Summary
Interacting proteins typically coevolve, and the identification of coevolving amino acids can pinpoint residues required for interaction specificity. This approach often assumes that an interface-disrupting mutation in one protein drives selection of a compensatory mutation in its partner during evolution. However, this model requires a non-functional intermediate state prior to the compensatory change. Alternatively, a mutation in one protein could first broaden its specificity, allowing changes in its partner, followed by a specificity-restricting mutation. Using bacterial toxin-antitoxin systems, we demonstrate the plausibility of this second, promiscuity-based model. By screening large libraries of interface mutants, we show that toxins and antitoxins with high specificity are frequently connected in sequence space to more promiscuous variants that can serve as intermediates during a reprogramming of interaction specificity. We propose that the abundance of promiscuous variants promotes the expansion and diversification of toxin-antitoxin systems and other paralogous protein families during evolution.
Introduction
Many interacting proteins within the same cell, particularly signaling proteins, are members of large paralogous families which have expanded through duplication and divergence. To expand in number, paralogous interacting proteins typically must become specific after duplication to avoid unwanted cross-talk (Capra et al., 2012; Zarrinpar et al., 2003). The specificity determinants of protein-protein interactions remain poorly defined in most systems. Even in the cases where they have been identified, we lack a detailed understanding of how a new, insulated protein-protein interaction emerges during the course of evolution and, more generally, the mutational paths followed during protein evolution (DePristo et al., 2005).
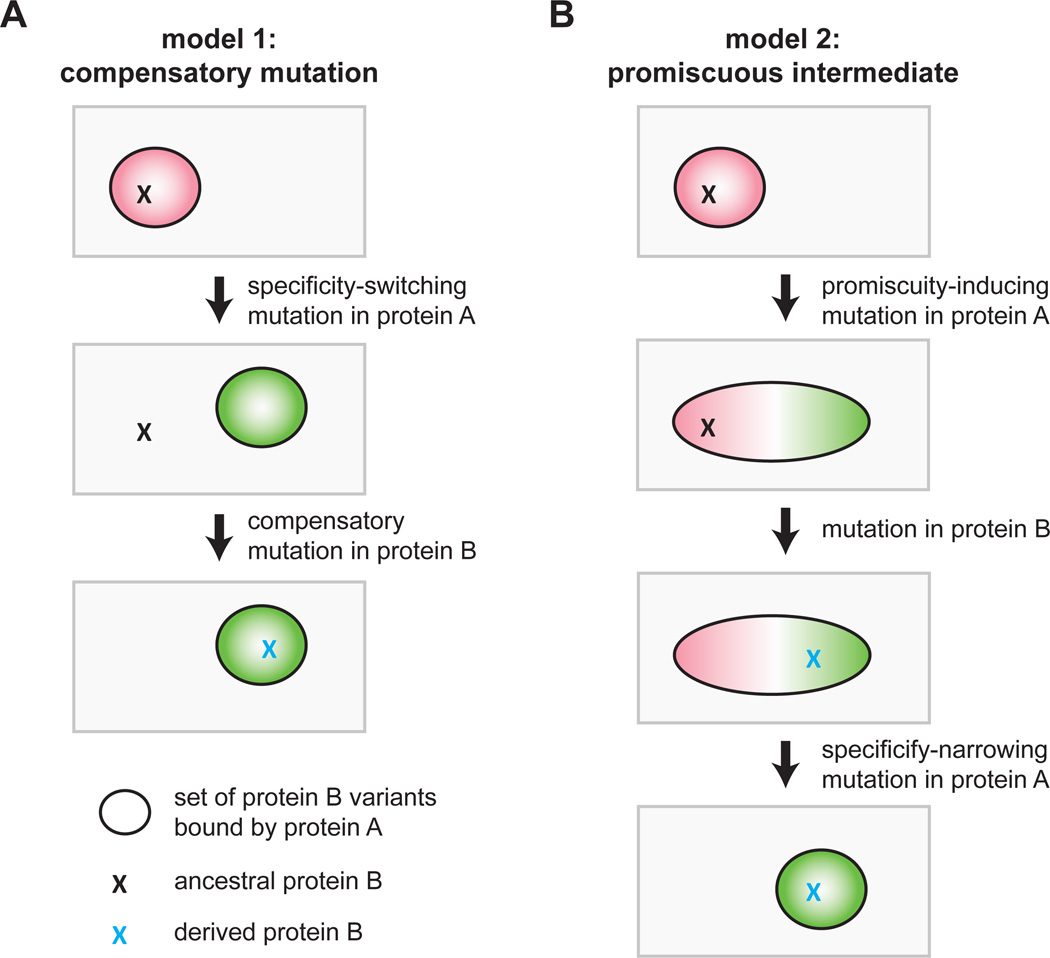
Computational studies demonstrate that interacting proteins often coevolve. Indeed, identification of coevolving residues has helped guide identification of the specificity determinants of many protein-protein interfaces (Ovchinnikov et al., 2014; Skerker et al., 2008). The implicit notion or underlying model behind these analyses is usually that an interaction-disrupting mutation in one protein can be rescued by a mutation in its partner (Fig. 1A). This model, which we call the compensatory mutation model, implies that the system passes through a non-functional or non-interacting state. However, such a state is highly unlikely, particularly for a protein-protein interaction that is critical for the viability of an organism. Alternatively, the specificity of a given protein-protein interaction could change, and become insulated from other paralogous systems, if one of the proteins passes through a promiscuous intermediate (Fig. 1B). In this model, an initial mutation in protein A would broaden its specificity, enabling its partner, protein B, to accumulate a mutation that would have disrupted its interaction with the original, ancestral form of protein A. A subsequent mutation in protein A would then narrow its specificity to include the derived, but not the ancestral, form of protein B. In this promiscuous intermediate model, the specificities of the interacting proteins change without ever transitioning through a nonfunctional intermediate state. Note that in both models A and B continue to interact through the same set of interfacial residues and do not evolve an alternative interface de novo (Kuriyan and Eisenberg, 2007).
Figure 1. Models for the evolution of new protein-protein interaction specificity.
(A) In a model of coevolution through compensatory mutations, an initial mutation in protein A that disrupts the A-B interaction is rescued by a compensatory mutation in protein B. Ovals represent the set of protein B variants that are bound by protein A, and Xs indicate particular protein B variants. Note that the intermediate state is a non-functional interaction.
(B) In an alternative model for protein coevolution, protein A first accumulates a mutation that broadens its specificity, followed by a second mutation in protein B that retains its interaction with the new form of A but that would have disrupted its interaction with the ancestral form of protein A. In a final step, protein A mutates to narrow its specificity to include the derived, and not ancestral, form of protein B.
Which of the two models in Figure 1 applies to most pairs of interacting proteins is unclear. In each case, the mutational trajectory involved would produce a signature of pairwise amino-acid coevolution in the phylogenetic record. However, only the latter, promiscuous intermediate model invokes the existence of mutations that are transiently introduced to broaden the specificity of one of the two proteins. The prevalence of such promiscuous states is unknown, as is whether they are easily reached from more specific, extant states.
Bacterial toxin-antitoxin (TA) systems provide an excellent model system for dissecting the coevolutionary dynamics of protein-protein interactions. Originally identified on plasmids, these systems are widely found in bacterial chromosomes, with many species encoding multiple, paralogous copies that share extensive similarity at the sequence and structural levels (Leplae et al., 2011). The biological function of TA systems is unclear, but they have been implicated in stress responses, resistance to phage, formation of persister cells, and bacterial pathogenicity (Yamaguchi et al., 2011). Typically, the toxin is a stable, globular protein that can inhibit cell growth or viability unless antagonized by a cognate antitoxin that directly binds and sequesters the toxin. Changes in the degradation rate or synthesis of the antitoxin can trigger release of the toxin. A toxin is typically encoded in the same operon as an antitoxin, and toxin-antitoxin paralogs frequently arise through operon duplications. An unresolved question is whether toxin-antitoxin systems interact in an exclusive one-to-one manner. Genetic data suggest that these interactions may be specific (Fiebig et al., 2010), and the growth inhibitory effects of a toxin are usually rescued only by expressing its co-operonic antitoxin (Hallez et al., 2010; Ramage et al., 2009). However, interaction specificity has only been directly tested in a limited number of cases, and some groups have suggested that toxins and antitoxins encoded in different operons are capable of interacting in vivo and in vitro, possibly forming large, promiscuous networks (Yang et al., 2010; Zhu et al., 2010).
