Abstract
Human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) is an important target for antiviral therapy against acquired immunodeficiency syndrome. However, the efficiency of available drugs is impaired most typically by drug-resistance mutations in this enzyme. In this study, we applied a nuclear magnetic resonance (NMR) spectroscopic technique to the characterization of the binding of HIV-1 RT to various non-nucleoside reverse transcriptase inhibitors (NNRTIs) with different activities, i.e., nevirapine, delavirdine, efavirenz, dapivirine, etravirine, and rilpivirine. 1H-13C heteronuclear single-quantum coherence (HSQC) spectral data of HIV-1 RT, in which the methionine methyl groups of the p66 subunit were selectively labeled with 13C, were collected in the presence and absence of these NNRTIs. We found that the methyl 13C chemical shifts of the M230 resonance of HIV-1 RT bound to these drugs exhibited a high correlation with their anti-HIV-1 RT activities. This methionine residue is located in proximity to the NNRTI-binding pocket but not directly involved in drug interactions and serves as a conformational probe, indicating that the open conformation of HIV-1 RT was more populated with NNRTIs with higher inhibitory activities. Thus, the NMR approach offers a useful tool to screen for novel NNRTIs in developing anti-HIV drugs.
Human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) plays an important role in HIV-1 replication by catalyzing the conversion of single-stranded RNA into double-stranded DNA. This enzyme is one of the most promising targets for anti-HIV drug development to suppress the production of new viral particles. The structure of HIV-1 RT consists of an asymmetric heterodimer of two subunits, a 66 kDa subunit (p66) containing both polymerase and RNase H domains, and a 51 kDa subunit (p51) containing only a polymerase domain1,2,3. Each polymerase domain is comprised of four subdomains: fingers, thumb, palm, and connection1,3. The p66 subunit carries the functional sites including the polymerase active site, the RNase H domain and the non-nucleoside binding site, whereas p51 provides the structural foundation4.
HIV-1 RT inhibitors can be divided into two classes, nucleoside reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs). NRTIs are nucleoside analogs lacking the 3′-OH group and acts as a chain terminator of DNA synthesis. NNRTIs are small molecules that bind to a hydrophobic pocket located in proximity to the polymerase active site on the p66 subunit5,6. It is expected that NNRTIs are able to circumvent the toxic side effects associated with nucleoside chain termination7. Accordingly, the NNRTI binding pocket is considered to be an important target for further development of novel anti-HIV-1 drugs. Five NNRTIs, nevirapine, delavirdine, efavirenz, etravirine, and rilpivirine, have currently been approved by the U.S. Food and Drug Administration8. However, the efficiencies of these inhibitors are impaired by mutations in HIV-1 RT9, requiring continuous development of novel NNRTIs capable of inhibiting both wild-type and mutated HIV-1 RT enzymes. Hence, a detailed knowledge about the interactions between this enzyme and NNRTIs in solution is crucial for antiviral therapy against acquired immunodeficiency syndrome.
Biophysical and structural approaches are useful for rapid, efficient development of small molecule inhibitors targeting HIV-1 RT. X-ray crystallography offers atomic images of the different binding modes of HIV-1 RT between NRTIs and NNRTIs5,6,8,10,11,12,13. The availability of these crystallographic structures has greatly facilitated the optimization of NNRTIs. Nuclear magnetic resonance (NMR) is also a useful method for studying HIV-1 RT binding to drugs. Although applying the NMR technique to analysis of large proteins remains challenging, this spectroscopic method provides valuable information regarding dynamic aspects of ligand binding. It has been reported that selective isotope labeling with 13C at the methyl side chain of methionine offers useful spectroscopic probes for investigating the structures and dynamics of larger proteins14,15,16,17,18. Zheng et al. previously reported heteronuclear single-quantum coherence (HSQC) spectra for observing signals from the methionine methyl groups of the HIV-1 RT p66 subunit in the absence and presence of nevirapine, with assignments based on the site-directed mutagenesis method16,17. In this study, the response of HIV-1 RT binding to its ligands in solution was probed with methyl 13C resonances.
