SUMMARY
MYC is a highly pleiotropic transcription factor whose deregulation promotes cancer. In contrast, we find that Myc haploinsufficient (Myc+/−) mice exhibit increased lifespan. They show resistance to several age-associated pathologies, including osteoporosis, cardiac fibrosis and immunosenescence. They also appear to be more active, with a higher metabolic rate and healthier lipid metabolism. Transcriptomic analysis reveals a gene expression signature enriched for metabolic and immune processes. The ancestral role of MYC as a regulator of ribosome biogenesis is reflected in reduced protein translation, which is inversely correlated with longevity. We also observe changes in nutrient and energy sensing pathways, including reduced serum IGF-1, increased AMPK activity, and decreased AKT, TOR and S6K activities. In contrast to observations in other longevity models, Myc+/− mice do not show improvements in stress management pathways. Our findings indicate that MYC activity has a significant impact on longevity and multiple aspects of mammalian healthspan.
INTRODUCTION
Myc is a helix-loop-helix leucine zipper transcription factor that is highly conserved among metazoans (Meyer and Penn, 2008). It was discovered as the transforming oncogene of the MC29 avian myelocytomatosis virus, and subsequently as the cellular proto-oncogene activated in Burkitt's lymphoma. Increased expression of the MYC protein strongly promotes cell proliferation, and has been documented as a frequent event in a wide variety of human cancers (Dang, 2012).
By interacting with partners such as MAX and ZBTB17 (MIZ1), MYC can either activate or repress transcription (Meyer and Penn, 2008). Much effort has been focused on understanding how MYC influences signaling networks, and it has emerged as a major regulatory hub. In addition to its role in cancer, it is also critically involved with many essential cellular processes, and the mouse knockout is embryonic lethal. By conservative estimates, 15–20% of all genes are directly regulated by MYC, including genes that play key roles in metabolism, ribosome biogenesis, cell cycle, apoptosis, differentiation, and stem cell maintenance (Dang, 2012).
While age does not have a significant effect on Myc expression in any mouse tissue examined (Zahn et al., 2007), many of the biological processes regulated by MYC have also been implicated in aging and age-associated diseases. MYC upregulates major biosynthetic pathways leading to cellular growth and proliferation, and enhances energy production through glycolysis and oxidative phosphorylation (Dang, 2012). In contrast, calorie restriction (CR) and reduction of insulin/IGF-1 signaling promote longevity (Gems and Partridge, 2013). MYC also increases protein synthesis by positively regulating ribosome biogenesis (Brown et al., 2008), while reducing translation can extend lifespan (Johnson et al., 2013).
MYC overexpression results in an increase in reactive oxygen species (ROS) and DNA damage (Vafa et al., 2002), which are believed to contribute to the progression of aging (Hoeijmakers, 2009). Stem cell populations decline in number and functionality with normal aging (Cho et al., 2008; Jang et al., 2011), and ectopic MYC expression depletes stem cell populations (Eilers and Eisenman, 2008). MYC may also affect the inflammatory state that accompanies aging, since it directly regulates expression of some cytokines (Whitfield and Soucek, 2012) and may influence the composition of the leukocyte population via its roles in proliferation and stem cell maintenance (Eilers and Eisenman, 2008; Wang et al., 2011a).
The overall trend suggested by this evidence is that increased MYC activity promotes several processes that have been connected with aging and age-associated diseases. To address the role of Myc in aging, given that a complete loss of Myc is embryonic lethal while overexpression promotes cancer, we established a partial loss of function (hypomorphic) model in the mouse. We previously found that cells knocked out for one copy of the Myc gene display a variety of mild but distinct phenotypes, including reduced rates of proliferation (Mateyak et al., 1997). Similarly, heterozygous (Myc+/−) mice appear normal and healthy, but have a 20% smaller body mass (de Alboran et al., 2001; Trumpp et al., 2001). We used this simple constitutive hypomorphic model to address the consequences of reduced MYC activity on aging.
RESULTS
Experimental Strategy
Mice with all coding exons of one copy of the Myc gene flanked by LoxP sites (de Alboran et al., 2001) were bred to mice expressing germline Cre recombinase, converting the floxed allele to a deletion, and subsequently backcrossed to C57BL/6 for 10 generations. The expected decreases of MYC mRNA and protein levels in Myc+/− mice were confirmed in tail fibroblasts and several tissues (Figure 1A; Figure S1A–C). MYC chromatin immunoprecipitation of liver extracts from Myc+/− animals reproducibly gave lower yields, indicating that in vivo a smaller fraction of DNA was bound by MYC (Figure S1D).
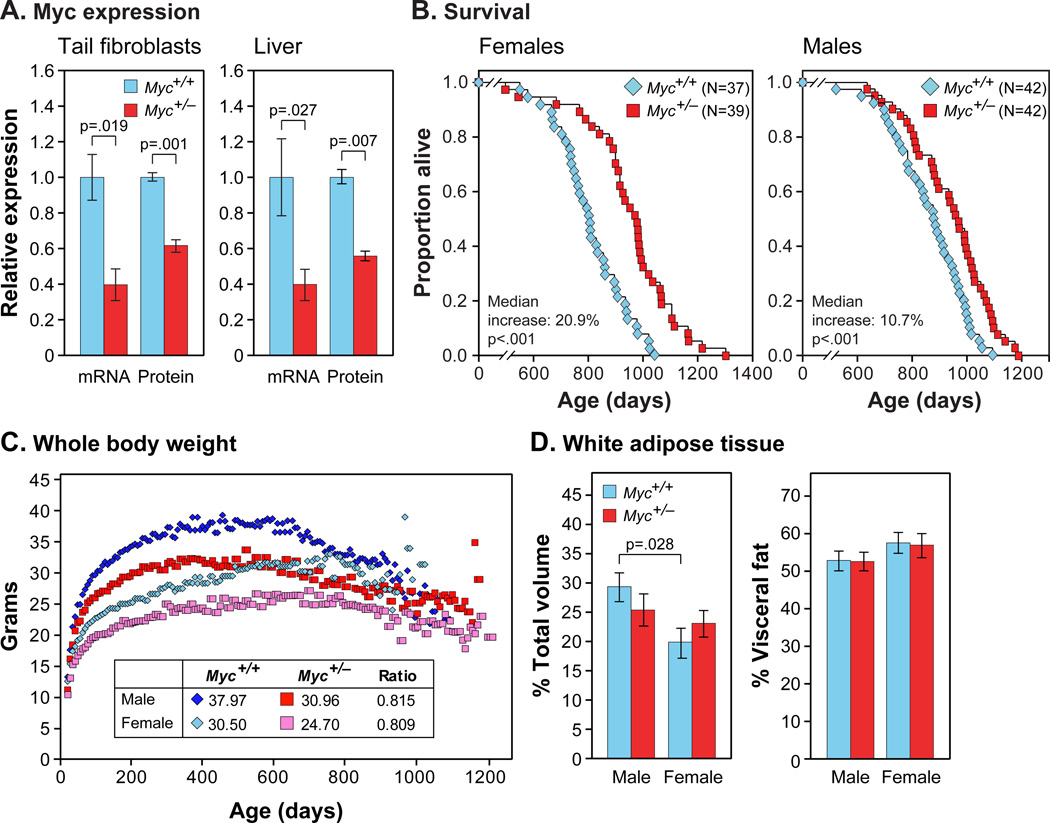
Figure 1. MYC Expression, Longevity, Body Mass and Composition.
(A) MYC mRNA and protein expression. mRNA was measured by qRT-PCR. Protein was measured by immunoblotting of tail fibroblasts or by IF of liver sections. Fibroblasts: N=3, 3 months, females. Liver: N=3–5, 5–9 months, both sexes.
(B) Survival of Myc+/+ (blue) and Myc+/− (red) mice. Each data point shows one animal (N, number of animals in each cohort).
(C) Lifelong trends of whole body weights. Median weights (3-week sliding window) of the aging cohorts in (B). Inset: weights at 500 days and relative weights of Myc+/− animals.
(D) Relative adiposity. Animals were scanned by micro-CT imaging. Left: volume of all white fat as percent of total volume of the animal. Right: volume of visceral fat as percent of total fat. N=6, 5 months.