Here, we systematically measure the binding preferences of twenty ParD-ParE TA family members and find that these toxins and antitoxins are highly specific, interacting almost exclusively with their partner from the same operon. This specificity is encoded by a small set of coevolving residues at the toxin-antitoxin interface, and mutations in these residues are sufficient to reprogram a ParD antitoxin to interact with non-cognate ParE toxins. Guided by these findings, we generated a library with ~104 variants of the key, specificity-determining residues in a ParD antitoxin and selected mutants that antagonize the cognate toxin, a non-cognate toxin, or both. Strikingly, we find that promiscuous variants that antagonize multiple toxins are easily obtained and are also highly connected in sequence space to specific variants. These results suggest that mutational paths leading to changes in toxin-antitoxin specificity are likely to involve promiscuous intermediates. Such paths enable the reprogramming of toxin-antitoxin specificity through the pairwise coevolution of interfacial residues, but without passing through an intermediate state that disrupts the protein-protein interaction. The abundance of promiscuous states likely facilitates the evolutionary expansion of these and other paralogous protein families following operon and whole genome duplications during evolution.
Results
Toxins and antitoxins from the ParDE family exhibit high interaction specificity
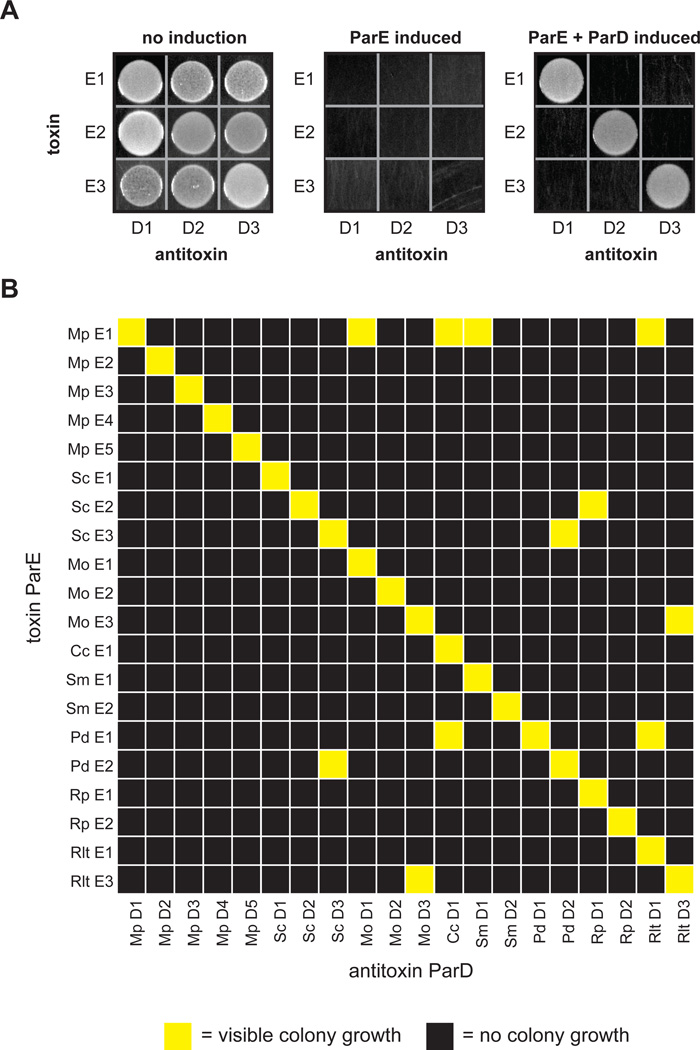
To systematically measure the interaction specificity of TA systems, we focused on the ParD-ParE family, which is often found in multiple copies on bacterial chromosomes (Fiebig et al., 2010; Leplae et al., 2011) (Fig. S1A). We initially cloned the three chromosomally-encoded ParD-ParE pairs from the α-proteobacterium Mesorhizobium opportunistum into vectors that allow for separate and inducible expression of the ParE toxin and ParD antitoxin. To measure the interaction specificity for these pairs, we then co-transformed all pairwise combinations of toxin and antitoxin plasmids into E coli, and assessed whether the induced expression of each ParD antitoxin rescues the growth arrest resulting from inducing each ParE. As a control, we first confirmed that inducing each ParE toxin inhibited growth of E. coli (Fig. 2A). Then, plating on a medium that induces both ParD and ParE, we observed growth for each of the three cognate ParD-ParE pairings (Fig. 2A). No growth was observed for the six non-cognate pairs, indicating that the ParD antitoxins from M. opportunistum can only neutralize their cognate ParE toxins.
Figure 2. Toxins and antitoxins from the ParD-ParE family exhibit high interaction specificity.
(A) Testing of interaction specificity for ParD antitoxins and ParE toxins from Mesorhizobium opportunistum. Plasmids harboring the toxins and antitoxins indicated were cotransformed into E. coli with ParD and ParE induced as indicated.
(B) Comprehensive testing of interaction specificity for 20 ParD and ParE pairs from eight different species. Cells containing each possible ParD-ParE pair were grown on plates that induce the toxin and antitoxin, respectively, and grown overnight at 37°C. Yellow, visible colonies following serial dilution; black, no visible colonies.
Also see Fig. S1.
We extended this analysis to the 20 chromosomally-encoded ParDE pairs from eight different bacteria, including the three pairs from M. opportunistum (Fig. S1B). For this 20×20 matrix of ParD and ParE pairs we observed strong interactions between all 20 co-operonic ParDE pairs, but only 11 of the 380 (or 3%) other possible pairings (Fig. 2B). Importantly, these cross-reactions were only observed between ParD and ParE proteins not encoded in the same species, indicating that the ParDE pairs within a given organism are typically insulated from one another. These results indicate that ParD antitoxins are highly specific for their cognate ParE toxins.
Identification of covarying residues in ParD and ParE
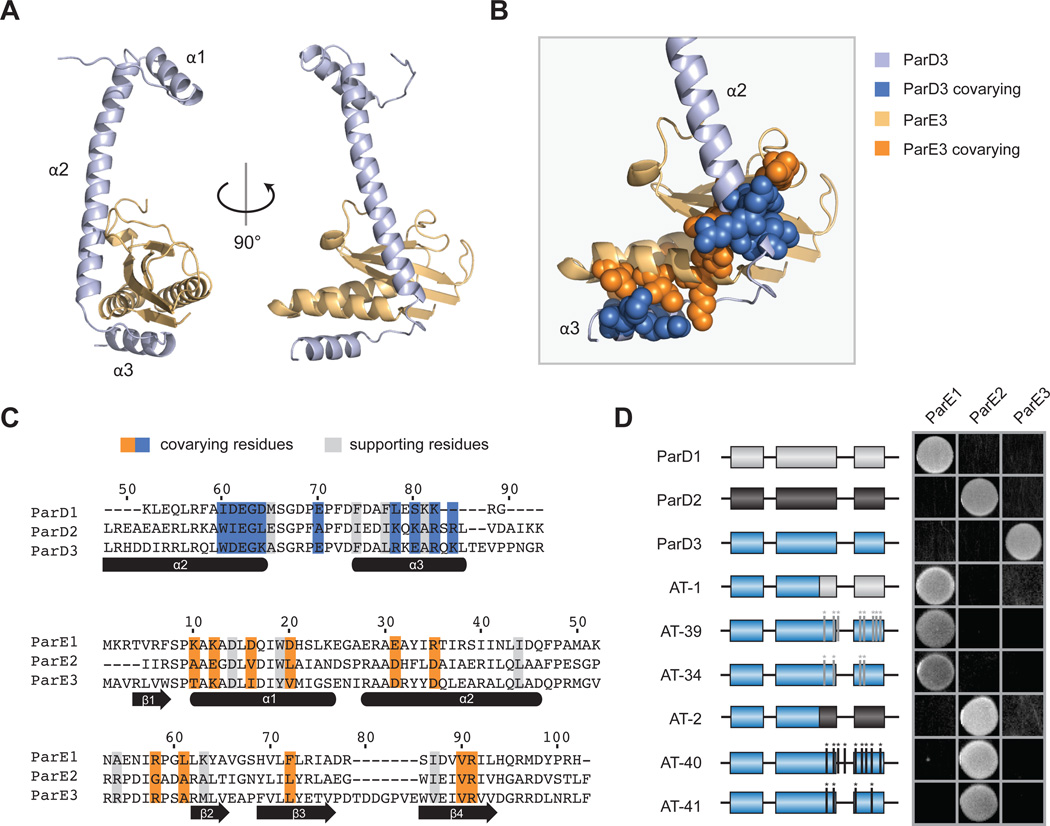
As a first step in understanding the molecular basis of specificity in ParD-ParE complexes, we solved a 1.59-Å cocrystal structure of the M. opportunistum ParD3 antitoxin bound to ParE3, its cognate toxin. This structure revealed a heterotetrameric asymmetric unit composed of ParD3 and ParE3 dimers (Fig. S2A), similar to a C. crescentus ParD-ParE structure (Dalton and Crosson, 2010). Crystal packing and an estimated mass of ~87 kDa in solution indicate that the biological assembly is composed of two tetramers (Fig. S2B–C). Within this complex, each ParD3 subunit makes extensive contacts with a neighboring ParE3 subunit primarily through its second and third alpha helices, with a total buried surface area of 1624 Å2 (Fig. 3A).
