Abstract
The protein kinase PKR is a sensor in innate immunity. PKR autophosphorylates in the presence of dsRNA enabling it to phosphorylate its substrate, eIF2α, halting cellular translation. Classical activators of PKR are long viral dsRNAs, but recently PKR has been found to be activated by bacterial RNA. The features of bacterial RNA that activate PKR are unknown, however. We studied the B. subtilis trp 5’-UTR, which is an indirect riboswitch with secondary and tertiary RNA structures that regulate gene function. Additionally, the trp 5’-UTR binds a protein, TRAP, which recognizes L-tryptophan. We present the first evidence that multiple structural features in this RNA, which are typical of bacterial RNAs, activate PKR in TRAP-free and TRAP/L-Trp-bound forms. Segments from the 5’-UTR, including the terminator, 5’-stem-loop, and Shine-Dalgarno blocking hairpins, demonstrated 5’-triphosphate and flanking RNA tail dependence on PKR activation. Disruption of long-distance tertiary interactions in the 5’-UTR led to partial loss in activation, consistent with highly base-paired regions in bacterial RNA activating PKR. One physiological change a bacterial RNA would face in a human cell is a decrease in the concentration of free magnesium. Upon lowering the magnesium concentration to human physiological conditions of 0.5 mM, the trp 5’-UTR continued to activate PKR potently. Moreover, total RNA from E. coli, depleted of rRNA, also activated PKR under these ionic conditions. This study demonstrates that PKR can signal the presence of bacterial RNAs under physiological ionic conditions and offers a potential explanation for the apparent absence of riboswitches in the human genome.
Keywords: PKR, bacterial RNA, riboswitch, innate immunity, RNA folding
Graphical abstract
Introduction
The first line of defense to pathogens is the innate immune system. One of the key ways the body discriminates self and non-self is through molecular patterns involving RNA. PKR is the RNA-activated protein kinase and it plays a key role in innate immunity [1–3]. In the presence of pathogenic RNA, which serve as a pathogen-associated molecular pattern (PAMP), PKR dimerizes and autophosphorylates, which activates it to phosphorylate translation initiation factor eIF2α, blocking viral replication and shutting down translation [4].
PKR is comprised of a C-terminal kinase domain and two N-terminal dsRNA-binding motifs (dsRBMs) [5]. Long stretches of viral dsRNA bind to the dsRBMs in a sequence-independent fashion and activate PKR [6]. In addition, we and others have shown that many other RNA motifs serve as PAMPs to activate PKR. In particular, complex tertiary structures including pseudoknots [7], IRESs [8], and UTRs [9] activate PKR, as well as a number of misfolded or dimerized functional RNAs [10–12]. Multiple RNA secondary structures have been reported to promote activation of PKR including stem-loops containing defects such as smaller bulges [13]. Moreover, certain unstructured RNA regions promote activation. In particular single-stranded regions with a 5’-triphosphate activate PKR, especially when fused to short stem-loops [14,15], and they do so without specificity to the 5’-base to offer broad-spectrum protection [16]. We also found that activation of PKR is abrogated by many covalent modifications of RNA, especially in single-stranded regions of RNA [17,18].
Recent reports indicate that bacterial RNAs activate PKR in human cells [19,20]. For instance, transfection of human cardiac myocytes with total RNA from E. coli and S. aureus leads to potent activation of PKR, while transfection with human RNAs does not. Similarly, others reported and we confirmed [14] that PKR from E. coli cells is purified in a phosphorylated form that must be dephosphorylated to make PKR responsive to RNA [21]. These observations support the ability of bacterial RNAs to activate PKR, although this could also arise from high PKR concentrations that may be present during expression[22]. However, the molecular features of bacterial RNA that activate PKR have not been identified, which motivated our present study.
The trp 5’-UTR from B. subtilis, which regulates the biosynthesis of tryptophan, is known to switch between several different conformations to regulate gene expression at the transcription and translational levels (Fig. 1). At the transcription control level, an antiterminator forms in the trp 5’-UTR. In the presence of excess tryptophan, TRAP binds L-Trp. The Trp-bound TRAP then binds the 11 GAG/UAG single-stranded triplet repeats to drive formation of an intrinsic terminator to promote attenuation (Fig. 1a). At the translation control level, a large hairpin forms in the 5’-UTR that allows ribosomes access to the Shine-Dalgarno (SD) sequence (Fig. 1b). Upon binding TRAP, the 5’-UTR refolds to form the SD blocking hairpin to inhibit translation (Fig. 1) [23]. In regulating expression at the transcription and translational levels, the trp 5’-UTR serves as an indirect riboswitch, sensing tryptophan levels through binding of L-Trp to TRAP protein. In addition, the trp 5’-UTR has a long-distance pseudoknot with a well-characterized Mg2+ response that helps promote translation (Fig. 1b, lower) [24]. Given the diversity of elements in the trp 5’-UTR—multiple stem-loops, a long-range RNA tertiary structure, extensive dsRNA and ssRNA regions, and an RNA-binding protein—it is an ideal system to investigate activation of PKR by bacterial RNAs. Moreover, some of these RNA elements are quite unique to bacteria, such as the terminator, SD blocking hairpin, and 5’-triphosphate, making our findings applicable to other bacterial RNAs.
Figure 1.
TRAP-free and TRAP–bound models of the RNA folds present in the B. subtilis trp 5’-UTR transcription and translation control mechanisms. (a) RNA folds of trp 5’-UTR present in the transcription control construct [23,24]. Left is the TRAP-free construct that forms in tryptophan-limiting conditions where TRAP is unable to bind, which leads to formation of an antiterminator resulting in transcription readthrough and eventually L-Trp biosynthesis. Right is the TRAP/L-Trp-bound construct wherein TRAP is able to bind to the GAG/UAG triplet repeats (bold) as they are synthesized. TRAP/L-Trp binding leads to the formation of the terminator (green) resulting in termination of transcription. (b) RNA fold of the full-length trp 5’-UTR present in the translation control construct [23,24]. The AUG (bold) start codon is the last three nucleotides shown. Left is the TRAP-free construct that forms in tryptophan-limiting conditions where the trp 5’-UTR transcript adopts a structure in which the SD sequence is single-stranded allowing the ribosome to bind and translation to occur. The structure below this one is a magnesium-dependent tertiary structure model where 24 nt from both the 5’- (magenta) and 3’-(blue) pyrimidine-rich internal loop base pair with upstream residues 36–60 [24]. Right is the TRAP/L-Trp-bound construct that forms a structure where the SD sequence is sequestered, which inhibits ribosome binding. In all structures, the 11 identical subunits of TRAP are shown in blue, yellow, red and orange, and L-Trp molecules are shown in green as per PDB 1C9S [51]. Numbering is from the start of transcription and specific numbers labeled include the boundaries for sequence truncations used in this study.
In the present study, we conducted PKR activation studies on the trp 5’-UTR in the presence and absence of TRAP and investigated the secondary structural features of trp 5’-UTR responsible for activating PKR. We also tested various RNA segments for 5’-triphosphate dependence to activation and investigated the contribution of the long-distance tertiary structure to activation. To generalize our findings, we conducted experiments under physiological ionic conditions and investigated the ability of total bacterial RNAs to activate PKR. Our study provides the first evidence that specific bacterial RNA motifs activate PKR very strongly. These results support the ability of bacterial RNAs to activate PKR under physiological conditions.
Results
The trp 5’-UTR activates PKR in both its TRAP-free and TRAP-bound states
In an effort to assess whether the trp 5’-UTR can activate PKR, we conducted activation assays with the RNA alone, the RNA with TRAP protein, or the RNA with TRAP plus L-Trp. We began with the full-length translation control trp 5’-UTR shown in Figure 1b, which we term ‘FL RNA (1–206)’. As shown in Figure 2, the trp 5’-UTR activated PKR with a bell-shaped dependence on RNA concentration. The level of activation in the lowest concentration of added RNA (0.02 µM) was similar to that in the negative control of no-added RNA, as was the level of activation in the highest concentration of added RNA (10 µM) (Fig. 2a). These data indicate that there is a low background overall and support the bell-shaped dependence of PKR activation by RNA, where high concentrations of RNA titrate PKR protein out into inactive monomers [25]. At intermediate concentrations of RNA (~1.0 µM), PKR activation occurred with a maximum value of ~130% the level of the reference activator, dsRNA-79 (Fig. 2d). The trp 5’-UTR is thus a potent activator of PKR in its ligand-free form.
