Abstract
Virus protein U (Vpu) from HIV-1, a small membrane protein composed of a transmembrane helical domain and two α-helices in an amphipathic cytoplasmic domain, down modulates several cellular proteins, including CD4, BST-2/CD317/tetherin, NTB-A, and CCR7. The interactions of Vpu with these proteins interfere with the immune system and enhance the release of newly synthesized virus particles. It is essential to characterize the structure and dynamics of Vpu in order to understand the mechanisms of the protein-protein interactions, and potentially to discover antiviral drugs. In this article, we describe investigations of the cytoplasmic domain of Vpu as well as full-length Vpu by NMR spectroscopy. These studies are complementary to earlier analysis of the transmembrane domain of Vpu. The results suggest that the two helices in the cytoplasmic domain form a U-shape. The length of the inter-helical loop in the cytoplasmic domain and the orientation of the third helix vary with the lipid composition, which demonstrate that the C-terminal helix is relatively flexible, providing accessibility for interaction partners.
Keywords: human immunodeficiency virus (HIV), viral protein U (Vpu), nuclear magnetic resonance (NMR), membrane protein, lipid bilayers
Graphical abstract
1. Introduction
Virus protein U (Vpu) is one of the accessory proteins encoded by HIV-1. Although it is dispensable for viral replication in vitro, it has multiple biological functions that are crucial for viral infectivity in vivo (1–3). Vpu removes newly synthesized CD4 receptors from the endoplasmic reticulum (ER), and causes their subsequent degradation. As a result, it prevents CD4 receptors from binding to the viral envelope (Env) precursor gp160 thereby enhancing viral infectivity. Vpu also enhances the release of newly formed virus particles by antagonizing the human immune restriction factor BST-2/CD317 (tetherin), which otherwise prevents their release (4–6). Recently Vpu has been shown to induce down modulation of natural killer (NK) cell receptor NK, T-cell, B-cell antigen (NTB-A) from the cell surface, and prevent HIV-Infected cells from degranulation and lysis by NK cells (7,8). Vpu also down regulates the chemokine receptor CCR7 on the cell surface, and subsequently reduces the ability of CD4+ T cells to migrate into the lymph nodes, where it helps with priming and assembly of immune responses (9).
Vpu is a small 81-residue type I membrane protein that has two distinct domains, a hydrophobic transmembrane (VpuTM) domain with helix-1, and an amphipathic cytoplasmic (VpuCyto) domain with helix-2 and helix-3. Loop-1 connects the VpuTM and the VpuCyto domains, and loop-2 connects the two helices in the VpuCyto domain and contains two conserved phosphorylation sites. These two domains are individually associated with biological activities mentioned above that contribute to the pathogenicity of HIV-1 infection. The structures and properties of three polypeptides are discussed in this article: TM domain (residues 2–31) of Vpu (VpuTM), Cyto domain (residues 28–81) of Vpu (VpuCyto), and full-length Vpu (VpuFull) (residues 2–81).
The interactions of Vpu with BST-2, NTB-A and CCR7 have been mapped to the transmembrane (TM) domains of each of the polypeptides. VpuTM is shown to employ the A10, A14, A18 and W22 helix face in its protein-protein interactions with both biological and spectroscopic studies (8–10). Helix-2 in Vpu, which is in the cytoplasmic domain, is required for CD4 binding (11); the conserved DSGXXS phosphorylation site in loop-2, which connects helix-2 and helix-3, acts as a molecular adaptor to recruit β-TrCP, linking CD4 and/or BST-2 to the ubiquitin ligase complex for degradation (2,6,12–17). The tryptophan in the C-terminal helix of VpuCyto has been identified as a key residue for enhancement of virus release, primarily due to the ability of Vpu to displace BST-2 from the virus assembly site (18–20).
It is essential for the samples to mimic the properties of biological membranes for structural, dynamic and functional studies of membrane proteins. Micelles, isotropic bicelles, nanodiscs, magnetically aligned bicelles, and liposomes have been widely applied in both solution NMR and solid-state NMR studies. The isotropic micelle is the most widely used membrane environment for solution NMR, since it reorients fast enough to give narrow line widths and at the same time it provides a hydrophobic interior environment to solubilize and stabilize membrane proteins. Although proteoliposomes more closely mimic membrane bilayers, their slow tumbling rates require the techniques of solid-state NMR in order to obtain high-resolution spectra. In addition, all the mentioned systems can be further altered by changing composition; pH, charge and hydration levels.
Solution NMR is well established as a method to characterize the structures and dynamics of proteins. Isotropic chemical shifts provide the empirical torsional angle restraints for the structure refinements, and residual dipolar couplings (RDCs) are available from samples in weakly aligned media as angular restraints. Distance restraints can be derived from homo-nuclear Overhauser effects (NOEs) and paramagnetic relaxation enhancements (PREs). PREs provide a source of long-range distance restraints for structure determination of helical membrane protein, which usually lack measurable long-range 1H-1H NOEs in micelle environments. The dynamics of local motions (usually within the picosecond to nanosecond range) can be monitored by T1 and T2 relaxation, and heteronuclear NOEs. Similarly, solid-state NMR can be applied to characterize structures of proteins with input of torsion angle restraints, angular restraints and distance restraints. In magic angle spinning (MAS) solid-state NMR, dipolar couplings and chemical shift anisotropy are averaged when the samples are spinning at 54.7°, the magic angle, and as a result the spectra have narrow resonance line widths. A complication is that MAS solid-state NMR experiments require recoupling pulse sequences to measure the dipolar couplings. The heteronuclear dipolar couplings measured from the unoriented proteins are uninformative, because the powder patterns and the bond lengths (N-H amide bonds or C-H bond at Cα) are essentially identical for all sites. However, when the proteins are undergoing fast rotational diffusion about the bilayer normal, the tensors are partially averaged, resulting in “reduced powder patterns” (21–23), whose breadths reflect the magnitudes of the dipolar couplings. In order to measure the angular-dependent heteronuclear dipolar couplings, it is necessary to find the condition of fast rotational diffusion for the proteins. This technique is known as rotationally aligned (RA) solid-state NMR. The symmetric pulse sequence, R1871 can recouple heteronuclear dipolar couplings, which are used as angular restraints in the structure refinements, and long mixing intervals during homonuclear correlation experiments and PREs provide distance restraints.
Solid-state NMR experiments have been applied to Vpu full-length in phospholipid bilayers (24–26). Marassi et al showed that the TM and Cyto domains of Vpu have distinct orientations with the TM helix approximately parallel to the bilayer normal and Cyto domain perpendicular to the bilayer normal in DPPC/DPPG bilayers aligned on glass plates (24). Similarly, with MAS experiments, the results of Do et al reflect membrane insertion of the TM domain, and the water accessible Cyto domain in POPC bilayers (26). While the three-dimensional structure of Vpu full-length has not been determined previously, the structures of various constructs of the TM domain of Vpu have been well studied in various environments including TFE (27) and detergent micelles (28) by solution NMR, and also in phospholipid bicelles (28), and phospholipid bilayers (29–31) by solid-state NMR. All the results in phospholipid bilayers suggest that the TM domain of Vpu forms a single bilayer spanning α-helix with a tilt angle between 15° and 30° relative to the bilayer normal. The Cyto domain has proven to be more difficult to study largely because of its conformational flexibility. Previously, the structure of the VpuCyto domain from HIV strain HV1S1 has been studied in 50% TFE (32), aqueous solution (33) and DPC micelles (34) by solution NMR. All of the results show that the Cyto domain contains two α-helices, and a C-terminal turn. In addition, VpuCyto alone has been shown to align parallel on membrane surface in lipid bilayers on glass plates (35). In addition to experimental studies, molecular dynamics (MD) simulations were applied independently as well as in combination with NMR results to several constructs of Vpu including VpuTM (36–38), the first two helices of Vpu (39,40) and full-length Vpu (41) in lipid environments.
Even though several biological studies have identified the interaction partners of Vpu and shown the key residues or motifs involving phosphorylation or protein-protein interactions, the details of structure and corresponding mechanisms at the molecular level remain of interest for investigations. For instance, determining the conformation of the Cyto domain is crucial for understanding the mechanism of phosphorylation and how it recruits other interaction partners. In addition, the conformation of full-length Vpu and its possible conformational changes upon interactions are of interest, since many of the interactions collectively utilize both the TM and Cyto domains, for example the down modulation of CD4 and the dysfunction of BST-2. Several key points need to be clarified with structural studies of Vpu, for example: (1) what is the relative orientation of the TM domain and the Cyto domain? (2) What is the relative orientation of the two helices in the Cyto domain? Are they linear or forming a U-shape conformation? (3) How do the helices associate with phospholipids? Are they lying on the phospholipid surface, partially or fully embed in the membrane, or exposed in the aqueous solution? (4) Is loop-2 structured or flexible? (5) How do loop-2 and terminal region associate with phospholipids?
NMR spectroscopy has the ability to answer these questions, and in this article, we focus on solving the structure of full-length Vpu, and describing both the structure and dynamics of VpuCyto in order to provide more insights in the biological functions at the molecular level.
