Abstract
Gene regulatory networks are multistable dynamical systems in which attractor states represent cell phenotypes. Spontaneous, noise-induced transitions between these states are thought to underlie critical cellular processes, including cell developmental fate decisions, phenotypic plasticity in fluctuating environments, and carcinogenesis. As such, there is increasing interest in the development of theoretical and computational approaches that can shed light on the dynamics of these stochastic state transitions in multistable gene networks. We applied a numerical rare-event sampling algorithm to study transition paths of spontaneous noise-induced switching for a ubiquitous gene regulatory network motif, the bistable toggle switch, in which two mutually repressive genes compete for dominant expression. We find that the method can efficiently uncover detailed switching mechanisms that involve fluctuations both in occupancies of DNA regulatory sites and copy numbers of protein products. In addition, we show that the rate parameters governing binding and unbinding of regulatory proteins to DNA strongly influence the switching mechanism. In a regime of slow DNA-binding/unbinding kinetics, spontaneous switching occurs relatively frequently and is driven primarily by fluctuations in DNA-site occupancies. In contrast, in a regime of fast DNA-binding/unbinding kinetics, switching occurs rarely and is driven by fluctuations in levels of expressed protein. Our results demonstrate how spontaneous cell phenotype transitions involve collective behavior of both regulatory proteins and DNA. Computational approaches capable of simulating dynamics over many system variables are thus well suited to exploring dynamic mechanisms in gene networks.
Introduction
Multistable dynamics in gene regulatory networks has been proposed as the basis for the existence of diverse cell types (1, 2, 3). In this view, the biochemical interactions encoding gene networks give rise to complex, nonlinear expression dynamics. Distinct gene expression states—cellular phenotypes—are self-stabilizing attractors of the dynamical system. Transitions between attractors correspond to critical cellular processes, including developmental fate decisions (4, 5), cellular reprogramming (3), phenotype switching (6), and carcinogenesis (7). As such, there is interest in characterizing the global dynamics of complex multistable gene networks to gain insight into the relative stability of cell states and the processes by which transitions between states can occur.
Over the past 20 years, the inherent noisiness of gene expression has been amply demonstrated. Stochastic biomolecular fluctuations can significantly impact dynamics of expression due to small-number effects (8, 9, 10). Positive feedback, which is common in regulatory networks, can amplify these fluctuations, giving rise to large-scale changes in gene expression programs. As such, intrinsic biomolecular noise is thought to underlie phenotypic variability in cell populations (11, 12, 13, 14). Stochastic processes can also allow for spontaneous noise-induced transitions between metastable gene expression states. This stochastic state switching can be advantageous by priming cells to diversify according to alternative developmental programs (15, 16) or by promoting survival of microorganisms or cancer cells in fluctuating environments (17, 18, 19). These findings highlight the need to understand how cellular networks achieve—or remain resistant to—noise-induced switching.
Common approaches to modeling gene networks formulate system dynamics as a set of stochastic biochemical reactions encompassing, for example, transcription-factor binding and unbinding to regulatory sites on DNA, gene transcription and translation, and degradation/dilution of mRNA and protein molecules. The associated master equations are amenable to Monte Carlo simulation (e.g., by the Gillespie algorithm (20)), which can exactly account for intrinsic stochastic fluctuations and molecular discreteness. Many studies of gene regulatory networks employ continuum approximations and dimensionality reduction to aid in the analysis of complex dynamics. For example, occupancies of binding sites on DNA are often assumed to be in quasiequilibrium with gene expression levels (i.e., concentrations of expressed proteins), which leads to nonlinear Hill function expressions for gene interactions (21, 22, 23, 24). This assumption stems from a separation of timescales, when rates of binding and unbinding of regulatory proteins to DNA sites are fast relative to the rates of protein synthesis and degradation. This kinetic regime has been termed the adiabatic limit, in analogy to electron transfer reactions (25).
Recent studies have focused on gene network dynamics in cases where this separation of timescales does not apply (4, 26, 27, 28, 29). In eukaryotic regulation, highly complex regulatory processes such as chromatin remodeling (in contrast to simple protein binding) drive fluctuations in gene expression. Such processes occur on relatively slow timescales; slow fluctuations in chromatin structure have been identified as a major source of gene expression noise in eukaryotes (9, 10). Although knowledge of the detailed biochemical reactions underlying such higher-order regulation is generally lacking, theoretical studies have explored how different kinetic regimes contribute to the stability of gene expression states. Notably, cell-states have been found to be most stable (most resistant to noise-induced switching) in the adiabatic limit, where fast kinetics at DNA regulatory sites allows rapid response of gene expression states to local concentrations of regulatory proteins (28, 30). This has led to the hypothesis that a weakly adiabatic kinetic regime allows for relatively frequent stochastic state switching, and thus developmental plasticity, in pluripotent stem cells (4, 26, 31).
In addition to exploring the principles governing the stability of gene expression states, recent theoretical studies have predicted transition paths (or reaction coordinates) of stochastic state switching in regulatory networks. These paths describe the most probable stepwise changes over multiple species in the network (e.g., the time-dependent changes in gene expression patterns) that occur as the system moves from one metastable state to another. Approaches for calculating transition paths have largely adopted analytical or numerical methods based on large deviation theory from chemical physics (32, 33, 34, 35, 36), related path integral approaches (4, 5, 27, 37), or Monte Carlo simulations (31, 38, 39). Valuable insights into the dynamics of noise-induced switching in gene networks have emerged from these studies, including the significance of nonequilibrium phenomena (e.g., the irreversibility of switching paths) in gene network dynamics.
Gaining quantitative insight into stochastic state switching in gene networks is mathematically and computationally challenging. Stochastic fluctuations, overlapping temporal scales, and large numbers of interacting species can preclude the use of analytical approaches. Approximation methods may not accurately predict switching mechanisms driven by intrinsic biochemical fluctuations. For example, mean-field approximations have been found to distort multistable landscapes in some biochemical networks (40, 41, 42, 43). On the other hand, brute-force Monte Carlo simulations can be plagued by inefficiency because state switching may be a rare event: long waiting times between transitions mean that simulations capture few, if any, switching events.
Numerical rare-event sampling algorithms provide an alternative approach by preferentially simulating events of interest without modifying the underlying system dynamics (44, 45, 46). Moreover, these algorithms aid interpretation of large amounts of noisy simulation data by providing automated means of extracting essential dynamical properties (47). These types of approaches have been adopted for the study of biochemical networks, including circadian rhythms (48), enzymatic cycles (46), and genetic switches (38). So-called string-based sampling algorithms (45, 49, 50) are especially well suited to discovery of transition paths for rare events in complex dynamical systems.