Myc+/− Mice Have Increased Longevity
Large cohorts of both sexes and genotypes were maintained in a barrier facility and allowed to die of natural causes. A highly significant increase in median lifespan was observed: 10.7% for males, 20.9% for females, and 15.1% for both sexes combined (Figure 1B; Tables S1, S2). We do not know the reason for the greater effect in females; we note, however, that control (Myc+/+) females were shorter lived than males (Figure S1E), an observation that has been reported in several colonies of C57BL/6 mice (Ladiges et al., 2009). Thus, while Myc+/+ males lived 8.8% longer than Myc+/+ females, Myc+/− males and females had equivalent lifespans. Maximum lifespans were commensurately increased, with nearly all of the mice surviving to the longest-lived decile being of Myc+/− genotype. The instantaneous mortality rate was lower for Myc+/− mice across all ages (Figure S1F), indicating that the health benefits are not limited to a particular age.
Myc+/− Mice are Smaller and Develop and Reproduce Normally
Myc+/− mice are healthy, robust and can be group-housed with their Myc+/+ littermates without any adverse consequences. Animals of both sexes are 15–20% smaller as adults (Figure 1C). These differences were apparent at weaning and the weight curves of Myc+/+ and Myc+/− mice were parallel over time. Mass has been noted as a predictor of longevity in mice, although this effect is strain specific (Anisimov et al., 2004). We found no correlation between longevity and the weight attained by individual mice within any cohort (Figure S1G).
The decreased size of Myc+/− mice is proportional across all parts of the body, with the mass ratio of major organs to total body weight not significantly different between genotypes (Figure S1H). Prior examination of Myc hypomorphic mice did not find significant changes in cell size among several organs, which we confirmed in our animals (Figure S1I). Of particular interest was adipose tissue, since in several long-lived mouse models reduced adiposity has been associated with longevity. Myc+/− and Myc+/+ mice have a similar proportion of adipose tissue relative to total body volume, and also have similar proportions of visceral and subcutaneous adipose depots (Figure 1D). Consistent with this, Myc+/− and Myc+/+ mice had similar levels of both leptin and adiponectin in serum at both 6 and 22 months, implying similar adipose tissue function (Figure 2A,B). The reduced size of Myc+/− mice may be due to their lower levels of serum IGF-1, which we documented in both young and old animals (Figure 2C).
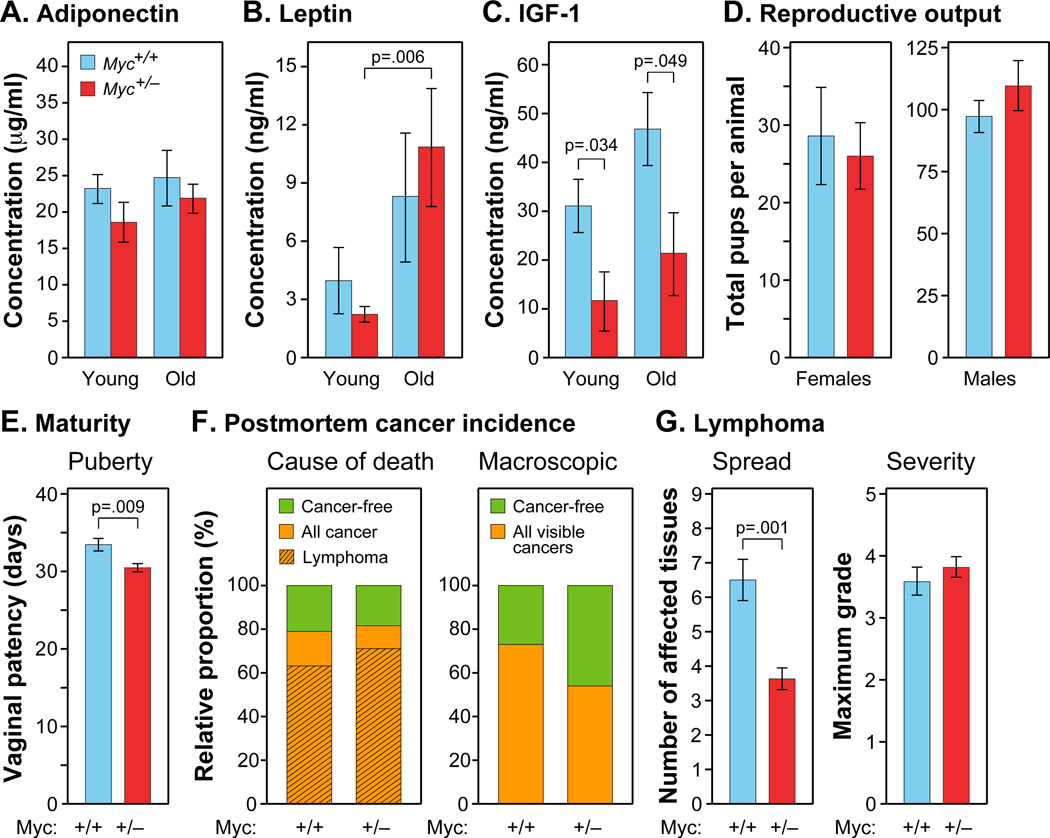
Figure 2. Metabolic Hormones, Fecundity and Cancer Incidence.
(A–C) Levels of adiponectin, leptin, and free IGF-1 in plasma. Blood samples were collected after overnight fasting. N=5–12, 5 and 22 months, females.
(D) Lifetime reproductive output. Myc+/− and Myc+/+ females (N=5) were bred with Myc+/+ males (left), and Myc+/− and Myc+/+ males (N=7–9) were bred with Myc+/+ females (right).
(E) Sexual maturation of females. N=12–19.
(F) Left: Cause of death determined by a veterinary pathologist. N=19 Myc+/+, 38 Myc+/−, both sexes. Right: Incidence of macroscopic cancer noted at time of autopsy (Myc+/+, 73.4%; Myc+/−, 53.7%; p=0.03). N=64 Myc+/+, 67 Myc+/−, both sexes.
(G) Histopathological analysis was used to determine the spread (left; number of affected tissues) and severity (right; maximum grade) of lymphoma. N=12 Myc+/+, 27 Myc+/−, both sexes.
The concept of a tradeoff between longevity and reproduction, namely that increased longevity may bear the cost of decreased efficacy or duration of reproductive ability, has been widely discussed. We tested both sexes for reproductive fitness and longevity and found no significant differences between the two genotypes (Figure 2D; Figure S2A–C). Myc+/− females displayed slightly accelerated vaginal patency (Figure 2E), indicating that sexual maturation is not retarded. The increased longevity of Myc+/− mice is thus not associated with either slower development or reduced fecundity. We also performed skin wound-healing assays and did not find statistically significant differences between the genotypes, even in old animals (Figure S2D).
The Major Cause of Death is Lymphoma in Both Genotypes
We performed autopsies on all adequately preserved mice at time of death and a subset of animals were subjected to a histopathological analysis of 18 tissues. 79% of Myc+/+ and 81% of Myc+/− mice were deemed to have died of cancers (of which 80% and 87%, respectively, were lymphomas; Figure 2F). The spectrum of pathologies was typical of the C57BL/6 strain of mice and very similar in both genotypes (Table S3). Hence, the increased longevity of Myc+/− mice is not due to fewer cancer-caused deaths.
We however also noted evidence for decreased progression of cancer in Myc+/− animals. First, careful macroscopic examination at time of autopsy revealed fewer tumors visible to the naked eye (Figure 2F). Second, histopathological analysis showed that by the time of death lymphoma spread to significantly fewer organs (Figure 2G). Third, while the severity (maximum grade) of lymphoma at time of death was similar in both genotypes, Myc+/− mice survived considerably longer. Others have reported that reducing cancer prevalence alone does not substantially extend lifespan (Matheu et al., 2004). Additional data from multiple organ systems presented below indicate that other effects of the Myc+/− genotype are likely to contribute to the extended lifespan.
Gene Expression Analysis Points to Changes in Metabolism, the Immune System and a Unique Expression Signature
Microarray expression profiling was performed on liver, skeletal muscle, and white adipose (gonadal) tissues in 5 and 24 month old animals. A principal component analysis showed that the first component was tissue, the second age, and the third genotype, representing 61.6%, 30.0%, and 2.9% of the total variability, respectively (Figure 3A; Table S4). The relatively small effect of the Myc+/− genotype is notable, accounting for 10-fold fewer changes than those due to age. For all three tissues, both the number of differentially expressed genes and magnitude of average change with age were lower in Myc+/− animals (Figure 3B). This reduction in the apparent aging of the transcriptome suggests that Myc+/− mice are long-lived because of widespread changes that affect multiple tissues.