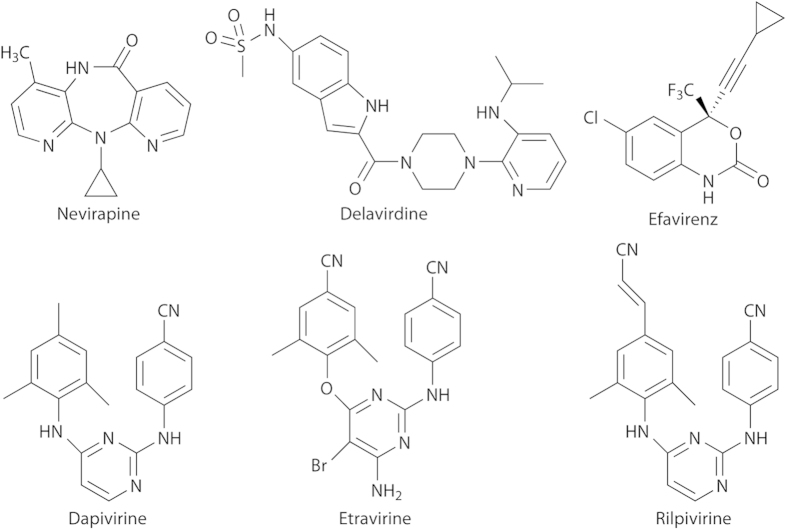
In the present study, we have applied the NMR technique to characterize the interactions of HIV-1 RT with various NNRTIs with different inhibitory activities, nevirapine, delavirdine, efavirenz, dapivirine, etravirine, and rilpivirine (Fig. 1). We found that the methyl 13C chemical shift of M230 in the p66 subunit, which is located in close proximity to the inhibitor binding pocket, serves as a useful indicator of the efficacy of these NNRTIs.
Figure 1. Structures of nevirapine, delavirdine, efavirenz, dapivirine, etravirine, and rilpivirine.
Results and Discussion
Spectral assignments of the apo form of HIV-1 RT with the 13C-labeled p66 subunit
In the present NMR study, HIV-1 RT complex composed of 13C-labeled p66 and unlabeled p51 was prepared by bacterial expression using [methyl-13C]methionine. The recombinant p66 subunit possesses six intrinsic methionine residues and an extra methionine residue at its N-terminus. The 1H-13C HSQC spectrum of the apo form of the 13C-labeled HIV-1 RT protein gave four peaks (supplementary Fig. S1). To assign each methyl resonance, six different mutants of HIV-1 RT were prepared, substituting each methionine in the p66 subunit with leucine16,17. The 1H-13C HSQC spectra of these mutants were compared with those of the wild type, thereby identifying peaks originating from M16, M184, and M357, because these peaks were missing in the spectra of the corresponding mutants (supplementary Fig. S1). The remaining mutants, i.e., M41L, M164L, and M230L, exhibited virtually identical 1H-13C HSQC spectra with those of the wild type, indicating that these methionine residues gave no observable peaks, presumably because their side chains are buried in the protein and thereby have low mobility. These observations are consistent with the 1H-13C HSQC spectral data reported previously16,17. We confirmed that the sharp peak (indicated by an asterisk) was derived from the N-terminal extra methionine because it was eliminated by treatment with methionine aminopeptidase (supplementary Fig. S2).