Figure 2.
Activation of PKR by the trp 5’-UTR translation control RNA (1–206) in the absence and presence of TRAP and L-Trp. PKR activation assays on trp 5’-UTR (1–206) (a) alone, (b) in the presence of TRAP, and (c) in the presence of TRAP/L-Trp. RNA was serially diluted ~2-fold from 10 to 0.02 µM. In each lane, TRAP was present in five-fold excess over the RNA, and L-Trp was present at a final concentration of 1 mM [26]. A buffer-only negative control is included and PKR activation is normalized to dsRNA-79 in each panel. Activation values are provided under each gel and the position of phosphorylated PKR is indicated as ‘p*PKR’. Gels shown are raw data from one representative trial. (d) Graphical representation of percent PKR activation from panels (a)-(c) as a function of the concentration of FL RNA (1–206). Plotted is the average of two independent trials and error bars represent the range of these two trials. The lines for FL RNA(1–206)/TRAP and FL RNA(1–206)/TRAP/L-Trp also average points immediately adjacent on the x-axis. All forms of the RNA activated PKR with a bell-shaped dependence on RNA concentration.
Next we tested whether TRAP (+/− L-Trp) altered the activation of PKR. When we added TRAP in 5-fold excess over RNA to the activation mixture in the absence of L-Trp, there was little to no effect on the activation profile. Maximal activation remained at ~125% (Fig. 2b and 2d). TRAP is known to be unable to bind the 5’-UTR in the absence of L-Trp [26], consistent with no measurable effect on PKR activation. As a control we also tested whether TRAP alone can activate PKR, and no activation was observed (Fig. S2), in agreement with the observations above.
When TRAP was added to the activation mixture in the same five-fold excess over the RNA but in the presence of 1 mM L-Trp, the maximum activation shifted to ~150% with a sharper profile (Fig. 2c and 2d). These conditions were chosen to assure saturation of the RNA with TRAP (see Materials and Methods). Overall, these data indicate that PKR is activated by the trp 5’-UTR both in its ligand-free and ligand-bound states. Furthermore, these results suggest that TRAP does not compete with PKR for binding to the RNA, which is consistent with the opposite RNA binding preferences of these two proteins—TRAP is known to bind to single-stranded segments of the RNA while PKR tends to bind double-stranded segments. We next investigated which of the structured regions of the trp 5’-UTR are capable of activating PKR.
Multiple secondary structural features in the trp 5’-UTR are capable of activating PKR
The previous section showed that PKR is activated potently by the trp 5’-UTR in both its TRAP-free and TRAP-bound states. There have been no investigations in the literature, however, of which bacterial RNA elements can activate PKR. We therefore did a systematic analysis to determine which elements of the trp 5’-UTR are capable of activating PKR. As described above, the full-length trp 5’-UTR activated PKR ~130% relative to dsRNA-79. Knowing that PKR generally prefers RNA substrates that are structured, we assessed whether the large hairpin or ‘LH’ (nt 61–188) in the TRAP-free 5’-UTR is capable of activating PKR. The results are summarized in Table 1 and the activation gels are provided in Figure S3. The LH (61–188) potently activated PKR at a maximum value ~95% the level of 0.1 µM dsRNA-79. Moreover, less of the LH (61–188) than full-length trp 5’-UTR was required to activate PKR, with the maximum activation occurring at ~0.625 µM RNA versus 1.0 µM. This gave a combined effect that was similar (1.2-fold) to full-length 5’-UTR (Table 1). The combined effect is calculated by multiplying maximal PKR activation and minimal RNA concentration to yield the overall potency of PKR activation, which accounts for the bell-shaped profile of PKR activation.
Table 1.
Multiple structural features of the trp 5’-UTR lead to activation of PKR
| RNA | Shorthand Notationa |
Maximal PKR Activation (%)b |
[RNA] at Max. Act. (µM)b |
Max. Act. Fold Effectc |
[RNA] Fold Effectd |
Combined Effecte |
ppp Contributionf |
|---|---|---|---|---|---|---|---|
| 1–206 | FL | 128 | 1.0 | 1.0 | 1.0 | 1.0 | 1.1 |
| 61–188 | LH | 95.0 | 0.625 | 0.7 | 1.6 | 1.2 | 1.1 |
| 89–159 | T + Tails | 88.9 | 0.625 | 0.7 | 1.6 | 1.1 | 4.7 |
| 89–133 | T + 5’ Tail | 12.6 | 10 | 0.1 | 0.1 | 0.01 | |
| 108–159 | T + 3’ Tail | 100 | 0.39 | 0.8 | 2.6 | 2.0 | |
| 108–133 | T | ~ 0 | N/A | N/A | N/A | N/A | N/A |
| 1–60 | 5’-SL + Tails | 69.1 | 0.156 | 0.5 | 6.4 | 3.2 | 1.7 |
| 142–206 | SD SL + Tails | 167 | 2.5 | 1.3 | 0.4 | 0.5 | 24.6 |
| 172–200 | SD SL | 6.1 | 6.25 | 0.05 | 0.2 | 0.01 | |
| dsRNA-79 | ds-79 | 100 | 0.1 | 0.78 | 10 | 7.8 |
Abbreviations for shorthand notation: FL: full-length, LH: long hairpin, T: terminator, SD: Shine-Delgarno, SL: stem-loop.
Each of the truncated trp 5’-UTRs tested for PKR activation are listed at the left and the effect in activating PKR is calculated in two ways: the highest percentage of activation relative to dsRNA-79 and the concentration of RNA at the max activation. All experiments were repeated at least twice.
An activation fold-effect is calculated for each truncation with respect to the full-length (1–206) RNA by dividing the maximal PKR activation of the corresponding truncation by that of the full-length.
An RNA concentration fold-effect is calculated for each truncation with respect to the full-length (1–206) RNA by dividing the RNA concentration at the maximum activation of the full-length by that of the corresponding truncation.
The combined effect is calculated by multiplying the activation and concentration fold effects together.
Triphosphate (ppp) fold-effects were calculated for the four RNAs tested for triphosphate dependence. The ppp-effects are calculated by dividing the 5’-ppp maximum activation by those of the 5’-OH values corresponding to the same RNA (rather than full-length). Actual quantities used to calculate the ppp contribution values can be found in figure 3.
Knowing that certain short hairpins with single-stranded tails can also activate PKR [14,15], we next investigated activation of PKR by the terminator hairpin, ‘T’ both with and without its flanking tail sequences, which we termed ‘tails’. The RNA segment from 89–159 contains the 11 bp terminator hairpin and both its 5’- and 3’-tails. This construct activated PKR potently, at a maximum value ~90% relative to that of dsRNA-79. Again, less of T + tails (89–159) RNA than full-length trp 5’-UTR was necessary to activate PKR at this maximum value, at only ~0.625 µM RNA. Together, the maximum activation and RNA requirement gave a combined effect of approximately equal to full-length trp 5’-UTR. Next, the dependence of activation on the tails flanking the terminator hairpin was determined, where construct 89–133, termed ‘T + 5’ tail’, contains the terminator hairpin and the 5’ tail, and construct 108–159, termed ‘T + 3’ tail’, contains the terminator hairpin and the 3’ tail. It was found that T + 5’ tail (89–133) only slightly activated PKR, at ~10% maximum activity, and that this occurred only at the highest RNA concentration of 10 µM, giving a very weak total combined effect of ~0.01-fold relative to the full-length trp 5’-UTR. Conversely, the T + 3’ tail (108–159) construct displayed a high maximum activation of PKR was at ~100% relative to dsRNA-79, and the concentration of RNA at this maximum activation was low ~0.39 µM. This leads to a total combined effect that was 2.6-fold greater than the full-length 5’-UTR. It was also determined that when both tails were removed from the terminator hairpin, in the 108–133 (T) construct, all PKR activity was lost (Table 1). Apparently, the 3’-tail is an important and essential contributor to activation by the terminator hairpin. Promotion of activation by a single stranded 3’-tail is consistent with previous observations [14], although preference for the 3’-tail is stronger in this bacterial RNA. This may be due to the stem being only 11 bp in the present study, while it was 16 bp in the earlier work.
