Abstract
Endo-β-N-acetylglucosaminidase isolated from B. infantis ATCC 15697 (EndoBI-1) is a novel enzyme that cleaves N-N′-diacetyl chitobiose moieties found in the N-glycan core of high mannose, hybrid, and complex N-glycans. These conjugated N-glycans are recently shown as a new prebiotic source that stimulates the growth of a key infant gut microbe, Bifi-dobacterium longum subsp. Infantis. The effects of pH (4.45–8.45), temperature (27.5–77.5°C), reaction time (15–475 min), and enzyme/protein ratio (1:3,000–1:333) were evaluated on the release of N-glycans from bovine colostrum whey by EndoBI-1. A central composite design was used, including a two-level factorial design (24) with four center points and eight axial points. In general, low pH values, longer reaction times, higher enzyme/protein ratio, and temperatures around 52°C resulted in the highest yield. The results demonstrated that bovine colostrum whey, considered to be a by/waste product, can be used as a glycan source with a yield of 20 mg N-glycan/g total protein under optimal conditions for the ranges investigated. Importantly, these processing conditions are suitable to be incorporated into routine dairy processing activities, opening the door for an entirely new class of products (released bioactive glycans and glycan-free milk). The new enzyme’s activity was also compared with a commercially available enzyme, showing that EndoBI-1 is more active on native proteins than PNGase F and can be efficiently used during pasteurization, streamlining its integration into existing processing strategies.
Keywords: N-glycans, endo-β-N-acetylglucosaminidase, whey
Introduction
Milk contains a variety of components, including proteins, peptides, lipids, carbohydrates, and minerals that contribute to the growth and development of newborns. Whey proteins represent 18–20% of total milk protein and have high nutritive and functional properties for the young.1 During biosynthesis in the mammary epithelial cell, some milk proteins are decorated with long carbohydrate chains (glycoproteins). Glycoproteins contain a variety of oligosaccharides (glycans) covalently attached to the polypeptide side chains.2 The most abundant bovine milk glycoproteins are lactoferrin, immunoglobulins, and α-lactalbumin.3 These proteins have interesting biological activities including antioxidant, antibacterial, and antiviral action, as well as immunomodulatory activity.4-7 These functions are supposedly modulated by both the polypeptide chain and the glycans. However, the function of glycoproteins’ N-glycans is not well elucidated. They may have similar functions to those described for free milk oligosaccharides from human milk, including inhibition of pathogen adhesion to intestinal epithelial cells and promoting the growth of beneficial commensal microbes in the infant intestine.8-11 In fact, we have recently shown that a key infant gut microbe, Bifidobacterium longum subsp. infantis, is able to consume these conjugated N-glycans as a sole carbon source while other less important microbes do not.
Glycans are linked to the protein through O-glycosidic or N-glycosidic bonds. O-linked glycans (O-glycans) are frequently attached to the polypeptide via N-acetylgalactosamine to a hydroxyl group of a serine or threonine residue. N-linked glycans (N-glycans) are linked, via N-acetylglucosamines (HexNAc), to an asparagine residue of proteins in the specific amino acid sequence AsN-X-Ser/Thr (where X can be any amino acid except proline). The N-glycan core is composed of two HexNAc and three mannose residues. This core is elongated by other monosaccharides, including fucose and sialic acid, via the actions of glycosyltransferases and glycosidases, which determine the degree of branching and the type of linkage.12 N-glycans are divided into three main classes: high mannose, complex, and hybrid.
Whey, a by-product of cheese production from bovine milk, is an attractive source of milk compounds with interesting biological properties. It contains 93–94% water, 4.5–5% lactose, 0.8–1% total protein, 0.5–0.7% minerals, and others.13 Whey accounts for 90% of the initial milk volume processed, which means that approximately 9 L of whey are produced from 10 L of bovine milk during cheese-making. Whey worldwide production is estimated to be 190,000,000 ton/year.14,15 The underutilization of the organic components in whey means that only 50% of the whey produced is used in food applications or for animal feed. The unused waste stream causes serious economic and environmental problems, as cheese processors have to dispose this highly organic into wastewater stream. In addition to containing proteins with high functionality, whey is also a potential source of N-glycans, with concentrations of ~4.5 g glycoproteins/L.16 Therefore, the recovery of whey bioactive compounds is an opportunity for the development of new high-value products for the dairy industry while reducing the industry’s environmental pollution.
Hydrazinolysis and alkali/reducing conditions (β-elimination) are commonly used techniques for deglycosylation of glycoproteins.17,18 However, the chemicals used for glycan release may result in the degradation of glycoproteins and are potentially hazardous.19-21 There are also commercially available enzymes (endoglycosidases, glycosamidases) that can be used for N-glycan release.19 However, commercial enzymes are affected by fucosylation of the N-glycan core resulting in limited glycan release. For example, peptidyl-N-glycosidase F (PNGase F), one of the most commonly used enzymes,22 catalyzes the cleavage between HexNAc and asparagine residues, but it is not able to cleave N-glycans from glycoproteins when an α-1-3 fucose residue is linked to the core HexNAc.23 Additionally, the release of N-glycans at large scale has been impaired by the price of commercial enzymes. We previously showed that a novel Endo-β-N-acetylglucosaminidase isolated from B. infantis ATCC 15697 cleaves the N-N′-diacetyl chitobiose moiety found in the N-glycan core of high mannose, hybrid, and complex N-glycans. Its activity is not affected by a fucosylated N-glycan core, and it was also shown to be heat resistant which enables its use during heat treatments at 95°C,24,25 a common practice in milk processing.