In this study, we employed a recently developed weighted-ensemble string (WE-string) simulation method (50) to study noise-induced switching in a set of genetic toggle-switch networks. By efficiently predicting transition paths involving collective changes over all species in the biochemical network, the approach uncovered new insights, to our knowledge, into how spontaneous switching depends on fluctuations of both protein number and DNA-binding occupancies. In particular, we found that the kinetics of protein binding and unbinding to DNA regulatory sites controls the mechanism of switching by determining whether the switching event is driven by these binding/unbinding events, or is instead driven by fluctuations in protein copy numbers. Our results potentially inform strategies to perturb gene network dynamics to stabilize or destabilize particular cellular states, or to drive a desired cellular transition.
Materials and Methods
Modeling and simulation approach
Toggle-switch models
To explore the capability of the WE-string simulation method to predict transition paths in gene networks, we applied it to a set of related chemical network models for the genetic toggle switch. In this ubiquitous motif, two genes mutually repress one another through the action of their protein products, giving rise to a bistable gene expression pattern. Described in detail for the lysis/lysogeny decision of bacteriophage λ (51), the mutual repression motif is recognized as underlying a wide variety of cellular decisions, from embryonic development (52) to hematopoiesis (53) to specialization of T-cell subsets (54). The discovery of this motif paved the way for theory-based approaches to the design of synthetic gene circuits (23).
Quasipotential landscape
The stability of cell states can be expressed by a potential landscape over the gene network state space (5, 55, 56). We use a definition for a quasipotential function based on the stationary probability distribution: the quasipotential φ is taken to be φ = −ln(PS), where PS is the stationary (steady-state) probability over the state space (Fig. 1 A). Defined in this way, areas of state space with low potential are most probable, and therefore most stable. However, this quasipotential does not completely describe dynamics in nonequilibrium open systems such as biochemical networks (5, 35, 45).
Figure 1.
(A) (Top) The quasipotential surface, φ = −ln(PS), for a bistable gene network. States with lower potential (bluer colors) are more stable (color scale applies to the whole figure). (Bottom) A single stochastic simulation trajectory that switches from the SA attractor basin (high expression of protein a encoded by gene A) to the SB basin (high expression of protein b by gene B). φ is projected onto subspaces representing a and b protein copy numbers. (B) The simulation method predicts the most probable transition path by utilizing a string connecting two states of interest to adaptively partition the state space and sample short stochastic trajectories initialized in each partition. (Top) The simulation is initialized with a string representing a guess path for the transition (the corresponding Voronoi partitions are plotted underneath). (Bottom) After the simulation has converged, the string (and corresponding partitions) indicates the most probable transition path. (C) The predicted transition path contains information on all network species; for the toggle-switch network, it contains information on protein copy numbers and gene activities (DNA occupancies). (Top) Transition path for the switch, projected onto a, b, and Aon subspaces, where Aon is the probability that the regulatory site for gene A is unbound, rendering it on (active). (Bottom) Same transition path projected onto a, b, and Bon subspaces, where Bon is the probability that the regulatory site for gene B is unbound.
Transition-path sampling algorithm
Predicting the most probable switching path by brute-force simulations is inefficient, because switching events are infrequent, and individual trajectories may be highly variable (Fig. 1 A and Figs. S1 and S2 in the Supporting Material). To predict the most probable path between two states of interest, the WE-string algorithm preferentially samples dynamics in the transition region (a low-probability area of state space), statistically merging information from many short trajectories. The algorithm achieves this by combining WE rare-event sampling (57) with a string-based adaptive discretization method for partitioning the state space (58). Adelman and Grabe recently introduced the method (50) and showed that it compares favorably to other path sampling methods in terms of efficiency, and could successfully sample dynamics in the space of many collective variables in an application to protein conformational change.
Rare-event sampling methods typically involve discretizing the state space of the system to access long-timescale kinetics on the basis of short-timescale trajectories in each region (59). The computational expense of discretizing a system evenly over N dimensions scales exponentially with N, which quickly becomes intractable for gene networks. The state space is naturally defined in terms of copy numbers and conformational/binding configurations of all biomolecular species, but even the smallest two-gene network (i.e., the toggle switch) has more than two species (including DNA promoter or regulatory binding sites, mRNA transcripts, protein products with multiple configurations, and so on, depending on the level of detail of the model). This challenge can be addressed by sampling along a single, relevant order parameter, such as a progress coordinate for the transition of interest, as pioneered by Allen and ten Wolde with the forward flux sampling (FFS) method (60). Alternatively, string-based methods adaptively partition space along a string: a one-dimensional path winding through a high-dimensional space, which connects two states of interest. This approach enables us to sample collective dynamics involving multiple system variables, without prior knowledge of an order parameter or the nature of dynamics in the transition region (61).
The string is defined by a set of Nstr evenly spaced nodes, or points in the full state space; these nodes define the centers of Nstr regions. All points in the space lie in a region that is defined by the nearest string node. Thus, the string nodes are the generating points of Voronoi polyhedra. The string is initialized as a guess path connecting two states (Fig. 1 B). For each iteration of the simulation, multiple weighted stochastic trajectories (replicas) in each region are simulated for a time τ. After each iteration, the WE method is used to statistically combine and duplicate weighted replicas as needed to ensure unbiased sampling of the partitioned space. After Tmove iterations of τ, the string position is updated by moving the string nodes toward the average position of replicas in each region collected from the last Tavg iterations of τ, using a procedure of moving, smoothing, and reparameterization. Details of simulation parameters and algorithm implementation are in the Methods section. A more detailed description of the general algorithm is in Adelman and Grabe (50).
Because the string is defined as a list of points in the system state space, the converged string contains information about the probable stepwise changes along all network dimensions over the course of the transition. For our model toggle-switch network (Fig. 2), this means that the predicted transition path includes information about both gene expression levels (copy numbers of protein products a and b) and gene activity states (i.e., likelihood of gene states Aon and Bon), which reflect the occupancy states of the DNA-binding sites (Fig. 1 C).
Figure 2.
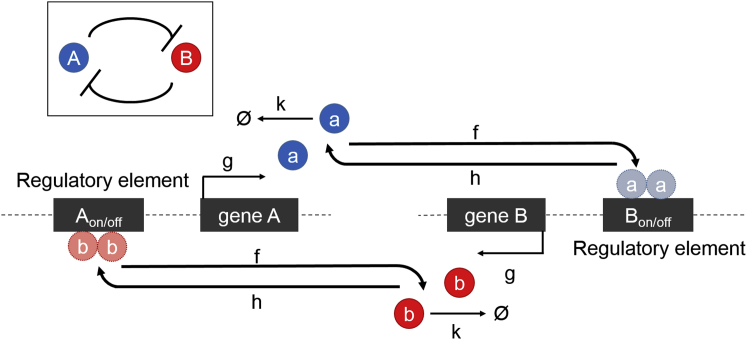
Schematic diagram of biochemical reactions in the genetic toggle switch network (the inset shows the basic network motif of two mutually repressing genes). Gene A codes for repressor protein a (blue) and gene B codes for protein b (red). Each gene is controlled by a regulatory element (e.g., the promoter). When the regulatory element is unbound, the gene is on (active), or free to express protein. When it is bound by a homodimer of the opposing repressor, the gene is off (inactive). To see this figure in color, go online.