Spectral changes upon drug binding to HIV-1 RT
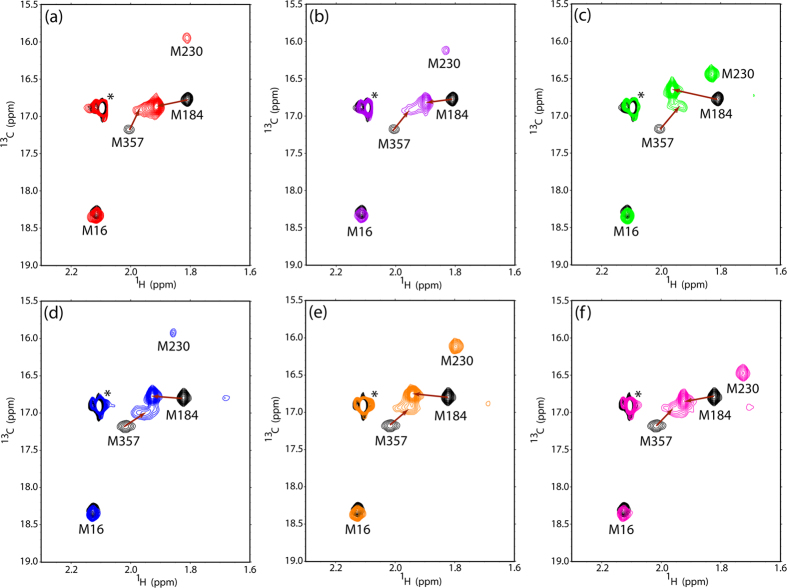
To examine the effects of NNRTIs bound to HIV-1 RT, 1H-13C HSQC spectral data of HIV-1 RT with the [methyl-13C]methionine-labeled p66 subunit were collected in the presence of six NNRTIs, nevirapine, delavirdine, efavirenz, dapivirine, etravirine, and rilpivirine, as shown in Fig. 2. Chemical shift changes of the methionine methyl resonances were observed upon the addition of these NNRTIs to HIV-1 RT. NNRTIs examined induced significant chemical shift changes for the resonances originating from M184 and M357. Moreover, the M230 peak became observable upon addition of these drugs. The chemical shift of the M16 resonance was little affected by drug binding, whereas the M41 and M164 resonances remained unobservable.
Figure 2. Superposition of the 1H- 13C HSQC spectra of apo HIV-1 RT (black) and HIV-1 RT complexed with (a) nevirapine, (b) efavirenz, (c) rilpivirine, (d) delavirdine, (e) dapivirine, and (f) etravirine.
The crystal structures of the apo form without a ligand and the complexed form with these inhibitors suggest that the binding of NNRTIs stabilizes the open conformation of HIV-1 RT. The thumb and fingers domains are more distant than in the closed conformation adopted by the apo form of HIV-1 RT, in which the thumb domain is in contact with the fingers domain with occlusion of the NNRTI-binding pocket1,19,20,21. M184 and M230 are located in the vicinity of this pocket but not directly involved in drug binding. In the drug-binding open conformation, M230 is more exposed to the solvent, in contrast to the apo closed conformation in which this methionine residue gave no observable peak because of its low mobility. Therefore, this methionine signal might serve as a conformational probe for characterizing NNRTI-binding to HIV-1 RT.
Spectral comparison among the six NNRTI-bound forms of HIV-1 RT indicated significant chemical variation of the M230 peak (Fig. 3a). The methyl 13C chemical shift of M230 exhibited a high correlation with anti-HIV-1 RT activity reported by Yang et al.22 (r2 = 0.91 and p = 0.012, *p < 0.05), as shown in Fig. 3b, demonstrating its utility for evaluating and even predicting the efficacies of NNRTIs.
Figure 3.
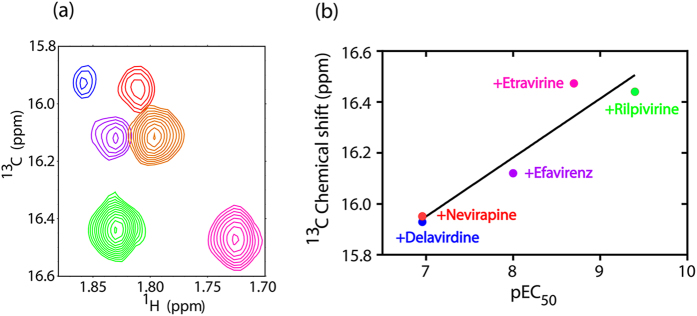
(a) Superposition of 1H-13C HSQC spectra showing the M230 peak of the p66 subunit in HIV-1 RT with nevirapine (red), efavirenz (purple) and rilpivirine (green), delavirdine (blue), dapivirine (orange), and etravirine (magenta). (b) Correlation between anti-HIV-1 RT activity reported by Yang et al.22 and the 13C chemical shift of M230 of the HIV-1 RT p66 subunit upon binding to these inhibitors. Linear regression analyses showed a significant correlation between the chemical shift and pEC50 values (r2 = 0.91, p = 0.012, *p < 0.05).