Improving the release of N-glycans is an essential step to enable the large-scale production of N-glycans. This will also enable a better understanding of their biological properties and source of new bioactive ingredients. In this work, the effects of pH, temperature, reaction time, and enzyme/protein ratio on the total release of N-glycans by EndoBI-1 were evaluated. In order to identify the optimal combination of pH, reaction time, temperature, and enzyme/protein ratio within the most likely processing range, a central composite rotatable design was employed including a four-factor, two-level full factorial design (24) along with four center points and eight axial points. Because it was previously shown that EndoBI-1 is a heat stable enzyme,26 we investigated how efficiently it can be applied within pasteurization (62°C, 30 min) so that no additional heating step would be required for deglycosylation. We also demonstrated how denaturation of substrates affected N-glycan release by EndoBI-1 and commercial enzyme PNGase F.
Materials and Methods
Bacteria, media, and substrates
The B. longum subsp. Infantis ATCC 15697 strain used in this study was obtained from the University of California Davis Viticulture and Enology Culture Collection (Davis, CA). Bifidobacterium was grown in Man–Rogose–Sharp (MRS) broth supplemented with 0.05% (w/v) l-cysteine (Sigma-Aldrich). The cells were anaerobically grown in a vinyl chamber (Coy Laboratory Products, Grass Lake, MI) at 37°C for 24 h, in an atmosphere consisting of 5% carbon dioxide, 5% hydrogen, and 90% nitrogen. The E. coli Bl21* strain used for protein expression was grown in Luria Broth (LB) media containing Carbenicillin (100 μg/mL). All incubations were carried out in an Innova 4000 shaker (New Brunswick Scientific, New Jersey) at 200 rpm and 37°C. Bovine lactoferrin and RNase B were obtained from Sigma-Aldrich (St. Louis, MO, USA). Gene cloning, expression, and purification and pilot-scale production of protein concentrate from bovine colostrum whey were performed as described before.27
N-Glycan release of RNase B
Twenty micrograms of Ribonuclease B (RNase B) (Sigma-Aldrich, San Diego, CA, USA) was reconstituted in 20 μL of 0.2 M Na2HPO4 (pH 5) and 200 μg of EndoBI-1 were added to the sample. PNGase F (New England BioLabs, Ipswich, MA, USA) was used as a positive control. Two microliters of 500,000 units/mL PNGase F were added to 20 μg of RNase B reconstituted in 20 μL of commercial denaturing buffer (New England BioLabs). The two mixtures were incubated 2 h at 37°C. The enzyme activity was analyzed by SDS-PAGE on 4–15% precast polyacrylamide gels.
Experimental design and statistical analysis
In order to find the optimal combination of pH, reaction time, temperature, and enzyme/protein ratio, a complete 24 factorial design of the central rotational type was established, with four central points and eight axial points, based on Response Surface Methodology.27 The effects of pH (4.45–8.45), temperature (27.5–77.5°C), reaction time (15–475 min), and enzyme/protein ratio (1:3,000–1:333) on the release of N-glycans from bovine colostrum whey by EndoBI-1 were tested. The independent variables (pH, reaction time, temperature, and enzyme/protein ratio) were evaluated according to coded levels (−α, −1, 0, +1, +α). Center points are located at the average of levels −1 and +1 for each factor, while axial points were determined by interpolation (α ±2.0) for each factor. Coded and uncoded levels and their corresponding independent variables are shown in Table 1. The dependent variable (i.e., evaluated response) was the release of N-glycans from bovine colostrum whey by EndoBI-1. Data were analyzed by Statistica version 8.0 software. The significance of the model was tested by Analysis of Variance (ANOVA).
Table 1.
Experimental Design for Optimizing the Release of N-Glycans from Bovine Colostrum Whey by EndoBI-1
| Treatments | Coded Levels
|
Uncoded Levels
|
Glycans Release (μg) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| pH (X1) |
Time (min) (X2) |
Temperature (°C) (X3) |
Enzyme/Protein Ratio (X4) |
pH (X1) |
Time (min) (X2) |
Temperature (°C) (X3) |
Enzyme/Protein Ratio (X4) |
||
| 1 | +1 | +1 | +1 | +1 | 7.00 | 360 | 65.0 | 1:500 | 9.35 |
| 2 | +1 | +1 | −1 | −1 | 7.00 | 360 | 40.0 | 1:1,500 | 3.85 |
| 3 | +1 | +1 | −1 | +1 | 7.00 | 360 | 40.0 | 1:500 | 9.85 |
| 4 | +1 | −1 | −1 | −1 | 7.00 | 130 | 40.0 | 1:1,500 | 3.97 |
| 5 | −1 | −1 | −1 | −1 | 5.30 | 130 | 40.0 | 1:1,500 | 12.87 |
| 6 | −1 | −1 | −1 | +1 | 5.30 | 130 | 40.0 | 1:500 | 23.33 |
| 7 | −1 | −1 | +1 | +1 | 5.30 | 130 | 65.0 | 1:500 | 17.62 |
| 8 | −1 | +1 | +1 | +1 | 5.30 | 360 | 65.0 | 1:500 | 25.84 |
| 9 | +1 | −1 | +1 | −1 | 7.00 | 130 | 65.0 | 1:1,500 | 3.15 |
| 10 | −1 | +1 | −1 | +1 | 5.30 | 360 | 40.0 | 1:500 | 54.75 |
| 11 | +1 | −1 | +1 | +1 | 7.00 | 130 | 65.0 | 1:500 | 3.98 |
| 12 | +1 | +1 | +1 | −1 | 7.00 | 360 | 65.0 | 1:1,500 | 5.50 |
| 13 | +1 | −1 | −1 | +1 | 7.00 | 130 | 40.0 | 1:500 | 5.60 |
| 14 | −1 | +1 | −1 | −1 | 5.30 | 360 | 40.0 | 1:1,500 | 24.60 |
| 15 | −1 | −1 | +1 | −1 | 5.30 | 130 | 65.0 | 1:1,500 | 8.47 |
| 16 | −1 | +1 | +1 | −1 | 5.30 | 360 | 65.0 | 1:1,500 | 13.32 |
| 17 | 0 | 0 | 0 | 0 | 6.15 | 245 | 52.5 | 1:1,000 | 27.47 |
| 18 | 0 | 0 | 0 | 0 | 6.15 | 245 | 52.5 | 1:1,000 | 27.29 |
| 19 | 0 | 0 | 0 | 0 | 6.15 | 245 | 52.5 | 1:1,000 | 27.36 |
| 20 | 0 | 0 | 0 | 0 | 6.15 | 245 | 52.5 | 1:1,000 | 27.48 |
| 21 | +2 | 0 | 0 | 0 | 7.85 | 245 | 52.5 | 1:1,000 | 4.78 |
| 22 | −2 | 0 | 0 | 0 | 4.45 | 245 | 52.5 | 1:1,000 | 36.13 |
| 23 | 0 | +2 | 0 | 0 | 6.15 | 475 | 52.5 | 1:1,000 | 38.32 |
| 24 | 0 | −2 | 0 | 0 | 6.15 | 15 | 52.5 | 1:1,000 | 4.03 |
| 25 | 0 | 0 | +2 | 0 | 6.15 | 245 | 77.5 | 1:1,000 | 5.64 |
| 26 | 0 | 0 | −2 | 0 | 6.15 | 245 | 27.5 | 1:1,000 | 8.43 |
| 27 | 0 | 0 | 0 | +2 | 6.15 | 245 | 52.5 | 1:333 | 43.49 |
| 28 | 0 | 0 | 0 | −2 | 6.15 | 245 | 52.5 | 1:3,000 | 6.43 |
Complete 24 factorial design parameters, with four independent variables in two levels, four repetitions in the central point and eight in the axial points.