Methods
Toggle-switch reaction network
We considered two previously studied variants of the toggle switch, which we term the base network (40) and the network with explicit dimerization (Fig. 2 and Eqs. 1–3) (38). In both variants, protein dimers act as repressors of the other competitor gene. The switch is symmetric with respect to the rate parameters governing the behavior of each of the two genes, A and B. Translation and transcription are subsumed into a single biochemical reaction for synthesis of the two encoded proteins, a and b. The two genes exist in one of two occupancy states, giving rise to binary regulation (Aon/off, Bon/off), where only the on states contribute to protein synthesis. We furthermore studied the general and exclusive versions of the switch; the exclusive switch assumes that the a2 and b2 repressors competitively bind to a single regulatory site that controls both genes; therefore, both genes cannot simultaneously be in the off state (i.e., the doubly repressed state is disallowed (equations are in the Supporting Material)).
Protein synthesis and degradation reactions are given by
| (1) |
In the base network, dimerization is neglected, and gene repression is represented by
| (2) |
These reactions give rise to an overall rate of gene inactivation of , where x is the repressor protein encoded by the competing gene. In the network with explicit dimerization, the above reactions are replaced by
| (3) |
Rate parameters for the reaction network are given in Table 1. The effect of DNA-binding kinetics was studied by varying the rates h and f (binding and unbinding, respectively, of transcription factors to DNA). The binding equilibrium constant was maintained at a constant value, such that the locations of the two stable states are preserved.
Table 1.
Rate parameters and calculated switching rates, kAB, for all studied genetic toggle-switch variants
| Network Parameters |
kAB [t−1] (General) |
kAB [t−1] (Exclusive) |
||||||
|---|---|---|---|---|---|---|---|---|
| Mean | Std. | Mean | Std. | |||||
| Base Network | ||||||||
| h [k−1] | f [k−1] | g [k−1] | k [t−1] | |||||
| I | 102 | 104 | 80 | 1 | 1.9 × 10−6 | 2 × 10−7 | 5.8 × 10−10 | 6 × 10−11 |
| II | 10−1 | 10 | 80 | 1 | 3 × 10−4 | 3 × 10−4 | 3 × 10−4 | 3 × 10−4 |
| III | 10−2 | 1 | 80 | 1 | 7 × 10−3 | 7 × 10−3 | 3 × 10−3 | 3 × 10−3 |
| Explicit Dimerization | ||||||||
| h [u−1] | f [u−1] | g [u−1] | k [u−1] | d [u−1] | u [t−1] | |||
| I | 8.89 × 103 | 105 | 80 | 8.944 | 1 | 1 | 3.3 × 10−10 | 7 × 10−11 |
Included are parameters for base and explicit-dimerization networks for both general and exclusive switch versions. Rate parameters correspond to the schematic in Fig. 2 (see Methods): h and f, binding and unbinding rates, respectively, of proteins to DNA; g, protein synthesis; k, protein degradation; d and u, binding and unbinding rates, respectively, of monomer proteins to form homodimers. All network parameters are reported in units of [k−1] (the inverse degradation rate) in the base network and [u−1] (the inverse dimer unbinding rate) in the explicit-dimerization network. Parameter set I corresponds to the adiabatic regime, where the propensities of protein binding to and unbinding from DNA ( and f, respectively) are large relative to g and k. Std., standard deviation.
Calculation of quasipotential
We calculated PS and φ by expressing the chemical master equation (CME) for the network in matrix form: , where A is the transition-rate matrix over a truncated state space , and solving for the eigenvector associated with the zero eigenvalue (62) using MATLAB (63). For parameter regimes giving relatively short waiting times between switching events (i.e., parameter sets II and III), we also obtained PS from a single long Gillespie simulation trajectory, using BioNetGen (version 2.2.2) (64). The quasipotential landscapes obtained from each method were indistinguishable.
Transition-path simulation
The WE-string sampling method was adapted from Adelman et al. (50). The procedure for string smoothing, parameterization, and separate sampling of forward and backward paths (by separating replicas according to their most recently visited basin) was adapted from Dickson et al. (48). String ends were fixed in the two stable attractor basins, SA and SB, corresponding to states with high gene A expression and high gene B expression, respectively. SA and SB were defined as hyperspheres with unit radius in N-dimensional state space (base network, N = 4 {a, b, Aon/off, Bon/off}; explicit-dimerization network, N = 6 {a, b, a2, b2, Aon/off, and Bon/off }). The basin centers were found by identifying the minima of the quasipotential for each parameter set. The number of string nodes, Nstr, was 20, and the number of replicas in each region (Nrep) was 150. Definitions and values of additional simulation parameters are given in Table S1. Switch progress is defined as the normalized distance along the converged string. That is, for the forward path, the progress at the ith string node is defined by , where Di is the Euclidean distance to the ith node from the center of SA, and DAB is the total distance along the string. The simulation was determined to have converged after the position of the string remained stable over >1000 string movements. Convergence was determined from the root mean-squared difference between the current node positions and the running average of the previous 100 positions (Fig. S3). Sampling code was written in MATLAB and Gillespie trajectories were simulated using BioNetGen (64).
Calculation of switching rates
Switching-rate constants, kAB, were estimated from brute-force Gillespie simulations when possible (the faster switching regime: parameter sets II and III) or WE sampling in cases where capturing switching events from brute-force simulations was intractable (the rare-switching regime: parameter set I). The network parameters are symmetric, such that . For the brute-force simulations, kAB was estimated by inverting the mean first-passage time of transitions from 1000 trajectories. The WE rate estimation followed (65), using a linear partitioning of state space along the progress coordinate λ with bins of unit length. λ is given by λ = na − nb, where na = a + 2Boff, nb = b +2Aoff (general switch) or na = a + 2a2 + 2Boff, nb = b + 2b2 + 2Aoff (exclusive switch); this is the same progress coordinate as used previously (38, 60). The simulation time step was chosen to be the same as that of the WE-string simulation for the corresponding parameter set; 150 replicas were simulated in each bin.