Figure 3. Covarying residues dictate interaction specificity in the ParD-ParE family.
(A) Structure of the M. opportunistum ParD3-ParE3 complex. Light orange, ParE3 monomer; light blue, ParD3 monomer.
(B) A section of the ParD3-ParE3 structure from (A) magnified; covarying residues shown in space-filling representation.
(C) Alignment of M. opportunistum ParD and ParE paralogs with coevolving residues highlighted in blue or orange for ParD or ParE, respectively. Supporting residues, which coevolve with the interfacial coevolving residues, are highlighted in grey.
(D) Mutations in the C-terminus of ParD3 can reprogram interaction specificity. The indicated ParD3 mutants were tested against each ParE homolog from M. opportunistum using the E. coli toxicity-rescue assay.
Previous work with bacterial two-component signaling systems demonstrated that their interaction specificity is controlled by a subset of residues at the protein-protein interface formed by a histidine kinase and response regulator (Skerker et al., 2008). These specificity-determining residues coevolve to maintain the interaction between cognate signaling proteins. Thus, to pinpoint the residues that contribute to the specificity of ParD-ParE interactions, we used GREMLIN, a pseudo-likelihood-based model for coevolution (Kamisetty et al., 2013; Ovchinnikov et al., 2014), to search for residues that strongly covary in a multiple sequence alignment of concatenated, co-operonic ParD and ParE proteins. This analysis identified 10 residues in ParD and 11 residues in ParE that coevolve most strongly. Hereafter, we call these 21 amino acids 'specificity' residues, as our work below indicates that they play the dominant role in determining partner specificity. Mapping these specificity residues onto the ParD3-ParE3 crystal structure indicated that they cluster into two groups at the primary molecular interface formed by these proteins (Fig. 3B–C). The first group sits at the base of the second alpha helix in ParD3 and covaries with residues in the three-stranded beta sheet in ParE3. The second group clusters in the third alpha helix in ParD3 and covaries with residues in the first and second alpha helices of ParE3. We also used GREMLIN to identify residues within each protein (four in ParD and six in ParE) that coevolve with the specificity residues (Fig. 3C, S3A). These 'supporting' residues may indirectly contribute to ParD-ParE interaction specificity by influencing the orientation or packing of the interfacial specificity residues.
Covarying residues dictate interaction specificity in the ParD-ParE family
To determine whether the coevolving residues identified are sufficient to dictate interaction specificity of the ParD-ParE family, we constructed a series of chimeric proteins in which different regions of the M. opportunistum ParD3 were replaced with the corresponding regions of ParD1 or ParD2 (Fig. S3B). Replacing the entire C-terminal region of ParD3 with the corresponding region of ParD1 or ParD2 produced a chimera that lost its ability to interact with ParE3 but gained the ability to interact with ParE1 or ParE2 (Fig. 3D). These chimeras involved both clusters of interfacial residues identified as coevolving between ParD and ParE proteins. Replacing only one of these clusters in the ParD3 C-terminus was sometimes sufficient to reprogram specificity, but depended on the toxin tested (Fig. S3C). These results indicate that the C-terminal region of ParD, which contains the specificity and supporting residues, is sufficient to dictate interaction specificity.
To pinpoint the residues required for interaction specificity, we focused additional mutagenesis on the coevolving residues identified computationally. We generated variants of ParD3 in which all of the specificity and supporting residues were replaced with the corresponding residues in ParD1 or ParD2, for a total of 8 or 9 substitutions, respectively. In each case, we found that these mutations were sufficient to reprogram ParD3 to interact with ParE1 or ParE2 and lose its ability to interact with ParE3 (Fig. 3D). Interestingly, ParD3 could be reprogrammed to interact with ParE1 or ParE2 with fewer substitutions. For example, we found sets of four substitutions that were sufficient to reprogram ParD3 to interact with ParE1 or ParE2 (Fig. 3D). Taken together, our results indicate that mutating the most highly coevolving residues in an antitoxin can be sufficient to reprogram its interaction specificity, and, in some cases, mutating only a subset of these residues allows a complete switch in partner specificity.
High-throughput mapping of interface mutant fitness
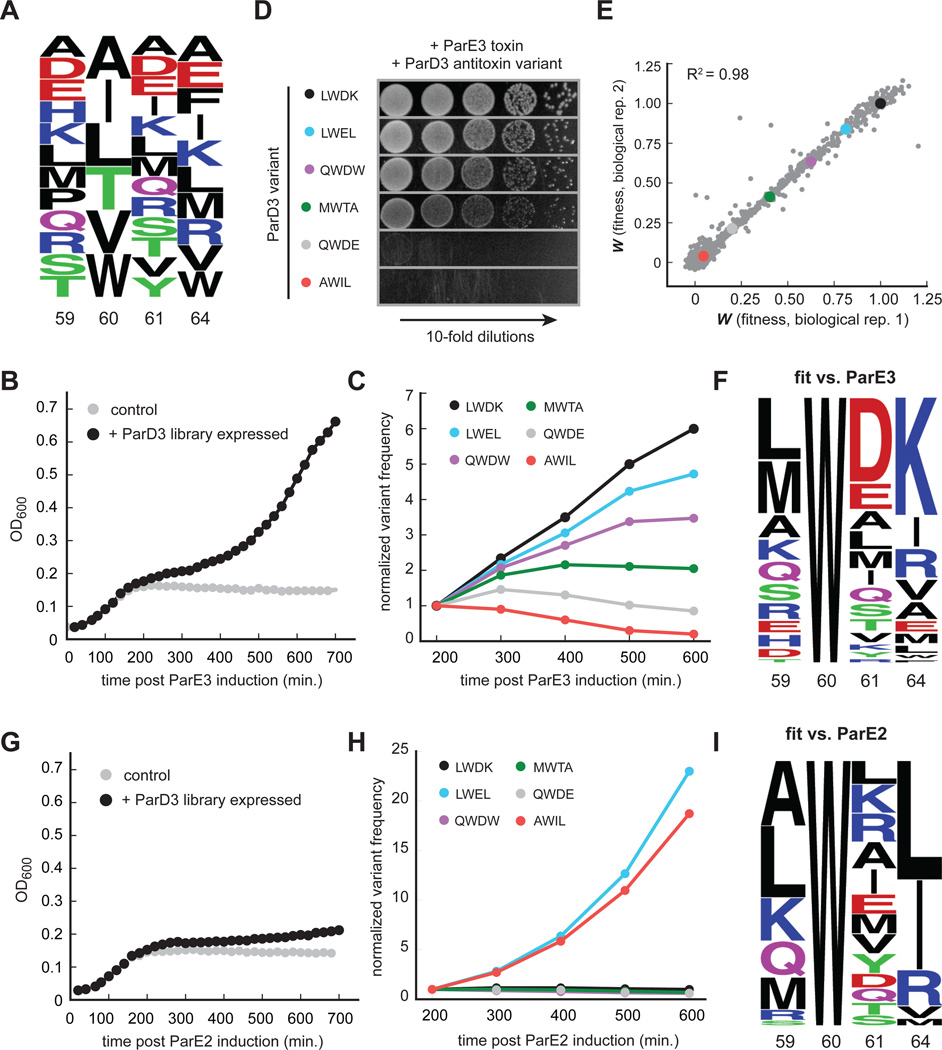
The results presented above indicate that antitoxin interaction specificity can be reprogrammed by changing just four residues. But how does specificity change as these four individual substitutions are introduced, and does the substitution order matter? Does the specificity of antagonizing one ParE toxin to another change abruptly, or are there promiscuous mutational intermediates? To answer these questions, we sought to generate a large library of ParD3 variants that included combinations of residues shown to be specific for antagonizing ParE3 or ParE2, as well as the mutational intermediates separating these specific states. To this end, we generated a library of mutants at four of the key interfacial positions in the ParD3 antitoxin, Leu59, Trp60, Asp61, and Lys64 (LWDK). To reduce the complexity of our library, we only allowed residues at each library position that are commonly found in naturally occurring ParD homologs (see Experimental Procedures). The resulting library has a theoretical diversity of 9,360 variants, with 12, 6, 13, and 10 possible residues encoded at the four respective positions of the library (Fig. 4A). Deep-sequencing of the relevant region in parD3 in the initial library revealed that >98% of the predicted variants were represented by at least 10 reads and >94% had at least 100 reads (Fig. S4A). Measurements of read numbers were highly reproducible between replicates (R2 > 0.99, Fig. S4B).