For each experiment, 450 μL of concentrated whey protein were diluted in 50 μL of 0.2 mM Na2HPO4. Each experiment was performed in duplicate. Five milliliters of cold pure ethanol were added into the samples to stop the reactions, precipitate proteins, and collect the released glycans. Samples were dried overnight by vacuum centrifugation. Samples were rehydrated in 100 μL of water.
N-Glycan release comparison of EndoBI-1 and PNGase F on denatured/native substrates and pasteurization conditions
Five hundred microliters of concentrated bovine colostrum whey, bovine lactoferrin (5 mg/mL), or RNase B (1 mg/mL) were used to examine how the denaturation state of substrates affects the deglycosylation efficiency of glycoproteins by EndoBI-1 and PNGase F. Samples were denatured at 95°C for 5 min in a 10 mM final concentration of DTT. Two hundred and fifty micrograms of EndoBI-1 and PNGase F were added into samples and enzymatic digestions were performed under the conditions described before by Garrido et al.26 and incubated for 8 h.
The N-glycan release comparison of EndoBI-1 and PNGase F on concentrated bovine colostrum whey under pasteurization conditions (62°C, 30 min) was also investigated. Two hundred and fifty micrograms of enzyme was added into 500 μL of whey when the temperature reached 62°C and incubated for 30 min. All the reactions were stopped with 1 M Na2CO3. Total released N-glycans by each enzyme were calculated after glycan purification.
Glycan quantification
A total carbohydrate Assay Kit, (Biovision Milpitas, CA, USA), was used to quantify released N-glycans. To create a standard curve, 0, 2, 4, 6, 8, and 10 μL of mannose standard (2 mg/mL) were added into a series of wells in a 96-well microplate (Supporting information Figure S1). This generated 0, 4, 8, 12, 16, and 20 μg/well of mannose standard. The volume of each sample was adjusted to 30 μL per well with water. Thirty microliters of sample was used. One hundred and fifty microliters of concentrated sulfuric acid (98%) were added to each well. Samples were mixed on a shaker for ~1 min and then incubated at 85°C for 15 min. After incubation, 30 μL of Developer (provided by the manufacturer) was added to each well. Samples were again mixed on the shaker for 5 min. OD of each sample was measured at 490 nm. Sample OD was applied to the mannose standard curve linear function to calculate the quantity of carbohydrate in the sample.
Glycan purification
Samples 9, 10, and 17 were loaded on PGC SPE plates (Glygen, Columbia, MD, USA) that were conditioned using 3 × 100 μL of 80% ACN (acetonitrile) containing 0.1% TFA in water, followed by 3 × 100 μL of water. After sample loading, wells were washed using 6 × 200 μL of water and N-glycans were eluted using 3 × 200 μL of 40% ACN containing 0.1% TFA (trifluoroacetic acid) in water. The enriched N-glycan fractions were dried overnight under vacuum. Samples were rehydrated in 50 μL of water, vortexed, and sonicated prior to mass spectrometry analysis. For each condition tested, the same quantity of total carbohydrate was analyzed by mass spectrometry.
N-Glycan analysis by Nano-LC-Chip-Q-TOF MS
N-glycans were analyzed using an Agilent 6520 accurate-mass Q-TOF LC/MS with a microfluidic nano-electrospray chip (Agilent Technologies, Santa Clara, CA, USA). N-glycans were separated using an HPLC-chip with a 40-nL enrichment column and a 43 mm × 75 μm analytical column, both packed with 5 μm porous graphitized carbon (PGC). The system was composed of a capillary and nano-flow pump, and both used binary solvents consisting of solvent A (3% ACN, 0.1% formic acid in water (v/v)) and solvent B (90% ACN, 0.1% formic acid in water (v/v)). Two microliters of sample were loaded with solvent A at a capillary pump flow rate of 4 μL/min. N-glycan separation was performed on a 65-min gradient delivered by the nanopump at a flow rate of 0.3 μL/min. The 65-min gradient followed this program: 0% B (0.0–2.5 min), 0–16% B (2.5–20.0 min), 16–44% B (20.0–30.0 min), 44–100% B (30.0–35.0 min), and 100% B (35.0–45.0 min). The gradient was followed by equilibration at 0% B (45.0–65.0 min). Data were acquired within the mass range of 450–3,000 m/z for N-glycans in the positive ionization mode with an acquisition rate of 2.01 spectra/s for N-glycans. An internal calibrant ion of 922.010 m/z from the tuning mix (ESI-TOF Tuning Mix G1969–85000, Agilent Technologies) was used for continual mass calibration. For tandem MS analysis, N-glycans were fragmented with nitrogen as the collision gas. Spectra were acquired within the mass range of 100–3,000 m/z. The collision energies correspond to voltages (Vcollision) that were based on the equation: Vcollision = m/z (1.5/100 Da) Volts—3.6 V; where the slope and offset of the voltages were set at (1.5/100 Da) and (−3.6), respectively. Acquisition was controlled by MassHunter Workstation Data Acquisition software (Agilent Technologies, Santa Clara, CA, USA).