Validation of transition-path simulation
Validation of switching mechanisms predicted by the transition-path simulations was done by comparison to brute-force switching trajectories (where possible), and by committor analysis (66). Successful switching trajectories harvested from brute-force simulations were binned according to the progress coordinate λ. Additional simulations assessed committor probabilities along the converged strings: these reflect the probability that simulation replicas in a given region will next commit to the final target state rather than returning to their most recently visited basin.
Results
Stability of phenotypes depends on DNA-binding kinetics
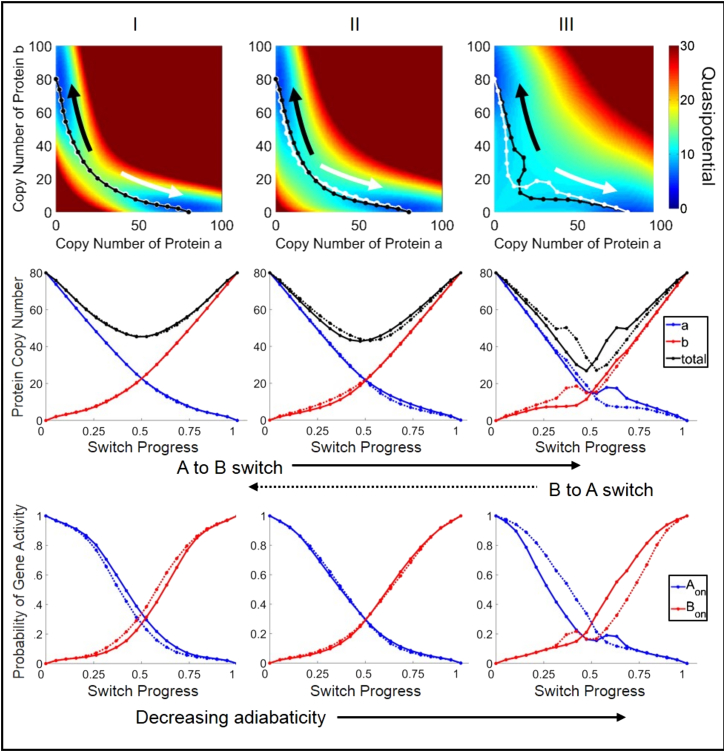
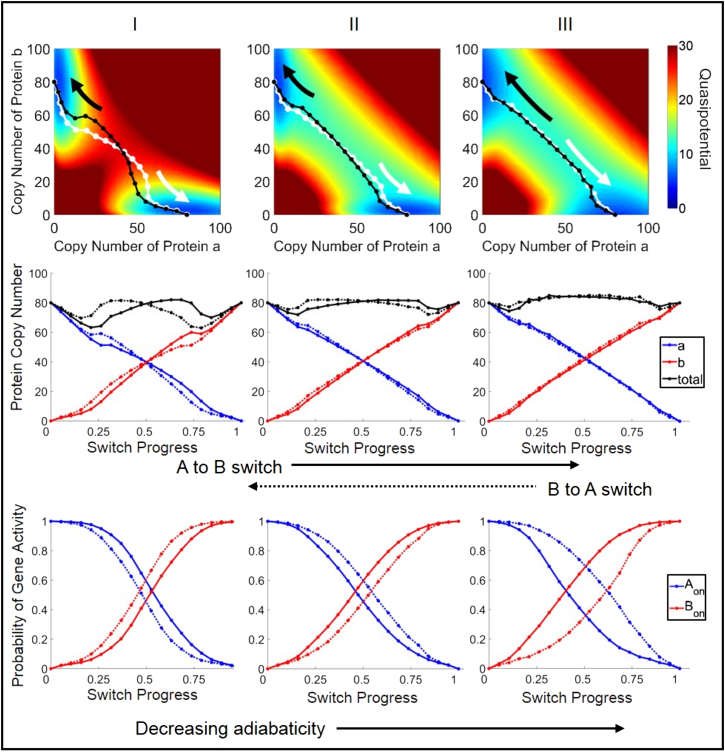
We calculated the quasipotential landscape for the genetic switch, and varied the parameters governing DNA-binding/unbinding kinetics between the adiabatic (fast kinetics (Figs. 3 and 4, parameter set I)) and nonadiabatic (slow kinetics (Figs. 3 and 4, parameter set III)) regimes. By varying rates of unbinding (f) and binding (h) while maintaining their constant ratio, , the bistability and locations of the attractor basin centers are preserved, but the barrier height separating the two states changes (Figs. S4 and S5). Our results show that the barrier height decreases with decreasing adiabaticity, in agreement with previous results (40). This difference is also reflected in the calculated rate constants, kAB, for spontaneous switching from the attractor state SA to SB (Table 1). The rate constant for spontaneous switching between metastable states has been identified as a measure of stability or robustness of gene expression states to noise. In agreement with previous studies (4, 26, 28, 30, 40), we find that the rate of switching slows with increasing adiabaticity, indicating that gene expression states are more stable with faster DNA-binding kinetics. This trend was preserved for all variants of the switch (Table 1). For parameter set I, kAB is much lower for the exclusive than for the general switch, whereas the switching rates for the two variants are of the same order of magnitude for sets II and III. Interestingly, the switching rates are nearly identical for parameter set II.
Figure 3.
Quasipotential surfaces and predicted transition paths for the base general-toggle-switch network. (Left to right) Network rate parameters with decreasing adiabaticity (slower DNA-binding kinetics) (see Table 1 for parameters). (Top). Transition paths are superimposed on the two-dimensional projection of the quasipotential surface. The black and white paths illustrate the forward and backward transitions, respectively. (Middle) Transition paths are plotted as protein copy numbers versus switch progress (protein a, blue; protein b, red; total protein number (a + b), black). Switch progress is defined as the normalized distance along the transition path (see Methods). Forward switching is displayed as solid lines and backward as dotted lines. (Bottom) Transition paths are plotted as gene activities versus switch progress. The activity of gene A (Aon, blue curve) is the probability of the regulatory site of gene A being unbound, which renders it active. This is similar for Bon (red curve).
Figure 4.
Quasipotential surfaces and predicted transition paths for the exclusive-toggle-switch network, with competitive binding of repressors to regulatory sites. All definitions are the same as in Fig. 3. (Left to right) Network rate parameters with decreasing adiabaticity (slower DNA-binding kinetics) (see Table 1 for parameters). (Top). Transition paths are superimposed on the two-dimensional projection of the quasipotential surface. (Middle) Transition paths are plotted as protein copy numbers versus switch progress. (Bottom) Transition paths are plotted as gene activities versus switch progress.