2. Materials and methods
2.1. Plasmids, Cells, and Reagents
Vpu expression plasmids for NL4-3 VpuFull and VpuCyto constructs were prepared similarly to those previously described (42). Synthesized DNA corresponding to VpuCyto (Integrated DNA Technologies) was ligated into phLV vector for expression as a Trp-leader fusion. Synthesized DNA for the VpuFull sequence was inserted into pET31b(+) vector (Agilent Technologies) for expression as a KSI fusion. For both constructs the methionines were mutated to leucines to assist protein purification, and were shown to have no effect on the properties of Vpu. Expression plasmids encoding mutants VpuCyto-L41C, VpuCyto-A74C, and VpuFull-L41C were constructed using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). The primers were synthesized by Integrated DNA Technologies. BL21-CodonPlus competent cells were obtained from Agilent, and Overexpress C41(DE3) competent cells were obtained from Lucigen. The chelated manganese was prepared by adding manganese sulfate (MnSO4) to ethylenediaminetetraacetic acid (EDTA) (0.5 M) as a 2:1 molar ratio in 20mM HEPES pH 7.3, and an MnEDTA precipitate was formed overnight. The precipitated complex was centrifuged, washed with water and methanol, and dried by lyophilization. The lipids 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) were purchased from Avanti Polar Lipids. MTSL, 5-DOXYL-stearic acid free radical (5DSA), ascorbic acid and deuterium oxide (D2O) were obtained from Sigma. 13C-glucose and (15NH4)2SO4 were obtained from Cambridge Isotope Laboratories. Tris-HCl buffer was obtained from Teknova. Dithiothreitol (DTT) and HEPES buffer were purchased from Invitrogen.
2.2. Expression, purification and sample preparation
The expression and purification of VpuFull, VpuCyto and mutants were performed similarly to the methods described previously (24,42). Briefly, the proteins were overexpressed in BL21-CodonPlus competent cells and Overexpress C41 (DE3) competent cells, respectively for VpuCyto and VpuFull in LB or minimal M9 growth media with (15NH4)2SO4 and/or 13C-glucose for preparation of unlabeled or isotopically labeled samples. The proteins were isolated using nickel affinity chromatography, and then separated from the fusion partner by cyanogen bromide cleavage, followed by reverse phase high performance liquid chromatography (HPLC) for final purification. The molecular weight of the purified protein was confirmed to be correct using SDS-PAGE and Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. Milligram amounts of the isotopically labeled proteins were obtained for the NMR experiments.
Protein samples for solution NMR were prepared by dissolving purified and lyophilized VpuCyto protein powder in either aqueous solution with 10%(V/V) D2O with the pH adjusted to 3.0, or aqueous solution containing 100 mM DHPC with 10% (v/v) D2O with the pH adjusted to 4.0. For RDC measurements, DHPC containing VpuCyto was weakly aligned in vertically stretched 6% polyacrylamide gels, as previously described (43). Proteoliposome samples for solid-state MAS experiments were prepared by first mixing purified lyophilized Vpu powder with DMPC powder (1:3.5 mass ratio) and dissolving in 10% SDS buffer to obtain a clear solution with 10mg/mL DMPC concentration. The sample was diluted 20-fold, and incubated at room temperature for 4 hours. Subsequently, SDS was removed from the solution by first dialysis against 20mM HEPES, pH 7.3 overnight, and then 5 days dialysis against 20 mM KCl in 20 mM HEPES, pH 7.3. After dialysis, the proteoliposomes were collected by ultracentrifugation at 235,000 ×g overnight at 15 °C. The concentrated proteoliposome sample was packed into a 3.2 mm MAS rotor for solid-state NMR experiments.
To measure PREs, the cysteines in each of the mutants were covalently linked to MTSL using the following protocol (44). Purified, lyophilized mutant protein powder was dissolved in 8 M urea in 20 mM Tris-HCl, pH 8.0 with 20-fold molar excess DTT and incubated for 2 hours. The protein solution was then passed through a G10 desalting column pre-equilibrated with urea buffer to remove DTT. A 10-fold excess of MTSL was added to the reduced protein, and the mixture was incubated at room temperature overnight. Free MTSL was separated from MTSL-labeled protein by passing the sample through a G10 desalting column followed by dialysis against water for a day. The MTSL-labeled protein was obtained by lyophilization. For solution NMR measurements, MTSL-labeled protein was refolded in 100 mM DHPC micelles with a final protein concentration of ~0.1 mM. For MAS experiments, MTSL-labeled protein was reconstituted into proteoliposomes with DMPC, as previously described. MTSL-labeled protein was reduced by addition of 10-fold excess of ascorbic acid in both micelles and proteoliposomes. 5SDA and/or MnEDTA were incorporated into either VpuCyto in 100 mM DHPC or VpuFull in DMPC proteoliposomes to a final concentration of 0.5 mM for 5SDA and 1 mM for MnEDTA respectively.
2.3. CD spectroscopy
VpuCyto in aqueous solution and in DHPC micelles were analyzed by CD spectroscopy in the far UV (176 – 260 nm) region. Lyophilized protein was dissolved in either H2O or 100mM DHPC to a final concentration of 1 mg/mL to obtain the spectra. Samples were transferred to a 0.1 mm path-length cell for data collection. (NSG Precision Cells) Data were collected on an AVIV model 2.98 spectrophotometer at 25 °C, in 1.0 nm wavelength intervals. Data were analyzed using an online server DichroWeb (45,46).
2.4. Solid-state NMR Experiments
For MAS solid-state NMR experiments, 13C-detected cross polarization (CP) spectra were acquired with a MAS spinning speed of 5 kHz over the temperature range 5 °C to 30 °C to determine the minimum temperature that allows VpuFull to undergo fast rotational diffusion about the bilayer normal. Two- and three-dimensional experiments were acquired with a MAS spinning speed of 10 kHz, at 25 °C. 13C/13C correlations were obtained with 50 ms mixing by proton-driven spin diffusion (PDSD) (47) and 200 ms mixing by dipolar-assisted rotational resonance (DARR) (48,49). Magnetization transfer from 15N to 13Cα was performed with SPECIFIC-CP (50). The 15N and 13Cα chemical shifts were assigned with three-dimensional NCACX and NCOCX experiments. The 1H-13C and 1H-15N dipolar couplings were assigned based on a combination of assigned chemical shifts and thee-dimensional separated local field (SLF) experiments including 1H-13C/N/Cα and 1H-15N/N/Cα, where R1871 pulse sequences (51) with 100 kHz 1H B1 fields, were implemented to measure heteronuclear dipolar couplings. Details of all the experiments are in Table S1. All the experiments were performed at a 1H resonance frequency of 700 MHz using a home-built triple-resonance MAS probe and a Bruker Avance II console.
2.5. Solution NMR experiments
Solution NMR experiments were performed on Bruker Avance 600 MHz and Varian 800 MHz spectrometers equipped with 5 mm z-axis pulse-field gradient, 1H/15N/13C triple resonance cryoprobes. All of the experiments were performed at 50 °C. Triple-resonance HNCA, HNCACB, HN(CO)CA and HNCO experiments were performed on uniformly 13C and 15N labeled protein for the backbone amide resonance assignments. The side-chain assignments were completed using HCC(CO)NH, CC(CO)NH and TOCSY-HSQC experiments. 15N-edited NOESY-HSQC (mixing time, 120 ms) and 13C-edited NOESYHSQC (mixing time, 120 ms) experiments were performed to measure distance restraints. 15N T1 measurements were recorded with relaxation delays of 20, 220, 420, 620, 820, 1020, 1320, and 1520 ms.15N T2 measurements were recorded with relaxation delays of 10, 30, 50, 70, 90, 130, and 170 ms. Heteronuclear 1H-15N NOEs were obtained using a gradient sensitivity enhanced 1H-15N NOE experiment, recorded with and without 1H saturation after a relaxation delay of 4 s. RDCs were measured with HSQC in-phase and anti-phase (IPAP) experiments.
2.6. NMR data analysis
All of the NMR data were processed using NMRPipe (52) and analyzed with Sparky. All of the figures were prepared using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). The resonance intensities and chemical shifts were extracted using Sparky. Both T1 and T2 relaxation time were fitted and calculated using SPARKY.
2.7. Structure calculation
Structure calculations for VpuFull were carried out with Xplor-NIH[16]. A total of 100 structures were generated in the refinement step, and the 20 structures with lowest-energy were used for analysis. For structure calculation of VpuFull in proteoliposome, a three-step process was employed, and the lowest energy structure was chosen as the initial structure for refinement in the following step. Dipolar couplings and torsion angle were used in the initial structure determination. In the second and third steps of the calculations distance restraints between the residues and the plane distance restraints (53) were added with kdih = 400 kcal mol−1 rad−2. 1H-15N krdc was ramped from 0.01 to 2 kcal mol−1 rad−2, 1H-13C krdc was ramped from 0.01 to 1 kcal mol−1 rad−2, know was ramped from 2 to 40 kcal mol−1 Å−2, and kplane was ramped from 0.01 to 5 kcal mol−1 Å−2.
Structure calculations for VpuCyto were carried out with Xplor-NIH (54,55) ab initio simulated annealing protocol with two steps: (1) high temperature simulated annealing, and (2) refinement. The initial structures were generated from an extended random coil based on dihedral angle restraints derived from TALOS+ (56) and distance restraints derived from the NOE cross-peaks in the simulated annealing. Residual dipolar couplings as angular restraints and paramagnetic relaxation data as distance restraints were incorporated during the refinement step. Simulated annealing was performed with a starting temperature of 3500 K and cooling to 25 K in steps of 12.5 K. kdih = 400 kcal mol−1 rad−2. Knoe was ramped from 2 to 50 kcal mol−1 Å−2. The refinement was performed with Xplor-NIH torsion angle molecular dynamics and experimental restraints included. The temperature was cooled to 50 K from an initial value of 2000 K in steps of 12.5 K. kdih = 400 kcal mol−1 rad−2, knoe = 40 kcal mol−1 A−2, krdc was ramped from 0.2 to 2 kcal mol−1 rad−2.