Released glycans were identified with MassHunter Qualitative Analysis software (version B.04.00 SP2, Agilent Technologies). Compounds were extracted using the Molecular Feature Extractor algorithm. The software generated extracted compound chromatograms (ECCs) in a range of 400–3,000 m/z with a ≥1000 ion count cut-off, allowing charge states of +1–3, a retention time from 5–40 min, and a typical isotopic distribution of small biological molecules. The resulting compounds were matched to a bovine milk N-glycan library28 using a mass error tolerance of 20 ppm. The N-glycans from the library were composed of hexose (Hex), HexNAc, Fuc, N-acetylneuraminic acid (NeuAc), and N-glycolylneuraminic acid (NeuGc).
Results
Production of EndoBI-1 and activity of the enzyme on RNase B
The production of poly-histidine-tagged EndoBI-1 was achieved with a yield of 1.7 mg/L using LB media under optimum induction conditions. The activity of recombinant EndoBI-1 was tested on RNase B, an N-glycosylated protein of 17 kDa that contains high mannose N-linked glycans,29 and compared to commercial enzyme, PNGase F (Figure 1). Cleavage of RNase B by an endoglycosidase results in a 14 kDa deglycosylated protein. This experiment was performed under nondenaturing conditions. After 2 h of incubation with the enzymes, the band corresponding to glycosylated RNase B disappeared. The protein was detected at lower molecular weight in presence of EndoBI-1 and PNGase F. The bands observed in lane 2 and 3 corresponded to the deglycosylated RNase B. These data indicate that the recombinant EndoBI-1 was as efficient as PNGase F in releasing N-glycans.
Figure 1. Enzymatic deglycosylation of RNase B by Endo-B-N-acetylglucosaminidase and PNGase F on 12% SDS-PAGE gel. Lane 1: glycosylated RNase B (17kDa). Lane 2: deglycosylated RNase B by Endo-β-N-acetyl-glucosaminidase (47kDa). Lane 3: deglycosylated RNase B by PNGase F (36kDa).

Optimization of the release of N-glycans from bovine colostrum whey by EndoBI-1
In Table 1, the effects of pH, incubation time, temperature, and enzyme/protein ratio on the release of N-glycans from bovine colostrum whey protein concentrate are presented. In general, deglycosylation activity was reduced by higher pH values regardless of the other operation parameters evaluated. The lowest values of N-glycan release (3.97–9.35 μg of N-glycans) were observed at pH 7.0 and 7.85, in agreement with the qualitative results presented by Garrido et al. (2012). The highest N-glycan release (54.7 μg) was observed at pH 5.3, the longer incubation time (360 min), a temperature of 40°C, and an enzyme/protein ratio of 1:500. Enzymatic activity reduction was observed when temperature was increased from 40 to 65°C. N-glycan release was reduced from 24.6 to 13.2 μg when temperature increased from 40 to 65°C, respectively (Treatments 14 and 16).
Statistical analysis
The estimated regression model and coefficient of determination (R2) for total release of N-glycans from bovine colostrum whey by EndoBI-1 was determined as described in Table 2. According to the model, higher glycan release will happen at lower pH values (−2 = 4.45); longer reaction time (+2 = 475 min); temperature in the center point (0 = 52.5°C); and higher enzyme/protein ratio (+2 = 1:333). Only parameters significant at p < 0.05 were used in the regression model. The coefficient of determination (R2) of the predictive model for release of N-glycans from bovine colostrum whey by EndoBI-1 was 0.74, indicating that the regression model was able to explain 74% of the total variation between the observed values for release of N-glycans from bovine colostrum whey by EndoBI-1.
Table 2.
Estimated Regression Model and Coefficient of Determination for the Release of N-Glycans from Bovine Colostrum Whey by EndoBI-1
| Estimated Regression Model* R2 | |
| Estimated release of N-glycans from whey by | 0.74 |
X1 = coded level corresponding to pH; X2 = coded level corresponding to time; X3 = coded level corresponding to temperature; X4 = coded level corresponding to enzyme/protein ratio.
Only parameters with a confidence level above 95% (P < 0.05) were considered as significant.
The analysis of variance of the estimated regression model is presented in Table 3. The regression was significant (Fcalculated > Ftable) and thus could be used for predictive goals in the range of parameters evaluated since the ratio Fcalculated/Ftable is greater than 3.27 Although the F-test for the lack of fit was statistically significant (Fcalculated > Ftable), the high lack of fit of the model was associated with the extremely low value of the pure error (0.026) likely due to the low degrees of freedom (3).
Table 3.
Analysis of Variance of the Estimated Model for the Release of N-Glycans from Bovine Colostrum Whey by EndoBI-1
| Source of Variation | Sum of Squares | Degrees of Freedom | Mean Squares | F-test |
|---|---|---|---|---|
| Regression | 3,968.56 | 4 | 992.14 | 16.33a |
| Residual | 1,397.249 | 23 | 60.75 | |
| Lack of fit | 1,397.223 | 20 | 69.86 | 8,123.2b |
| Pure error | 0.026 | 3 | 0.0086 | |
| Total | 5,365.809 |
Values in bold are statistically meaningful (P < 0.05).
Coefficient of determination: R2 = 0.74; F0.95–4,23 = 2.80; F0.95–20,3 = 8.66.