Sampling algorithm finds a transition path for the toggle switch
Transition paths of noise-induced switching between gene expression states were predicted by performing WE-string sampling of stochastic simulations. Transition paths were predicted for both forward and backward switching events and were compared to the numerically calculated quasipotential landscapes for the three parameter sets. Although the WE-string simulations require no foreknowledge of the underlying potential in the transition region, the paths converge to the relatively low-potential (i.e., high-probability density) transition tube connecting the two states (Figs. 3 and 4). In addition to mapping out the region of state space traversed by successful transitions, committor analysis (Figs. S9 and S10 and Methods) indicates that for most studied parameter sets, the string also accurately reflects the true reaction coordinate for the transition. Progress along the string corresponds to the most probable step-by-step changes undergone as the system progresses from one basin to another. However, slight discrepancies exist for the general switch in the nonadiabatic regime, likely due to a known limitation of string-based sampling methods in cases of wide transition tubes (58). Predicted transition paths were robust to differences in simulation initial conditions (guess paths) (Fig. S6).
Transition paths reveal additional dynamic features and nonequilibrium phenomena
The predicted transition paths also reveal that the system dynamics is not governed solely by the topography of the potential surface. The paths reveal additional features, including pathway oscillations and nonoverlapping forward and reverse paths, in agreement with previous studies (5, 38). These features can be seen by comparison to calculated minimum energy paths (Figs. S7 and S8), which reflect only gradient dynamics over the projected quasipotential surface. The degree to which the forward and reverse paths diverge depends both on the parameters and on the choice of subspace on which the system dynamics is projected. For example, when projected onto the protein copy number subspace, the paths show more divergence in the nonadiabatic regime than in the adiabatic regime for the general switch (Fig. 3), whereas the opposite is true for the exclusive switch (Fig. 4). For the exclusive switch in the nonadiabatic regime (Fig. 4, parameter set III), there is clear divergence of the forward and reverse transition paths in the subspace of DNA-occupancy states (gene states), yet the transition paths are nearly superimposed in the protein subspace (Fig. 4.).
Protein number oscillations are observed in the adiabatic regime for the exclusive switch (Fig. 4, parameter set I). This motion is not echoed in the DNA binding occupancies, suggesting that it results solely from the birth-death reactions for protein expression. In the nonadiabatic regime for the general switch (Fig. 3, parameter set III), oscillatory motion in the proteins is echoed in the DNA-binding occupancies (gene activities), suggesting that it could result from previously described eddy currents in gene networks, where slow transcription factor binding/unbinding events drive cyclic dynamics of protein expression in the nonadiabatic limit (26, 30, 37). However, as discussed above, the detailed features of the string for the nonadiabatic general switch may not reflect dynamics along the true reaction coordinate.
Influence of DNA-binding kinetics on the switching mechanism
The predicted transition paths for the toggle switch reveal that the switching mechanism is altered by the DNA-binding kinetic parameters. This is seen by considering the switch progress separately in terms of either protein copy numbers or gene activities: the switch progress in these subspaces is not synchronized, and the degree to which they are asynchronous depends on the DNA-binding kinetics. In this symmetric system, when the system reaches the separatrix at (a = b), the transition can be considered to be half-complete. This crossing corresponds closely to the half-distance point along the transition path (switch progress = 0.5) (Figs. 3 and 4). Under a quasiequilibrium assumption, the gene activities depend directly on the levels of protein expression according to and . As such, when protein expression from the two competing genes reaches equivalence, the genes would then also have equal probabilities of being in the active (unbound) states, giving (Aon = Bon). However, the transition paths reveal that this crossing is not necessarily reached simultaneously in both subspaces. Similar trends are seen in the parameter dependence for the different switch variants: in the adiabatic regime, the gene activities reach (Aon = Bon) after the protein numbers reach (a = b) (Figs. 3 and 4, left column), whereas in the nonadiabatic regime, the gene activities equalize before the proteins (Figs. 3 and 4, right column). In other words, when DNA-binding kinetics is slow, stochastic binding and unbinding events play a larger role in driving the switch toward completion, and protein expression follows. Conversely, when DNA-binding kinetics is fast, birth-death fluctuations in protein numbers drive the switch. This trend was preserved for additional studied parameter sets (Figs. S11 and S12) and also appeared in averaging of switching trajectories harvested from brute-force simulations (Figs. S1 and S2).
The simulations predict that the exclusive and general switch variants (see Methods) progress through different transition states. Our results are in agreement with the previous finding (38) that the general switch transitions through a state in which both genes are repressed, which is also reflected in the decrease in overall expression of proteins (Fig. 3, total expression curves). For the exclusive switch variant, the transition progresses through a region where each gene has a nearly equal probability of being on or off. Additionally, the total protein number when the switch is half-complete (switch progress = 0.5) remains high after an initial decrease (Fig. 4, total). These transition states clearly reflect the difference in the DNA occupancy states available to each toggle-switch variant. In the general switch, all combinations of bound and unbound states at the regulatory sites of the two genes are allowed (i.e., Aon/Bon, Aon/Bon, Aoff/Bon, Aoff/Boff), whereas in the exclusive switch, the Aoff/Boff state is not accessible due to competitive binding in the case of overlapping regulatory sequences. This scenario is the limiting case of a reduced binding rate of a repressor if the competing repressor is already bound to the DNA.
Advantage of the transition-path simulation method for multidimensional networks with rare switching
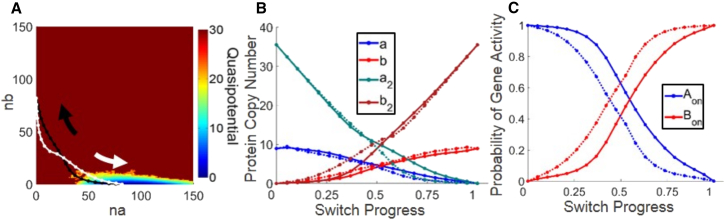
We applied the sampling algorithm to the extended network with explicit dimerization (Methods). The parameters of the extended network were chosen such that the base and extended networks give identical total protein expression at steady state, in terms of the variables na and nb (see Methods and the Supporting Material). The predicted transition paths showed qualitatively similar switching mechanisms for the base and extended networks. In particular, the trend of asynchronous switching in protein numbers and gene activities was preserved in the network with explicit dimerization, with gene activities lagging behind protein numbers in the adiabatic regime (Fig. 5). For this parameter set, it was not possible to access the full quasipotential surface: because the size of the state space increases exponentially with dimensionality (i.e., number of species), the addition of two more molecular species (the dimers a2 and b2) renders the transition rate matrix too large for straightforward numerical calculations (see Methods). This curse of dimensionality does not limit Gillespie simulations; however, brute-force simulation of 109 time steps failed to capture a single switching event (Fig. 5 A) due to the low probability of switching (kAB = 3.3 × 10−10, Table 1). Our results thus demonstrate that the WE-string sampling method is particularly advantageous for studying transition paths for networks with several or more molecular species and long waiting times between events of interest.