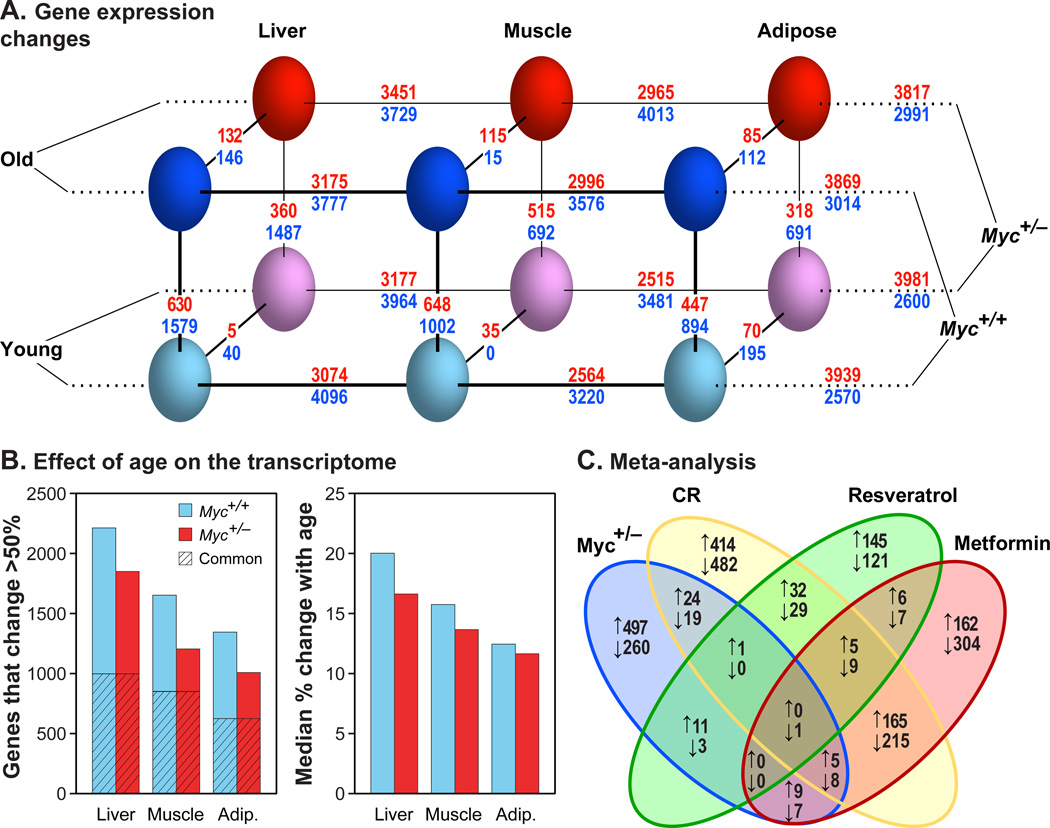
Figure 3. Transcriptome Analysis.
(A) Gene expression changes. Three parameters were compared: genotype (Myc+/+, Myc+/−), age (5, 24 months) and tissue (liver, muscle, adipose). Genotype, age, and tissue comparisons are connected with diagonal, vertical and horizontal lines, respectively. The number of genes changing expression is shown above and below each line. Red numbers: genes upregulated in Myc+/+ versus Myc+/− animals (MYC activated genes); blue numbers: genes upregulated in Myc+/− versus Myc+/+ animals (MYC repressed genes). A 1.5-fold cutoff and a FDR threshold of <5% were used. N=5–8 male animals.
(B) Effect of age on the transcriptome. Left panel: number of genes whose expression changes with age by more than 50% in either direction; hatched area represents genes in common between the two genotypes. Right panel: the median change in expression with age (expressed as %) across all expressed genes.
(C) Meta-analysis. Differentially expressed genes in Myc+/− versus Myc+/+ animals were compared with genes similarly recovered in studies of calorie restriction, metformin, or resveratrol treatment (Martin-Montalvo et al., 2013; Pearson et al., 2008). Upward arrows indicate genes upregulated in the long-lived condition (and conversely for downward arrows). Expression in liver of old male mice was compared. Skeletal muscle showed the same trends.
Differentially expressed genes were enriched in pathways related to the immune system and metabolism, especially that of lipids (Figure S3A). We also evaluated which upstream regulators could explain this pattern of transcription (Table S5). When sorted by significance for the effect of genotype in old animals, regulators of lipid metabolism and the immune system were prominently enriched. Transcription factors that promote lipid biosynthesis and adipogenic fate were predicted to become more active with age in tissues in which adipogenesis and lipid accumulation are considered pathogenic, such as the liver and skeletal muscle, and less active with age in adipose tissue. These changes were reversed in old Myc+/− tissues. The effects of numerous immune system regulators (including interferons, interleukins, colony stimulating factors and NF-κB) were strongly predicted to increase with age in all three tissues, and this effect was predicted to be counteracted in muscle of Myc+/− mice.
Expression of xenobiotic metabolism enzyme (XME) genes increases with age and is further elevated in male mice that are long-lived due to genetic, pharmacologic, or dietary interventions (female mice express these genes at an elevated and mostly constant level (Li et al., 2013). The XME gene expression signature in male Myc+/− mice did not resemble the signatures observed in several mouse longevity models (Table S6). This analysis is consistent with the results of stress tests performed in cell culture with tail fibroblasts established from adult Myc+/− mice (Figure S3D), which did not display increased resistance to any of the stressors previously shown to be counteracted by cells from a variety of long-lived mice. We also note that compared to other longevity models, some of which display very pronounced changes in XME gene expression, the changes in our animals were very modest (Table S6).
In a broader meta-analysis, we compared our gene expression datasets with those of other lifespan and healthspan extending interventions: CR and treatments with resveratrol or metformin. While some of these interventions show considerable overlap with one another in differentially expressed genes, especially CR and metformin (Martin-Montalvo et al., 2013), none were similar to the Myc+/− signature (Figure 3C; Figure S3B). A meta-analysis based on Gene Ontology (GO) pathways also failed to indicate substantial overlap (Figure S3C). As before, CR and metformin showed much greater similarity to each other than either with Myc+/−. The few universally enriched GO terms, however, further implicated metabolism and inflammation; for example, in muscle the only commonly enriched GO term was immune response, while in liver high density lipoprotein binding and hormone activity were two of only five enriched GO terms.
Myc+/− Mice Show Evidence of Improved Healthspan in Multiple Tissues
The pathological effects of aging in several organ systems were found to be attenuated in Myc+/− mice. Cardiac fibrosis increases with age and in diabetes and obesity, and has been found to be reduced in several long-lived mouse models (Dai et al., 2009). Myc+/− mice had less cardiac fibrosis in old age than Myc+/+ animals (Figure 4A). An important component of healthspan in females is osteoporosis (Syed and Melim, 2011). In Myc+/+ females, both bone volume and trabecular number declined with age, whereas trabecular spacing increased (Figure 4B,C). Using all these parameters, old Myc+/− mice were indistinguishable from young Myc+/+ animals, indicating that Myc+/− females do not develop osteoporosis by 22 months of age. Musculoskeletal and neurological performance also decline with age, and the rotarod test is a frequently used measure of motor coordination that reflects these parameters. Old Myc+/− mice were able to remain on the rotarod nearly twice as long as Myc+/+ animals of the same age (Figure 4G), indicating an attenuation of the effects of aging on motor function.