Based on the estimated regression model generated, response surfaces were built to express the release of N-glycans from bovine colostrum whey by EndoBI-1. According to the estimated regression model and Figure 2, the release of N-glycans is favored by increased reaction time and low pH values. The highest release of N-glycans was observed in the axial points or both variables: 4.45 pH and 475 min. Based on the estimated regression model and Figure 2, EndoBI-1 activity was favored by temperature values ranging from 40 to 65°C, with the highest activity observed at the temperature of the central point (52.5°C) and lowest pH values. Reduction of EndoBI-1 activity when moving further away from the center point is likely due to some protein denaturation effects at higher temperature (+1, 65°C) and to temperatures that are below its optimum value (−1, 40°C).
Figure 2. Effects of temperature vs time, temperature vs pH, and time vs pH on the release of N-glycans from bovine colostrum whey by EndoBI-1.
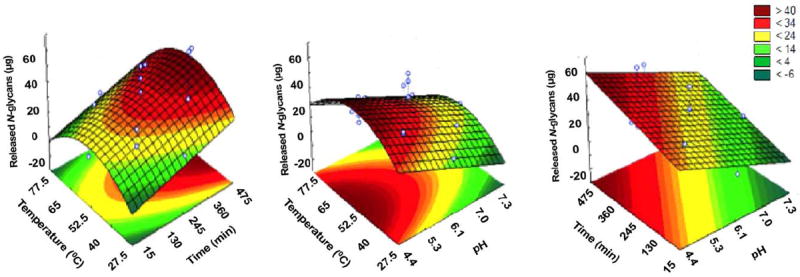
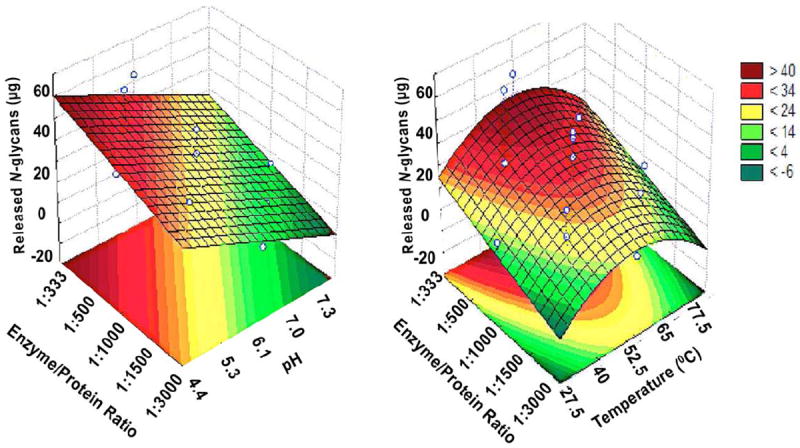
According to the estimated regression model and Figure 3, EndoBI-1 activity increases when enzyme/protein ratio increases from −2.0 to +2.0 (1/3,000 to 1/333), with highest values at lowest pH (−2.0, 4.45 pH). Based on the estimated regression model and Figure 2, temperature in the central point (52.5°C) and longer extraction times (+1–2.0, 360–475 min) did favor EndoBI-1 activity, with highest glycan release observed at 52.5°C and 475 min of reaction time. According to the estimated regression model and Figure 3, EndoBI-1 activity was favored by temperature in the central point (52.5°C) and higher enzyme/protein ratio (+1–2.0, 1/500–1/333 min), with the highest glycan release observed at 52.5°C and 1/333 enzyme/protein ratio. Overall, for the conditions tested, optimal conditions for glycan release were observed for low pH, a temperature of 52.5°C, a high enzyme/protein ratio, and longer reaction times.
Figure 3. Effects of enzyme/protein ratio vs pH and enzyme/protein ratio vs temperature on the release of N-glycans from bovine colostrum whey by EndoBI-1.
N-Glycan release comparison of EndoBI-1 and PNGase F on denatured/native substrates and pasteurization conditions
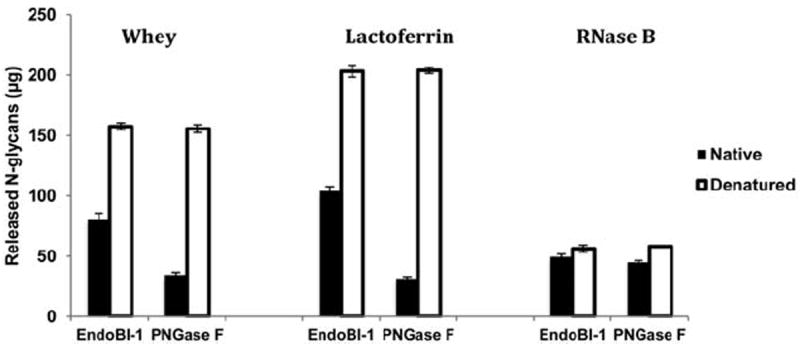
The yield of N-glycans released by EndoBI-1 and PNGase F from native or denatured bovine colostrum whey, bovine lactoferrin, or RNase B is shown in Figure 4. The treatment of native whey, lactoferrin, and RNase B by EndoBI-1 resulted in 80, 103, and 49 μg N-glycans, respectively, while the yield after denaturation was 157, 203, and 56 μg, respectively. In comparison, the yield of N-glycans from native whey, lactoferrin, and RNase B after treatment with PNGase F was found to be only 34, 31, and 44 μg, respectively. After denaturation, treatment with PNGase F produced 155, 203, and 58 μg N-glycans, respectively.
Figure 4. Denaturation effect of concentrated bovine colostrum whey, bovine lactoferrin, and RNase B on N-glycan release by EndoBI-1 and PNGase F.