The 13C chemical shift of the methionine methyl group can refer to side chain conformations, associated with χ3 values: the 13C chemical shifts of 16 ppm and 19 ppm correspond to gauche and trans conformations, respectively, whereas 17 ppm indicates flexibly exchanging trans and gauche conformations23. In the closed apo structure of HIV-1 RT (PDB codes 1DLO and 3DLK), M230 is buried, exclusively exhibiting the gauche conformation, while it occasionally adapted a trans conformation in the rilpivirine-bound open form (PDB code 3MEE), presumably with increased mobility. Under this circumstance, 13C chemical shift of M230 reflects drug-induced population shifts of the open and closed conformations. The intensity of the M230 resonance in the rilpivirine- and etravirine-bound forms of HIV-1 RT was higher than that in the nevirapine-, delavirdine- and efavirenz-bound forms. This indicates that diarylpyrimidine-based inhibitors (DAPYs) such as rilpivirine and etravirine fix the open conformation most tightly, rendering M230 exposed with its highest mobility (with a partial trans conformation) in comparison with nevirapine, delavirdine, and efavirenz. In other words, fixation in open conformation by nevirapine, delavirdine, and efavirenz are not complete. Consistent with this interpretation, the enzyme bound to efavirenz remains under dynamic equilibrium between the open and closed forms24. On the basis of these data, we conclude that the open conformation of HIV-1 RT was more populated in complexes with NNRTIs with higher inhibitory activities.
In summary, we applied NMR spectroscopy to probe NNRTI binding to HIV-1 RT using the methionine methyl 13C resonances from the p66 subunit. We confirmed that the compounds examined in this study share the same binding pocket. Moreover, our NMR data revealed that the methyl 13C chemical shift of M230 can be used as an indicator of the efficacy of NNRTI, offering a useful tool in screening for novel inhibitors in developing anti-HIV drugs.
Materials and Methods
Materials
Powdered rilpivirine, delavirdine, dapivirine, and etravirine were purchased from MedChem Express. Nevirapine and efavirenz were synthesized by Dr. Supanna Techasakul (Department of Chemistry, Faculty of Science, Kasetsart University).
Preparation of HIV-1 RT variants by site-directed mutagenesis
Site-directed mutagenesis was performed to prepare six variants of the p66 subunit of HIV-1 RT, M16L, M41L, M164L, M184L, M230L, and M357L. The recombinant plasmids containing HIV-1 RT genes, pGEX3X 25 for p66 and pET33B26 for p51, were previously constructed. The pGEX3X vector carrying the wild-type p66 subunit of HIV-1 RT was used as a template for site-directed mutagenesis. PCR was performed with two synthetic oligonucleotide primers containing the desired mutation using the KOD-plus DNA polymerase (TOYOBO). PCR products were digested by DpnI (TOYOBO) and transformed into the Escherichia coli strain DH5α. The mutations were confirmed by DNA sequencing using an ABI 3130xl genetic analyzer (Applied Biosystems).
Protein expression and purification of wild-type and mutated HIV-1 RT
Wild-type and mutated p66 subunit proteins were individually expressed in the E. coli BL21(DE3)-RIL strain (Agilent Technologies) in M9 medium containing 50 μg/ml of ampicillin with L-[methyl-13C]methionine (Cambridge Isotope Laboratories) using previously described protocols27,28. The wild-type p51 subunit was expressed separately using M9 medium with 15 μg/ml of kanamycin. Production of recombinant proteins was induced by the addition of 0.5 mM isopropyl β-D-thiogalactopyranoside at 16 °C for 16–18 h. The harvested cells of p51 and p66 HIV-1 RT were suspended and mixed into 50 mM Tris-HCl (pH 7.5) containing 0.5 mM EDTA, 50 mM NaCl, 1 mM DTT, 5% (w/v) glycerol, 0.5% (w/v) Triton X-100, and protease inhibitor cocktail (Sigma-Aldrich) and then disrupted by sonication. The HIV-1 RT protein was sequentially purified from cell lysates with a DEAE cellulose column (Whatman), a phosphocellulose P11 column (Whatman), a Chelating Sepharose Fast Flow (GE Healthcare) charged with nickel sulfate, and a RESOURCE S column (GE Healthcare). Finally, the HIV-1 RT protein was purified by Superdex-200 (HiLoad 16/60) gel filtration using a FPLC column (GE Healthcare). All purification steps of HIV-1 RT were performed at 4 °C with buffer containing protease inhibitors.