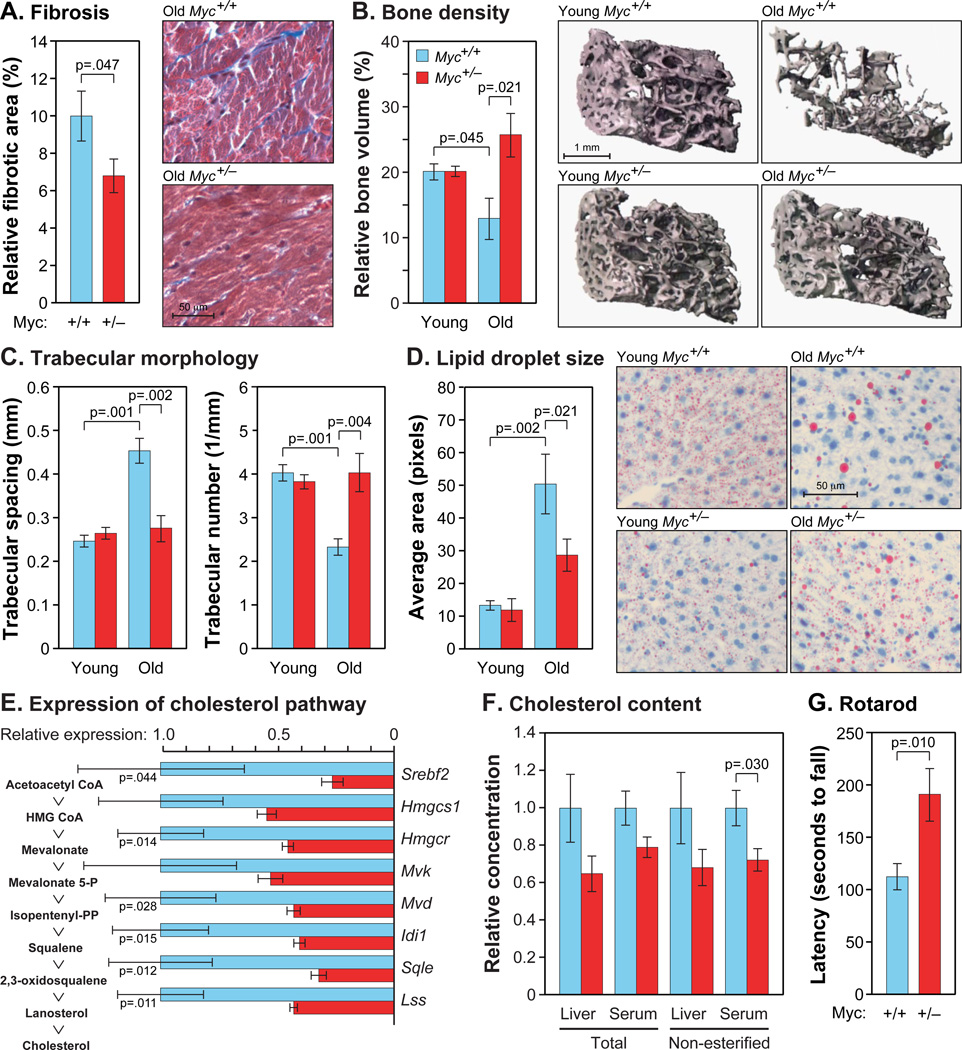
Figure 4. Amelioration of Age-Associated Phenotypes.
(A) Cardiac fibrosis was scored in ventricular cross sections using Masson’s trichrome stain. N=11–14, 22–24 months, both sexes.
(B) Osteoporosis in females was assessed using micro-CT analysis. N=3–7, 5 and 22 months.
(C) Trabecular spacing and number were scored by micro-CT, as above.
(D) Liver sections were stained with Oil Red O. N=6, 5 and 24 months, males.
(E) Gene expression in liver was measured by qRT-PCR. Intermediates in the cholesterol biosynthetic pathways are shown on the left, and the corresponding genes on the right. Data are normalized to Myc+/+ for each comparison. N=4, 24 months, males.
(F) Total and non-esterified cholesterol in liver extracts and serum. Normalized to Myc+/+. N=5–6, 24 months, males.
(G) Animals of average weight were chosen for rotarod tests, and their performance was corrected for their weight. N=3–4, 24 months, males.
See also Figure S4.
Lipid metabolism is strongly affected by aging. Hepatic lipid droplets (LDs) are small membrane-bound organelles that store fatty acids and cholesterol and regulate triglyceride metabolism (Fujimoto and Parton, 2011). LD activities occur mainly at their surface, and hence their size rather than aggregate volume is a relevant measure of functional capacity. We found that while total hepatic LD area decreased ~2-fold in Myc+/+ animals with age (Figure S4A), average LD size increased by over 3.5-fold (Figure 4D). In contrast, total LD content in Myc+/− mice did not change, and LD size increased to a lesser extent. The relative preservation of LD surface area in Myc+/− animals is also consistent with increased expression of genes involved in their biogenesis (Plin2, Cidec, Fitm1).
Another important healthspan parameter of lipid metabolism is cholesterol. Cholesterol levels in mouse liver increase with age, and interventions that promote longevity, such as CR, result in decreased cholesterol synthesis (Tsuchiya et al., 2004). In our microarray datasets the expression of cholesterol biosynthetic genes increased with age in Myc+/+ mice. In contrast, almost all genes in this pathway showed lower expression in Myc+/− animals, which we confirmed by qRT-PCR (Figure 4E; this included the rate limiting enzyme, Hmgcr, and the master transcriptional regulator, Srebf2). Furthermore, both esterified and non-esterified cholesterol was reduced in serum of Myc+/− mice, as well as in liver, where it is synthesized and stored (Figure 4F).
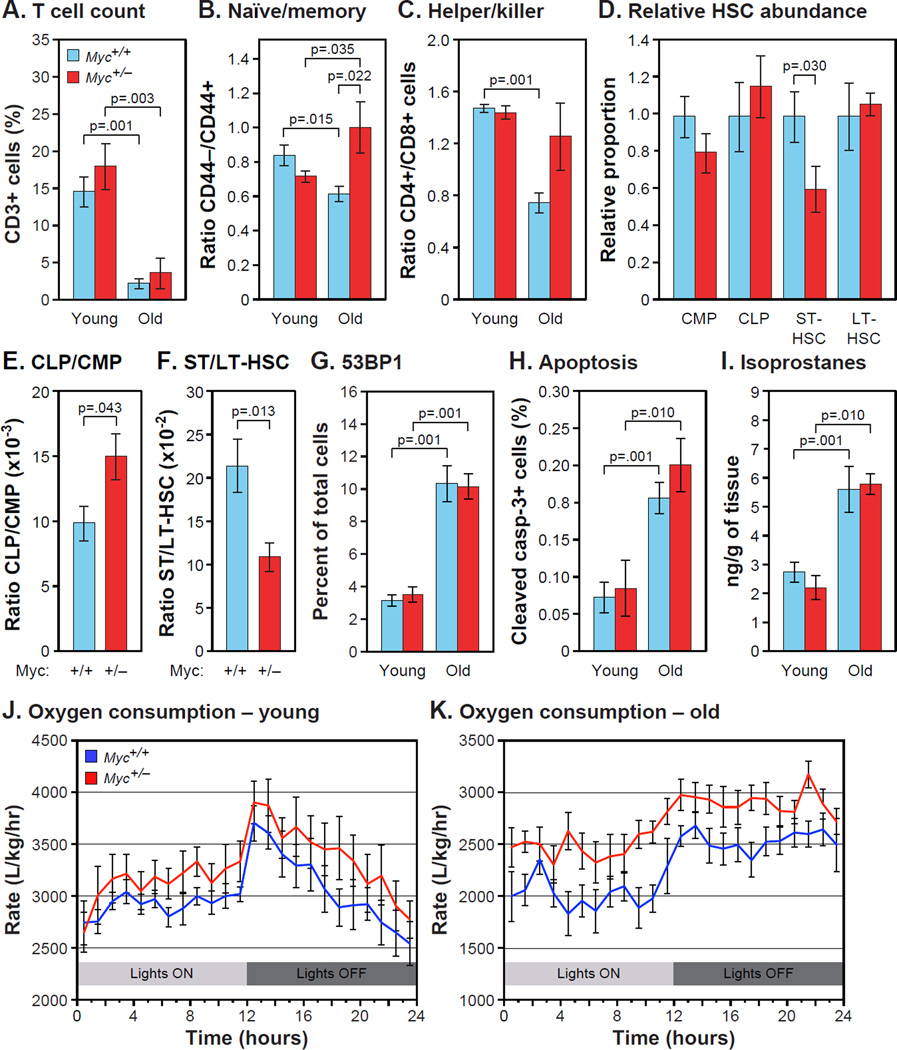
Immunosenescence results in part from thymic involution as well as replicative senescence of some T cell populations (Akbar and Henson, 2011). Since few T cells are produced by the thymus in older animals, the T cell pool is maintained by the expansion of existing memory T cells, which is greater for cytotoxic CD8+ than regulatory CD4+ T cells (Goronzy and Weyand, 2005). Naïve T cells do not proliferate. As previously reported, the ratios of both CD4+ to CD8+ T cells and of naïve to memory T cells declined with age in normal (Myc+/+) mice, but both were significantly elevated in Myc+/− animals at 24 months of age (Figure 5B,C). We found no effect of genotype on total T cell levels (Figure 5A), the presence of macrophages in several tissues (Figure S4B), or total white blood cell counts (Figure S4C). These results indicate that Myc+/− animals can maintain younger and presumably healthier proportions of T cell populations.
Figure 5. Immunosenescence, Stress Defenses and Metabolic Activity.
(A) Total T cells (CD3+) were analyzed by flow cytometry as % of total peripheral lymphocytes. N=8–12, 5 and 24 months, males.
(B) Ratio of naive to memory T cells (CD44−/CD44+), and (C) ratio of helper to killer T cells (CD4+/CD8+) were measured in the same samples.