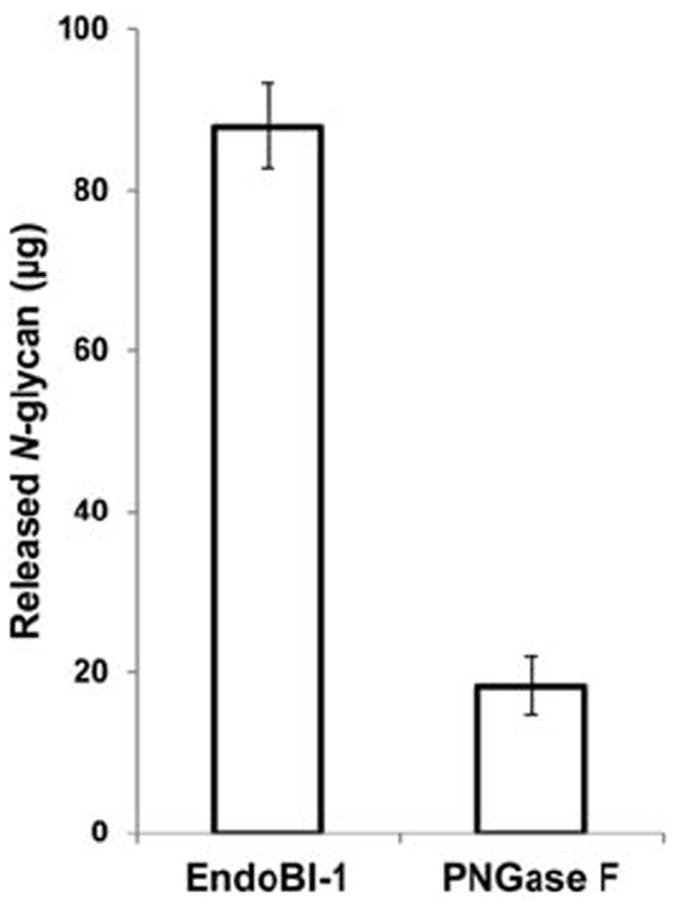
Similarly, the N-glycan yield of EndoBI-1 and PNGase F on 500 μL concentrated bovine whey under pasteurization conditions (62°C, 30 min) showed that EndoBI-1 yield was 87 μg, while the PNGase F yield was only 18 μg (Figure 5).
Figure 5. N-glycan release efficiency comparison of EndoBI-1 and PNGase F on concentrated bovine whey colostrum under pasteurization conditions (62°C, 30 min).
N-Glycan analysis by Nano-LC-Chip-Q-TOF MS
After digestion by EndoBI-1, released N-glycans from the bovine milk glycoproteins were purified by solid-phase extraction and analyzed by nano-LC-Chip–Q-TOF MS for a sample from experimental treatment 10 (Table 1). The PGC chip was able to separate N-glycans based on their structures and their monosaccharide compositions.
Figure 6 shows the extracted ion chromatogram (EIC) for one neutral and one sialylated oligosaccharide. For each of them, the composition was determined based on the mass and data on bovine milk N-glycans from the literature.26 EIC of these N-glycans shows 2 peaks at different retention times for the same mass. These results suggest two structural isomers or α or β anomers for the same composition.
Figure 6. Extracted ion chromatogram (EIC) of two isomers or anomers for neutral N-glycans with 638.73 m/z (z = 1) and sialylated N-glycan with 703.23 m/z (z = 1).
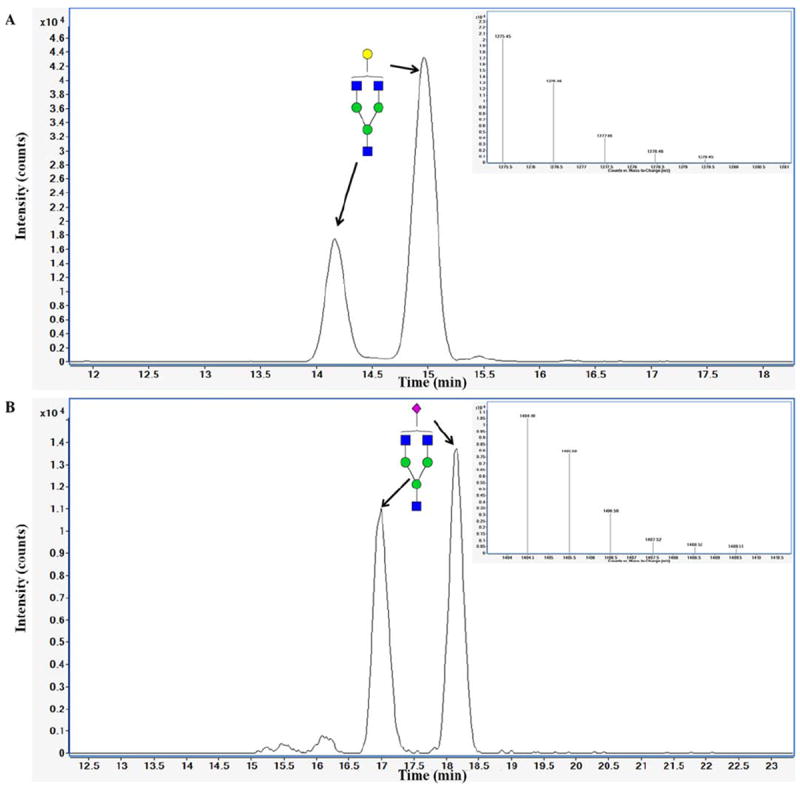
Deconvoluted CID Endo-B-N-acetylglucosaminidase was incubated with bovine colostrum whey protein concentrate at pH 5.30, 360 min, 40°C, enzyme/protein ratio 1/500. N-glycans were purified by solid-phase extraction and analyzed by Nano-LC-Chip-Q-TOF MS. Green circles, yellow circles, blue squares, red triangles, and purple diamonds represent mannose, galactose, HexNAc, fuc, and NeuAc residues, respectively.