Figure 5.
Transition paths predicted for the exclusive switch with explicit dimerization, parameter set I (see Table 1), and incomplete quasipotential surface. (A) Predicted transition paths for the (black) and (white) transitions. The two-dimensional projection shows the quasipotential surface obtained from a brute-force Gillespie simulation of 109 time steps initialized in SA. The long simulation fails to produce a single switching event, giving rise to a one-sided, incomplete quasipotential surface. Despite the rarity of switching for this network (kAB = 3.3 × 10−10 (Table 1)), the simulation method remains capable of predicting the most probable transition path. (B) Transition paths are plotted as protein copy numbers versus switch progress. (C) Transition paths are plotted as gene activities versus switch progress.
Discussion
In this work, we applied recently developed stochastic simulation methods to the study of gene regulation dynamics to predict switching mechanisms in a ubiquitous bistable gene network. Our simulations recapitulate several general aspects of gene network switching, including the increased stability of cell states to noise-induced switching with fast DNA-binding/unbinding kinetics and evidence of nonequilibrium phenomena in switching dynamics, in agreement with previous studies (4, 5, 28, 30, 38, 40). Our simulations also uncovered new insights, to our knowledge, into stochastic phenotype switching. In particular, we found that switching progresses asynchronously in the separate subspaces of protein copy numbers or DNA occupancies (i.e., gene activities). We observed a trend wherein the gene activities lagged behind the protein numbers during the switching transition in the fast-DNA-binding (adiabatic) kinetic regime and switched ahead of the proteins in the slow-DNA-binding (nonadiabatic) regime. Thus, our results reveal that the detailed mechanisms (and overall rates, kAB) of spontaneous switching depend on the network parameters. Our findings suggest that protein copy-number fluctuations play a more significant role than DNA-site-occupancy fluctuations in driving the switch toward completion in the adiabatic regime, whereas the converse is true in the nonadiabatic regime. Our study also demonstrates that rare-event sampling algorithms, in particular string-based methods (49, 50), are well-suited to the discovery of transition paths that link metastable states in gene networks.
Stochastic state switching in gene networks is thought to be a mechanism driving phenotypic heterogeneity in genetically identical cell populations. This type of nongenetic heterogeneity induced by transient switching events appears to play a role in diverse biological contexts, including development and disease. In stem cell networks, probabilistic differentiation has been linked to stochastic switching of pluripotent cells between substates, distinguished by different levels of expression of key transcription factors (67, 68, 69, 70). Stochastic state switching is also thought to allow bacteria to switch into and out of antibiotic-resistant states (17), virus-infected cells to switch into and out of latency (12), and cancer cells to switch into and out of chemotherapy-resistant states (19). Identification of the mechanisms by which these switching events occur can provide a route to discovery of novel strategies for cellular engineering (e.g., stem cell reprogramming) and drug treatment. However, mechanistic studies of switching can be experimentally challenging, particularly when only a fraction of cells in a population is poised to transition between states (71). As such, quantitative models coupled with the computational approaches presented here could provide important insight into underlying mechanisms, e.g., by predicting patterns of gene expression associated with cells that are in transit between phenotypic states.
Chromatin remodeling contributes to gene expression noise by stochastically transitioning individual genes between on (active) and off (inactive) states (9, 10). These transitions can occur on timescales that are of the same order as or longer than mRNA and protein lifetimes (72). Though the toggle-switch model studied here is highly simplified, the slow-DNA-binding (weakly adiabatic or nonadiabatic) kinetic regime broadly captures this complex epigenetic regulation. The relative instability of phenotypic states in this regime has been hypothesized to underlie developmental plasticity of pluripotent stem cells (4, 26). Our results suggest that stochastic fluctuations at the level of the individual promoter sites, rather than changes in local regulatory protein concentration, play a critical role in driving the spontaneous, global switching events that poise pluripotent cells to choose probabilistically between alternative lineages.
The simulation method employed in this study simultaneously tackles two challenges encountered in modeling gene network dynamics: the rare-event problem (switching between gene states may occur rarely, giving rise to impossibly long simulation convergence times) and the curse of dimensionality (switching requires coordinated changes involving many species in the network). By applying the WE-string method to different toggle-switch variants in multiple kinetic regimes, we demonstrate its potential flexibility for capturing stochastic dynamics in different gene network systems. We found that a major advantage of the method is its ability to simultaneously track dynamics of both protein products and DNA occupancy states (i.e., promoter states), which together control global gene network dynamics.
In previous studies of transition paths in gene networks where DNA occupancies were treated as being in quasiequilibrium with protein numbers, it was implicitly assumed that the dynamics in the two network subspaces are identical (32, 33, 34, 35, 36).Our results show that even in the adiabatic regime, where a separation-of-timescales assumption is justified, the switching dynamics of the gene activities and protein numbers have distinct features. Theoretical methods have been developed recently for studying noise-induced transitions in gene networks, which can account for nonadiabaticity (slow DNA binding) by path integral approaches based on approximations to the CME (4, 37). In this study, we explore an alternative approach in direct sampling, which circumvents the need for approximations to the CME, can be used in conjunction with available stochastic simulation software packages (64), and potentially scales more easily to other types of biochemical networks with many species.
The application of rare-event sampling techniques to study switching dynamics in gene networks was pioneered previously with the FFS method (60). Morelli et al. (38) carried out a detailed study of the toggle switch by FFS; our network model is largely based on theirs, although we studied parameter regimes commensurate with those of Sasai et al. (25), producing larger average protein numbers. The transition states we found for the general versus exclusive switch variants are in qualitative agreement with that study (38), although direct quantitative comparison of our results to theirs is not possible because of the different parameter regimes and because their study did not focus explicitly on the mechanistic dependence on adiabaticity.
In principle, FFS and string-based sampling methods can access the same information; the advantages and disadvantages of each method, making them potentially complementary, have been discussed previously (61). FFS samples dynamics along a single order parameter, which must be defined a priori, and which tracks progress between initial and final states. The choice of this parameter is nontrivial, and a poor choice can lead to computational inefficiency, although statistical methods to optimize the order parameter have been developed (73). Use of a single order parameter necessarily results in loss of information about dynamics along separate system coordinates; the progress coordinate λ is much more sensitive to protein copy numbers than to DNA-occupancy states. Adaptive string-based methods were developed to circumvent this problem and access collective dynamics in spaces of many order parameters (45). Because progress through a transition is measured by successive string nodes, each of which represents a configuration in the full state space, this approach offers a more intuitive description of dynamics in gene networks with many species. Moreover, the approach enabled us to directly isolate separate contributions of protein numbers and DNA-occupancy states to the switching mechanism, which in turn revealed the mechanistic dependence on adiabaticity.