(D) Proportions of common myeloid and lymphoid progenitors (CMP, CLP), and short-term and long-term hematopoietic stem cells (ST-HSC, LT-HSC) were scored as % of Lin− cells in bone marrow (tibia and femur). N=6, 16 months, females. Normalized to Myc+/+ for each comparison.
(E) Ratio of CLP to CMP, and (F) ratio of ST-HSC to LT-HSC in the same samples.
(G) 53BP1-positive cells were visualized by IF in liver sections. N=5–6, 5 and 25 months, males.
(H) Apoptotic cells in liver were identified by IF with an antibody to cleaved caspase-3. N=6–8, 5 and 22 months, females.
(I) F2 isoprostane levels were measured in liver extracts using gas chromatography and mass spectrometry. N=4–7, 5 and 23–27 months, males.
(J) O2 consumption by young animals over a 24 hour period. Statistical significance in was computed using two-way ANOVA (time, genotype). Genotype factor was significantly different; p<0.001. N=8, 5 months, both sexes.
(K) O2 consumption by old animals. P(genotype) <0.001. N=7–8, 17–22 months, males.
See also Figure S5.
We next examined age related changes in the bone marrow and thymus where T cells develop and mature. In C57BL/6 mice the thymus begins to involute at 10 months of age and very little remains by 20 months. Thymic mass was slightly decreased in Myc+/− mice although the rate of involution was not changed; adiposity and the proportion of senescent cells were also not affected by genotype (Figure S4D,E). Hence, the younger T cell profiles in Myc+/− animals are unlikely to be caused by reduced thymic involution.
Hematopoietic stem cells (HSC) differentiate through several subpopulations: long term HSC (LT-HSC) differentiate into short term HSC (ST-HSC), which can then differentiate into either common myeloid progenitors (CMP) or common lymphoid progenitors (CLP). With normal aging, the CLP/CMP ratio decreases by about 3-fold (Cho et al., 2008). Flow cytometry of HSC showed that 16 month old Myc+/− mice had a higher CLP/CMP ratio relative to Myc+/+ animals (Figure 5D,E). This is further evidence for decelerated aging of the hematopoietic system, and might be responsible for the observed changes in T cell proportions. Consistent with this, we found that Myc+/− mice have a lower ratio of ST-HSC to LT-HSC (Figure 5F), suggesting that LT-HSC can persist in greater numbers and/or functional capacity into old age.
Myc+/− Mice Do Not Show Changes in Pathways Linked to Macromolecular Damage
A commonly invoked cause of aging is the accumulation of damage to macromolecules, particularly from oxidative stress. Genotoxic stress, manifested as various forms of DNA damage, increases dramatically with age (Hoeijmakers, 2009). We examined the frequency of DNA damage foci, and while we saw clear evidence of the age-associated increase, the Myc genotype had no effect (Figure 5G). Genotoxic stress and other forms of damage can lead to apoptosis, which has been found to rise with age in several tissues (Kujoth et al., 2005). While we observed the increase with age, the changes were equivalent in Myc+/+ and Myc+/− mice (Figure 5H). Genotoxic stress is also a major trigger of cellular senescence. As with apoptosis, however, Myc genotype did not affect the age-associated increase in cellular senescence (Figure S4E).
The cyclin-dependent kinase inhibitors p21 (Cdkn1a) and p16 (Cdkn2a) are important regulators of cellular senescence. As expected, both p21 and p16 were upregulated with aging (Figure S4F,G). The expression of p21, which responds strongly to DNA damage and ROS. was unaffected by Myc genotype in five tissues. The response of p16, whose regulation is not well understood, varied between tissues: it was unaffected in Myc+/− relative to Myc+/+ mice in heart, reduced in liver and spleen, and increased in lung. F2-isoprostanes, products of lipid peroxidation, are a sensitive and accurate biomarker of oxidative status. As expected, we found that levels of F2-isoprostanes rose with age, but the changes were the same in Myc+/+ and Myc+/− animals (Figure 5I). Hence, Myc+/− animals do not seem to be protected from age-associated increases in ROS, or from the consequences of this and other forms of stress, such as DNA damage, apoptosis and senescence.
Effects on Metabolic Pathways that Regulate Aging
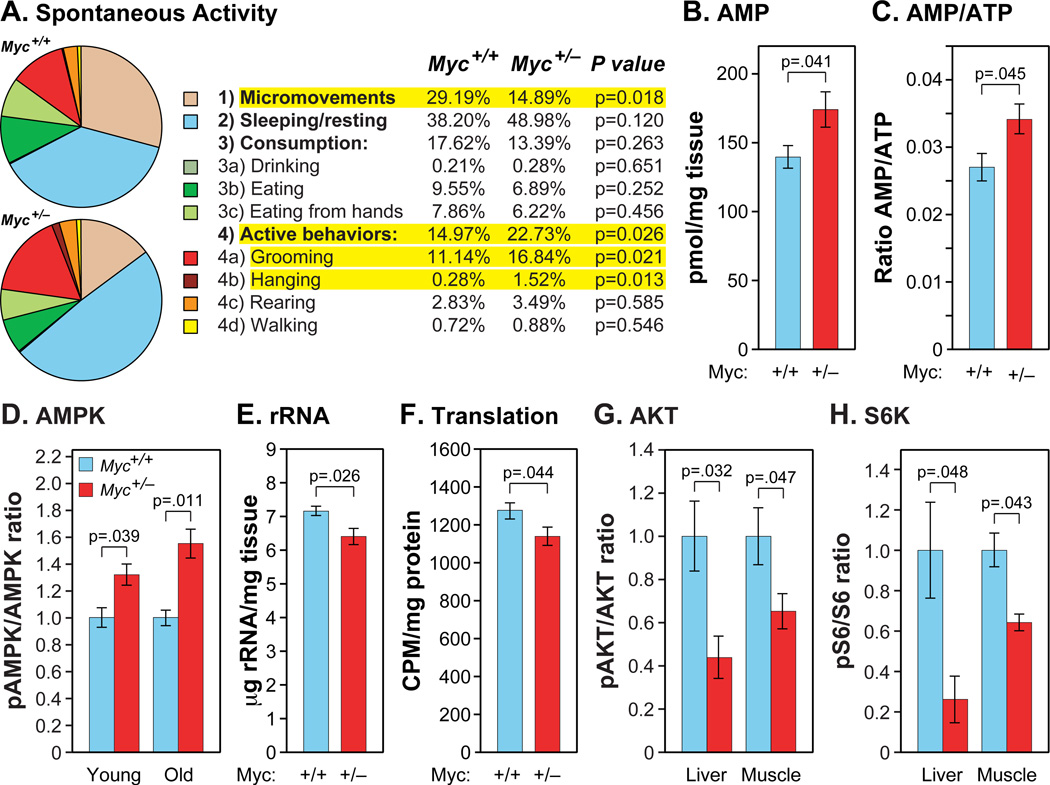
Metabolic rate, measured by O2 consumption and CO2 production, declines during normal aging, and is increased by CR and in the long lived Ames and GHR-KO dwarf mice (Bartke and Westbrook, 2012). Interestingly, Myc+/− mice have a significantly higher metabolic rate than Myc+/+ animals (Figure 5J,K; Figure S5A,B). Although smaller in magnitude, this trend is already apparent in young Myc+/− mice. Myc+/− animals also have higher food and water consumption (Figure S5C,D). Animals of both genotypes had equivalent body temperatures (Figure S5E). The higher metabolism of Myc+/− mice might be needed to thermoregulate their smaller body mass, which would be expected to predispose to hypothermia. A small but significant increase in mitochondrial copy number was found in skeletal muscle (Figure S5F). We also assessed spontaneous activity using a home-cage monitoring system based on automated computer analysis of continuous video recordings (Jhuang et al., 2010), and found that 16–18 month old Myc+/− mice displayed a higher level of active behaviors and a lower level of micromovements (Figure 6A; Figure S6A–D). Of the active behaviors, hanging from the wire cage roof was notable as being almost completely absent in Myc+/+ animals. The large difference in this high-energy behavior is consistent with the notably improved rotarod performance (Figure 4G) and lack of osteoporosis (Figure 4B,C) in older Myc+/− animals.
Figure 6. Spontaneous Activity, Energy Metabolism and Signaling Pathways.
(A) Spontaneous home cage activity. Of the 4 categories of behaviors (micromovements, sleeping, consumption, and active behaviors), micromovements and active behaviors were statistically different. N=6, 16–18 months, males.
(B) AMP concentrations in extracts of muscle. N=5–7, 25–30 months, both sexes.
(C) AMP to ATP ratio in the same samples (B).