Figure 4. High-throughput mapping of mutant fitness at co-evolving interface.
(A) Composition of the ParD3 antitoxin library at the four variable positions.
(B) Library growth following ParE3 toxin induction.
(C) Frequency changes over time for the indicated ParD3 variants following ParE3 induction.
(D) Testing of individual variants from (C) using the toxicity rescue assay. 10-fold serial dilutions were plated from cultures expressing the ParD3 variant indicated and the ParE3 toxin.
(E) Two biological replicates of fitness measurements derived from screening the ParD3 library against the ParE3 toxin.
(F) Frequency logo for ParD3 library variants with high fitness against ParE3 (WE3 > 0.5).
(G) Library growth following induction of the non-cognate ParE2 toxin.
(H) Frequency changes over time for the indicated ParD3 library variants.
(I) Frequency logo for ParD3 library variants with high fitness against ParE2 (WE2 > 0.5).
Also see Fig. S4.
To assess the ability of each ParD3 variant to bind and antagonize ParE3, we co-transformed E. coli with the ParD3 library and an inducible ParE3 vector. When cultured in conditions that do not induce ParD3, cell growth arrested within 200 minutes after inducing the ParE3 toxin (Fig. 4B). In contrast, when the ParD3 library was expressed, growth slowed after inducing the toxin but eventually resumed, suggesting that some fraction of the population could neutralize ParE3 toxicity (Fig. 4B). To determine which mutants neutralized ParE3 and hence were enriched during the course of this experiment, we harvested samples every 100 minutes and deep-sequenced the relevant region of parD3. We observed large changes in the frequency of individual variants over this time course (Fig. S4C). For example, the variant containing the wild-type ParD3 residues (LWDK) was enriched ~6-fold, whereas variants with frameshift mutations in parD3, which are presumably non-functional, were depleted ~7-fold (Fig. S4C). To validate the functionality of variants inferred from this competitive growth assay, we isolated six mutants that exhibited different frequency dynamics following toxin induction (Fig. 4C). We tested these six mutants individually using our toxicity-rescue assay and found clear agreement between the change in the frequency of each variant in the library and its individual plating efficiency (Fig. 4D).
To quantify differences in variant behavior during competitive growth, we generated a linear fit to the frequencies of each mutant as a function of time, and then calculated the log-fold expansion of each mutant relative to the rest of the population, producing a raw fitness value (Wraw) for each mutant. We then transformed these raw fitness values such that the W value for frameshift variants was 0 and the W value for the wild-type (LWDK) sequence was 1; the resulting distribution of W values ranged from −0.04 to 1.13 and was highly reproducible between biological replicates (Fig. 4E, R2 = 0.98). We found a total of 252 variants with W values > 0.5, representing 2.7% of the total (Fig. 4F). This set included the wild-type combination of residues (LWDK) and 31 single, 189 double, and 31 triple mutants relative to the wild-type sequence (Fig. S4D). There were no quadruple mutants, as position 60 was invariantly tryptophan. The most common residues in this set as a whole were wild type. However, the identification of 252 variants that can effectively antagonize ParE3 indicates a substantial degree of functional degeneracy in the ParD3 interfacial residues.
Next, to assess the ability of each ParD3 variant to antagonize the non-cognate toxin ParE2, we repeated the competitive growth experiment but co-transformed E. coli with our ParD3 library and an inducible ParE2 vector. As before, we observed growth rescue following ParD3 library expression with large changes in the frequency of individual variants over time (Fig. 4G, S4E). However, the frequency changes observed here differed from those observed against the cognate toxin ParE3. For example, a variant containing the specificity residues found in the native ParD2 antitoxin, AWIL, was enriched in the ParD3 library screened against ParE2 but was depleted when screened against ParE3 (Fig. 4C, 4H). We quantified variant fitness as before and found a total of 151 variants (1.6% of the total) capable of antagonizing ParE2 with W values > 0.5 (Figure 4I, S4F). The most common residues were Ala59, Trp60 (invariant), Leu61, and Leu64. However, we noted important differences between variants reactive against ParE2 and ParE3, particularly at the last two variable positions in our library. ParE2-specific variants tended to have small hydrophobic or positively-charged residues at position 61, whereas ParE3-specific variants favored negatively-charged residues at this position (Fig. 4F, 4I). Additionally, ParE2-specific variants were more likely to contain small hydrophobic residues at position 64, whereas ParE3-specific variants tended to have positively-charged residues (Fig. 4F, 4I).
Mutational paths that reprogram specificity tend to involve promiscuous variants
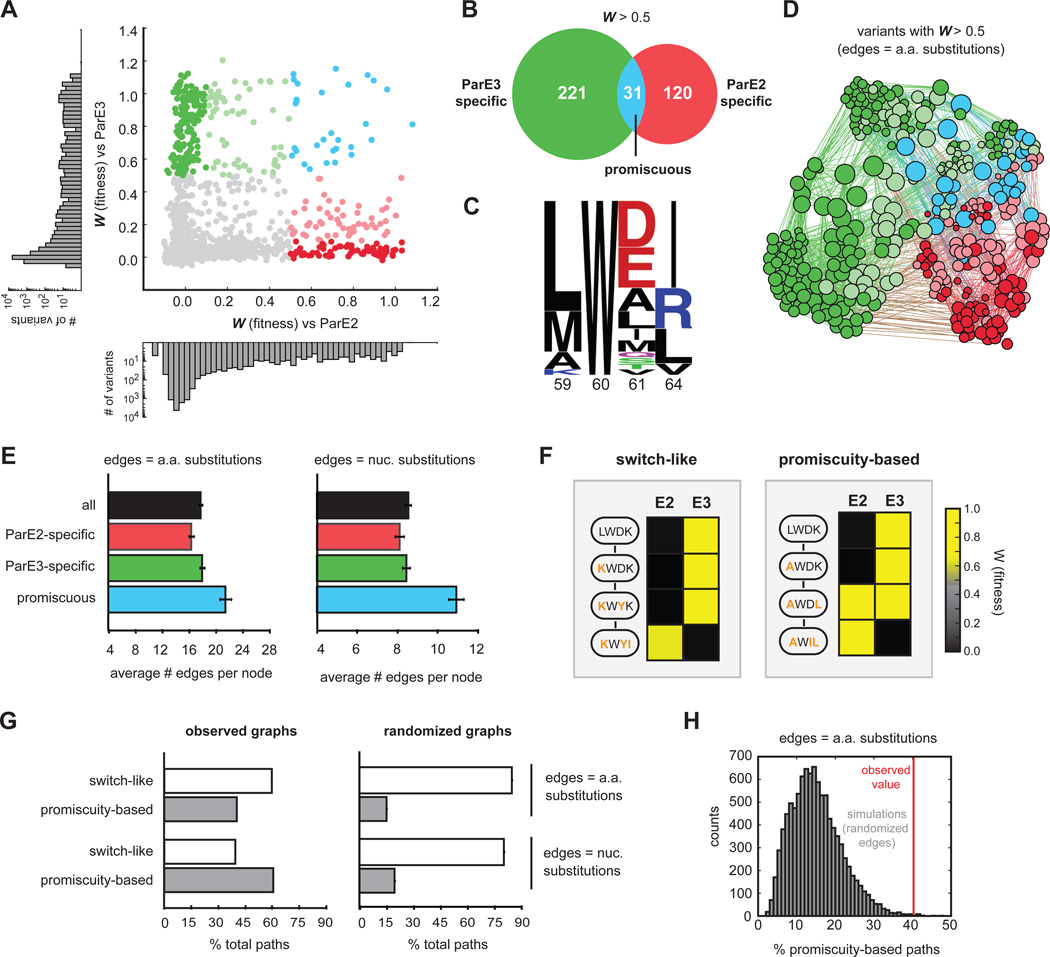
To more systematically probe the sequence space governing the specificity of ParD3, we generated a scatterplot of ParD3 variant fitness when screened against the ParE2 or ParE3 toxin (Fig. 5A). This analysis revealed variants spanning all ranges of fitness, including those capable of antagonizing ParE2, ParE3, or both toxins simultaneously. We identified a total of 31 promiscuous variants (W > 0.5 for both toxins), which represents a subset of the 252 ParE3-reactive and 151 ParE2-reactive variants (Fig. 5B). We then grouped variants by specificity class (Fig. S5A) and found that the promiscuous variants, such as LWEL, tended to harbor sequence elements from both ParD3 and ParD2, often with negatively charged residues at position 61 (ParD3-like) and aliphatic residues at position 64 (ParD2-like) (Fig. 5C).