3. Results
3.1. CD spectroscopy and secondary structures of VpuFull and VpuCyto
The secondary structures of VpuCyto in aqueous solution and in DHPC micelles were characterized by CD spectroscopy. All of the CD spectra show a maximum around 191nm, and local minima near 208nm and 222nm, which are indicative of the presence of helical secondary structure. The comparison of the CD spectrum of VpuCyto in aqueous solution (Figure 1B) with that of VpuCyto in DHPC micelles (Figure 1A indicates that VpuCyto is more helical in the presence of DHPC micelles. This indicates that the cytoplasmic domain is affected by the presence of DHPC, and is not freely soluble in the aqueous portion of the sample. Our CD spectra provide evidence of structure in VpuCyto, even in aqueous solution, which differs somewhat from the findings of Wittlich et al (34), who found predominantly disordered Vpu38-81 in aqueous solution. However, the results are in agreement in showing an increase in secondary structure in the presence of micelles.
Figure 1. CD spectra of VpuCyto.
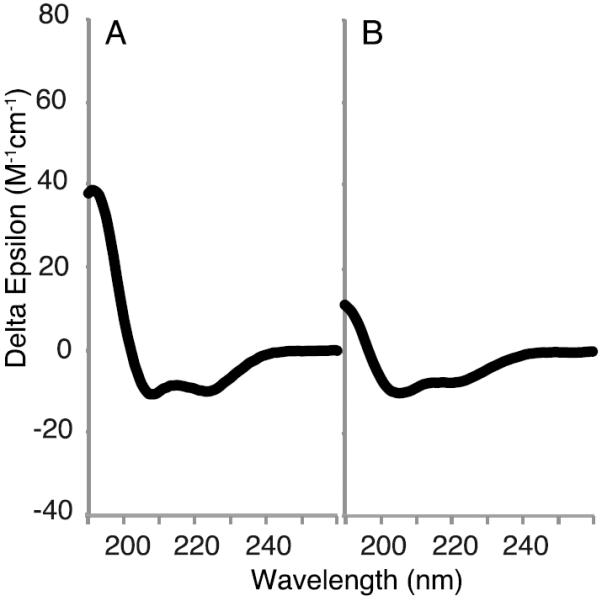
A. CD spectrum of VpuCyto in 100 mM DHPC micelles. B. CD spectra of VpuCyto in aqueous solution.
3.2. Solution NMR spectroscopy and resonance assignments
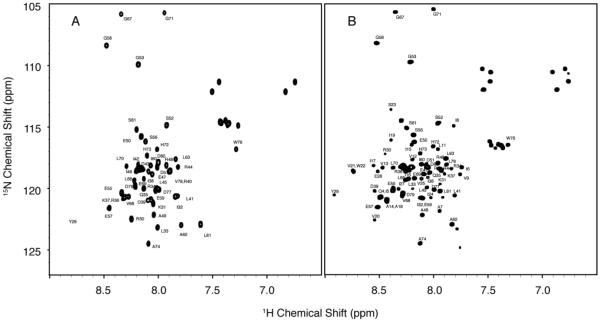
Both uniformly 15N labeled VpuCyto and VpuFull solubilized in DHPC micelles gave well-resolved two-dimensional HSQC solution NMR spectra (Figure 2). The two spectra overlap to a large extent, indicating that the structure of VpuCyto alone is representative of the cytoplasmic domain in full-length Vpu. In addition, the resonances could be fully assigned to the residues in the polypeptides (Figure 2).
Figure 2. 1H/15N HSQC NMR spectra of VpuCyto and VpuFull.
A. Assigned 1H/15N HSQC spectrum of VpuFull in 100 mM DHPC micelles. B. Assigned 1H/15N HSQC spectrum of VpuCyto in 100 mM DHPC micelles.
3.3. Vpu undergoes fast rotational diffusion in proteoliposomes
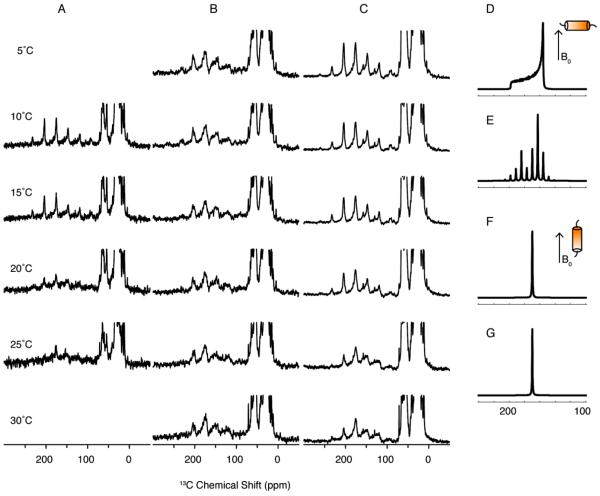
The series of one-dimensional 13C-detected cross-polarization solid-state NMR spectra shown in Figure 3 were acquired over a range of temperatures in order to characterize the rotational diffusion of three Vpu constructs in phospholipid bilayers. Relatively slow magic angle spinning, at 5 kHz, was used in order to observe the sideband patterns of the 13C' signals. The results show that VpuTM undergoes fast rotational diffusion when the temperature is higher than 25°C (Figure 3A). By contrast, no powder pattern is found even when VpuCyto is at 5°C (Figure 3B). At high temperatures, the ratio of the first left sideband to the center band of the carbonyl carbon of VpuCyto is 0.5, which is relatively high compared to VpuTM. The orientation effects were simulated under the conditions that the principal values of C' δ11, δ22, and δ22 are 244 ppm, 170.8 ppm, and 95 ppm, respectively, where δ22 is 3.2° away from the carbonyl bond (57). For α-helices, carbonyl bonds are roughly parallel to the helical axes (22), and therefore, we simplified the system in the simulations so that δ22 represents the axial directions, which are either parallel or perpendicular to the bilayer normal.
Figure 3. 13C solid-state NMR spectra of three constructs of Vpu in DMPC proteoliposomes.
A. VpuTM. B. VpuCyto. and C. VpuFull. All the spectra were acquired at 5 kHz MAS at various temperatures. VpuTM is static below 15 °C and undergoing fast rotational diffusion above 25 °C. D. Simulated C' powder pattern of helix undergoing fast rotational diffusion with the helix axis perpendicular to the bilayer normal. E. Simulated C' powder pattern of helix undergoing fast rotational diffusion with the helix axis perpendicular to the bilayer normal under 5 kHz MAS. F. Simulated C' powder pattern of helix undergoing fast rotational diffusion with the helix axis parallel to the bilayer normal. G. Simulated C' powder pattern of helix undergoing fast rotational diffusion with the helix axis parallel to the bilayer normal under 5 kHz MAS.
Comparison of simulated powder patterns of helices with different orientations with experimental spectra of VpuTM and VpuCyto shows that the orientation of the TM domain is approximately parallel to the bilayer normal, while the orientation of cytoplasmic domain is approximately perpendicular to the bilayer normal. By comparing the spectra of VpuTM, VpuCyto, and VpuFull, we find that the spectra of VpuFull are combinations of those from VpuTM and VpuCyto. The range of reduced dipolar powder patterns observed in 1H-13C/13C and 1H-15N/13C SLF experiments (Figure 4) provide direct evidence that Vpu is undergoing fast rotational diffusion at 25 °C.
Figure 4. 1H-13C/13C and 1H-15N/13C SLF spectra of VpuFull in DMPC proteoliposomes.
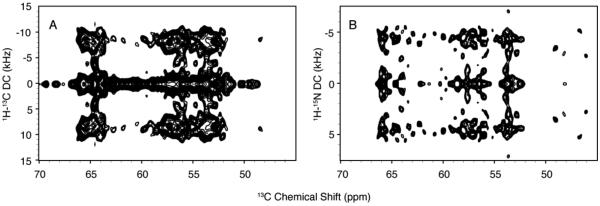
A. 1H-13C/13C SLF spectrum. B. 1H-15N/13C SLF spectrum. The various dipolar couplings obtained from both experiments show that VpuFull is undergoing fast rotational diffusion, and single-site resolution is achievable when it is incorporated in proteoliposomes at 25 °C.