There are several other hairpins that can form in the trp 5’-UTR. Upon binding of Trp-bound TRAP, the full-length trp 5’-UTR can switch its conformation between two very different folds (Figure 1b). In this conformational switch, the large hairpin (61–188) region refolds to release a complement to the Shine-Dalgarno (‘anti-SD’) that sequesters the SD sequence to form an 11 bp stem-loop termed the ‘SD-blocking hairpin’ or ‘SD SL’. We first tested an RNA segment that spans from 142–206 that contains the SD blocking hairpin, a long 5’-tail (30 nt) and a shorter 3’ tail (6 nt), termed ‘SD SL + tails’. This RNA potently activated PKR at a maximal level of ~170% that of dsRNA-79. The relative amount of this RNA necessary to activate PKR was 2.5 µM, however, which was one of the higher values of all the RNAs tested. This led to a slightly diminished combined effect of 0.5-fold that of the full-length 5’-UTR. One possibility is that such an element could be a potent activator of PKR if it accumulates to high concentrations. We also tested the contribution of the tails to activation of PKR by the SD blocking hairpin. The construct SD SL (172–200) has both the 5’- and 3’-tails removed. This RNA was an exceptionally poor activator of PKR with a maximal activation level of just ~6% and a very high RNA requirement at 6.25 µM. The inability of the SD blocking hairpin alone (172–200) to activate PKR is similar to the inability of the terminator hairpin alone (108–133) to activate PKR.
The final hairpin that we studied is the 5’-stem-loop (5’-SL). The 5’-SL is comprised of 9 bp and resides at the very 5’-end of the transcript. One of its functions is to increase the affinity of the interaction between TRAP-Trp and the 5’-UTR [27]. We prepared an RNA segment from 1–60 that contains the 5’-SL and 28 downstream nucleotides, termed ‘5’-SL + tails’. This RNA activated PKR very well, at a maximum value that was ~70% that of dsRNA-79. The relative amount of 5’-SL + tails (1–60) RNA necessary to activate PKR at this maximum value was the lowest of any trp 5’-UTR segments at just ~0.16 µM. These two values lead to a combined effect of ~3.2-fold greater than that of the full-length 5’-UTR, which is the most potent of the trp 5’-UTR segments studied. Furthermore, this combined effect for the 5’-SL + tails (1–60) RNA is within 2.5-fold the combined effect of dsRNA-79.
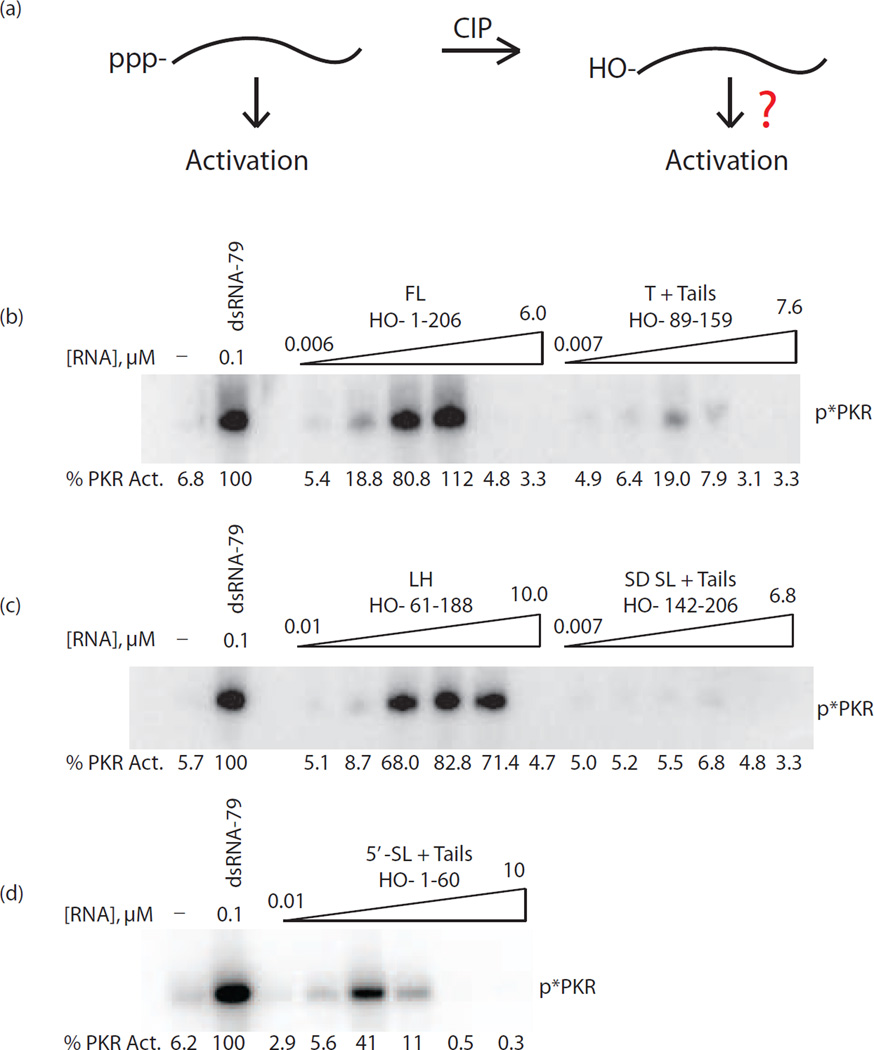
Various trp 5’-UTR segments display a 5’-triphosphate dependence for PKR activation
We previously reported that short hairpins with single-stranded tails activate PKR in a 5’-triphosphate dependent fashion [15]. Given that several of the PKR-activating trp 5’-UTR segments discussed in the preceding section are hairpins that depend on single-stranded tails for activation, we tested whether these also activate PKR in a 5’-triphosphate dependent fashion. The terminator with both 5’- and 3’-tails, ‘T + tails (89–159)’, was prepared with and without treatment by calf intestinal phosphatase (CIP), which removes the 5’-triphosphate (Fig. 3a) [28,15], and tested for PKR activation. As shown in Figure 3b, upon treatment with CIP, T + tails (89–159) became a poor activator of PKR, with a ~5-fold loss in activation. Even larger effects were obtained with the SD blocking hairpin with 5’- and 3’-tails, SD SL + tails (142–206), in which activation diminished by nearly 25-fold upon CIP treatment (Fig. 3c, Table 1). A much more subtle triphosphate effect was seen with the 5’-SL and tails (1–60) where the CIP-treated construct still activated PKR at around 40% of untreated, which yielded only a 1.7-fold loss in activation of PKR (Fig. 3d, Table 1).
Figure 3.
Dependence of activation of PKR on 5’-triphosphate for full-length and truncated trp 5’-UTRs. (a) Model of activation by 5’-triphosphate RNA (left) and 5’-hydroxyl RNA (right). Calf-intestinal phosphatase (‘CIP’) is used to remove the 5’-triphosphate and leave a 5’-hydroxyl. In all RNA constructs tested, the 5’-triphosphate form activated PKR, but activation after CIP treatment was dependent on the size and structure of the RNA, as indicated by the question mark. (b–d) PKR activation assays of CIP-treated trp 5’-UTR constructs (b) FL (1–206) and T + tails, (89–159), (c) LH (61–188) and SD SL + tails (142–206), and (d) 5’-SL + tails (1–60). A buffer-only negative control is included and PKR activation is normalized to dsRNA-79 in each panel. Activation values are provided under each gel and the position of phosphorylated PKR is indicated as ‘p*PKR’. All assays were performed twice.
We previously reported that RNA activators with extensive double-stranded structure do not show dependence on a 5’-triphosphate [15]. For instance, activation of PKR by dsRNA-79 is not diminished upon CIP-treatment [25]. Consistent with this, the more structured activating RNA constructs of full-length trp 5’-UTR (1–206) and large hairpin construct (61–188) continued to activate PKR potently after CIP treatment (Fig. 3b,c and Table 1). Importantly, observation of full activation by CIP-treated 1–206 and 61–188 RNA indicates that loss of activity upon CIP treatment in the 89–159 and 142–206 constructs is not due to inhibition of PKR activity by CIP treatment. Overall, the above results indicate that the 5’-UTR of a bacterial RNA can activate PKR both with and without a 5’-triphosphate.