Figure 5. Specificity-reprogramming paths are highly enriched for promiscuous variants.
(A) Fitness of ParD3 variants against ParE2 and ParE3. Green, specific for ParE3; blue, capable of antagonizing both ParE2 and ParE3; red, specific for ParE2. Histograms of fitness values against ParE2 and ParE3 are shown.
(B) Venn diagram of ParD3 variants reactive against ParE3, ParE2, or both.
(C) Frequency logo of promiscuous ParD3 variants (WE2 > 0.5, WE3 > 0.5).
(D) Force-directed graph of all ParD3 variants reactive against ParE3 or ParE2 (W > 0.5). Nodes represent individual variants and edges represent single amino acid substitutions. Node size scales with increasing degree and color corresponds to the specificity classes in (A).
(E) Average number of edges per node for the indicated categories of ParD3 variants. Error bars indicate SEM.
(F) Examples of ‘switch-like’ and ‘promiscuity-based’ mutational paths from an E3-specific variant to an E2-specific variant with the fitness against each variant color-coded based on the scale shown.
(G) Left, percentage of ‘switch-like’ and ‘promiscuity-based’ paths from the wild-type ParD3 sequence (LWDK) to each of the 66 ParE2-specific variants (WE2 > 0.5, WE3 < 0.1). Right, same as left panel but for 10,000 simulations in which the graph edges were randomly shuffled while keeping the total edge count and degree distribution constant. Error bars represent SEM.
(H) Histogram representing percentage of ‘promiscuity-based’ paths in 10,000 edge shuffling simulations; red line indicates percentage for the observed amino acid graph.
Also, see Fig. S5.
To visualize the connectivity of functional variants in sequence space, we created a force-directed graph where individual nodes represent functional variants with lines connecting variants that differ by a single amino acid (Fig. 5D). Node sizes increase with greater connectivity and node colors represent the specificity class of a given variant (Fig. 5D). The resulting graph was densely interconnected but generally grouped variants based on their specificity. The average number of edges per node, or degree, was 17.8 and ranged from 7 to 31. However, we noted that the average number of edges per node was 23% higher for promiscuous variants than for variants specific for ParE2 or ParE3 (Fig. 5E). We also generated a force-directed graph in which edges represent variants that differ by a single nucleotide substitution, following the standard genetic code (Fig. S5B). For this graph, promiscuous variants were, on average, 31% more connected to other nodes than their ParE2- or ParE3-specific counterparts (Fig. 5E). This increased connectivity of promiscuous variants was highly significant for both amino acid and nucleotide graphs, as it was lost when the edges of each graph were randomly shuffled (p < 10−4, Fig. S5C–D). The high connectivity of promiscuous variants was even more pronounced with a more stringent definition of specificity (Fig. S5E).
The dense connectivity of promiscuous variants suggested that mutational paths that change ParD3 specificity (from ParE3-specific to ParE2-specific, or vice versa) tend to travel through promiscuous intermediates. To test this hypothesis, we first defined two types of specificity-reprogramming paths. Note that for the following analysis, we exclude paths in which ParD3 fails to interact with both ParE3 and ParE2 (also see Discussion). The first class of paths are “switch-like” and only involve intermediates that are specific for ParE2 or ParE3, whereas the second class of paths are "promiscuity-based" and travel through at least one intermediate that can inhibit both ParE2 and ParE3 (Fig. 5F). To determine whether paths that change the interaction specificity of ParD3 tend to be switch-like or promiscuity-based, we identified all shortest mutational paths from the wild-type ParD3 variant (LWDK) to each of the 66 variants that are highly specific for ParE2 (WE2 > 0.5, WE3 < 0.1; Fig. S5A); for this analysis, each mutational step involved a single amino-acid substitution. We found a total of 370 shortest paths, of which 40% involved a promiscuous intermediate (Fig. 5G). The percentage of paths via promiscuous intermediates increased to 61% when considering only paths that involve single nucleotide substitutions (Fig. 5G).
To determine whether the number of paths that involve promiscuous variants is greater than would be expected by chance, we generated graphs in which the edges were randomly shuffled, and again calculated the percentage of each class of paths from ParD3 (LWDK) to the ParE2 highly specific variants. For these graphs with randomized edges, the percentage of paths involving promiscuous intermediates dropped to 15% for the amino acid neighbor graph and 20% for the nucleotide neighbor graph (Fig. 5G–H). Thus, the enrichment of promiscuity-based paths in the observed graphs is significant (p < 0.005) (Fig. 5G–H, S5F). Collectively, our results demonstrate the dense connectivity of functional variants in the sequence space governing ParD-ParE interaction specificity and reveal that specificity-reprogramming paths are highly enriched for those that involve promiscuous variants, which may facilitate the evolution of ParD-ParE systems with new specificities.
Epistasis: Mutational order dictates specificity class of intermediate variants
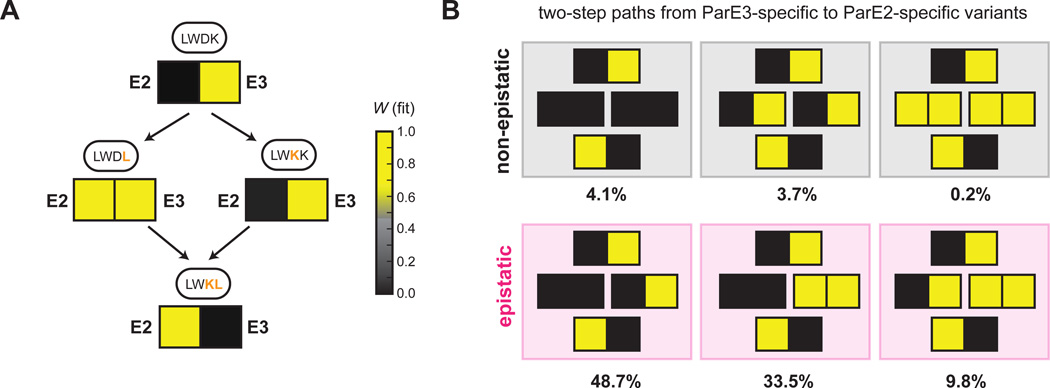
Inspection of the paths connecting ParD3 variants with different specificities indicated that the third and fourth library positions, residues 61 and 64 in ParD3, contribute significantly to the insulation of the ParD-ParE system. For instance, the wild-type residue combination in ParD3, LWDK, renders it specific for binding to ParE3, whereas the double-mutant variant LWKL is specific for ParE2. Strikingly, however, the two possible paths connecting LWDK and LWKL are in different classes (Fig. 6A). A single ParD3 substitution (K64L in LWDL) resulted in promiscuous binding to ParE2 and ParE3, whereas a second substitution in this background (D61K in LWKL) resulted in specificity for ParE2 (Fig. 6A). In contrast, incorporating these substitutions in the reverse order, D61K and then K64L, resulted in a switch-like change in specificity in which the initial D61K substitution retained specificity for ParE3, but then enabled the subsequent K64L substitution to produce a ParE2-specific antitoxin (Fig. 6A). These results underscore how a small number of mutations can fully reprogram protein-protein interaction specificity and demonstrate that the order of mutations can strongly affect whether the path to a new specificity state involves a promiscuous intermediate or a rapid switch.