3.4. Solid-state NMR spectroscopy and assignment
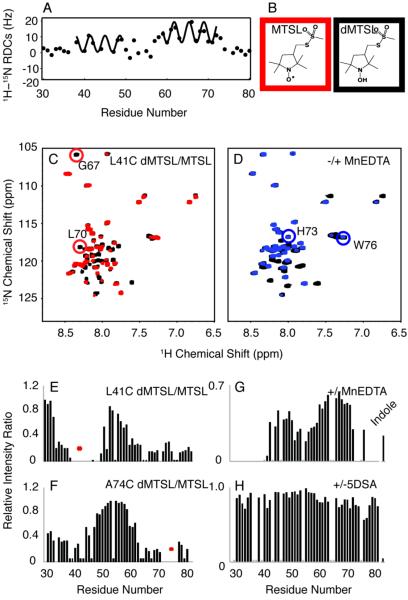
It is possible to obtain high-resolution two-dimensional 13C/13C and 13C/15N correlation spectra under these sample conditions (Figure 4). Spectral resolution is further improved in three-dimensional experiments. Importantly, resonances from the mobile regions of the Vpu polypeptide are characterized by TOBSY (total through-bond correlation spectroscopy) (58,59) (Figure 5B). This J-coupling-based 13C/13C correlation experiment is only feasible for residues that are mobile enough to average out the dipolar couplings. Signals from about 30 residues are found in the TOBSY spectrum. Partial assignments are available by considering the dynamics studies of VpuCyto in micelles. Most of the mobile residues are found in loop-2 and both termini, suggesting that these regions are also highly dynamic in proteoliposomes. Complementary dipolar based NCACX and NCOCX experiments identify the structured parts of VpuFull. This allows the sequential assignment of the chemical shifts of 15N, 13Cα and 13C' resonances. Signals from 43 immobile residues were assigned with NCACX and NCOCX experiments (Figure S1), and their dipolar couplings are measured from 1H-15N/15N/13C and 1H-13C/15N/13C three-dimensional SLF experiments. The measured values of chemical shifts and heteronuclear dipolar couplings are listed in Table 1. Dipolar couplings provide valuable information about secondary structure, especially α-helix, in a protein. The resulting 1H-15N dipolar coupling values for the backbone amides in an ideal helix display a sinusoidal wave pattern referred to as a Dipolar Wave. Three Dipolar Waves are found in the plot of dipolar couplings versus residue numbers (Figure 6A), which indicates the existence of three helices in VpuFull. The tilt angles estimated according to the mean values and the amplitudes of dipolar waves, are 20°, 80° and 80° from the bilayer normal for helices-1, -2 and -3, respectively.
Figure 5. Correlation spectra of VpuFull in DMPC proteoliposomes.
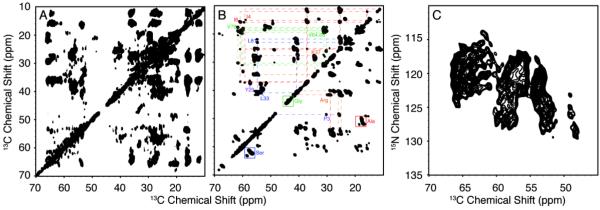
A. 13C/13C correlation spectrum obtained with 50 ms PDSD mixing. B. 13C/13C TOBSY spectrum; the J-coupling based 13C/13C correlation spectrum indicates the mobile residues of VpuFull. C. 15N/13C HETCOR spectrum, where the 15N/13C transfer is obtained from SPECIFIC-CP. All of the experiments show that single-site resolution is achievable when VpuFull is incorporated in DMPC proteoliposomes at 25 °C.
Table 1.
List of chemical shifts and dipolar couplings of VpuFull in proteoliposomes.
| Residue | 15N CS (ppm) | 13Cα CS (ppm) | 13C' CS (ppm) | 1H-15N DC (kHz) | 1H-13CDC (kHz) |
|---|---|---|---|---|---|
| I8 | 119.52 | 65.30 | 175.90 | 8.5 | |
| V9 | 119.93 | 66.80 | 176.69 | 8.5 | 18.1 |
| A10 | 116.54 | 55.39 | 175.62 | 15.2 | |
| L11 | 119.26 | 58.53 | 176.15 | 7.7 | 17.7 |
| V12 | 119.70 | 66.77 | 175.68 | 10.6 | 6.9 |
| V13 | 120.64 | 66.66 | 175.57 | 4.5 | 12.9 |
| A14 | 121.48 | 54.52 | 176.03 | 3.5 | 14.5 |
| I15 | 115.91 | 63.85 | 176.20 | 7.4 | 17.6 |
| I16 | 122.12 | 65.29 | 176.00 | 9.2 | 18.6 |
| I17 | 121.84 | 65.05 | 176.14 | 4.3 | 16.4 |
| A18 | 121.44 | 54.31 | 177.07 | 9.5 | 5.9 |
| I19 | 116.00 | 64.61 | 176.98 | 10.3 | 18.3 |
| V20 | 122.70 | 66.74 | 175.87 | 4.8 | 17.1 |
| V21 | 120.20 | 66.56 | 175.67 | 4.5 | 14.6 |
| W22 | 121.45 | 56.54 | 177.27 | 8.2 | 15.3 |
| S23 | 118.8 | 60.19 | 175.99 | 9.1 | 15.0 |
| I24 | 115.36 | 64.66 | 176.99 | 14.9 | |
| V25 | 121.10 | 66.96 | 176.59 | 6.0 | 16.9 |
| I26 | 120.79 | 64.54 | 176.03 | 8.4 | |
| I27 | 122.04 | 66.56 | 175.75 | 8.6 | |
| Q35 | 120.84 | 60.81 | 174.68 | 7.6 | |
| R36 | 116.76 | 60.34 | 177.83 | 15.3 | |
| K37 | 120.94 | 60.81 | 174.94 | 7.8 | |
| I38 | 118.10 | 63.39 | 176.31 | 19.6 | |
| D39 | 119.20 | 56.44 | 176.81 | ||
| R40 | 121.63 | 54.80 | 177.05 | 7.8 | 14.8 |
| L41 | 118.82 | 58.25 | 177.62 | 9.2 | 2.4 |
| I42 | 118.20 | 63.42 | 176.63 | 8.5 | |
| D43 | 119.59 | 56.96 | 176.55 | 6.3 | 13.5 |
| R44 | 121.76 | 57.08 | 176.97 | 8.1 | 18.6 |
| L45 | 120.35 | 57.78 | 175.88 | 8.6 | 16.6 |
| I46 | 119.07 | 65.05 | 175.97 | 8.9 | 15.4 |
| E47 | 122.35 | 60.12 | 177.29 | 8.0 | 7.3 |
| R48 | 122.48 | 56.98 | 176.83 | 6.7 | 9.2 |
| E59 | 117.57 | 58.99 | 177.28 | ||
| I60 | 119.07 | 65.55 | 176.01 | 7.4 | 16.2 |
| S61 | 117.32 | 58.05 | 175.87 | 8.6 | 15.8 |
| A62 | 120.23 | 52.83 | 175.95 | 5.9 | 3.4 |
| L63 | 115.86 | 56.65 | 176.98 | 16.0 | |
| V64 | 119.73 | 66.96 | 176.86 | 8.6 | 17.8 |
| E65 | 118.47 | 58.28 | 176.50 | 5.3 | |
| L66 | 115.81 | 58.05 | 175.97 | 8.0 | 13.9 |
| G67 | 103.05 | 46.51 | 173.83 | 1.3 | |
| V68 | 115.55 | 66.44 | 175.66 | 6.5 | |
| E69 | 120.24 | 59.42 | 176.01 | 7.2 | 18.2 |
| L70 | 125.55 | 60.04 | 174.02 | 9.2 | 13.4 |
Figure 6. Solid-state NMR measurements for structure calculation of VpuFull in DMPC proteoliposomes.
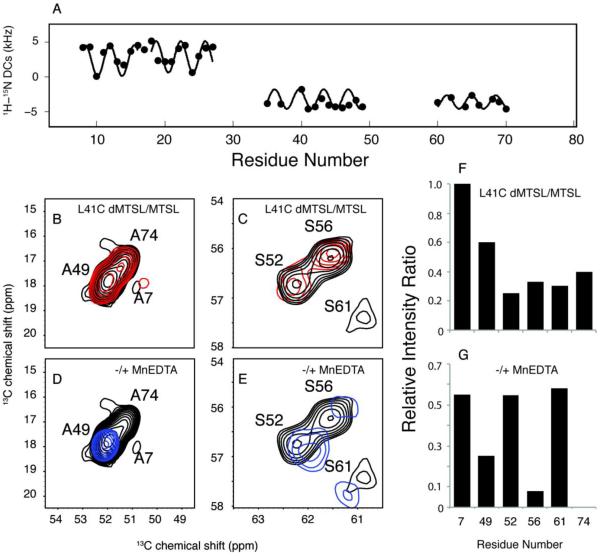
A. Plot of 1H-15N dipolar couplings versus residue numbers, indicating the helical regions in VpuFull. B. Overlap of 13C/13C TOBSY spectrum of L41C-VpuFull with MTSL (red) and its reduced diamagnetic form (black) at the alanine region. C. Overlap of 13C/13C TOBSY spectrum of L41C-VpuFull with MTSL (red) and its reduced diamagnetic form (black) at the serine region. D. Overlap of 13C/13C TOBSY spectrum of VpuFull with (blue) and without (black) MnEDTA present at the alanine region. E. Overlap of 13C/13C TOBSY spectrum of VpuFull with (blue) and without (black) MnEDTA present at the serine region. F. Plot of residues intensity change caused by L41C-MTSL. G. Plot of residues intensity change caused upon addition of MnEDTA solution.