(D) The ratio of phosphorylated (Thr172) to total AMPKα in muscle was determined by immunoblotting. Data are normalized to Myc+/+ for each comparison. N=4, 9–11 months, females; N=3, 25–30 months, both sexes.
(E) Ribosomal RNA content of liver. N=4, 23–25 months, female.
(F) Translation rates in live animals (liver) were assessed using 3H-phenylalanine incorporation into total protein. Muscle showed the same trend. N=5, 5–7 months; males.
(G) The ratio of phosphorylated (Ser473) to total AKT in liver and muscle. N=4, 9–11 months, female (same samples as in D). Normalized to Myc+/+.
(H) The ratio of phosphorylated (Ser235/236) to total S6 ribosomal protein in liver and muscle (same samples as in G. Normalized to Myc+/+.
See also Figure S6.
To further investigate these metabolic changes, we measured the levels of the major energy metabolites ATP, ADP and AMP. We found a small but significant increase in AMP levels and the AMP to ATP ratio in muscle of old Myc+/− mice (Figure 6B,C), and the same trends were observed in young animals (Figure S6E,F). There was also a trend towards higher ADP levels (Figure S6G), and similar overall trends in liver (Figure S6I,J). Although small in magnitude, in aggregate these changes are suggestive of a lower energy status in Myc+/− animals, a condition known to activate AMP dependent kinase (AMPK). In agreement, we observed a significantly higher ratio of Thr172-phosphorylated AMPKa to total AMPKa (Figure 6D), indicative of its activation.
MYC is a major regulator of ribosome biogenesis (Brown et al., 2008). Indeed, analysis of our microarray data suggested that genes encoding ribosomal proteins are downregulated in Myc+/− tissues (Figure S6K). We verified this effect by biochemical measurements of rRNA content which showed a small but significant decrease in Myc+/− mice (Figure 6E). Finally, to directly query the rate of translation, we measured the in vivo incorporation of a radioactive amino acid into total protein, and also found a significant decrease in Myc+/− mice (Figure 6F).
Myc+/− mice have reduced levels of circulating IGF-1 (Figure 2C). To more directly examine signaling through this pathway we assessed the activation status of protein kinase B (AKT). We observed a significantly lower ratio of Ser473-phosphorylated AKT to total AKT in liver and muscle of both young and old Myc+/− mice (Figure 6G), indicative of reduced activity. Both AMPK and AKT regulate mTOR, and increased AMPK and decreased AKT activity would promote reduced mTOR activity. We assessed S6 protein kinase activity, a downstream target of mTOR in the protein translation pathway, by measuring the phosphorylation state of ribosomal S6 protein. We found a lower level of phosphorylation in both liver and muscle of adult Myc+/− mice (Figure 6H).
DISCUSSION
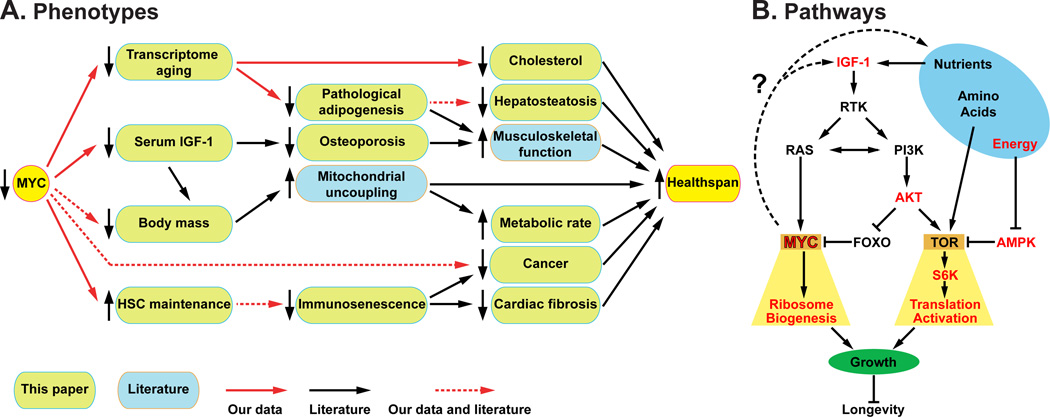
Myc+/− mice have significantly extended median and maximum lifespans in both sexes and a reduced mortality rate across all ages relative to their normal Myc+/+ littermates. Myc haploinsufficiency in Drosophila also extends lifespan, pointing to a deep conservation of the underlying processes (Greer et al., 2013). Myc+/− mice display ameliorated aging phenotypes across a variety of pathophysiological processes in multiple organs, the breadth of which suggests that their increased longevity is not attributable to the prevention of a specific fatal disease, but rather to a broadly increased healthspan.
Aging Myc+/− mice have healthier lipid and cholesterol metabolism, stronger bones, less fibrosis, reduced cancer progression, a higher metabolic rate, improved motor control, and reduced immunosenescence compared to their normal littermates. All of these phenotypes oppose the effects of normal aging and are shared with many other long-lived mouse models (Figure 7, Table S7). For example, CR and Ames dwarf mice also have an increased metabolic rate (Bartke and Westbrook, 2012); CR and metformin-treated mice have lower cholesterol (Martin-Montalvo et al., 2013; Tsuchiya et al., 2004); Ames dwarf, CR and rapamycin or metformin administration ameliorate cancer (Ikeno et al., 2003; Martin-Montalvo et al., 2013; Neff et al., 2013); and CR, Ames dwarf, and metformin-treated mice have better motor coordination (Brown-Borg et al., 2012; Lanza et al., 2012; Martin-Montalvo et al., 2013). All of these long-lived models show reduced body mass. However, Myc hypomorphic mice also exhibit contrasting phenotypes; for example, rapamycin administration increases serum cholesterol and does not improve motor coordination (Neff et al., 2013), Ames dwarf mice have decreased bone density (Heiman et al., 2003), and CR reduces fecundity (Lee and Longo, 2011). Compared to most other longevity extending interventions, many of which compromise some aspects of healthy physiology, we have to date not found significant physiological deficits in Myc+/− mice.
Figure 7. The Effects of Myc on Healthspan and Longevity.
(A) Phenotypes of Myc+/− mice demonstrating their interconnectedness and impact on healthspan (they key under the drawing applies to this panel only).
(B) Pathways affected by Myc hypomorphism and their relationship to increased longevity. The components investigated in this report are highlighted in red.
How can MYC coordinate these effects? It took several decades of work to elucidate the complex spectrum of genes regulated by this transcription factor. In addition to directly binding the promoters of its target genes and upregulating them, MYC also exerts strong secondary effects. For example, it can upregulate the expression of other transcription factors, such as TFAM (Gomes et al., 2013). It can modulate gene expression elicited by other factors, such as ZBTB17 (MIZ1) by directly binding to them (Herkert and Eilers, 2010), or by displacing some factors, such as FOXO3A from their promoters (Peck et al., 2013). Finally, MYC regulates a considerable number of miRNA genes, with widespread effects on gene expression (Dang, 2012).
Many MYC targets, direct or indirect, are involved in anabolic and energy producing processes, and their upregulation is believed to be the major mechanism by which MYC promotes cancer (Dang, 2012). The largest category of genes directly upregulated by MYC are involved in protein translation, and regulation of the ribosome biogenesis (RiBi) regulon is believed to be the core and most ancient function of MYC, extending back to primitive holozoans, the first appearance of the MYC-MAX complex in evolution (Brown et al., 2008). We found that the expression of Rpl and Rps genes was coordinately reduced in Myc+/− tissues. In addition, levels of mature rRNA and in vivo translation rates were decreased. Reducing translation, by a variety of means and in multiple species, is well known to extend lifespan. For example (among many others), yeast and nematodes with mutated ribosomal protein genes, Drosophila overexpressing TSC1 or TSC2, and mice fed rapamycin or mutated in S6 kinase 1 are all long lived (Johnson et al., 2013). Given the major and direct role of MYC on RiBi regulon expression, it is likely that at least part of the longevity of Myc+/− mice can be attributed to effects on translation.
TOR is a central hub in the networks that sense nutrients and energy status, and MYC plays a key downstream role in these pathways (Figure 7). For example, in Drosophila MYC was reported to mediate the protein biosynthetic capacity in response to insulin signaling (Teleman et al., 2008). Both TOR and MYC are activated by receptor tyrosine kinase signaling, through PI3K, RAS and AKT, with TOR providing an acute response by post-translationally regulating its targets, and MYC enabling a long-term response though transcriptional regulation. Given the downstream position of MYC, it is remarkable that Myc+/− mice display changes throughout this nutrient and energy sensing network, all of which have been documented to promote longevity: increased AMPK activity, decreased AKT and S6K activities, and even reduced circulating levels of IGF-1. None of the genes encoding components of these pathways (including IGF-1 binding proteins) are regulated by MYC in our expression datasets. AMPK is likely to be regulated secondarily by the reduced energy status elicited by the higher metabolic rate and activity of Myc+/− animals. Increased AMPK activity would then result in reduced TOR and S6K activities. The PI3K and TOR pathways also contain feedback loops that can be affected by MYC, for example, acute deletion of Myc was found to impair TOR signaling during T cell activation (Wang et al., 2011b).