Figure 6. Mutational order dictates specificity class of intermediate variants.
(A) Mutational paths from LWDK to LWKL for ParD3 with fitness of each variant against ParE2 and ParE3 shown as a heatmap: yellow, high fitness; black low fitness.
(B) The six path types that reprogram ParD3 specificity in two mutational steps. Percentage of mutational paths in each category is indicated for a threshold of 0.5 used to define a positive interaction.
Also see Fig. S6.
Our finding that changes in specificity can depend strongly on the order of substitutions represents a form of epistasis, broadly defined as cases where the functional effect of individual substitutions is context-dependent rather than additive and independent (Lehner, 2011). To more broadly quantify this epistasis for the ParD3 interfacial residues, we first defined six types of specificity-reprogramming paths that involve two amino-acid substitutions (Fig. 6B). Three of the six path types are epistatic with the two intermediates having different specificities, implying that substitution order influences changes from ParE3- to ParE2 specificity. We quantified the path type for each case in which two substitutions reprogram ParD3 from being specific for ParE3 (WE3 > 0.5, WE2 < 0.5) to being specific for ParE2 (WE3 < 0.5, WE2> 0.5) and found a total of 2,653 such cases, of which 92% were epistatic (Fig. 6B). The percentage of epistatic paths was robust to the threshold used for defining positive interactions (Fig. S6A–B). Taken together, our results highlight the pervasive effects of epistasis on ParD function. Although studies of epistasis typically consider the interdependence of individual substitutions with respect to protein folding or a single protein function (Kondrashov and Kondrashov, 2015; Lehner, 2011), our findings indicate that epistasis can also manifest at the level of interaction specificity. This form of epistasis may significantly impact the evolution of new ParD-ParE systems. Promiscuous intermediates enable a change in protein-protein interaction specificity without passing through a non-functional state, in which a liberated toxin would suppress growth and proliferation (Fig. 1A). Thus, the epistasis documented here may fundamentally restrict mutational trajectories during evolution to those involving promiscuous intermediates.
Mutational trajectories to an orthogonal ParD3-ParE3 pair
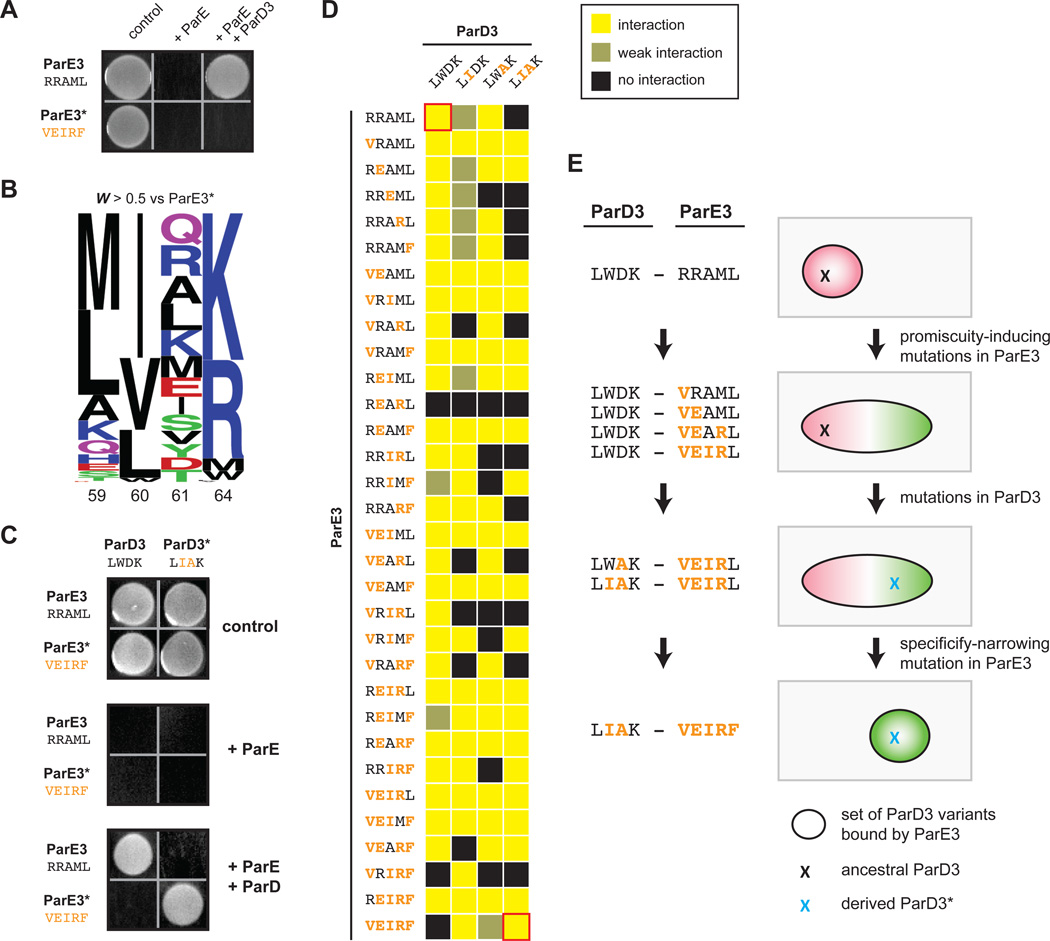
Thus far, we have considered changes to one side of the ParD-ParE interface. To probe how the interaction specificity of a ParD-ParE protein pair coevolves, we sought to generate a variant of the toxin ParE3 that does not interact with ParD3, and then select ParD3 variants from our library that can neutralize this novel toxin. To this end, we generated a variant of the toxin, called ParE3*, that retains toxicity but is incapable of binding to the ParD3 antitoxin. In particular, we mutated five ParE3 positions (Arg54, Arg58, Ala61, Met63, and Leu72, or RRAML) that strongly covary with the specificity residues in ParD3. We mutated RRAML➔VEIRF, as each individual variant residue was frequently observed in ParE3 homologs and was chemically different from the corresponding wild-type residue (Fig. S7A). As expected, we found that ParE3* retained toxicity but was no longer neutralized by ParD3 (Fig. 7A).
Figure 7. Mutational trajectories to an orthogonal ParD3*-ParE3* pair.
(A) ParE3* is insulated from antitoxin ParD3. A plasmid containing either ParE3 or ParE3* was co-transformed into E. coli with a plasmid expressing ParD3, and cells were plated on medium that induces or represses expression of the toxin and antitoxin.
(B) Frequency logo for ParD3 library variants with high fitness against ParE3* (WE3* > 0.5).
(C) ParE3*-ParD3* is insulated from the wild-type ParD3-ParE3 pair.
(D) Toxicity-rescue interaction assays for all ParD3 and ParE3 mutant combinations. Top left, wild-type ParD3-ParE3 pair; bottom right, orthogonal ParD3*-ParE3* pair. Promiscuous ParE3 intermediates are those capable of interacting with both ParD3 and ParD3*.
(E) Example of a series of single substitutions that lead to the insulated ParE3*-ParD3* system while retaining the toxin-antitoxin interaction at each step by first expanding the specificity of ParE3, followed by changes in ParD3, and finally by restricting the specificity of ParE3.
Also see Fig. S7.