Tertiary structure in the trp 5’-UTR contributes to PKR activation
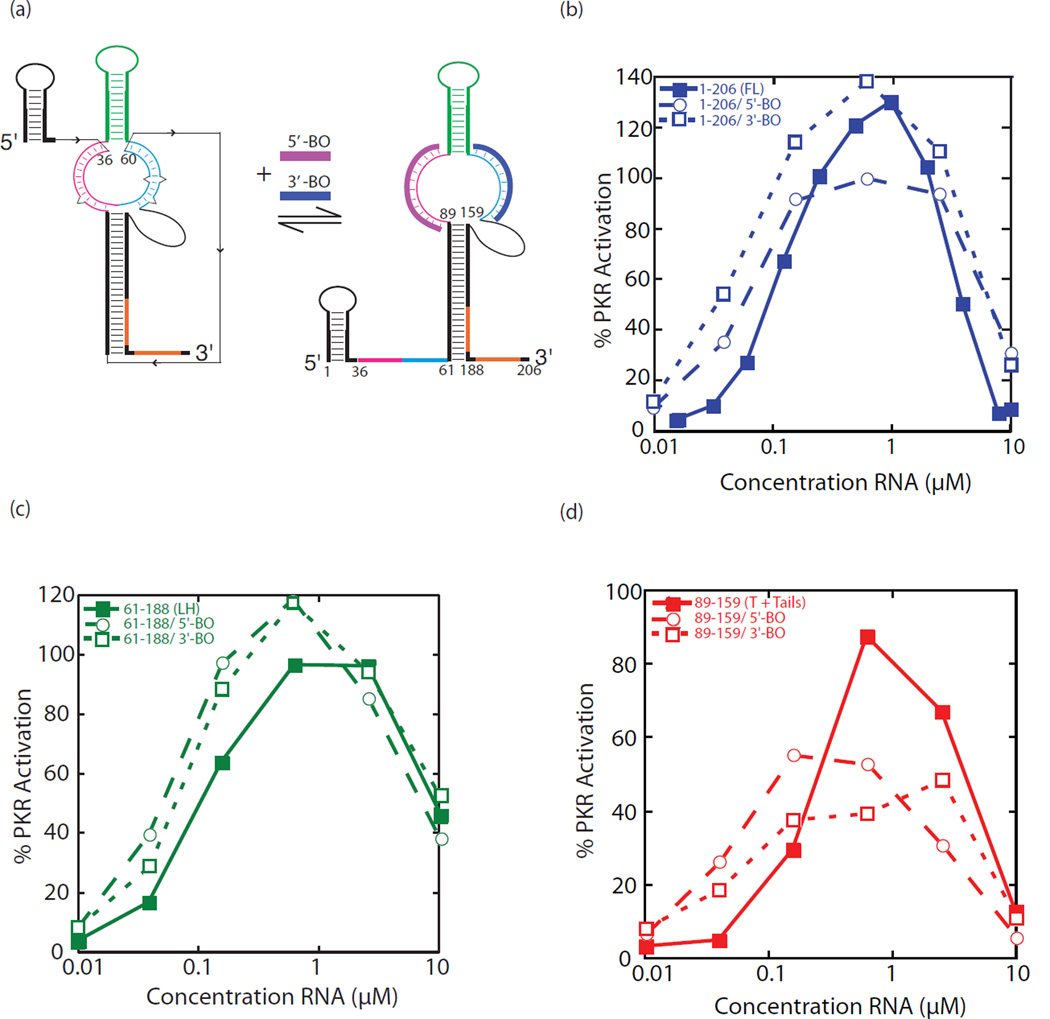
Prior investigations from our lab identified a long-distance Mg2+-dependent pseudoknot in the trp 5’-UTR that functions to limit binding of TRAP [24]. The 5’-half of the purine-rich single-stranded triplet repeat region (nt 36–60) was shown to base pair with both strands of the pyrimidine-rich internal loop in the large hairpin (Fig. 4a). The base pairing of these triplet repeats is more extensive with the 5’-strand of the internal loop than the 3’-strand. Pairing with the 5’-strand of the internal loop consists of 13 base pairs with just one single-nucleotide bulge, while pairing with the 3’-strand has only 11 base pairs and these are interrupted by two defects— a single-nucleotide bulge and a 1×1 internal loop (Fig. 4a). We also demonstrated that this tertiary structure could be disrupted by DNA blocking oligonucleotides (BOs) directed to either the 5’- or 3’-pyrimidine-rich internal loops, with more potent disruption by the 5’-BO, as expected given the stronger long-distance base pairing of this element [24]. Controls clearly showed binding of both BOs to the trp 5’-UTR [24]. Given that this long-distance tertiary structure substantially increases the number of base pairs in the trp 5’-UTR, it seemed possible that activation of PKR would increase upon its formation.
Figure 4.
Activation of PKR by the trp 5’-UTR in the presence and absence of a known longdistance tertiary interactions. (a) Model of the full-length (1–206) trp 5’-UTR in the TRAP-free form. (Left) RNA-fold with long-distance tertiary interaction between residues 36–60 and both the 5’- and 3’- regions of the downstream internal loop. (Right) Addition of either a 5’- or 3’-chimeric DNA/LNA blocking oligonucleotide (5’-BO and 3’-BO) disrupts these tertiary interactions. (b–d) Percent PKR activation as a function of RNA concentration for trp 5’-UTR constructs (b) FL (1–206), (c) LH (61–188), and (d) T + tails (89–159) with and without 5’- or 3’-BOs (legends provided in figures). Blocking oligonucleotides are in two-fold excess over the trp 5’-UTR. The average of three independent trials is plotted, connected by trend lines. The average standard deviation was ~20%PKR activation.
We began by investigating the effect of the BOs on the full-length trp 5’-UTR. Representative activation gels are provided in Figure S4 and the average data are plotted in Figure 4. Addition of the 3’-BO had little to no effect on activation of PKR (Fig. 4b). This is consistent with our earlier studies that the 3’-BO is not highly effective in disrupting the pseudoknot. Addition of the 5’-BO, on the other hand, led to a flattened profile with a ~40% reduction in activation at the RNA concentration corresponding to the activation maximum (Fig. 4b). Because the 5’-BO fully disrupts the pseudoknot [24], this finding suggests that the longdistance tertiary structure aids in activating PKR.
Next, we describe control experiments for any direct effects of the BOs on PKR activation. Addition of either the 3’- or 5’-BO’s to PKR in the absence of any RNA did not activate PKR (Fig. S2). Furthermore, addition of either BO to just the large hairpin structure (61–188) led to minimal effects on activation, with perhaps slight enhancement rather than the loss of activation seen with the full-length trp 5’-UTR (Fig. 4c). These controls suggest that the loss of activation upon addition of 5’-BO to the full-length trp 5’-UTR is not due to direct effects of the BO with PKR.
Lastly, we tested the effect of the BOs on the terminator hairpin plus its 5’- and 3’- tails (89–159). As shown in Figure 4d, addition of either BO led to a ~50% reduction in activation. As presented above, activation of PKR by the terminator is dependent on single-stranded tails. Observation of substantial loss in activation upon addition of the BOs is consistent with this single-stranded tail requirement along with possible blockage of 5’-triphosphate accessibility by the 5’-BO. Moreover, addition of blocking oligonucleotides leads to RNA-DNA hybrids in the tail regions, and such hybrids have been shown to not activate PKR [29].
Bacterial RNAs activate PKR under physiological magnesium concentrations
The concentration of Mg2+ in human cells, where PKR would be exposed to bacterial RNAs, is relatively low at ~0.5 to 1.0 mM [30,31]. Standard PKR activation assays in the literature have been typically conducted over a range of 4–10 mM Mg2+[14,22,32]. This is much higher than the human physiological concentration of Mg2+ as well as bacterial physiological concentration of Mg2+, which is ~2 mM [33,34]. It is known that many structured RNAs refold with Mg2+1/2 values in the broad range of 0.5 to 10 mM [35–37,24,38,39]. We thus wanted to test whether PKR could be activated at biological Mg2+ concentrations.