3.5. Solid-state NMR measurements of VpuFull in DMPC proteoliposomes
Rotationally averaged dipolar coupling and chemical shift anisotropy powder patterns provide angular restraints and are essential for structure determination. However, we would obtain degeneracies from structural calculations only relying on the angular restraints. For instance, the two helices in the cytoplasmic domain could be linear or form a U-shape based on the analysis of only dipolar couplings. Distance restraints help characterize the relationships between the secondary structure elements. In general, in solid-state NMR, the distance restraints could be extracted from homonuclear spin diffusion with long mixing times. However, since no significant correlations are found from 13C/13C correlation experiments with DARR mixing up to 200 ms for VpuFull, two PRE approaches were applied to obtain long-range restraints. (1) (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl) methanesulfonate (MTSL) was attached to L41C VpuFull to probe the distances of nearby nuclei. (2) MnEDTA, a bulky paramagnetic reagent that is restricted to the aqueous environment, was utilized to distinguish between residues exposed to the solution or embedded in the phospholipids, which helps determine how the cytoplasmic domain of the full-length Vpu interacts with phospholipids. The intensities of the resonances from TOBSY spectra vary in the PRE experiments; therefore we can assign the resonances for the mobile region more specifically. For instance, A7, A49 and A74 are found in the TOBSY spectra (Figure 6B), yet they are ambiguous without assignment experiments. The intensities of both A49 and A74 are reduced by L41C-MTSL, suggesting that helix-2 and helix-3 form a U-shape conformation, and these resonances are assigned according to the intensity ratios in the spectra of L41C-MTSL and its reduced form (Figure 6B). On the other hand, the resonance from A74 disappears with the addition of MnEDTA (Figure 6D), which indicates A74 is exposed to the solvent. Similarly, we can assign serines according to the changes of intensities assuming the Cyto domain is U-shaped (Figure 6C). S61 is less affected by MnEDTA, suggesting that S61 in helix-3 may face the phospholipids or be slightly embedded into the phospholipids. S52 and S56 are more exposed in the solution due to their presence in the loop; however, S56 is substantially more affected by MnEDTA than S52, suggesting that two serines adopt different orientations, with S52 more exposed to the aqueous environment. (Figure 6E).
3.6. Structure of VpuFull in proteoliposomes
During the structure calculation of VpuFull in DMPC liposomes, torsion angle restraints were obtained from TALOS+ with chemical shift input of N, CA, CB and C'. Orientational restraints were obtained from 1H-15N and 1H-13C dipolar couplings. In addition, two types of distance restraints obtained from L41C-MTSL VpuFull and MnEDTA were incorporated to further refine the structure. Refined structures based only on dipolar couplings give many possible conformations (Figure S2A) when the relative orientations between helix-2 and helix-3 are allowed to form either a linear or U-shape. By considering the distance restraints obtained from L41C-MTSL VpuFull, helix-3 is found to be folded back, yet the relative orientation between the cytoplasmic domain and membrane surface is still ambiguous (Figure S2B). Helix-3 is further confined to the bilayer surface by introducing the plane distance restraint obtained from MnEDTA experiments (Figure S2C). The RMSD of the ten lowest energy structures shows that all the helices are well defined, with RMSDs of 0.48 Å, 0.31 Å, and 0.23 Å for helix-1, helix-2, and helix-3, respectively. Most of the variations come from the loop regions (loop-1 is 1.92 Å and loop-2 is 2.74 Å), and the RMSD of the entire structural region (7–70) converges at 5.18 Å. The relatively high overall RMSD mainly reflects the dynamics of helix-3.
3.7. Solution NMR determines secondary structure and dynamics of VpuCyto
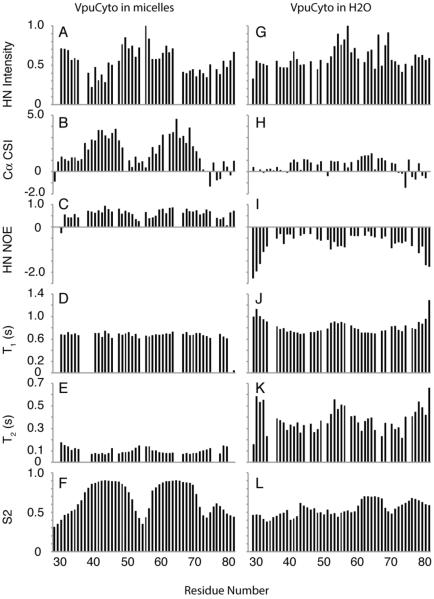
The secondary structure of VpuCyto in both types of samples can also be confirmed by solution NMR experiments (Figure 6). VpuCyto in aqueous solution has a relatively short T1 (between 690 ms and 1 s) and long T2 (between 210 ms and 600 ms), indicating that the protein is undergoing rapid reorientation, and the backbone is highly mobile. Also the negative 1H-15N heteronuclear NOE values throughout the polypeptide suggest that the internal motions of VpuCyto in aqueous media are rapid. However, T1, T2 and NOE have a non-uniform distribution, where the terminal and inter-helical loop region (51–58) have longer T1, longer T2 and more negative 1H-15N heteronuclear NOE than the helical regions. The variation in T1, T2 and H-15N heteronuclear NOE agrees with the CD data, suggesting that there is a structural distinction between the helical regions and loop region in aqueous solution, and that the structure is not fully disordered. In contrast, the Cα chemical shift index has values around +1 ppm, which cannot be categorized as helical. In addition, TALOS+ only predicts residues 62–70 as helix, and fails to predict any secondary structure for the rest of the protein due to the low order parameters. VpuCyto in aqueous solution may be in an intermediate state between well structured and fully disordered, with the helix-3 somewhat more structured.
In DHPC micelles, plots of the relative intensities of resonances in the HSQC NMR spectrum, and the Cα chemical shift index (Figure 7G and 7H) indicate two distinct helical regions connected by a mobile inter-helical loop. These helical regions are also consistent with the RDC data acquired in a 6% polyacrylamide stretched gel. When incorporated into DHPC micelles, the mobility is drastically reduced, as shown by the significant decrease of T2 relaxation (70 ms to 170 ms). In addition, the same trends were observed for T1, T2 and 1H-15N heteronuclear NOE, suggesting a significant difference in dynamics between the helical regions and the loop region. Overall, helix-3 and helix-2 exhibit similar restricted backbone dynamics, with similar values of 1H-15N heteronuclear NOEs, order parameters and 1H-15N HSQC peak intensities, in DHPC micelles. While residues at the N-, C-terminus and loop are significantly more flexible, with smaller values of the 1H-15N NOE and greater peak intensities.
Figure 7. Secondary structure and dynamics of VpuCyto in aqueous solution and DHPC micelles.
A.-F. are obtained from VpuCyto in aqueous solution. G.-L. are obtained from VpuCyto in 100 mM DHPC micelles. A. 1H/15N HSQC resonance intensity plot. B. Cα chemical shift index. C. 1H-15N heteronuclear NOE. D. T1 relaxation. E T2 relaxation. F. Order parameters derived from TALOS+. G. 1H-15N HSQC resonance intensity plot. H. Cα chemical shift index. I. 1H-1N heteronuclear NOE. J. T1 relaxation. K. T2 relaxation. L. Order parameters derived from TALOS+.
3.8. Solution NMR measurements for VpuCyto in DHPC micelles
Residual dipolar coupling measurements not only provide reliable information on the secondary structure of the protein, but also yield relative orientations of the helical segments in the protein. Residues in both N- and C-termini and the loop regions have close to 0 Hz RDCs (Figure 8A), which is an indication of the mobile properties of these regions. Two separate Dipolar Waves with very different amplitudes, indicating a considerable angle difference between the helices, fit the RDC values of the helical region. These RDC values can be used as angular restraints in structure calculations (60–62). The PRE values were used as distance restraints. The secondary structure and motion information were considered before determining the optimal placement of the spin labels. Residues at three positions 41, 62 and 74, located at helix-2, helix-3 and the C-terminal region respectively, were mutated to cysteines individually for MTSL attachment. 1H-15N HSQC NMR spectra were collected on the samples L41C-MTSL VpuCyto, S61C-MTSL VpuCyto, and A74C-MTSL VpuCyto. In both the L41C and A74C HSQC NMR spectra, residues near the mutation site showed small chemical shift changes compared to spectra of the wild type protein, which indicates that the mutation and the spin label did not change the protein structure dramatically. On the other hand since the S61C sample showed quite significant changes, it was not used for PRE measurements. The 1H-15N HSQC NMR spectra (Figure 8C) illustrate the overlapped spectra of the L41C-MTSL VpuCyto in red and its reduced form in black. Residues close to the mutation site L41C were broadened beyond detection. Observable reductions in intensity due to PRE were found for residues 67 and 70, even though they are not close to L41C in the sequence; this indicates that they are proximate to L41 in the folded polypeptide. The normalized PRE intensity ratios versus residue numbers for L41C VpuCyto and A74C VpuCyto are plotted in Figure 8E and 8F, and it is obvious that when the spin label is added to L41C, not only residues near 41, but also those in helix-3 and near the C-terminus are broadened. Similarly, for MTSL-A74C, not only are the residues in the C-terminal region broadened, but also the intensities of residues at helix-2 are significantly reduced. To ensure that the PRE effect is not intermolecular instead of intramolecular, protein concentrations were limited to 0.1mM during data acquisition; additionally, the intensity plot did not change significantly upon addition of unlabeled VpuCyto to the MTSL labeled sample, indicating that no intermolecular PRE effect are observed.
Figure 8. Solution NMR measurements for structure calculation of VpuCyto in DHPC micelles.
A. Plot of residual dipolar couplings as a factor of residue numbers obtained from weakly aligned sample in 6% polyacrylamide stretch gel. B. Chemical structure of MTSL (red box) and its reduced form (black box). C. Overlap of 1H-15N HSQC spectra of L41C VpuCyto with MTSL label (red) and its reduced diamagnetic label (black). D. Overlap 1H-15N HSQC spectra of VpuCyto with MnEDTA (blue) and without MnEDTA (black). E. Intensity plot shows PRE intensity ratio (Ipara/Idia) for L41C VpuCyto. F. Intensity plot shows PRE intensity ratio (Ipara/Idia) for A74C VpuCyto. G. Intensity change measured before and after addition of MnEDTA. H. Intensity change measured before and after addition of 5SDA to VpuCyto in DHPC micelles.