Considering the major longevity models, no set of phenotypes or pathways emerges as a unifying correlation. In other words, extended lifespan can be achieved by amelioration of some age-associated pathophysiologies even if others are left intact. Aging is increasingly being viewed as a segmental process (Wu et al., 2013), and the phenotypes of Myc hypomorphic animals are consistent with this notion. In agreement, a meta-analysis of gene expression showed limited overlap between Myc+/− mice and CR, metformin or resveratrol. The same analysis readily revealed previously reported connections between CR and metformin treatment (Martin-Montalvo et al., 2013), and underscores the apparently unique nature of the Myc-elicited lifespan extension.
How can this observation be reconciled with the prominent position of MYC in nutrient sensing pathways? First, while MYC and TOR are both major regulatory hubs, their downstream outputs are only partially overlapping. For example, while TOR activity opposes autophagy (Johnson et al., 2013), MYC upregulates it (Dey et al., 2013). Second, we have not detected significant changes in stress levels, stress management pathways, or stress responses in Myc+/− animals. These differences resulted in the low overlap scores returned by the meta-analysis, although these methods clearly detected commonalities in pathways related to metabolism and the immune system.
Production of ROS is well known to increase with age. Although manipulation of these pathways has not yet achieved robust lifespan extension in mammals (Salmon et al., 2010), a stress resistance signature is a component of several important longevity models (CR, reduced somatotrophic signaling, metformin treatment, Table S7). The XME pathway, which promotes the removal of ingested or endogenous toxic metabolites, has been strongly linked with longevity in invertebrate models (Gems and Partridge, 2013), and to a more limited extent in the mouse (Li et al., 2013). Although the expression of some XME genes in liver increased less with age in male Myc+/− than Myc+/+ mice, this effect was of much smaller magnitude than in other long-lived models. Furthermore, fibroblasts from Myc+/− mice did not display increased resistance to a variety of chemical stressors. We looked for changes in the production of ROS (F2 isoprostanes), as well as some notable consequences of stress exposure (DNA double strand breaks, apoptosis, cellular senescence, p21 and p16 induction). Although we observed the expected age-associated increases, we did not observe any significant or consistent effects of the Myc+/− genotype. In aggregate, these results indicate that Myc+/− mice can achieve impressive gains in longevity without significant improvements in the efficacy of stress management pathways.
Although it is possible that the phenotypes of Myc+/− mice predominantly originate from one cell type or tissue and are mediated in an endocrine manner, given the diverse and widespread nature of these effects we think this is unlikely. Furthermore, cell-autonomous effects are clearly apparent in cell culture. As floxed Myc alleles are available (de Alboran et al., 2001; Trumpp et al., 2001), investigating the effects of tissue-specific reduction of MYC activity would be of considerable interest. Similarly, the relationship between lifespan and healthspan benefits and the level of MYC activity is not known. We tested a heterozygous (Myc+/−) condition that resulted in ~50% levels of MYC mRNA and protein. Given that Myc hypomorphs down to ~25% expression are viable, it would be interesting to ask whether these animals continue to accrue healthspan benefits.
A general theme emerging from aging studies in multiple species is that "less is better": less food, less somatotrophic/IGF-1 signaling, less anabolic activity, less protein translation, etc., and MYC certainly fits well in this paradigm. The majority of genes studied for their promotion of longevity act in signaling pathways. MYC, as a transcription factor, stands out because of the vast size of its regulome. The Myc gene is strongly upregulated by mitogens, and by virtue of its widespread targets constitutes the "gas pedal" that allows a cell to double its mass every 18–24 hours. It is thus not surprising that MYC is necessary during embryonic development, and that its deregulated expression strongly promotes cancer. Our discovery that long-term reduction of MYC activity robustly extends lifespan greatly extends our understanding of the biology regulated by this fascinating gene. Given the variety of other physiological benefits, further studies of MYC and its targets should be an interesting avenue to explore in human medicine.
EXPERIMENTAL PROCEDURES
Use and Treatment of Animals
Mice were produced and housed in a specific pathogen-free AAALAC-certified barrier facility. All procedures were approved by the Brown University IACUC committee. For longevity studies animals were allowed to die naturally. Both females and males in the longevity study were virgins. Animals of both genotypes and the same sex were housed together.
Statistical and Demographic Analysis
Data are shown as means with SEM (unless stated otherwise). N indicates the number of animals per test group; age and sex are also noted. Student's t-test (unpaired, two-tailed, equal variance) was used for all pairwise comparisons. All relevant p values are shown in the figures; if not shown, the values were >0.05. Demographic data were processed with JMP software to compute mean and median lifespans, SEM, percent increase of the median, and p values (log-rank test) for each cohort. Mortality rate was calculated as log (−log (survival)). Maximum lifespans were calculated as the proportion of each cohort still alive when the total population reached 90% mortality, using Fisher’s exact test to determine statistical significance.
Procedures Performed on Live Animals
To score vaginal patency, mice were examined daily from weaning until vaginal opening was observed. Reproductive longevity and output of males were determined by continuously housing young males of either genotype with Myc+/+ females until pregnancies ceased. Females were replaced every 6 months. For female fecundity, young females of either genotype were continuously mated with one Myc+/+ male. For rotarod tests the apparatus (MedAssociates, Inc., St. Alban, VT) was accelerated continuously from 4 to 40 revolutions per min. O2 consumption, CO2 production, food and water intake were measured using the comprehensive lab animal monitoring system (CLAMS, Oxymax Open Circuit Calorimeter, Columbus Instruments). Spontaneous home cage activity was monitored using fully automated computer vision analysis of continuous video recordings. Wound-healing experiments were performed by introducing one 6 mm diameter full thickness skin punch on the back between the shoulder blades.
Histological Analysis
Mice were euthanized in the morning (8–10 AM) by isofluorane anesthesia followed by cervical dislocation. The dissection was performed rapidly (<3 min) by several trained staff members working in concert on one mouse. Paraffin-embedded specimens were stained with H&E. OCT-embedded specimens were used for the determination of fibrosis (Masson's trichrome stain), lipid content (Oil Red O stain), and cellular senescence (senescence-associated β-galactosidaase stain). Immunofluorescence microscopy was used to quantify MYC protein, cleaved caspase-3, 53BP1 foci and macrophages.
Analytical Procedures
Individual gene expression was measured by qRT-PCR (SYBR Green system, ABI 7900 Fast Sequence Detection instrument). Total RNA was extracted from cells or tissue samples (20–50 mg, stored at −80°C) with Trizol reagent (Invitrogen), purified using the RNeasy Mini kit (QIAGEN), and reverse transcribed using the Taqman kit (Applied Biosystems). Expression profiling was performed using Mouse 1.0 Gene ST arrays (Affymetrix), and analyzed using Expression Console software (Affymetrix) and Ingenuity Pathway Analysis software (Ingenuity Systems). rRNA was purified by affinity capture using the RiboMinus kit (Life Technologies). Chromatin immunoprcipitation was performed with the Magna ChIP kit (Millipore) on nuclei isolated from intact frozen liver tissue. Immunoblotting of proteins was performed using SDS-PAGE of whole cell extracts, electro-transfer onto Immobilon-P membranes (Millipore), and staining with the indicated antibodies. Signals were detected using the LI-COR Odyssey (LI-COR Biosciences) infrared imaging system. Bone density and adipose tissue were assessed using a Scanco Medical Micro-CT 40 instrument on whole animals immediately after euthanasia. Lipids were extracted using the Folch method and cholesterol was measured with the Amplex Red Cholesterol kit (Invitrogen). F2 isoprostane levels were measured using gas chromatography and mass spectrometry. Leptin and adiponectin were quantified in plasma using the Luminex magnetic bead platform (Millipore) with the mouse adipokine panel and the adiponectin single-plex assay. Free IGF-1 in plasma was measured with an ELISA kit from Abcam. Lymphoid and myeloid cells in blood or bone marrow were quantified by flow cytometry. Blood was harvested by cardiac puncture into heparinized tubes, cells were collected by centrifugation, and erythrocytes were lysed with FACS lysing solution (Becton Dickinson). Bone marrow was flushed out of the femur and tibia. In both cases the cell suspensions were stained with the indicated fluorescently-conjugated antibodies and analyzed using a Becton Dickinson FACSAria instrument. To measure AMP, ADP and ATP, frozen tissue specimens were homogenized (Fisher Scientific, PowerGen Model 125), extracted in perchloric acid, and neutralized with K2CO3 on ice. Ion-paired HPLC was run on an Agilent 1200 series instrument using a ZORBAX Eclipse XDB-C18 column (Agilent). To assess protein translation in live animals, L-3H-phenylalanine (Perkin Elmer, 100–140 Ci/mmol) was combined with unlabeled phenylalanine (135 mM), and injected at 1 ml/100g body weight into the tail vein. Animals were kept under anesthesia for the entire duration (0–30 min), sacrificed by cervical dislocation, and tissues were snap frozen in liquid nitrogen. Incorporation of 3H-phenylalanine was measured by TCA precipitation of total protein and scintillation counting. Detailed protocols are provided in the Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant R37 AG016694 to J.M.S. J.W.H. was supported in part by grant F30 AG035592. J.M.S. was a Senior Scholar of the Ellison Medical Foundation and a recipient of the Glenn Award for Research on the Biological Mechanisms of Aging. The Brown Molecular Pathology and Genomics core facilities were supported by NIH grants P42 ES013660 and P30 GM103410. RdC is funded by the Intramural Research Program of the National Institute on Aging, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
GEO accession number for microarray data is GSE55272.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, 6 figures, 7 tables and associated references.