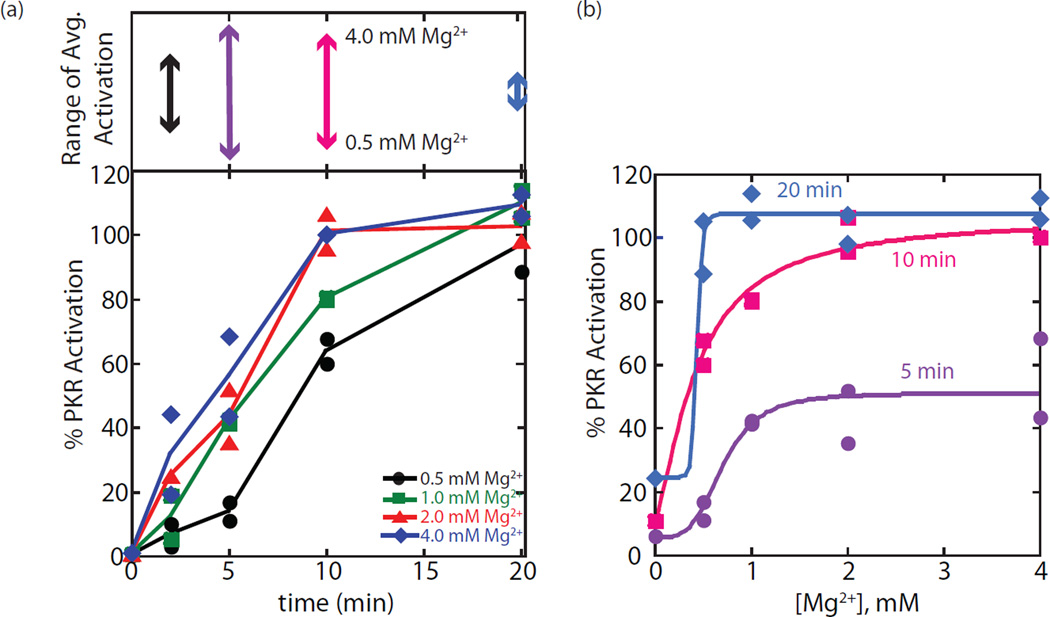
We began by testing the Mg2+- and time-dependence of PKR activation by dsRNA-79. Activation assays are provided in Figure S5 and the data are plotted in Figure 5. As shown in Figure 5a (bottom), PKR underwent autophosphorylation in Mg2+ concentrations ranging from 0.5 to 4 mM. Under our standard PKR activation assay condition of 4 mM Mg2+, maximal activation was reached in 10 min, which remained at a plateau out to 20 min. As the Mg2+ concentration was lowered to 2.0, 1.0, and 0.5 mM, the level of activation lowered. The difference in extent of activation between lowest and highest Mg2+ concentrations changed as a function of time, increasing and then decreasing (Figure 5a, top). By the longest time point of 20 min, there was little difference in extent of PKR activation between the lowest and highest Mg2+ concentrations, as sufficient time had elapsed to allow the slower reactions at low Mg2+ concentrations to catch up with the 4 mM Mg2+ reaction.
Figure 5.
Dependence of activation of PKR on time and Mg2+ concentration. (a) PKR activation by dsRNA-79 was monitored at various concentrations of free Mg2+ as a function of time. The upper panel displays the range of PKR activation between 0.5 and 4 mM Mg2+ as a function of time where it can be seen that the range of activation is much larger for the 5 and 10 min time points. Spacing along the y-axis is the same in the lower and upper panels. (b) The 5, 10 and 20 min assays are plotted versus Mg2+ concentration. At 20 min, activation is approximately the same for all Mg2+ concentrations tested. In both panels, two individual trials are plotted. For panel (a), lines are drawn through the average of these two trials, and for panel (b) curves are from fits to a Hill equation. Colors of time match for panel (a) upper and panel (b).
We replotted the data from Figure S5 as a function of Mg2+ concentration, parametric in time (Fig. 5b). As expected, the plot at 5 min shows a stronger dependence of the extent of reaction on Mg2+ concentration, with 4 mM Mg2+ giving ~4-fold more phosphorylated product than 0.5 mM Mg2+. However, by 20 min, there was almost no dependence of the reaction on Mg2+ concentration between 0.5 and 4 mM. These plots led to an apparent Hill constant that increases with time. Given that 20 min was sufficient to allow full extent of PKR phosphorylation independent of Mg2+ concentration, we conducted subsequent assays at this time.
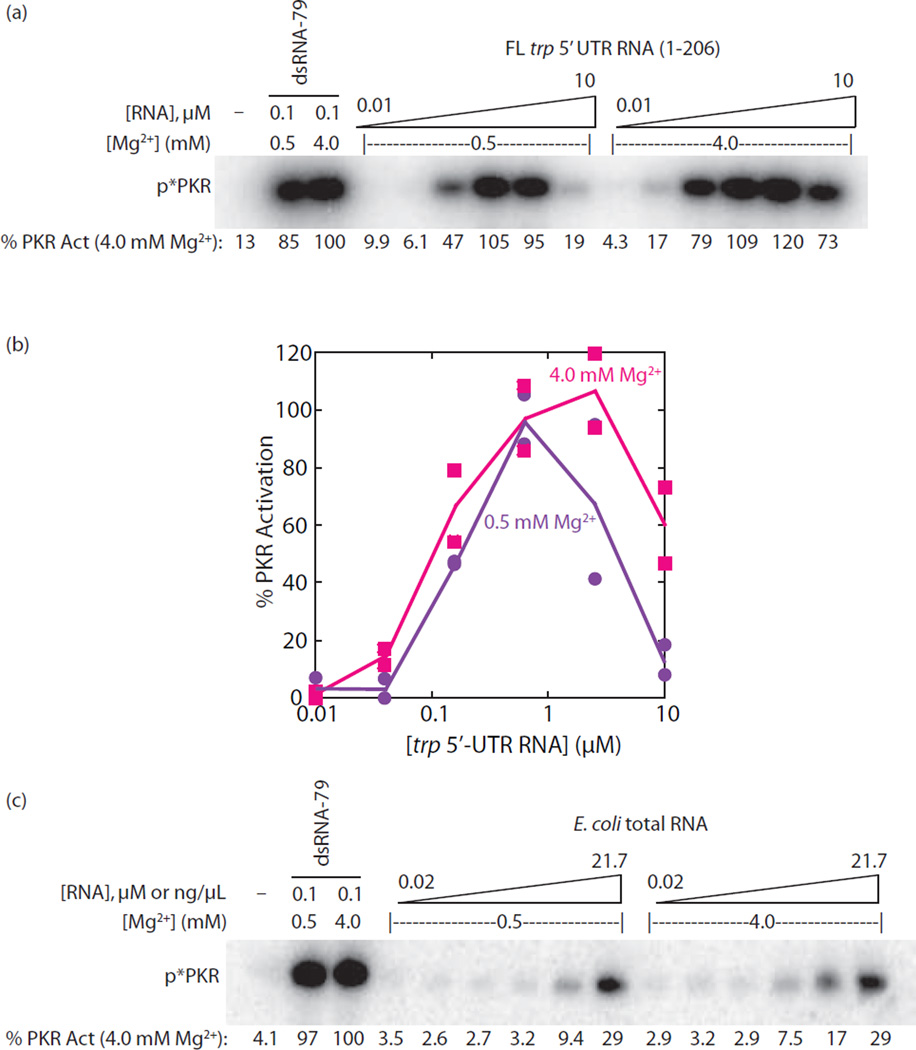
Next, the dependence of PKR activation by the FL trp 5’-UTR (1–206) on Mg2+ concentration was tested. Given that PKR autophosphorylates to a similar extent in both 0.5 and 4 mM Mg2+ concentrations at 20 min, this experiment reports on the ability of different Mg2+-dependent folds of the trp 5’-UTR to activate PKR. Previous studies from our lab revealed that the long-distance tertiary structure of the trp 5’-UTR has a Mg2+1/2 of ~0.5 mM and Hill constant of ~1.5 for folding[24]. Thus the trp 5’-UTR should be only ~50% folded in 0.5 mM Mg2+ but fully formed at 4 mM Mg2+. As shown in Figure 6a,b, PKR was potently activated by the trp 5’-UTR at both 0.5 and 4 mM Mg2+ concentrations, with small differences in extent of activation at low and high Mg2+ proportionate to small differences seen in the control lanes with dsRNA-79 at these two Mg2+ concentrations. These data are consistent with observations above that the trp 5’-UTR potently activates PKR in the presence and absence of the long-distance tertiary contacts (Fig. 4). Importantly, these data indicate that bacterial RNA can activate PKR under physiological ionic conditions.