To determine whether variants in the ParD3 library neutralized ParE3*, we performed a competitive growth experiment following co-transformation. As before, we converted changes in variant frequencies to fitness values, which were highly reproducible (R2 = 0.96, Fig. S7B). Sequence analysis of the high-fitness mutants (W > 0.5) revealed large differences in amino-acid preferences at positions 60 and 61 relative to those shown above (Fig. 4F, 7B). In particular, for the ParD3 variants that neutralized ParE3*, the invariant Trp60 was replaced by Ile/Val/Leu and the strong preference for a negatively charged residue at position 61 was replaced by positively-charged or neutral residues (Fig. 4F, 7B). One of the high-fitness variants with specificity residues LIAK, renamed ParD3*, no longer neutralized ParE3 but robustly interacted with ParE3* (Fig. 7C). Taken together, our results indicate that mutations in the specificity residues of ParD3 and ParE3 are sufficient to create an orthogonal, interacting protein pair.
Our results indicate that mutational paths leading to a change in ParD specificity tend to pass through promiscuous intermediates (Fig. 5). Thus, we wanted to determine whether mutational paths between the wild-type ParD3-ParE3 and the orthogonal ParD3*-ParE3* systems also pass through promiscuous intermediates, thereby changing the specificity of both proteins without disrupting their interaction. We therefore generated variants of ParE3 containing all possible subsets of the substitutions in ParE3* (32 mutants) and variants of ParD3 containing all possible subsets of the substitutions in ParD3* (4 mutants). We then co-transformed each possible pairing of ParD3 and ParE3 variants (128 pairs total) into E. coli and assessed interaction using the toxicity-rescue assay (Fig. 7D). Interestingly, 90 of the 128 pairs of ParD3 and ParE3 variants were capable of interacting, likely because most (17 of 32) of the ParE3 variants were promiscuous, which we define as interacting strongly with both ParD3 and ParD3* (Fig. 7D).
To determine whether paths between the wild-type and insulated ParD-ParE pairs tend to pass through promiscuous intermediates, we first enumerated the total number of trajectories between these systems. Assuming one residue is changed per step and no reversions are considered, there are 5,040 paths from ParD3-ParE3 to the orthogonal ParD3*-ParE3* pair; of these paths, 1,030 retain functionality at each intermediate step. Strikingly, we found that all of these 1,030 functional paths passed through at least one promiscuous intermediate of ParE3 with an average of five promiscuous ParE3 intermediates per path (Fig. S7C). The prevalence of these promiscuous states may enable the ParD-ParE system to readily evolve a new interaction specificity. An initial broadening of ParE3 specificity enables the movement of ParD3 in sequence space, followed by a narrowing of ParE3 specificity in the final step (Fig. 7E). By contrast, mutational paths in which a substitution in either ParD or ParE yields a "switch-like" change in specificity would, by definition, be broken until a second substitution restores the interaction. Thus, our results support the notion that the coevolution and expansion of the ParD-ParE family occurs through promiscuous intermediates.
Discussion
Mutational trajectories and the coevolution of protein-protein interactions
Interacting proteins coevolve, and the identification of coevolving amino acids in two proteins can often help to pinpoint the residues that mediate their interaction. Such analyses are typically predicated on the idea that a mutation in one protein that disrupts an interaction then drives selection of a compensatory mutation in the partner, thereby restoring the interaction (Fig. 1A). However, this model implies that organisms tolerate (at least transiently) a non-functional, or less functional, interaction, which seems unlikely if the protein-protein interaction is essential for viability. Our results provide a solution to this conundrum, demonstrating experimentally how interacting proteins can coevolve and acquire new specificity by having one of the proteins pass through a promiscuous intermediate (Fig. 1B). For instance, a mutation in an antitoxin can initially broaden its specificity; the toxin can then accumulate a mutation that moves it in sequence space but retains its interaction with the antitoxin. A subsequent substitution in the antitoxin can then narrow its specificity to include the mutated toxin and exclude the original form. The net result is a change in specificity without disruption of the protein-protein interaction, which is critical as a disruption at any step would liberate a toxin that prevents growth and proliferation. This model for protein coevolution involves a minimum of three instead of two mutations but means that the protein-protein interaction is functional at each step. Thus, such mutational trajectories could be entirely neutral but importantly would retain a pairwise-coevolution signature in multiple sequence alignments.
Our systematic identification of ParD3 variants that can antagonize ParE3, ParE2, or both revealed an abundance of promiscuous variants in sequence space that are, on average, more highly connected to other functional variants than are specific variants. Consequently, the mutational trajectories that reprogram the specificity of ParD3 frequently involve promiscuous intermediates (Fig. 5F–G). The high frequency of mutational paths involving promiscuous intermediates was seen when considering transitions in ParD3 from being specific for ParE3 to specific for ParE2, and even more so when considering mutations on both sides of the interface. We assessed the complete set of mutational trajectories between the wild-type ParD3-ParE3 and the orthogonal ParD3*-ParE3* by testing 128 pairwise interactions between all possible ParD3 and ParE3 mutational intermediates. Strikingly, 17 of the 32 ParE3 intermediate variants were promiscuous, or capable of interacting with both the ParD3 and ParD3* variants (Fig. 7). Consequently, all of the functional paths between ParD3-ParE3 and ParD3*-ParE3* involved at least one promiscuous intermediate, with most involving more than five (Fig. 7). Our results thus suggest that promiscuous variants of ParD and ParE are abundant in sequence space and that promiscuity-enabling mutations can facilitate the evolution of new interaction specificities while still using the same set of interfacial residues.
A similar principle may apply to other protein-protein interactions throughout biology, even those not involving toxic proteins. The disruption of a given protein-protein interaction could prevent the execution of an essential cellular function or lead to an unwanted, detrimental interaction with another protein, thus favoring coevolutionary trajectories that retain function at each step. This same principle may also underlie the coevolution of transcription factors and their DNA binding sites. The evolutionary history of a steroid hormone receptor and its recognition element was recently reconstructed including the analysis of a possible ancestral state of the steroid receptor and mutational intermediates separating it from extant states (Anderson et al., 2015). Several of the intermediates were promiscuous and may have facilitated coevolution of the receptor and its recognition element toward a new specificity without disrupting the interaction. However, that study only considered mutational intermediates containing residues present in the ancestral or derived states, and our analyses of the ParD-ParE interface suggest that promiscuous intermediates can also involve substitutions that appear in neither the ancestral nor the derived states.
Like many protein families, toxin-antitoxin systems can expand through duplication and divergence. The duplication of a toxin-antitoxin system could allow one of the protein pairs to wander unconstrained in sequence space toward a new interaction specificity via switch-like paths that involve non-functional intermediates. After a duplication, one antitoxin could accumulate interaction-disrupting substitutions while its toxin is still inhibited by the other antitoxin. The toxin could then subsequently mutate to restore an interaction with the derived antitoxin. However, this scenario assumes that the evolving antitoxin does not, in the intermediate state, interact inappropriately with other proteins, and it assumes that the other antitoxin is produced at sufficiently high levels to inhibit twofold more toxin, i.e. that there is normally a significant excess of free antitoxin, which may not be the case. Determining whether and when switch-like or promiscuous paths are followed will require careful reconstructions of toxin-antitoxin evolution.
High-throughput mapping of protein interaction specificity
Deep mutational scanning via next-generation sequencing is a relatively new approach for interrogating the relationship between protein sequence and function, including folding, enzymatic activity, or the binding of a target protein or RNA (Fowler and Fields, 2014). These studies have begun to reveal the functional degeneracy of proteins by examining all, or nearly all, possible single mutants of a given protein. Similar approaches have also been used to probe subsets of all possible double and higher-order mutants (Melamed et al., 2013), or to systematically probe all possible mutants at a limited set of positions (Podgornaia and Laub, 2015).