The MTSL samples provide 72 PRE restraints that were particularly useful in defining distances and orientations between helix-2 and helix-3 in VpuCyto. Distance restraints were calculated based on the measured peak intensity ratios (Ipara/Idia) as described by Wagner (44) and Liang (63). The global correlation time of each residue (5 ns – 10 ns) was calculated based on T1 and T2 of the wild type VpuCyto in micelles (64,65). The calculated PRE distances were incorporated into the structure calculations with a lower bound of 4 Å and upper bound of 4 Å as compensation for inaccuracy in correlation time and other measurements.
3.9. Structure calculation of VpuCyto in DHPC micelles
During the structure calculation of VpuCyto in DHPC micelles, torsion angle restraints were obtained from TALOS+ with chemical shift input of HN, N, CA, CB, C' and HA resonances. Angular restraints were obtained from one RDC dataset with Da, Rh calculated by Xplor-NIH. Distance restraints were provided by 15N NOESY-HSQC NMR spectra, mainly for short range, and by PRE, mainly for long range. Although torsion angles, RDCs, and NOEs provide sufficient information to anneal the VpuCyto to two helices, they are not sufficient to obtain a converged set of structures. The inclusion of long-range distance restraints from PRE data was essential to produce three-dimensional structures. Ensembles of the 20 lowest energy structures calculated with only dihedrals and NOEs (Figure S3A), dihedral, NOEs and RDCs (Figure S3B), and dihedral, NOEs, RDCs and PREs (Figure S3C) with decreasing ensemble RMSDs, are observed. VpuCyto consists of two helices exhibiting a U-shape structure in DHPC micelles that are relatively well defined with an RMSD of 2.95 Å over all backbone atoms for a family of 20 VpuCyto structures.
3. 10. VpuCyto and DHPC micelles association
The depth of micelle insertion of VpuCyto backbone amide sites is reflected in the distance-dependent broadening effects of MnEDTA and 5SDA (Figure 8G, 8H). When MnEDTA was added to VpuCyto incorporated in DHPC micelles, signals of residues that are accessible to MnEDTA in an aqueous environment are broadened, and the observed intensities are reduced; residues associated with the hydrophobic region of the micelles are less affected. Signals from residues that are broadened by MnEDTA are colored red (Figure S4 A). Addition of MnEDTA results in complete disappearance of signals from both N- and C-termini due to their exposure in the aqueous solvent and MnEDTA. In addition, signals from residues 34–40 in helix-2 completely disappear, suggesting that the N terminal half of helix-1 is located outside the micelles. Notably, in the completely exposed C-terminal region, resonance signals of W76 amide, its indole nitrogen and H73 amide are not completely eliminated by MnEDTA, suggesting partial insertion in the micelles. The aromatic side chains of tryptophan and histidine are frequently found inserted in membranes as a result of the dipole moment of their aromatic ring interacting with the phosphatidylcholine head group. Moreover, the loop region, which is believed to be highly mobile, is less broadened by MnEDTA than the terminal region. This suggests that the loop is more restricted in mobility as a result of partial micelle association, compared to the solvent exposed terminal region, and this finding agrees with what we observe in the structure calculations. The line broadening effects were not observed equally for both helices, and those in helix-2 are less affected, indicating that it is more structured and embedded in the center of the micelles. 5SDA is a fatty acid with an 18-carbon hydrophobic chain that has a nitroxide group labeled at the fifth carbon position, close to the head group. When incorporated into the DHPC micelles, the 5SDA partitions to the hydrophobic interior of the micelles and the spin label is near the center of the micelles. Thus, it can be used to probe the portion of protein embedded in the micelles. Signals of residues that are broadened by 5SDA are colored yellow, and those less affected are colored blue (Figure S4 B). Opposite effects were observed compared to MnEDTA. The residues at helix-3 were slightly more affected than those of helix-2, and the C-terminal region seemed to be more affected than the N-terminal region, and W76 and especially its indole signals are the most broadened among all the residues. This is evidence that W76 is embedded in the micelles. Overall, line broadening was invariant for VpuCyto in DHPC micelles with 5SDA compared to MnEDTA. The possible explanations are: (1) both DHPC and 5SDA are dynamic and undergoing rapid rearrangement, and it is possible that at the nitroxide group is not always inserted the center of the micelles but sometimes located near the head group region; (2) due to the amphipathic nature of VpuCyto, it is highly mobile and is not firmly embedded in the micelle, but more likely loosely associated with the micelles.
4. Discussion
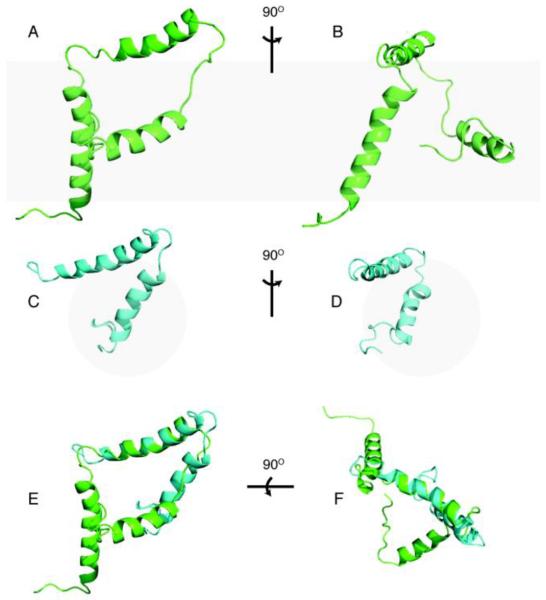
The structure ensemble of VpuCyto in DHPC micelles, disregarding the disordered terminal regions, has a well-defined helix-2-loop-helix-3 motif with 1.24 Å RMSD. The modest variations in the helices suggest that they are relatively rigid. The inter-helical loop itself appears to have RMSD of 0.7 Å. Along with a number of NOEs observed in residue 50–59 in the loop region, surprisingly it suggests that, the loop adopts a well defined structure. All the charged residues occur on either the end of the helices and the terminal region or were restricted to the outer surface of the U-shape structure (Figure S4), presumably exposed to the aqueous media. The U-shaped set of two helices also renders the more hydrophobic phase in the core of the structure. Both VpuFull (Protein data bank ID: 2N28) and VpuCyto (Protein data bank ID: 2N29) constructs have well-defined U-shaped helix-2 and helix-3 (Figure 9). The lengths of both helices are slightly longer in VpuCyto in DHPC micelles than those in VpuFull in DMPC liposomes; however, the helical lengths in both constructs are longer than those in Vpu 39–81 obtained from DPC micelles, presumably due to their truncation in the middle of helix-2 (34). The tilt angles of the average structure of ten lowest energy structures of VpuFull are approximately 25°, 89°, and 79° from the bilayer normal for helix-1, -2, and -3, respectively, and this suggests that the helices in the Cyto domain are located on the bilayer plane, approximately parallel to the bilayer surface. The 25° tilt angle of the TM domain is quite similar to the tilt angle of the VpuTM construct obtained in DMPC liposomes, which is 28° (66), and in DMPC bicelles, which is 30° (28). The slight difference may arise from the missing Cyto domain in the VpuTM construct. The angles between helix-2 and helix-3 of VpuFull and VpuCyto are approximately 42° and 36°, which shows that the orientations of Cyto domain are similar, but the spacing between helices is larger for VpuFull, and it results from VpuFull having a longer loop-2. As a result, helix-3 in VpuFull was observed to have a high degree of flexibility, which may be attributed to fewer structural restraints imposed in the calculation or its intrinsic dynamic properties. Loop-2 of VpuCyto in micelles is shorter and partially structured, and this may be caused by the interactions with the short-chain lipids. Both the effects on loop-2 and the helical lengths suggest that micelles alter the structures slightly from the bilayer environment. Thus, caution is necessary in the interpretation of structures of membrane proteins (or domains) determined in micelles because of the potential for structural distortion compared to the structures determined in phospholipid bilayer environments.
Figure 9. Structure comparison of VpuCyto and VpuFull.
A. Cartoon representation of VpuFull in DMPC proteoliposomes. B.90° rotation of A about the x axis. C. Cartoon representation of VpuCyto in DHPC micelles. D.90° rotation of C about the x axis. E. Alignment of VpuFull and VpuCyto structures (side view with respect to bilayer normal). F. Alignment of VpuFull and VpuCyto structures (top view with respect to bilayer normal). The structure of full length Vpu has PDB ID 2N28, and that of the cytoplasmic domain of Vpu has PDB ID 2N29.
It is challenging for solid-state NMR to quantify the dynamics of proteins, yet we still can investigate the dynamic properties with J-coupling based experiments, such as TOBSY. The signals from TOBSY require the motions of the residues to be faster than the μs range in order to average out the dipolar couplings, and this dynamics range is close to the lower-limit of measurements of T1, T2, and heteronuclear NOE in solution NMR. Based on the assignments from TOBSY spectrum, it suggests that both loops and C-terminus are mobile, and the results are similar with the dynamics studies in solution NMR.