The analytical platform used enables measurement of the presence of N-glycans in a complex mixture, bovine colostrum whey protein concentrate, with high resolution and separation of isomeric forms. The N-glycan composition was identified by an automated data analysis using a library for bovine milk glycoproteins. A complete N-glycan profiling was performed using LC-MS and LC-MS/MS on samples 9, 10, and 17 in another study to evaluate the effect of pH, reaction time, temperature, and enzyme/protein ratio on the N-glycan release diversity.27
Discussion
In this study, we investigate how different reaction conditions affect the efficiency of N-glycan release by EndoBI-1 from concentrated bovine colostrum whey glycoproteins. These released glycans are structurally similar to human milk oligosaccharides (HMO) that are shown to stimulate the development of a beneficial infant intestinal microbiota, in particular, enriching for Bifidobacterium.30,31 However, the large-scale isolation of these glycans is not feasible. Therefore, their use in both research and commercial applications is limited.
Currently, N-glycans are not commercially available, despite their biological activity, in large part because of the difficulty in producing them. Available technologies for releasing N-glycans from glycoproteins relies on the use of PNGase F, an enzyme derived from Flavobacterium,19 whose activity is limited by core-fucosylation, is heat-sensitive, active at a pH higher than that of most physiological systems, denaturation is required for activity, and the enzyme is expensive. These limitations restrict its use in the production of N-glycans from common sources, including whey protein.
Protein denaturation, especially, before enzymatic treatment plays an important role for efficient enzymatic release.32 However, the native environment of B. infantis from which EndoBI-1 was isolated26 likely contains few denatured glycoproteins and as a result, evolutionary pressures have shaped this enzyme’s activity to act efficiently on native-state glycoproteins. To investigate this possibility, we compared the ability to release N-glycans from native proteins, in comparison to PNGase F. The results show that both enzymes were more active on denatured proteins compared to native proteins. PNGase F showed 4.55, 7.41, and 1.31-fold reduced activity on native whey, lactoferrin, and RNase B, respectively, compared to denatured conditions, while the yield of N-glycans released from EndoBI-1 treatment of native whey, bovine lactoferrin, and RNase B was only 1.9, 1.95, and 1.14-fold reduced under native conditions, respectively. These results suggest that denaturation of substrates for N-glycan release by EndoBI-1 is not critical for N-glycan release, compared to PNGase F. Importantly, these results demonstrates that EndoBI-1 can be used in dairy industry to efficiently release N-glycans from milk proteins.
Bovine milk contains a large amount of protein (32 g/L), and a small proportion of free milk glycans, whose structural similarity to human milk oligosaccharides has been previously discussed.33 The biological activity of these milk oligosaccharides are chiefly responsible for the ability of milk to promote the growth of Bifidobacterium in the neonatal gut, and contribute to infant health.30,31 We have recently shown that N-glycans purified from milk glycoproteins bear structural similarity to the free glycans found in milk. These N-glycans also possess similar bifidogenic potential and could be used to improve human health, but these glycans are currently not widely available.34
The effect of temperature, pH, reaction time, and enzyme/protein ratio was examined to identify the optimum conditions under which a novel enzyme, EndoBI-1 is active on whey protein. In general, a higher glycan yield was obtained at lower pH values (4.45), longer reaction times (475 min), and moderate temperatures in the range studied (52.5°C), and a higher enzyme/protein ratio (1:333). A yield of 0.12 g L−1 is possible under optimum conditions, which is significantly higher than that obtained with PNGase F under its optimum conditions. Also, unlike PNGase F, these data also demonstrate that EndoBI-1 is a versatile enzyme, active under a wider variety of incubation time, temperature, and pH combinations that can be used in large-scale N-glycan production strategies. For example, a protein ratio of 1:1,000 enzyme/protein and 18 h incubation time was predicted to result in approximately the same yield as the optimum conditions (1/500 ratio and 360 min).
As a conclusion, the large-scale N-glycan production can be achieved with a novel enzyme EndoBI-1 and processing technique. These N-glycans will enable us to study their biological functions. Moreover, the use of whey as an N-glycan source provides added value for a former waste product, contributing to improved economic and environmental sustainability with the added benefit of the production of a beneficial bioactive food additive.
Supplementary Material
Acknowledgments
This work has been supported by the UC Davis RISE program, the Bill and Melinda Gates Foundation, National Institutes of Health awards R01AT008759, R01AT007079, and the Peter J. Shields Endowed Chair in Dairy Food Science.
Contract grant sponsor: University of California Discovery Grant Program, the UC Davis RISE program, the Bill and Melinda Gates Foundation, National Institutes of Health awards; Contract grant number: R21AT006180, R01AT007079.
Contract grant sponsor: Peter J. Shields Endowed Chair in Dairy Food Science.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Contributor Information
Sercan Karav, Dept. of Food Science and Technology, University of California, One Shields Avenue, Davis, CA 95616.
Juliana Maria Leite Nobrega De Moura Bell, Dept. of Food Science and Technology, University of California, One Shields Avenue, Davis, CA 95616.
Annabelle Le Parc, Dept. of Food Science and Technology, University of California, One Shields Avenue, Davis, CA 95616.
Yan Liu, Dept. of Food Science and Technology, University of California, One Shields Avenue, Davis, CA 95616.
David A. Mills, Dept. of Food Science and Technology, University of California, One Shields Avenue, Davis, CA 95616 Foods for Health Institute, University of California, One Shields Avenue, Davis, CA 95616; Dept. of Viticulture and Enology, University of California, Davis, CA 95616.
David E. Block, Dept. of Viticulture and Enology, University of California, Davis, CA 95616 Dept. of Chemical Engineering and Materials Science, University of California, Davis, CA 95616.
Daniela Barile, Dept. of Food Science and Technology, University of California, One Shields Avenue, Davis, CA 95616; Foods for Health Inst., University of California, One Shields Avenue, Davis, CA 95616.