To confirm the existence of the extra N-terminal methionine, HIV-1 RT protein was incubated with methionine aminopeptidase (Clontech) at 37 °C for 12 h, and checked by NMR.
NMR measurements
All NMR measurements were made using an AVANCE800 (Bruker BioSpin) spectrometer equipped with a cryogenic probe. The probe temperature was set at 25 °C. The wild-type or mutated HIV-1 RT was dissolved at a concentration of 27 μM in 10 mM Tris-HCl-d11 (pD 7.6) containing 200 mM KCl, 1.5 mM sodium azide, and 4 mM MgCl2. A 20 mM stock solution of each inhibitor was prepared in dimethylsulfoxide-d6 and was added in five-fold molar excess to the protein. Spectra were processed and analyzed with the programs Topspin and SPARKY29.
Additional Information
How to cite this article: Thammaporn, R. et al. NMR characterization of HIV-1 reverse transcriptase binding to various non-nucleoside reverse transcriptase inhibitors with different activities. Sci. Rep. 5, 15806; doi: 10.1038/srep15806 (2015).
Supplementary Material
Acknowledgments
The authors thank Dr. Mathew Paul Gleeson for valuable discussion and comments. This work was supported by the Thailand Research Fund (RTA5380010). R.T. is grateful to the Royal Golden Jubilee PhD Program (Grant No. 3.C.KU/51/B.1) for a scholarship, the Faculty of Science, Kasetsart University (Outbound Research Student Exchange, ORSE) for supporting, and the Institute for Molecular Science (IMS), Japan for a research fellowship. The authors are grateful to Kasetsart University Research and Development Institute (KURDI), National Nanotechnology Center (NANOTEC), Laboratory of Computational and Applied Chemistry (LCAC), the Commission on Higher Education, and Ministry of Education [through the “National Research University Project of Thailand (NRU)” and “National Center of Excellence for Petroleum, Petrochemical Technology and Advanced Materials (NCEPPAM)”] for research facilities. This work was supported, in part, by ORION project, the Nanotechnology Platform Project, and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology in Japan. This work was also supported by Functional Genomics Facility, NIBB Core Research Facilities in Japan.
Footnotes
Author Contributions S.H. and K.K. conceived and designed the experiments; R.T., M.Y-U., P.B. and T.Y. prepared protein samples and performed the NMR experiments; K.C. performed the original expression of HIV-1 RT; S.T. synthesized compounds; R.T., M.Y-U., T.Y., P.B., K.C., P.S., K.K. and S.H. analyzed the data; and all authors wrote and reviewed the manuscript.