However, string methods also have potential limitations. Convergence of the string to the most probable transition path may not occur in systems with highly complex quasipotential landscapes with several or more local minima, so extension of the method to more complex gene networks remains to be explored. The transition path predicted by the converged string may not correspond exactly to the true chemical reaction coordinate for some networks (as defined by committor probabilities (66)) due to the difficulty of accurately partitioning wide transition tubes (58). Our results show that despite these potential limitations, the WE-string method was able to resolve switching dynamics over a wide range of parameters. Even in the nonadiabatic limit, the major mechanistic predictions (regarding the nature of the transition state and the asynchronous switch progression in the protein versus gene activity subspaces) were found to be robust by comparison to brute force trajectories. Our findings suggest that the method may be particularly useful for exploring dynamics of gene networks where little information (such as a suitable progress coordinate) is available a priori and may prove particularly powerful in combination with other, complementary rare-event sampling approaches.
The code used to generate the data in this article will be accessible from https://github.com/Read-lab/WE-string-GRN.git.
Acknowledgments
We thank the administrators of the University of California, Irvine High-Performance Computing cluster.
Editor: Leah Edelstein-Keshet.
Footnotes
Supporting Materials and Methods, twelve figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00870-X.
Supporting Citations
Reference (74) appears in the Supporting Material.
Supporting Material
References
- 1.Kauffman S. Control circuits for determination and transdetermination. Science. 1973;181:310–318. doi: 10.1126/science.181.4097.310. [DOI] [PubMed] [Google Scholar]
- 2.Huang S., Eichler G., Ingber D. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 2005;94:128701. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- 3.Huang S. Reprogramming cell fates: reconciling rarity with robustness. BioEssays. 2009;31:546–560. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B., Wolynes P. Stem cell differentiation as a many-body problem. Proc. Natl. Acad. Sci. USA. 2014;111:10185–10190. doi: 10.1073/pnas.1408561111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J., Zhang K., Wang E. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc. Natl. Acad. Sci. USA. 2011;108:8257–8262. doi: 10.1073/pnas.1017017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong T., Xing J., Tyson J. A mathematical model for the reciprocal differentiation of T helper 17 cells and induced regulatory T cells. PLOS Comput. Biol. 2011;7:e1002122. doi: 10.1371/journal.pcbi.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang S., Kauffman S. How to escape the cancer attractor: Rationale and limitations of multi-target drugs. Semin. Cancer Biol. 2013;23:270–278. doi: 10.1016/j.semcancer.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Beard D.A., Qian H. Cambridge University Press; Cambridge, United Kingdom: 2008. Chemical Biophysics: Quantitative Analysis of Cellular Systems. [Google Scholar]
- 9.Raj A., van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller-Jensen K., Dey S., Arkin A. Varying virulence: epigenetic control of expression noise and disease processes. Trends Biotechnol. 2011;29:517–525. doi: 10.1016/j.tibtech.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Arkin A., Ross J., McAdams H. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1997;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberger L.S., Burnett J.C., Schaffer D.V. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Hume D.A. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood. 2000;96:2323–2328. [PubMed] [Google Scholar]
- 14.Choi P.J., Cai L., Xie X.S. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322:442–446. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H.H., Hemberg M., Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietrich J.-E., Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 17.Balaban N., Merrin J., Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 18.Acar M., Mettetal J.T., van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 19.Sharma S., Lee D., Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillespie D. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- 21.Kepler T., Elston T. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys. J. 2000;81:3116–3136. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasty J., Pradines J., Collins J. Noise-based switches and amplifiers for gene expression. Proc. Natl. Acad. Sci. USA. 2000;97:2075–2080. doi: 10.1073/pnas.040411297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner T., Cantor C., Collins J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 24.Segal E., Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat. Rev. Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasai M., Wolynes P.G. Stochastic gene expression as a many-body problem. Proc. Natl. Acad. Sci. USA. 2003;100:2374–2379. doi: 10.1073/pnas.2627987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasai M., Kawabata Y., Terada T. Time scales in epigenetic dynamics and phenotypic heterogeneity of embryonic stem cells. PLOS Comput. Biol. 2013;9:e1003380. doi: 10.1371/journal.pcbi.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C., Wang J. Quantifying Waddington landscapes and paths of non-adiabatic cell fate decisions for differentiation, reprogramming and transdifferentiation. J. R. Soc. Interface. 2013;10:20130787. doi: 10.1098/rsif.2013.0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng H., Han B., Wang J. Landscape and global stability of nonadiabatic and adiabatic oscillations in a gene network. Biophys. J. 2012;102:1001–1010. doi: 10.1016/j.bpj.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge H., Qian H., Xie X.S. Stochastic phenotype transition of a single cell in an intermediate region of gene state switching. Phys. Rev. Lett. 2015;114:078101. doi: 10.1103/PhysRevLett.114.078101. [DOI] [PubMed] [Google Scholar]
- 30.Walczak A.M., Onuchic J.N., Wolynes P.G. Absolute rate theories of epigenetic stability. Proc. Natl. Acad. Sci. USA. 2005;102:18926–18931. doi: 10.1073/pnas.0509547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng H., Wang J. A new mechanism of stem cell differentiation through slow binding/unbinding of regulators to genes. Sci. Rep. 2012;2:550. doi: 10.1038/srep00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M., Onuchic J., Ben-Jacob E. Construction of an effective landscape for multistate genetic switches. Phys. Rev. Lett. 2014;113:078102. doi: 10.1103/PhysRevLett.113.078102. [DOI] [PubMed] [Google Scholar]
- 33.Roma D.M., O’Flanagan R.A., Mukhopadhyay R. Optimal path to epigenetic switching. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;71:011902. doi: 10.1103/PhysRevE.71.011902. [DOI] [PubMed] [Google Scholar]
- 34.Aurell E., Sneppen K. Epigenetics as a first exit problem. Phys. Rev. Lett. 2002;88:048101. doi: 10.1103/PhysRevLett.88.048101. [DOI] [PubMed] [Google Scholar]