Figure 6.
Activation of PKR by bacterial RNAs in different concentrations of Mg2+. (a) PKR activation assays by full-length trp 5’-UTR (1–206) in low (0.5 mM) and standard (4 mM) Mg2+ concentrations. The RNA was incubated in 0.5 or 4 mM Mg2+ at 30˚C for 5 min followed by a 20 min incubation with PKR. A buffer-only negative control is included and PKR activation is normalized to dsRNA-79 at each Mg2+ concentration. Activation values are provided under the gel and are normalized to dsRNA-79 activation under 4 mM Mg2+ conditions. The position of phosphorylated PKR is indicated as ‘p*PKR’. (b) Graphical representation of percent PKR activation from panel (a) as a function of full-length trp 5’ UTR RNA (1–206) concentration. Plotted are two independent trials of each set of data where lines are connecting the averages. (c) PKR activation assays by E. coli total RNA in low (0.5 mM) and normal (4 mM) Mg2+ concentrations. The E. coli total RNA was rRNA depleted and incubated in 0.5 mM or 4 mM Mg2+ at 30˚C for 5 min and then incubated with PKR for 20 min. A buffer-only negative control is included and PKR activation is normalized to dsRNA-79 under 4 mM Mg2+ conditions‥. The position of phosphorylated PKR is indicated as ‘p*PKR’. Experiments were performed twice and a representative gel is provided here.
Lastly, the ability of a mixture of E. coli RNAs to activate PKR was tested under both 0.5 and 4 mM Mg2+ concentrations. We isolated total RNA from MG1655 E. coli and depleted rRNAs using standard laboratory procedures (see Materials and Methods). As shown in Figure 6c, total E. coli RNA activated PKR under both 0.5 and 4 mM Mg2+ conditions and did so to a similar extent, with ~30% activation (relative to 0.1 µM dsRNA-79) at a total bacterial RNA concentration of 22 ng/µL. This activation is within ~10-fold that of dsRNA-79: the concentration of RNA is ~4-fold greater than 0.1 µM dsRNA-79 and the activation level is ~3.3-fold lower. We note that this modest level of activation could indeed be significant because the E. coli RNA preparation consists of a vast number of RNAs, present at various levels and sizes, some of which are likely potent activators of PKR. Thus, a mixture of bacterial RNAs has the potential to evoke an innate immune response through PKR.
Discussion
The innate immune system is the body’s first line of defense against pathogens. Factors in this pathway recognize general PAMPs, some of which are comprised of RNA features that differ between pathogens and humans. Indeed, distinction between self and non-self is a key feature of innate immunity and perception of RNA signatures plays a key role. Two proteins that play key roles in innate immunity at the level of RNA recognition are 2’,5’- oligoadenylate synthase (OAS) and PKR. It is well known that 2’,5’- OAS is activated by the viral PAMP of long dsRNA to synthesize 2’, 5’- linked adenylate oligonucleotides [40]. The oligoadenylates activate latent RNase L which then degrades viral and cellular RNAs to block viral infection. In addition, dsRNAs activate PKR to undergo autophosphorylation, which then phosphorylates eIF2α, thereby inhibiting translation and blocking viral infection [41]. Two additional proteins that serve as receptors in the innate immune response to RNA are RIG-I and MDA5, which are helicases that bind dsRNA and are expressed and functional in the presence of viral or bacterial infection [42].
Recent reports indicate that bacterial RNAs activate PKR in mice [19,20]. We found that the trp 5’-UTR, which has structured RNA elements representative of many bacterial mRNAs such as a terminator, 5’-stem-loop, SD hairpin, and Mg2+-dependent tertiary structures is a potent activator of PKR both in the absence and presence of bound TRAP. Multiple secondary structures drive activation of PKR including the full-length trp 5’-UTR as well as its large hairpin, and furthermore that these RNA elements do so at a maximal level similar to that of perfect dsRNA-79 (Table 1). The fact that activation of these pieces adds up to more than activation by full length 5’-UTR may be due to the elements overlapping in their PKR binding sites. What is most significant is that any of several elements can lead to activation, indicating that if one element is absent, due to say protein binding or conformational switching, another can still activate PKR. We found that all of the bacterial hairpins that we examined required flanking tails to activate PKR, with a preference for a 3’-tail. Most bacterial RNAs are found in this context, however, especially those at the 5’-end of RNA, supporting the general ability of bacterial mRNAs to activate PKR.
Observation that a high level of PKR activation occurs when TRAP/L-Trp binds the 5’-UTR is consistent with TRAP binding primarily to single-stranded RNA regions, and TRAP/L-Trp-bound trp 5’-UTR still having multiple helical regions of RNA available for recognition by PKR. Additionally, the pseudoknot that forms in the 5’-UTR in the absence of bound TRAP promotes activation of PKR, which is supported by partial loss of activation upon disruption of the long-distance base pairing by the blocking oligonucleotides (Fig. 4). This observation is agrees with earlier reports that base pairing within RNA pseudoknots can promote PKR activation [7]. In addition to the large hairpin, the terminator and SD blocking hairpins potently activated PKR in the presence of single-stranded tails and a 5’-triphosphate. These findings indicate that the mode of PKR activation involving ssRNA tails and a 5’-triphosphate [14,15] is important in sensing bacterial RNA.
The 5’-SL construct activated PKR the strongest. On the basis of earlier studies from our lab, this likely arises from this construct having the longest single-stranded tail [14]. From a biological standpoint, this observation indicates that the very 5’ end of the 5’-UTR can activate PKR. Moreover, the 5’-SL continued to activate PKR at a very high level after CIP treatment indicating activation of PKR both with a 5’-triphosphate and a 5’-OH. It has been reported that a 5’-triphosphate is sometimes processed to a 5’-monophosphate in bacteria by action of a pyrophosphatase [43]. We previously showed that 5’-OH, -p, and –pp are recognized similarly by PKR [15]. The ability of the 5’-SL construct to activate PKR both with and without a 5’-triphosphate supports PKR’s ability to recognize a bacterial RNA as foreign no matter its modification status. Given that most cellular RNAs have a m7G cap at their 5’-ends, activation of PKR by an RNA with both a 5’-OH and a 5’-triphosphate supports broad spectrum recognition of this bacterial RNA element as foreign. From a molecular standpoint, the mild 5’-triphosphate dependence of the 5’-SL may be due to interactions of PKR with the very long 28 nt 3’ tail.
We also investigated the effect of using physiological Mg2+ concentration on PKR activation using a variety of RNAs. The concentration of Mg2+ in human cells is ~0.5 mM, which is much lower than in classical PKR activation assays or in bacteria. It thus seemed possible that the ability of bacterial RNAs to activate PKR is altered in the cytoplasm of human cells. We found, however, that lowering the concentration of Mg2+ had little effect on PKR activation either in the presence of the classical activator of dsRNA-79 or with the full-length trp 5’-UTR. Lastly, we found that a rRNA-depleted preparation of E. coli total RNA activated PKR under both 0.5 and 4 mM Mg2+ conditions. These data indicate that bacterial RNA can activate PKR under the Mg2+ conditions found in the cell.