The loop-2 region is the most highly conserved in Vpu, which contains S52 and S56 as phosphorylation sites that are crucial to recruit ubiquitin ligase complex to lead CD4 and BST-2 to degradation. NMR studies performed on Vpu 39–81 in DPC micelles (67) and Vpu 27–57 in mechanically aligned POPC bilayers (68) suggest that phosphorylation causes no drastic structural change in Vpu except an elongation of helix-3, reduction in helicity at helix-2 and reduction in mobility of the loop-2 region in micelles. While both NMR and MD simulation performed by Girault et al. (69–71) show that loop-2 becomes structured and helix-3 becomes unstructured upon phosphorylation. The lowest energy structure of VpuFull shows that S56 faces toward to the aqueous solution and S52 faces down to the lipid surface, and loop-2 roughly lies on the membrane, which might suggest that S56 is easier to be phosphorylated by CK2. The different orientations of the two serines were derived from the substantial difference in PRE caused by MnEDTA, suggesting that two serines have distinct orientations and the loop-2 adopts a defined structure. This agrees with the VpuCyto structure determined in DHPC micelles. The partially structured loop-2 may result from the association with lipid, which has also been shown by Sun with molecular dynamics (41). Even though the conformation of loop-2 might be hindered for the phosphorylation by CK2, it could be compromised by the dynamics of loop-2, which provides more opportunities for exposure to the aqueous solution. Once S52 and S56 are phosphorylated, the recruiting of β-TrCP is sterically more viable because it involves loop-2 and the loops in WD domain from β-TrCP, and both segments are relatively flexible.
Biological studies have revealed several interaction partners of Vpu including CD4, BST-2, NTB-A and CCR7, etc., which are mainly membrane proteins. To down regulate BST-2, NTB-A, and CCR7, Vpu directly interacts with them through the TM domain and employs the A10, A14, A18 and W22 face in the interaction. In the lowest energy structure obtained for VpuFull in DMPC proteoliposomes, it is shown that A10, A14, A18 and W22 are aligned, and face the opposite direction of the Cyto domain. This open space appears to favor the approach of its interaction partners.
In the case of interaction with CD4, helix-2 and 3 and the C-terminus of Vpu were shown to be involved as well as the membrane proximal region in CD4. Based on the CD4 structure obtained in DPC micelles, the Vpu binding residues 402–420 are lying on the surface of micelles (72). The structures of both constructs also show that the cytoplasmic domain is closely associated with the membrane surface, which provides feasibility of interaction with CD4 (72–74)
When Vpu antagonizes BST-2, the W76 at the C terminal region is found critical for counteraction of the restricted virion release, and the phenotype is attributed to the ability of W76 to displace BST-2 from the site on the plasma membrane where virion assembly is taking place. NMR experiments performed on VpuCyto in both DHPC and aqueous media suggested that this might due to anchoring property of W76 to the plasma membrane. Here, the VpuFull structure in DMPC proteoliposomes shows that helix-3 is closer to the membrane surface than helix-2, and in most structures in the ensemble, W76 is facing the head group, which supports the previous membrane anchoring model. In addition, no signals were found in TOBSY spectra from W76, although it is in the very mobile C-terminal region. This may indicate that W76 is immobilized due to interaction with the head group. In addition, the observation of P75 in 13C/13C DARR spectra indicates that P75, which is next to W76, is also immobilized in proteoliposomes.
Supplementary Material
Highlights.
Structure of Vpu from HIV-1 has been solved.
Structure of Cytoplasmic domain of Vpu has been solved.
Cytoplasmic domain of Vpu forms a U-shape and lies on the surface of membrane.
Acknowledgements
This research was supported by NIH grants P41EB002031, R01GM099986, R01GM066978, and R01AI065361. It utilized the Biotechnology Resource Center for NMR Molecular Imaging of Proteins. We are grateful to Yan Wang for performing the assignment of VpuCyto in DHPC micelles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dube M, Bego MG, Paquay C, Cohen EA. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology. 2010;7:114. doi: 10.1186/1742-4690-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy N, Pacini G, Berlioz-Torrent C, Janvier K. Mechanisms underlying HIV-1 Vpu-mediated viral egress. Front Microbiol. 2014;5:177. doi: 10.3389/fmicb.2014.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neil SJD, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 5.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, Barker E. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe. 2010;8:397–409. doi: 10.1016/j.chom.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sowrirajan B, Barker E. The natural killer cell cytotoxic function is modulated by HIV-1 accessory proteins. Viruses. 2011;3:1091–1111. doi: 10.3390/v3071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramirez PW, Famiglietti M, Sowrirajan B, DePaula-Silva AB, Rodesch C, Barker E, Bosque A, Planelles V. Downmodulation of CCR7 by HIV-1 Vpu results in impaired migration and chemotactic signaling within CD4(+) T cells. Cell Rep. 2014;7:2019–2030. doi: 10.1016/j.celrep.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skasko M, Wang Y, Tian Y, Tokarev A, Munguia J, Ruiz A, Stephens EB, Opella SJ, Guatelli J. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem. 2012;287:58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiganos E, Yao XJ, Friborg J, Daniel N, Cohen EA. Putative alpha-helical structures in the human immunodeficiency virus type 1 Vpu protein and CD4 are involved in binding and degradation of the CD4 molecule. J Virol. 1997;71:4452–4460. doi: 10.1128/jvi.71.6.4452-4460.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerkau T, Bacik I, Bennink JR, Yewdell JW, Hunig T, Schimpl A, Schubert U. The human immunodeficiency virus type 1 (HIV-1) Vpu protein interferes with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules. J Exp Med. 1997;185:1295–1305. doi: 10.1084/jem.185.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 14.Miyagi E, Andrew AJ, Kao S, Strebel K. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc Natl Acad Sci U S A. 2009;106:2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goffinet C, Homann S, Ambiel I, Tibroni N, Rupp D, Keppler OT, Fackler OT. Antagonism of CD317 restriction of human immunodeficiency virus type 1 (HIV-1) particle release and depletion of CD317 are separable activities of HIV-1 Vpu. J Virol. 2010;84:4089–4094. doi: 10.1128/JVI.01549-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tervo HM, Homann S, Ambiel I, Fritz JV, Fackler OT, Keppler OT. beta-TrCP is dispensable for Vpu's ability to overcome the CD317/Tetherin-imposed restriction to HIV-1 release. Retrovirology. 2011;8:9. doi: 10.1186/1742-4690-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokarev AA, Munguia J, Guatelli JC. Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J Virol. 2011;85:51–63. doi: 10.1128/JVI.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNatt MW, Zang T, Bieniasz PD. Vpu binds directly to tetherin and displaces it from nascent virions. PLoS Pathog. 2013;9:e1003299. doi: 10.1371/journal.ppat.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jafari M, Guatelli J, Lewinski MK. Activities of transmitted/founder and chronic clade B HIV-1 Vpu and a C-terminal polymorphism specifically affecting virion release. J Virol. 2014;88:5062–5078. doi: 10.1128/JVI.03472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewinski MK, Jafari M, Zhang H, Opella SJ, Guatelli J. Membrane Anchoring by a C-terminal Tryptophan Enables HIV-1 Vpu to Displace Bone Marrow Stromal Antigen 2 (BST2) from Sites of Viral Assembly. J Biol Chem. 2015;290:10919–10933. doi: 10.1074/jbc.M114.630095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell RF, Meirovitch E, Freed JH. Slow-motional NMR line shapes for very anisotropic rotational diffusion. Phosphorus-31 NMR of phospholipids. Journal of Physical Chemistry. 1979;83:525–533. [Google Scholar]
- 22.Lewis BA, Harbison GS, Herzfeld J, Griffin RG. NMR structural analysis of a membrane protein: bacteriorhodopsin peptide backbone orientation and motion. Biochemistry. 1985;24:4671–4679. doi: 10.1021/bi00338a029. [DOI] [PubMed] [Google Scholar]
- 23.Hong M, Doherty T. Orientation Determination of Membrane-Disruptive Proteins Using Powder Samples and Rotational Diffusion: A Simple Solid-State NMR Approach. Chem Phys Lett. 2006;432:296–300. doi: 10.1016/j.cplett.2006.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marassi FM, Ma C, Gratkowski H, Straus SK, Strebel K, Oblatt-Montal M, Montal M, Opella SJ. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc Natl Acad Sci U S A. 1999;96:14336–14341. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kochendoerfer GG, Jones DH, Lee S, Oblatt-Montal M, Opella SJ, Montal M. Functional characterization and NMR spectroscopy on full-length Vpu from HIV-1 prepared by total chemical synthesis. J Am Chem Soc. 2004;126:2439–2446. doi: 10.1021/ja038985i. [DOI] [PubMed] [Google Scholar]
- 26.Do HQ, Wittlich M, Gluck JM, Mockel L, Willbold D, Koenig BW, Heise H. Full-length Vpu and human CD4(372–433) in phospholipid bilayers as seen by magic angle spinning NMR. Biol Chem. 2013;394:1453–1463. doi: 10.1515/hsz-2013-0194. [DOI] [PubMed] [Google Scholar]