AUTHOR CONTRIBUTIONS
Conceived study: J.M.S. Designed experiments: J.M.S., J.W.H., X.Z., M.D.C. Performed experiments and analysis: J.W.H., X.Z., M.D.C., A.L.P., L.P., J.M., G.B.H., Y.I., Y.Z., B.F., X.L., T.S., W.Q., K.G.B., J.M.S. Contributed to supervision, interpretation and manuscript: Y.I., T.S., H.V.R., R.A.M., K.G.B., R.D.C., H.X., N.N., J.M.S. Wrote the manuscript: J.M.S., J.W.H., X.Z.
REFERENCES
- Akbar AN, Henson SM. Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nat. Rev. Immunol. 2011;11:289–295. doi: 10.1038/nri2959. [DOI] [PubMed] [Google Scholar]
- Anisimov VN, Arbeev KG, Popovich IG, Zabezhinksi MA, Rosenfeld SV, Piskunova TS, Arbeeva LS, Semenchenko AV, Yashin AI. Body weight is not always a good predictor of longevity in mice. Exp. Gerontol. 2004;39:305–319. doi: 10.1016/j.exger.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Bartke A, Westbrook R. Metabolic characteristics of long-lived mice. Front. Genet. 2012;3:288. doi: 10.3389/fgene.2012.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Cole MD, Erives AJ. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics. 2008;9:442. doi: 10.1186/1471-2164-9-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Johnson WT, Rakoczy SG. Expression of oxidative phosphorylation components in mitochondria of long-living Ames dwarf mice. Age (Dordr) 2012;34:43–57. doi: 10.1007/s11357-011-9212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RH, Sieburg HB, Muller-Sieburg CE. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood. 2008;111:5553–5561. doi: 10.1182/blood-2007-11-123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DF, Santana LF, Vermulst M, Tomazela DM, Emond MJ, MacCoss MJ, Gollahon K, Martin GM, Loeb LA, Ladiges WC, et al. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Dey S, Tameire F, Koumenis C. PERK-ing up autophagy during MYC-induced tumorigenesis. Autophagy. 2013;9:612–614. doi: 10.4161/auto.23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc's broad reach. Genes. Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 2011:3. doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 2013;75:621–644. doi: 10.1146/annurev-physiol-030212-183712. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy JJ, Weyand CM. T cell development and receptor diversity during aging. Curr. Opin. Immunol. 2005;17:468–475. doi: 10.1016/j.coi.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Greer C, Lee M, Westerhof M, Milholland B, Spokony R, Vijg J, Secombe J. Myc-dependent genome instability and lifespan in Drosophila. PLoS One. 2013;8:e74641. doi: 10.1371/journal.pone.0074641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman ML, Tinsley FC, Mattison JA, Hauck S, Bartke A. Body composition of prolactin-, growth hormone, and thyrotropin-deficient Ames dwarf mice. Endocrine. 2003;20:149–154. doi: 10.1385/ENDO:20:1-2:149. [DOI] [PubMed] [Google Scholar]
- Herkert B, Eilers M. Transcriptional repression: the dark side of myc. Genes Cancer. 2010;1:580–586. doi: 10.1177/1947601910379012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Ikeno Y, Bronson RT, Hubbard GB, Lee S, Bartke A. Delayed occurrence of fatal neoplastic diseases in ames dwarf mice: correlation to extended longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2003;58:291–296. doi: 10.1093/gerona/58.4.b291. [DOI] [PubMed] [Google Scholar]
- Jang YC, Sinha M, Cerletti M, Dall'Osso C, Wagers AJ. Skeletal muscle stem cells: effects of aging and metabolism on muscle regenerative function. Cold Spring Harb. Symp. Quant. Biol. 2011;76:101–111. doi: 10.1101/sqb.2011.76.010652. [DOI] [PubMed] [Google Scholar]
- Jhuang H, Garrote E, Mutch J, Yu X, Khilnani V, Poggio T, Steele AD, Serre T. Automated home-cage behavioural phenotyping of mice. Nat Commun. 2010;1:68. doi: 10.1038/ncomms1064. [DOI] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Ladiges W, Van Remmen H, Strong R, Ikeno Y, Treuting P, Rabinovitch P, Richardson A. Lifespan extension in genetically modified mice. Aging Cell. 2009;8:346–352. doi: 10.1111/j.1474-9726.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- Lanza IR, Zabielski P, Klaus KA, Morse DM, Heppelmann CJ, Bergen HR, 3rd, Dasari S, Walrand S, Short KR, Johnson ML, et al. Chronic caloric restriction preserves mitochondrial function in senescence without increasing mitochondrial biogenesis. Cell Metab. 2012;16:777–788. doi: 10.1016/j.cmet.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Longo VD. Fasting vs dietary restriction in cellular protection and cancer treatment: from model organisms to patients. Oncogene. 2011;30:3305–3316. doi: 10.1038/onc.2011.91. [DOI] [PubMed] [Google Scholar]
- Li X, Bartke A, Berryman DE, Funk K, Kopchick JJ, List EO, Sun L, Miller RA. Direct and indirect effects of growth hormone receptor ablation on liver expression of xenobiotic metabolizing genes. Am. J. Physiol. Endocrinol. Metab. 2013;305:E942–E950. doi: 10.1152/ajpendo.00304.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, et al. Metformin improves healthspan and lifespan in mice. Nat. Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- Matheu A, Pantoja C, Efeyan A, Criado LM, Martin-Caballero J, Flores JM, Klatt P, Serrano M. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18:2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Neff F, Flores-Dominguez D, Ryan DP, Horsch M, Schroder S, Adler T, Afonso LC, Aguilar-Pimentel JA, Becker L, Garrett L, et al. Rapamycin extends murine lifespan but has limited effects on aging. J. Clin. Invest. 2013;123:3272–3291. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck B, Ferber EC, Schulze A. Antagonism between FOXO and MYC Regulates Cellular Powerhouse. Front Oncol. 2013;3:96. doi: 10.3389/fonc.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB, Richardson A, Perez VI. Update on the oxidative stress theory of aging: does oxidative stress play a role in aging or healthy aging? Free Radic. Biol. Med. 2010;48:642–655. doi: 10.1016/j.freeradbiomed.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed FA, Melim T. Rodent models of aging bone: an update. Curr. Osteoporos. Rep. 2011;9:219–228. doi: 10.1007/s11914-011-0074-z. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Dhahbi JM, Cui X, Mote PL, Bartke A, Spindler SR. Additive regulation of hepatic gene expression by dwarfism and caloric restriction. Physiol. Genomics. 2004;17:307–315. doi: 10.1152/physiolgenomics.00039.2004. [DOI] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- Wang J, Geiger H, Rudolph KL. Immunoaging induced by hematopoietic stem cell aging. Curr. Opin. Immunol. 2011a;23:532–536. doi: 10.1016/j.coi.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011b;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JR, Soucek L. Tumor microenvironment: becoming sick of Myc. Cell. Mol. Life Sci. 2012;69:931–934. doi: 10.1007/s00018-011-0860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4:913–920. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.