002; 130:1770-5; PMID:12481060; http://dx.doi.org/ 10.1104/pp.007625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Penfield S, Hall A. A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 2009; 21:1722-32; PMID:19542296; http://dx.doi.org/ 10.1105/tpc.108.064022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arana MV, Marín-de la Rosa N, Maloof JN, Blazquez MA, Alabadi D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci U S A 2011; 108:9292-7; PMID:21576475; http://dx.doi.org/ 10.1073/pnas.1101050108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 2014; 26:1557-69; PMID:24781117; http://dx.doi.org/ 10.1105/tpc.114.123794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hedden PG, JE Inhibition of Gibberellin Biosynthesis by paclobutrazol in cell-free homogenates of Curcurba maxima endosperm and Maluz pumila embryos. J Plant Growth Regul 1985; 4:111-22; http://dx.doi.org/ 10.1007/BF02266949 [DOI] [Google Scholar]
- 56. Yamashino T, Nomoto Y, Lorrain S, Miyachi M, Ito S, Nakamichi N, Fankhauser C, Mizuno T. Verification at the protein level of the PIF4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signal Behav 2013; 8:e23390; PMID: 23299336; http://dx.doi.org/ 10.4161/psb.23390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M . Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One 2012; 7:e36210; PMID:22590525; http://dx.doi.org/ 10.1371/journal.pone.0036210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nomoto Y, Kubozono S, Miyachi M, Yamashino T, Nakamichi N, Mizuno T. A circadian clock- and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol 2012; 53:1965-73; PMID:23037004; http://dx.doi.org/ 10.1093/pcp/pcs141 [DOI] [PubMed] [Google Scholar]
- 59. Hauvermale AL, Ariizumi T, Steber CM. Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 2012; 160:83-92; PMID:22843665; http://dx.doi.org/ 10.1104/pp.112.200956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. . Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008; 451:475-9; PMID:18216856; http://dx.doi.org/ 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 2001; 13:1555-66; PMID:11449051; http://dx.doi.org/ 10.1105/tpc.13.7.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 2002; 14:57-70; PMID:11826299; http://dx.doi.org/ 10.1105/tpc.010319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 2003; 15:1120-30; PMID:12724538; http://dx.doi.org/ 10.1105/tpc.010827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. . Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 2003; 299:1896-8; PMID:12649483; http://dx.doi.org/ 10.1126/science.1081077 [DOI] [PubMed] [Google Scholar]
- 65. Weller Jea. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Natl Acad Sci U S A 2012; 109:21158-63; PMID:23213200; http://dx.doi.org/ 10.1073/pnas.1207943110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 2011; 156:357-72; PMID:21430186; http://dx.doi.org/ 10.1104/pp.111.172684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Frigerio M, Alabadi D, Perez-Gomez J, Garcia-Carcel L, Phillips AL, Hedden P, Blázquez MA. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 2006; 142:553-63; PMID:16905669; http://dx.doi.org/ 10.1104/pp.106.084871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reid JD, Davidson SE; Ross JJ. Auxin cts independently of DELLA proteins in regulating gibberellin levels. Plant Signal Behav 2010; 6:406-8; PMID:21358281; http://dx.doi.org/ 10.4161/psb.6.3.14352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 2007; 5:e222; PMID:17683202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim WY, Hicks KA, Somers DE. Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol 2005; 139:1557-69; PMID:16258016; http://dx.doi.org/ 10.1104/pp.105.067173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thines BC, Youn Y, Duarte MI, Harmon FG. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J Exp Bot 2014; 65:1141-51; PMID:24574484; http://dx.doi.org/ 10.1093/jxb/ert487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Blazquez MW, D. Independent Regulation of Flowering by Phytochrome B and GIbberellins in Arabidopsis. Plant Physiol 1999; 120:1025-32; PMID:10444085; http://dx.doi.org/ 10.1104/pp.120.4.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 2008; 20:337-52; PMID:18252845; http://dx.doi.org/ 10.1105/tpc.107.052142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schafer E, Quail PH. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 2007; 19:3915-29; PMID:18065691; http://dx.doi.org/ 10.1105/tpc.107.051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lilly ST, Drummond RSM, Pearson MN; MacDiarmid RM. Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol Plant Microbe Interact 2011; 24:294-304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