References
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A. & Steitz T. A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256, 1783–1790 (1992). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc. Natl. Acad. Sci. USA 91, 7242–7246 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A. et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90, 6320–6324 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos S. G., Das K., Hughes S. H. & Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 14, 716–730 (2004). [DOI] [PubMed] [Google Scholar]
- Sarafianos S. G. et al. Structure and Function of HIV-1 Reverse Transcriptase: Molecular Mechanisms of Polymerization and Inhibition. J. Mol. Biol. 385, 693–713 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Arias L. Molecular basis of human immunodeficiency virus type 1 drug resistance: Overview and recent developments. Antiviral Res. 98, 93–120 (2013). [DOI] [PubMed] [Google Scholar]
- Merluzzi V. J. et al. Inhibition of HIV-1 replication by a nonnucleoside reverse transcriptase inhibitor. Science 250, 1411–1413 (1990). [DOI] [PubMed] [Google Scholar]
- Esposito F., Corona A. & Tramontano E. HIV-1 Reverse Transcriptase Still Remains a New Drug Target: Structure, Function, Classical Inhibitors, and New Inhibitors with Innovative Mechanisms of Actions. Mol. Biol. Int. 2012, 586401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. & Stammers D. K. Structural basis for drug resistance mechanisms for non-nucleoside inhibitors of HIV reverse transcriptase. Virus Res. 134, 157–170 (2008). [DOI] [PubMed] [Google Scholar]
- Sluis-Cremer N. & Tachedjian G. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 134, 147–156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents 33, 307–320 (2009). [DOI] [PubMed] [Google Scholar]
- Menéndez-Arias L. Molecular basis of human immunodeficiency virus drug resistance: An update. Antiviral Res. 85, 210–231 (2010). [DOI] [PubMed] [Google Scholar]
- Nikolenko G. N., Kotelkin A. T., Oreshkova S. F. & Ilyichev A. A. Mechanisms of HIV-1 drug resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. Mol Biol 45, 93–109 (2011). [PubMed] [Google Scholar]
- Bose-Basu B. et al. Dynamic Characterization of a DNA Repair Enzyme: NMR Studies of [methyl-13C]Methionine-Labeled DNA Polymerase β. Biochemistry 43, 8911–8922 (2004). [DOI] [PubMed] [Google Scholar]
- DellaVecchia M. J. et al. NMR analysis of [methyl-13C]methionine UvrB from Bacillus caldotenax reveals UvrB-domain 4 heterodimer formation in solution. J. Mol. Biol. 373, 282–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Mueller G. A., DeRose E. F. & London R. E. Solution characterization of [methyl-13C]methionine HIV-1 reverse transcriptase by NMR spectroscopy. Antiviral Res. 84, 205–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Mueller Geoffrey A., DeRose Eugene F. & London Robert E. Protein-Mediated Antagonism between HIV Reverse Transcriptase Ligands Nevirapine and MgATP. Biophys. J. 104, 2695–2705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. et al. Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation. Nucl. Acids Res. 42, 5361–5377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers D. W. et al. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 92, 1222–1226 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiou Y. et al. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure 4, 853–860 (1996). [DOI] [PubMed] [Google Scholar]
- Alcaro S. et al. Molecular and structural aspects of clinically relevant mutations related to the approved non-nucleoside inhibitors of HIV-1 reverse transcriptase. Drug Resist. Updates 14, 141–149 (2011). [DOI] [PubMed] [Google Scholar]
- Yang S. et al. Molecular design, synthesis and biological evaluation of BP-O-DAPY and O-DAPY derivatives as non-nucleoside HIV-1 reverse transcriptase inhibitors. Eur. J. Med. Chem. 65, 134–143 (2013). [DOI] [PubMed] [Google Scholar]
- Kofuku Y. et al. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 3, 1045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. W., Sadiq S. K., De Fabritiis G. & Coveney P. V. Thumbs Down for HIV: Domain Level Rearrangements Do Occur in the NNRTI-Bound HIV-1 Reverse Transcriptase. J. Am. Chem. Soc. 134, 12885–12888 (2012). [DOI] [PubMed] [Google Scholar]
- Silprasit K., Thammaporn R., Hannongbua S. & Choowongkomon K. Cloning, Expression, Purification, Determining Activity of Recombinant HIV-1 Reverse Transcriptase. Kasetsart J. (Nat. Sci.) 42, 231–239 (2008). [Google Scholar]
- Silprasit K. Recombinant HIV-1 Reverse Transcriptase and Its Mutant Study; Cloning, Expression, Purification and Preliminary Crystallization for X-ray Crystallography. Ph. D. Thesis, Kasetsart University, Bangkok (2010).
- Nishida N. et al. Probing dynamics and conformational change of the GroEL-GroES complex by 13C NMR spectroscopy. J. Biochem 140, 591–598 (2006). [DOI] [PubMed] [Google Scholar]
- Yagi-Utsumi M., Matsuo K., Yanagisawa K., Gekko K. & Kato K. Spectroscopic Characterization of Intermolecular Interaction of Amyloid β Promoted on GM1 Micelles. Int. J. Alzheimers Dis 2011, 925073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T. D. & Kneller D. G. Sparky 3 University of California, San Francisco. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.