- 35.Zhou J., Aliyu M., Huang S. Quasi-potential landscape in complex multi-stable systems. J. R. Soc. Interface. 2012;9:3539–3553. doi: 10.1098/rsif.2012.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P., Song C., Xing J. Epigenetic state network approach for describing cell phenotypic transitions. Interface Focus. 2014;4:20130068. doi: 10.1098/rsfs.2013.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K., Sasai M., Wang J. Eddy current and coupled landscapes for nonadiabatic and nonequilibrium complex system dynamics. Proc. Natl. Acad. Sci. USA. 2013;110:14930–14935. doi: 10.1073/pnas.1305604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morelli M., Tanase-Nicola S., ten Wolde P. Reaction coordinates for the flipping of genetic switches. Biophys. J. 2008;94:3413–3423. doi: 10.1529/biophysj.107.116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strasser M., Theis F.J., Marr C. Stability and multiattractor dynamics of a toggle switch based on a two-stage model of stochastic gene expression. Biophys. J. 2012;102:19–29. doi: 10.1016/j.bpj.2011.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz D., Walczak A.M., Wolynes P.G. Extinction and resurrection in gene networks. Proc. Natl. Acad. Sci. USA. 2008;105:19165–19170. doi: 10.1073/pnas.0810366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipshtat A., Loinger A., Biham O. Genetic toggle switch without cooperative binding. Phys. Rev. Lett. 2006;96:188101. doi: 10.1103/PhysRevLett.96.188101. [DOI] [PubMed] [Google Scholar]
- 42.Ma R., Wang J., Liu H. Small-number effects: a third stable state in a genetic bistable toggle switch. Phys. Rev. Lett. 2012;109:248107. doi: 10.1103/PhysRevLett.109.248107. [DOI] [PubMed] [Google Scholar]
- 43.Artyomov M.N., Mathur M., Chakraborty A.K. Stochastic bimodalities in deterministically monostable reversible chemical networks due to network topology reduction. J. Chem. Phys. 2009;131:195103. doi: 10.1063/1.3264948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen R., Valeriani C., ten Wolde P. Forward flux sampling for rare event simulations. J. Phys. Condens. Matter. 2009;21:463102. doi: 10.1088/0953-8984/21/46/463102. [DOI] [PubMed] [Google Scholar]
- 45.Dickson A., Warmflash A., Dinner A.R. Nonequilibrium umbrella sampling in spaces of many order parameters. J. Chem. Phys. 2009;130:074104. doi: 10.1063/1.3070677. [DOI] [PubMed] [Google Scholar]
- 46.Donovan R., Sedgewick A., Zuckerman D. Efficient stochastic simulation of chemical kinetics networks using a weighted ensemble of trajectories. J. Chem. Phys. 2013;139:115105. doi: 10.1063/1.4821167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schütte C., Noé F., Vanden-Eijnden E. Markov state models based on milestoning. J. Chem. Phys. 2011;134:204105. doi: 10.1063/1.3590108. [DOI] [PubMed] [Google Scholar]
- 48.Dickson A., Warmflash A., Dinner A.R. Separating forward and backward pathways in nonequilibrium umbrella sampling. J. Chem. Phys. 2009;131:154104. doi: 10.1063/1.3244561. [DOI] [PubMed] [Google Scholar]
- 49.Vanden-Eijnden E., Venturoli M. Revisiting the finite temperature string method for the calculation of reaction tubes and free energies. J. Chem. Phys. 2009;130:194103. doi: 10.1063/1.3130083. [DOI] [PubMed] [Google Scholar]
- 50.Adelman J., Grabe M. Simulating rare events using a weighted ensemble-based string method. J. Chem. Phys. 2013;138:044105. doi: 10.1063/1.4773892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ptashne M. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. A Genetic Switch. [Google Scholar]
- 52.Niwa H., Toyooka Y., Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 53.Zhang P., Behre G., Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong T., Xing J., Tyson J. A simple theoretical framework for understanding heterogeneous differentiation of CD4+ T cells. BMC Syst. Biol. 2012;6:66. doi: 10.1186/1752-0509-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waddington C., Kacser H. Routledge; London: 1957. The Strategy of the Genes. [Google Scholar]
- 56.Huang S. The molecular and mathematical basis of Waddington’s epigenetic landscape: a framework for post-Darwinian biology? BioEssays. 2012;34:149–157. doi: 10.1002/bies.201100031. [DOI] [PubMed] [Google Scholar]
- 57.Huber G., Kim S. Weighted-ensemble Brownian dynamics simulations for protein association reactions. Biophys. J. 1996;70:97–110. doi: 10.1016/S0006-3495(96)79552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren E.W.W., Vanden-Eijnden E. Transition pathways in complex systems: reaction coordinates, isocommittor surfaces, and transition tubes. Chem. Phys. Lett. 2005;413:242–247. [Google Scholar]
- 59.Zwier M.C., Chong L.T. Reaching biological timescales with all-atom molecular dynamics simulations. Curr. Opin. Pharmacol. 2010;10:745–752. doi: 10.1016/j.coph.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Allen R.J., Warren P.B., ten Wolde P.R. Sampling rare switching events in biochemical networks. Phys. Rev. Lett. 2005;94:018104. doi: 10.1103/PhysRevLett.94.018104. [DOI] [PubMed] [Google Scholar]
- 61.Dickson A., Dinner A.R. Enhanced sampling of nonequilibrium steady states. Annu. Rev. Phys. Chem. 2010;61:441–459. doi: 10.1146/annurev.physchem.012809.103433. [DOI] [PubMed] [Google Scholar]
- 62.Van Kampen N.G. 3rd ed. Elsevier; Amsterdam: 2007. Stochastic Processes in Physics and Chemistry. [Google Scholar]
- 63.MATLAB and Parallel Computing Toolbox Release. 2012b. The MathWorks, Natick, MA.
- 64.Faeder J.R., Blinov M.L., Hlavacek W.S. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol. Biol. 2009;500:113–167. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- 65.Zwier M.C., Adelman J.L., Chong L.T. WESTPA: An interoperable, highly scalable software package for weighted ensemble simulation and analysis. J. Chem. Theory Comput. 2015;11:800–809. doi: 10.1021/ct5010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma A., Dinner A.R. Automatic method for identifying reaction coordinates in complex systems. J. Phys. Chem. B. 2005;109:6769–6779. doi: 10.1021/jp045546c. [DOI] [PubMed] [Google Scholar]
- 67.Yamanaka Y., Lanner F., Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 68.Singh A.M., Hamazaki T., Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 69.Chambers I., Silva J., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 70.Kalmar T., Lim C., Martinez Arias A. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buganim Y., Faddah D.A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mariani L., Schulz E., Höfer T. Short-term memory in gene induction reveals the regulatory principle behind stochastic IL-4 expression. Mol. Syst. Biol. 2010;6:359. doi: 10.1038/msb.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Borrero E.E., Escobedo F.A. Reaction coordinates and transition pathways of rare events via forward flux sampling. J. Chem. Phys. 2007;127:164101. doi: 10.1063/1.2776270. [DOI] [PubMed] [Google Scholar]
- 74.Ren E.W.W., Vanden-Eijnden E. String method for the study of rare events. Phys. Rev. B. 2002;66:052301. doi: 10.1021/jp0455430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.