Literature Cited
- 1.Pelegrine DHG, Gomes MTMS. Analysis of whey proteins solubility at high temperatures. Int J Food Eng. 2012;8:1–8. [Google Scholar]
- 2.Sasaki N, Toyoda M. Glycoconjugates and related molecules in human vascular endothelial cells. Int J Vasc Med. 2013;2013 doi: 10.1155/2013/963596. 963596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahan-Mihan A, Luhovyy BL, El Khoury D, Anderson GH. Dietary proteins as determinants of metabolic and physiologic functions of the gastrointestinal tract. Nutrients. 2011;3:574–603. doi: 10.3390/nu3050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley WL, Theil PK. Perspectives on immunoglobulins in colostrum and milk. Nutrients. 2011;3:442–474. doi: 10.3390/nu3040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legrand D, Elass E, Carpentier M, Mazurier J. Lactoferrin: a modulator of immune and inflammatory responses. Cell Mol Life Sci. 2005;62:2549–2559. doi: 10.1007/s00018-005-5370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orsi N. The antimicrobial activity of lactoferrin: current status and perspectives. Biometals. 2004;17:189–196. doi: 10.1023/b:biom.0000027691.86757.e2. [DOI] [PubMed] [Google Scholar]
- 7.Seganti L, Di Biase AM, Marchetti M, Pietrantoni A, Tinari A, Superti F. Antiviral activity of lactoferrin towards naked viruses. Biometals. 2004;17:295–299. doi: 10.1023/b:biom.0000027708.27142.bc. [DOI] [PubMed] [Google Scholar]
- 8.Gopal PK, Gill HS. Oligosaccharides and glycoconjugates in bovine milk and colostrum. Br J Nutr. 2000;84(Suppl 1):S69–S74. doi: 10.1017/s0007114500002270. [DOI] [PubMed] [Google Scholar]
- 9.Tao N, DePeters EJ, Freeman S, German JB, Grimm R, Lebrilla CB. Bovine milk glycome. J Dairy Sci. 2008;91:3768–3778. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- 10.Ruhaak LR, Lebrilla CB. Analysis and role of oligosaccharides in milk. BMB Rep. 2012;45:442–451. doi: 10.5483/BMBRep.2012.45.8.161. [DOI] [PubMed] [Google Scholar]
- 11.Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv Nutr. 2012;3:450S–455S. doi: 10.3945/an.112.001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Galvao CM, Silva AF, Custodio MF, Monti R, Giordano RL. Controlled hydrolysis of cheese whey proteins using trypsin and alpha-chymotrypsin. Appl Biochem Biotechnol. 2001;91-93:761–776. doi: 10.1385/abab:91-93:1-9:761. [DOI] [PubMed] [Google Scholar]
- 14.Shahani KM, Friend BA. Fuel alcohol production from whey and whey–grain mixtures. Abstr Pap Am Chem S. 1980;180(Aug):53–FUEL. [Google Scholar]
- 15.Halasz A, Zalan Z. Biochemical principles of the use of yeast biomass and lab starter cultures in food production. Acta Aliment Hung. 2009;38:71–85. [Google Scholar]
- 16.de Wit JN. Marschall Rhone-Poulenc Award Lecture. Nutritional and functional characteristics of whey proteins in food products. J Dairy Sci. 1998;81:597–608. doi: 10.3168/jds.s0022-0302(98)75613-9. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 18.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microb. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triguero A, Cabrera G, Royle L, et al. Chemical and enzymatic N-glycan release comparison for N-glycan profiling of monoclonal antibodies expressed in plants. Anal Biochem. 2010;400:173–183. doi: 10.1016/j.ab.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 20.Peter-Katalinic J. Methods in enzymology: O-glycosylation of proteins. Method Enzymol. 2005;405:139–171. doi: 10.1016/S0076-6879(05)05007-X. [DOI] [PubMed] [Google Scholar]
- 21.Zellner M, Winkler W, Hayden H, et al. Quantitative validation of different protein precipitation methods in proteome analysis of blood platelets. Electrophoresis. 2005;26:2481–2489. doi: 10.1002/elps.200410262. [DOI] [PubMed] [Google Scholar]
- 22.Tarentino AL, Plummer TH., Jr Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 1994;230:44–57. doi: 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- 23.Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1—3 to the asparagine-linked N-acetylglucosamine residue. Eur J Biochem/FEBS. 1991;199:647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 24.Garrido D, Nwosu C, Ruiz-Moyano S, et al. Endo-beta. N-acetylglucosaminidases from infant gut-associated Bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteomics. 2012;11:775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karav S, Parc AL, Moura Bell MLNJd, et al. Kinetic characterization of a novel endo-B-N-acetylglucosaminidase on concentrated bovine colostrum whey to release bioactive glycans. Enzyme Microbial Technol. 2015;77:46–53. doi: 10.1016/j.enzmictec.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrido D, Nwosu C, Ruiz-Moyano S, et al. Endo-beta. N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteomics: MCP. 2012;11:775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karav S, Parc LA, Bell MLNMJ, et al. A novel endo-β-N-acetyl-glucosaminidase releases specific N-glycans depending on different reaction conditions. Submitted to Biotechnol Prog. 2015 doi: 10.1002/btpr.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nwosu CC, Aldredge DL, Lee H, et al. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res. 2012;11:2912–2924. doi: 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prien JM, Ashline DJ, Lapadula AJ, Zhang H, Reinhold VN. The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J Am Soc Mass Spectrom. 2009;20:539–556. doi: 10.1016/j.jasms.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 31.Marcobal A, Barboza M, Froehlich JW, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hustoft HK, Malerod H, Wilson SR, Reubsaet L, Lundanes E, Greibrokk T. A critical review of trypsin digestion for LC-MS based proteomics. Integr Proteomics. 2012;73:73–91. [Google Scholar]
- 33.Haug A, Harstad OM. Bovine milk in human nutrition--a review. Lipids Health Dis. 2007;6:25. doi: 10.1186/1476-511X-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karav S, Parc LA, Bell MLNMJ, et al. Unpublished data. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