In sum, we observed potent activation of PKR by a representative bacterial mRNA from B. subtilis and a total RNA preparation from E. coli at human physiological Mg2+ concentrations. Many of the features that PKR recognizes in the B. subtilis trp 5’-UTR are general and present in many bacterial RNAs. It thus appears that activation of PKR by bacterial RNAs is general and applies to Gram-positive and Gram-negative bacteria. Interestingly, activation of PKR by this mixture of bacterial RNA is within ~10-fold that of perfectly dsRNA-79. During bacterial infection of human cells, bacteria are known to lyse and release their RNA contents. Before this step, certain bacteria are known to secrete factors into human cells [44]. Recent reports that Listeria activates RIG-I and MDA5 by secreting RNA and DNA into mice are consistent with this notion. Thus, it is possible that PKR is also activated by RNA during various stages of bacterial infections.
Several approaches for examining RNA structure transcriptome-wide have been developed recently, including ones that assay RNA folds in vivo [45–48]. Furthermore, studies from the Weissman lab suggest that human RNAs are significantly less structured in vivo than previously thought. Low amounts of structured, protein-free RNA in healthy human cells may be key to understanding how bacterial RNAs, which are generally quite structured, are potent activators of PKR and the innate immune system, and may suggest an explanation for the recently noted absence of riboswitches in the human genome [49]. Future experiments are needed on a transcriptome-wide level on bacterially infected human cells to determine the identity and structure of bacterial RNAs that can activate PKR.
Materials and Methods
Design of PCR Primers, RNAs and Blocking Oligonucleotides
The 79 bp control RNA, dsRNA-79, was prepared by transcribing opposing strands of a portion of the pUC19 vector using two separate hemi-duplex templates. The transcripts were purified by denaturing PAGE and annealed to yield a perfectly double-stranded 79 bp dsRNA, which we previously used as a positive control for PKR activation [15].
All trp 5’-UTRs were transcribed from a template prepared by PCR. The region of interest was amplified from a plasmid that contained the full-length 1–206 sequence, inserting a T7 promoter on the 5’- end. PCR primers used for the trp 5’-UTR truncations are provided below, where the T7 promoter is underlined and any transcribed G’s not present in the native trp 5’UTR sequence in bold font. Such G’s were added to enhance transcription efficiency. Numbers correspond to the first and last nucleotides of the resultant transcript.
1–206 (FL)
TS primer (1): 5′GAAATTAATACGACTCACTATAGGAGCTTAGAAATACACAAGAGTGTG
BS primer (206): 5’CATTGCTCTCACTCCTTATGGC
61–188 (LH)
TS primer (61): 5’GAAATTAATACGACTCACTATAGGTAGCAGAGAATGAGTTTA
BS primer (188): 5’GGCAAGGAGAATGAGAAGATGGC
89–159 (T + tails)
TS primer (89): 5’GAAATTAATACGACTCACTATAGGAGACATTATGTTTATTCTA
BS primer (159): 5’AGGATAAAATACTATATAACAAATAAACCC
142–206 (SD SL + tails)
TS primer (142): 5’GAAATTAATACGACTCACTATAGGATAGTATTTTATCCTCTCATGCC
BS primer (206): see above
1–60 (5’-SL + tails)
TS primer (1): see above
BS primer (60): 5’CTATTCTCTAACTCAACTCATTC
108–159 (T + 3’ tail)
TS primer (108): 5’GAAATTAATACGACTCACTATAGGACCCAAAAGAAGTCTTTC
BS primer (159): see above
108–133 (T)
TS primer (108): see above
BS primer (133): 5’ACCCAAAAGAAAGACTTCTTTTGG
Due to inefficient transcription from the PCR products, trp 5’-UTR truncations 172–200 and 89–133, were transcribed from hemi-duplex templates, where the bottom strand is complementary (underlined) to the T7 promoter. Bold C’s denote where G’s were added to the nascent transcript in order to enhance transcription.
172–200 (SD SL)
BS template (172–200) 5’CATTGCTCTCACTCCTTATGGCAAGGAGAATGAGACCTATAGTGAGTCGTATTAATTTC
89–133 (T + 5’ tail)
BS template (89–133) 5’ACCCAAAAGAAAGACTTCTTTTGGGTAGAATAAACATAATGTCTCCTATAGTGAGTCGATTAATTTC
Sequences of the DNA/LNA chimeric blocking oligonucleotides (BOs) are provided below, where the LNA (locked nucleic acid) substitutions, which were added to strengthen binding, are in bold and DNA bases are non-bold (Exiqon). The LNA content was designed as recommended by the manufacturer.
5’BO: 5’AGAATAAAACATAATG
3’BO: 5’GGATAAAATACTATATAACAAATAA
RNA preparation and purification
In vitro T7 transcription reactions of ~500 µL were carried out on the purified templates described above. Reactions were fully submerged in a water bath at 37˚C for 3–4 h, quenched with an equal volume of 2x formamide loading buffer (95% v/v formamide, 20 mM EDTA), and fractionated on a 10% denaturing PAGE gel. The RNA bands were visualized via UV-shadowing, excised with a razor blade, and extracted overnight into 1x TEN250 [10 mM Tris (pH 7.5), 1 mM EDTA, 250 mM NaCl]. RNA was then ethanol precipitated and resuspended in 1x TE [10 mM Tris (pH 7.5), 1 mM EDTA] and stored at −20 °C. Concentrations of RNA were determined using a Nanodrop spectrophotometer. As appropriate, dephosphorylated RNAs (‘HO-RNAs’) were prepared by treatment with calf-intestinal phosphatase ‘CIP’ (New England Biolabs, NEB). RNAs were then phenol-chloroform extracted, ethanol precipitated, and otherwise handled as described above.
For PKR activation assays, the highest concentration of RNA tested was prepared in 1xTEN100 and renatured at 90 ˚C for 1 min followed by room temperature for 10 min. If LNA blocking oligonucleotides were present, they were included in this renaturation in 2-fold excess over the trp 5’-UTR. For assays that included TRAP (+/− L-Trp), TRAP and L-Trp were added after renaturation and incubated at room temperature for 60 min, which is sufficient to allow complete binding [24], followed by serial dilution. TRAP was present in five-fold excess over the RNA concentration, and L-Trp was present at a final concentration of 1 mM. The Kd between L-Trp-saturated TRAP and the trp 5’-UTR is 93 nM under conditions of 4 mM Mg2+ and 100 mM K+[24], which is similar to the ionic conditions in the standard PKR activation assay. These reaction conditions thus assured that the RNA was saturated with L-Trp-bound TRAP throughout the activation assays; the only exception was the lowest concentration of RNA (0.02 µM) where L-Trp-bound TRAP was present at 0.1 µM, which is equal to the Kd. TRAP in the absence of L-Trp does not bind to RNA [26].
Protein expression and purification
Full-length wtPKR containing an N-terminal (His)6 tag was cloned into pET-28a vector and transformed into E. coli BL21 (DE3) Rosetta cells (Novagen) as previously described [29,14]. Cells were sonicated and purified on a Ni2+ -NTA agarose column (Qiagen) run over a high-salt imidazole gradient on a Bio-Rad FPLC. Purified protein was dialyzed into storage buffer [10 mM Tris (pH 7.6), 50 mM KCl, 2 mM Mg(OAc)2, 10% glycerol and 7 mM β-mercaptoethanol]. Protein concentration was determined spectrophotometrically, aliquotted, flash-frozen, and stored at −80˚C. TRAP was a gift from Prof. Paul Babitzke’s lab, which was purified as previously described [50].