Deep mutational scans have been focused primarily on how mutations alter a single function or protein interaction. One study examined the ability of a PDZ domain to interact with both a cognate and non-canonical peptide ligand (McLaughlin et al., 2012), but only queried single point mutants. However, the interaction specificity of a protein is a distributed property of multiple amino acids, and the prevalence of epistasis means that the behavior of multiple mutations is difficult to infer from the properties of the corresponding single mutants. We queried a diverse library of ParD3 variants harboring multiple mutations of key specificity residues against two separate proteins: the cognate toxin ParE3 and the non-cognate toxin ParE2. This focused library approach was possible as the specificity of ParD is largely determined by a small number of interfacial residues (Fig. 3). Our approach yielded a high-density map of the sequence space of ParD3 that underpins its substrate interaction specificity (Fig. 5A–D). From these data, we uncovered the residues in ParD3 most responsible for its selective binding of one toxin over another (Fig. 4F, 4I). We found that three positions (60, 61 and 64) primarily dictate specificity, with substitutions at two sites (61 and 64) sufficient to switch ParD3 from antagonizing ParE3 to ParE2, and substitutions at an overlapping set of sites (60 and 61) sufficient to switch ParD3 from antagonizing ParE3 to ParE3*. As noted, our results also demonstrated the existence of many residue combinations that promote a promiscuous state of ParD3 or ParE3. Mutations that render proteins more promiscuous, with respect to catalytic activities or binding partners, has been noted anecdotally (Aharoni et al., 2005; Bloom and Arnold, 2009), but the prevalence of such states and, importantly, their accessibility from more specific, wild-type states has never been mapped in a comprehensive manner.
By building and screening libraries harboring multiple mutations, our work also sheds new light on protein epistasis and the non-additive relationship of individual substitutions. Epistasis has been well documented but is typically assessed with respect to a single protein function. By contrast, the epistasis documented here for ParD3 pertains to its specificity and interaction with two different proteins, revealing interdependencies that would be missed when considering only a single function. For instance, consider the example in Fig. 6A where ParD3 transitions from the E3-specific residues LWDK to the E2-specific residues LWKL. With respect to antagonizing the toxin ParE3, the two single mutants, LWDL and LWKK, are each functional. However, with respect to toxin ParE2, LWDL is functional whereas LWKK is not, reflecting a non-additive relationship between the two substitutions leading to the double mutant LWKL. This type of epistasis may, like other forms of epistasis, restrict the evolution of ParD-ParE systems, which likely follows mutational paths that involve promiscuous states, as discussed above.
Interaction specificity of toxin-antitoxin systems
The specificity of interactions in bacterial toxin-antitoxin systems had previously been unclear, with some reports indicating that these protein-protein interactions are specific (Fiebig et al., 2010) and others suggesting that TA systems form large, cross-reactive networks (Yang et al., 2010; Zhu et al., 2010). Here, by performing a systematic assessment of interaction specificity for a TA family, we found that ParD antitoxins typically exhibit an exquisite preference for binding to their co-transcribed ParE toxins, forming exclusive, cognate pairs. Of 180 non-cognate pairings tested, we found cross-talk in only 11 cases (Fig. 2) and, importantly, no cross-talk was observed for non-cognate pairs present in the same species.
The high degree of protein-protein interaction specificity observed for the ParD-ParE family is similar to that observed for other large, paralogous protein families (Newman and Keating, 2003; Skerker et al., 2008; Stiffler et al., 2007; Zarrinpar et al., 2003). The specificity of many of these paralogous families has been attributed to selection against detrimental cross-talk (Capra et al., 2012; Zarrinpar et al., 2003), raising the possibility that the ParD-ParE family may be under similar selective pressures. However, the biological rationale for maintaining the specificity of TA systems is unclear, and will require a deeper understanding of the function of these systems in bacterial physiology.
Final Perspective
In sum, our work provides a rationale and molecular basis for how protein interaction specificity can change and how two proteins can coevolve without involving non-functional intermediates. Mutations that produce promiscuity have been described for a variety of proteins, but the frequency of such mutations and their accessibility from more specific states had been unclear. Our results indicate that, at least for ParD3 and likely other proteins, promiscuous mutants are prevalent and easily reached from the wild-type sequence through a single mutation. The prevalence of promiscuous intermediates may facilitate the expansion of toxin-antitoxin systems and, more broadly, other paralogous protein families.
Experimental Procedures
ParD3-ParE structure analysis
For details on the structural analysis of M. opportunistum ParD3 and ParE3, see Extended Experimental Procedures.
Identification of coevolving residues
Coevolving residues in the ParDE family were identified using GREMLIN at http://gremlin.bakerlab.org. Input sequences were ParD3 and ParE3 from M. opportunistum, and we set the number of iterations to four and the E-value cutoff to 1E-04. To identify specificity residues, we isolated all residue pairings that had a scaled coupling score greater than 1.25. To identify supporting residues, we performed the following iterative procedure using a score cutoff of 1.25: (1) identify residues within ParD or ParE that covary with the specificity residues; (2) identify residues within ParD or ParE that covary with either the specificity residues or the supporting residues identified in step (1); (3) repeat step (2) until no new supporting residues are identified.
ParD3 library construction and analysis
For details on construction of the ParD3 library, see the Extended Experimental Procedures. To assess the ability of each ParD3 variant to antagonize different ParE toxins, E. coli cells harboring the ParD3 plasmid library were electroporated with a plasmid containing an arabinose-inducible copy of the ParE toxin. Cells were grown out overnight in 200 ml M9L supplemented with 0.4% glucose and antibiotics. The following day, cells were spun down, washed in 50 mL of M9L, and re-suspended at an OD of 0.03 in 500 mL of M9L supplemented with 100 µM IPTG (to induce the ParD3 library) and antibiotics. Cells were grown out at 37°C with shaking for 100 min, and then ParE toxin expression was induced by the addition of 0.2% arabinose. Cell density was measured every 20 min and samples (50 ml) were taken every 100 min, pelleted, and frozen at −20°C. Competitive liquid growth assays were performed in duplicate. Plasmid DNA was extracted and used as template for PCR (20 cycles) with custom barcoded primers containing Illumina flowcell adaptor sequences. Samples were sequenced on an Illumina HiSeq and then filtered, counted, and converted to fitness values as described in the Extended Experimental Procedures.
Supplementary Material
Acknowledgements
We thank R. Sauer, A. Murray, and the Laub laboratory for discussions and comments on the manuscript. We acknowledge S. Ovchinnikov and C. Bahl for valuable discussions on GREMLIN. This work supported by a National Institutes of Health grant (5R01GM082899) to M.T.L. who is also an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aharoni A, Gaidukov L, Khersonsky O, Mc QGS, Roodveldt C, Tawfik DS. The 'evolvability' of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Anderson DW, McKeown AN, Thornton JW. Intermolecular epistasis shaped the function and evolution of an ancient transcription factor and its DNA binding sites. eLife. 2015;4 doi: 10.7554/eLife.07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Arnold FH. In the light of directed evolution: pathways of adaptive protein evolution. PNAS. 2009;(106 Suppl 1):9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Perchuk BS, Skerker JM, Laub MT. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell. 2012;150:222–232. doi: 10.1016/j.cell.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton KM, Crosson S. A conserved mode of protein recognition and binding in a ParD-ParE toxin-antitoxin complex. Biochemistry. 2010;49:2205–2215. doi: 10.1021/bi902133s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- Fiebig A, Castro Rojas CM, Siegal-Gaskins D, Crosson S. Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin-antitoxin systems. Mol Microbiol. 2010;77:236–251. doi: 10.1111/j.1365-2958.2010.07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallez R, Geeraerts D, Sterckx Y, Mine N, Loris R, Van Melderen L. New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol Microbiol. 2010;76:719–732. doi: 10.1111/j.1365-2958.2010.07129.x. [DOI] [PubMed] [Google Scholar]
- Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. PNAS. 2013;110:15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov DA, Kondrashov FA. Topological features of rugged fitness landscapes in sequence space. Trends Genet. 2015;31:24–33. doi: 10.1016/j.tig.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- Lehner B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011;27:323–331. doi: 10.1016/j.tig.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D, Young DL, Gamble CE, Miller CR, Fields S. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA. 2013;19:1537–1551. doi: 10.1261/rna.040709.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgornaia AI, Laub MT. Protein evolution. Pervasive degeneracy and epistasis in a protein-protein interface. Science. 2015;347:673–677. doi: 10.1126/science.1257360. [DOI] [PubMed] [Google Scholar]
- Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- Yang M, Gao C, Wang Y, Zhang H, He ZG. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS ONE. 2010;5:e10672. doi: 10.1371/journal.pone.0010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Park SH, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003;426:676–680. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- Zhu L, Sharp JD, Kobayashi H, Woychik NA, Inouye M. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J Biol Chem. 2010;285:39732–39738. doi: 10.1074/jbc.M110.163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.