- 27.Wray V, Kinder R, Federau T, Henklein P, Bechinger B, Schubert U. Solution structure and orientation of the transmembrane anchor domain of the HIV-1-encoded virus protein U by high-resolution and solid-state NMR spectroscopy. Biochemistry. 1999;38:5272–5282. doi: 10.1021/bi982755c. [DOI] [PubMed] [Google Scholar]
- 28.Park SH, De Angelis AA, Nevzorov AA, Wu CH, Opella SJ. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys J. 2006;91:3032–3042. doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, Montal M, Opella SJ. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J Mol Biol. 2003;333:409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- 30.Sharpe S, Yau WM, Tycko R. Structure and dynamics of the HIV-1 Vpu transmembrane domain revealed by solid-state NMR with magic-angle spinning. Biochemistry. 2006;45:918–933. doi: 10.1021/bi051766k. [DOI] [PubMed] [Google Scholar]
- 31.Lu JX, Sharpe S, Ghirlando R, Yau WM, Tycko R. Oligomerization state and supramolecular structure of the HIV-1 Vpu protein transmembrane segment in phospholipid bilayers. Protein Sci. 2010;19:1877–1896. doi: 10.1002/pro.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Federau T, Schubert U, Flossdorf J, Henklein P, Schomburg D, Wray V. Solution structure of the cytoplasmic domain of the human immunodeficiency virus type 1 encoded virus protein U (Vpu) Int J Pept Protein Res. 1996;47:297–310. doi: 10.1111/j.1399-3011.1996.tb01359.x. [DOI] [PubMed] [Google Scholar]
- 33.Willbold D, Hoffmann S, Rosch P. Secondary structure and tertiary fold of the human immunodeficiency virus protein U (Vpu) cytoplasmic domain in solution. Eur J Biochem. 1997;245:581–588. doi: 10.1111/j.1432-1033.1997.t01-1-00581.x. [DOI] [PubMed] [Google Scholar]
- 34.Wittlich M, Koenig BW, Stoldt M, Schmidt H, Willbold D. NMR structural characterization of HIV-1 virus protein U cytoplasmic domain in the presence of dodecylphosphatidylcholine micelles. FEBS J. 2009;276:6560–6575. doi: 10.1111/j.1742-4658.2009.07363.x. [DOI] [PubMed] [Google Scholar]
- 35.Marassi FM, Ma C, Gesell JJ, Opella SJ. Three-dimensional solid-state NMR spectroscopy is essential for resolution of resonances from in-plane residues in uniformly (15)N-labeled helical membrane proteins in oriented lipid bilayers. J Magn Reson. 2000;144:156–161. doi: 10.1006/jmre.2000.2036. [DOI] [PubMed] [Google Scholar]
- 36.Jo S, Im W. Transmembrane helix orientation and dynamics: insights from ensemble dynamics with solid-state NMR observables. Biophys J. 2011;100:2913–2921. doi: 10.1016/j.bpj.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Im W, Jo S, Kim T. An ensemble dynamics approach to decipher solid-state NMR observables of membrane proteins. Biochim Biophys Acta. 2012;1818:252–262. doi: 10.1016/j.bbamem.2011.07.048. [DOI] [PubMed] [Google Scholar]
- 38.Bond PJ, Holyoake J, Ivetac A, Khalid S, Sansom MS. Coarse-grained molecular dynamics simulations of membrane proteins and peptides. J Struct Biol. 2007;157:593–605. doi: 10.1016/j.jsb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Fischer WB, Sramala I, Cordes F, Angsuthanasombat C, Sansom MSP, Watts A. Molecular dynamics simulations on up to the first 52 amino acids from HIV-1's Vpu. BIOPHYSICAL JOURNAL, Biophysical Society. 2002 [Google Scholar]
- 40.Sramala I, Lemaitre V, Faraldo-Gomez JD, Vincent S, Watts A, Fischer WB. Molecular dynamics simulations on the first two helices of Vpu from HIV-1. Biophys J. 2003;84:3276–3284. doi: 10.1016/S0006-3495(03)70052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun F. Molecular dynamics simulation of human immunodeficiency virus protein U (Vpu) in lipid/water Langmuir monolayer. J Mol Model. 2003;9:114–123. doi: 10.1007/s00894-003-0123-3. [DOI] [PubMed] [Google Scholar]
- 42.Ma C, Marassi FM, Jones DH, Straus SK, Bour S, Strebel K, Schubert U, Oblatt-Montal M, Montal M, Opella SJ. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 2002;11:546–557. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DH, Opella SJ. Weak alignment of membrane proteins in stressed polyacrylamide gels. J Magn Reson. 2004;171:258–269. doi: 10.1016/j.jmr.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 44.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 45.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 47.Bloembergen N. On the interaction of nuclear spins in a crystalline lattice. Physica. 1949;15:386–426. [Google Scholar]
- 48.Takegoshi K, Nakamura S, Terao T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical Physics Letters. 2001;344:631–637. [Google Scholar]
- 49.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-driven 13C-13C recoupling without 13C rf irradiation in nuclear magnetic resonance of rotating solids. Journal of Chemical Physics. 2003;118:2325–2341. [Google Scholar]
- 50.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Molecular Physics. 1998;95:1197–1207. [Google Scholar]
- 51.Zhao X, Edén M, Levitt MH. Recoupling of heteronuclear dipolar interactions in solid-state NMR using symmetry-based pulse sequences. Chemical physics letters. 2001;342:353–361. [Google Scholar]
- 52.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Gong XM, Ding Y, Yu J, Yao Y, Marassi FM. Structure of the Na,K-ATPase regulatory protein FXYD2b in micelles: implications for membrane-water interfacial arginines. Biochim Biophys Acta. 2015;1848:299–306. doi: 10.1016/j.bbamem.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 55.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor–NIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 56.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartzell CJ, Whitfield M, Oas T, Drobny G. Determination of the nitrogen-15 and carbon-13 chemical shift tensors of L-[13C]alanyl-L-[15N]alanine from the dipole-coupled powder patterns. Journal of the American Chemical Society. 1987;109:5966–5969. [Google Scholar]
- 58.Lesage A, Bardet M, Emsley L. Through-bond carbon-carbon connectivities in disordered solids by NMR. Journal of the American Chemical Society. 1999;121:10987–10993. [Google Scholar]
- 59.Leppert J, Ohlenschlager O, Gorlach M, Ramachandran R. Adiabatic TOBSY in rotating solids. J Biomol NMR. 2004;29:167–173. doi: 10.1023/B:JNMR.0000019248.48726.ff. [DOI] [PubMed] [Google Scholar]
- 60.Prestegard JH, al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 61.Zweckstetter M, Bax A. Prediction of Sterically Induced Alignment in a Dilute Liquid Crystalline Phase: Aid to Protein Structure Determination by NMR. Journal of the American Chemical Society. 2000;122:3791–3792. [Google Scholar]
- 62.Mesleh MF, Opella SJ. Dipolar Waves as NMR maps of helices in proteins. J Magn Reson. 2003;163:288–299. doi: 10.1016/s1090-7807(03)00119-8. [DOI] [PubMed] [Google Scholar]
- 63.Liang B, Bushweller JH, Tamm LK. Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR spectroscopy. J Am Chem Soc. 2006;128:4389–4397. doi: 10.1021/ja0574825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 65.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 66.Das BB, Zhang H, Opella SJ. Dipolar Assisted Assignment Protocol (DAAP) for MAS solid-state NMR of rotationally aligned membrane proteins in phospholipid bilayers. J Magn Reson. 2014;242:224–232. doi: 10.1016/j.jmr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wittlich M, Koenig BW, Willbold D. Structural consequences of phosphorylation of two serine residues in the cytoplasmic domain of HIV-1 VpU. J Pept Sci. 2008;14:804–810. doi: 10.1002/psc.1004. [DOI] [PubMed] [Google Scholar]
- 68.Henklein P, Kinder R, Schubert U, Bechinger B. Membrane interactions and alignment of structures within the HIV-1 Vpu cytoplasmic domain: effect of phosphorylation of serines 52 and 56. FEBS Lett. 2000;482:220–224. doi: 10.1016/s0014-5793(00)02060-3. [DOI] [PubMed] [Google Scholar]
- 69.Coadou G, Evrard-Todeschi N, Gharbi-Benarous J, Benarous R, Girault J-P. HIV-1 encoded virus protein U (Vpu) solution structure of the 41–62 hydrophilic region containing the phosphorylated sites Ser 52 and Ser 56. International Journal of Biological Macromolecules. 2002;30:23–40. doi: 10.1016/s0141-8130(01)00184-2. [DOI] [PubMed] [Google Scholar]
- 70.Coadou G, Gharbi-Benarous J, Megy S, Bertho G, Evrard-Todeschi N, Segeral E, Benarous R, Girault JP. NMR studies of the phosphorylation motif of the HIV-1 protein Vpu bound to the F-box protein beta-TrCP. Biochemistry. 2003;42:14741–14751. doi: 10.1021/bi035207u. [DOI] [PubMed] [Google Scholar]
- 71.Evrard-Todeschi N, Gharbi-Benarous J, Bertho G, Coadou G, Megy S, Benarous R, Girault JP. NMR studies for identifying phosphopeptide ligands of the HIV-1 protein Vpu binding to the F-box protein beta-TrCP. Peptides. 2006;27:194–210. doi: 10.1016/j.peptides.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 72.Singh SK, Mockel L, Thiagarajan-Rosenkranz P, Wittlich M, Willbold D, Koenig BW. Mapping the interaction between the cytoplasmic domains of HIV-1 viral protein U and human CD4 with NMR spectroscopy. FEBS J. 2012;279:3705–3714. doi: 10.1111/j.1742-4658.2012.08732.x. [DOI] [PubMed] [Google Scholar]
- 73.Hill MS, Ruiz A, Schmitt K, Stephens EB. Identification of amino acids within the second alpha helical domain of the human immunodeficiency virus type 1 Vpu that are critical for preventing CD4 cell surface expression. Virology. 2010;397:104–112. doi: 10.1016/j.virol.2009.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruiz A, Guatelli JC, Stephens EB. The Vpu protein: new concepts in virus release and CD4 down-modulation. Curr HIV Res. 2010;8:240–252. doi: 10.2174/157016210791111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.