PKR Activation Assays
RNAs were tested for their ability to mediate PKR autophosphorylation. PKR is purified from E. coli in a phosphorylated form so it was first dephosphorylated by lambda protein phosphatase λPP (NEB). PKR and λPP were incubated at 30˚C for 1 h followed by inhibition of the λPP by freshly made 2 mM sodium orthovanadate[21,14]. Subsequently, PKR was incubated at a final concentration of 0.8 µM with various concentrations of RNA, RNA/BO, or RNA/TRAP (+/− L-Trp). Reactions were in PKR activation buffer [20 mM HEPES (pH 7.5), 4 mM MgCl2, 50 mM KCl] and 1.5 mM DTT, 100 µM ATP (Ambion) plus 15 µCi [γ-32P]-ATP. Standard incubation conditions were 30 ˚C for 10 min, followed by quenching with SDS loading buffer. Samples were fractionated on a 10% Bis-Tris (Novex) SDS PAGE gel, dried, and exposed to a storage PhosphorImager screen (Molecular Dynamics). Phosphorylated PKR bands were detected using a Typhoon PhosphorImager and quantified with ImageQuant. Each activation assay gel contains one negative control, a sample containing 1x TEN100 instead of RNA, as well as a positive control, a sample containing 0.1 µM dsRNA-79, which was used to normalize all phosphorylation activities. Each new preparation of dsRNA-79 was titrated to find the optimal concentration for activating PKR, which was 0.1 µM dsRNA-79 (Fig. S1). For activation assays performed at varying Mg2+ concentrations and reaction times, the appropriate concentration of free Mg2+ was added to the RNA alone and incubated at 30˚C for 5 min prior to the addition of PKR activation buffer (Mg2+ omitted) and PKR. For varying time assays, PKR was added to the RNA reaction mixture and aliquots were removed and quenched after 2, 5, 10 or 20 min.
Preparation of total E. coli mRNA
Wild-type MG1655 E. coli cells (a gift from Prof. Paul Babitzke’s lab) were grown overnight in LB broth and pelleted. Cells were treated with the RNAprotect bacterial reagent (Qiagen) to help prevent RNA degradation and then purified by the RNEasy Qiagen kit. A Ribozero kit (Epicentre) was used to deplete rRNAs, which was confirmed by a Bioanalyzer run (Agilent). Ribosomal RNA was depleted to avoid introducing naked rRNA, which is non-physiological.
Supplementary Material
Highlights.
Bacterial mRNA 5’-UTR activates the innate immune sensor PKR
Multiple secondary structural features in the 5’-UTR of trp mRNA from B. subtilis activate PKR
Various bacterial RNA segments display a 5’-triphosphate dependence for PKR activation
Tertiary structure in the 5’-UTR contributes to PKR activation
Bacterial RNAs activate PKR under physiological magnesium concentrations
Acknowledgments
We thank Paul Babitzke, Smarajit Mondal, and Alexander Yakhnin for providing bacterial reagents. We also thank Paul Babitzke, Rebecca Toroney, and members of the Bevilacqua Lab for critically reading the manuscript. Supported by National Institutes of Health Grant R01GM110237.
Abbreviations
- dsRBD
dsRNA binding domain of PKR
- eIF2α
eukaryotic initiation factor 2α
- IFN
interferon
- PAMP
pathogen-associated molecular pattern
- PKR
protein kinase R
- p*PKR
phosphorylated form of PKR
- TRAP
tryptophan RNA-binding attenuation protein
- 79 bp
dsRNA with 79 bp derived from pUC19
- λpp
lambda phosphatase
- 5’-SL
5’-stem-loop
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 2.Nallagatla SR, Toroney R, Bevilacqua PC. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struc Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munir M, Berg M. The multiple faces of proteinkinase R in antiviral defense. Virulence. 2013;4:85–89. doi: 10.4161/viru.23134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green SR, Mathews MB. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 6.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 8.Toroney R, Nallagatla SR, Boyer JA, Cameron CE, Bevilacqua PC. Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A. J Mol Biol. 2010;400:393–412. doi: 10.1016/j.jmb.2010.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis S, Watson JC. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3’ untranslated regions of human alpha-tropomyosin. Proc Natl Acad Sci U S A. 1996;93:508–513. doi: 10.1073/pnas.93.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, Bevilacqua PC. RNA dimerization promotes PKR dimerization and activation. J Mol Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinicke LA, Bevilacqua PC. Activation of PKR by RNA misfolding: HDV ribozyme dimers activate PKR. RNA. 2012;18:2157–2165. doi: 10.1261/rna.034744.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nallagatla SR, Jones CN, Ghosh SK, Sharma SD, Cameron CE, Spremulli LL, Bevilacqua PC. Native tertiary structure and nucleoside modifications suppress tRNA’s intrinsic ability to activate the innate immune sensor PKR. PLoS One. 2013;8:e57905. doi: 10.1371/journal.pone.0057905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinicke LA, Nallagatla SR, Hull CM, Bevilacqua PC. RNA helical imperfections regulate activation of the protein kinase PKR: effects of bulge position, size, and geometry. RNA. 2011;17:957–966. doi: 10.1261/rna.2636911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5’-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 16.Toroney R, Hull CM, Sokoloski JE, Bevilacqua PC. Mechanistic characterization of the 5’-triphosphate-dependent activation of PKR: lack of 5’-end nucleobase specificity, evidence for a distinct triphosphate binding site, and a critical role for the dsRBD. RNA. 2012;18:1862–1874. doi: 10.1261/rna.034520.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5’-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5:140–144. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleiblo F, Michael P, Brabant D, Ramana CV, Tai T, Saleh M, Parrillo JE, Kumar A, Kumar A. Bacterial RNA induces myocyte cellular dysfunction through the activation of PKR. J Thorac Dis. 2012;4:114–125. doi: 10.3978/j.issn.2072-1439.2012.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bleiblo F, Michael P, Brabant D, Ramana CV, Tai T, Saleh M, Parrillo JE, Kumar A, Kumar A. JAK kinases are required for the bacterial RNA and poly I:C induced tyrosine phosphorylation of PKR. Int J Clin Exp Med. 2013;6:16–25. [PMC free article] [PubMed] [Google Scholar]
- 21.Matsui T, Tanihara K, Date T. Expression of unphosphorylated form of human double-stranded RNA-activated protein kinase in Escherichia coli. Biochem Biophys Res Commun. 2001;284:798–807. doi: 10.1006/bbrc.2001.5039. [DOI] [PubMed] [Google Scholar]
- 22.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Babitzke P, Stults JT, Shire SJ, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994;269:16597–16604. [PubMed] [Google Scholar]
- 24.Schaak JE, Yakhnin H, Bevilacqua PC, Babitzke P. A Mg2+-dependent RNA tertiary structure forms in the Bacillus subtilis trp operon leader transcript and appears to interfere with trpE translation control by inhibiting TRAP binding. J Mol Biol. 2003;332:555–574. doi: 10.1016/s0022-2836(03)00969-0. [DOI] [PubMed] [Google Scholar]
- 25.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babitzke P, Yanofsky C. Structural features of L-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 1995;270:12452–12456. doi: 10.1074/jbc.270.21.12452. [DOI] [PubMed] [Google Scholar]
- 27.McGraw AP, Bevilacqua PC, Babitzke P. TRAP-5’ stem loop interaction increases the efficiency of transcription termination in the Bacillus subtilis trpEDCFBA operon leader region. RNA. 2007;13:2020–2033. doi: 10.1261/rna.719507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2 edit. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 30.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. 3rd edit. Garland, New York: 1994. [Google Scholar]
- 31.Feig AL, Uhlenbeck O. The Role of Metal Ions in RNA Biochemistry. In the RNAWorld. Cold Spring Harbor Press; 1999. [Google Scholar]
- 32.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truong DM, Sidote DJ, Russell R, Lambowitz AM. Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribozyme core. Proc Natl Acad Sci U S A. 2013;110:E3800–E3809. doi: 10.1073/pnas.1315742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang X, Pan T, Sosnick TR. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
- 36.Shelton VM, Sosnick TR, Pan T. Applicability of urea in the thermodynamic analysis of secondary and tertiary RNA folding. Biochemistry. 1999;38:16831–16839. doi: 10.1021/bi991699s. [DOI] [PubMed] [Google Scholar]
- 37.Misra VK, Draper DE. A thermodynamic framework for Mg2+ binding to RNA. Proc Natl Acad Sci U S A. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J Am Chem Soc. 2005;127:8272–4273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge density of divalent metal cations determines RNA stability. J Am Chem Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silverman RH. Viral encounters with 2’,5’-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samuel CE. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 42.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piton J, Larue V, Thillier Y, Dorleans A, Pellegrini O, Li de la Sierra-Gallay I, Vasseur JJ, Debart F, Tisne C, Condon C. Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates. Proc Natl Acad Sci U S A. 2013;110:8858–8863. doi: 10.1073/pnas.1221510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 46.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci. 2015 doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the Trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 2000;275:4519–4524. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]
- 51.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.