Summary
Somatic hypermutation (SHM) of immunoglobulin (Ig) genes appears to involve the generation of double-strand DNA breaks (DSBs) and their error-prone repair. Here we show that DSBs occur at a high frequency in unrearranged (germline) Ig variable (V) genes, BCL6 and c-MYC. These DSBs are blunt, target the mutational RGYW/RGY hotspot, and would be resolved through nonhomologous end-joining, as indicated by the presence of Ku70/Ku86 on these DNA ends. Upon CD40-induced expression of activation-induced cytidine deaminase (AID), DSBs increase in frequency and are resected to yield 5′- and 3′-protruding ends in hypermutating rearranged V genes, BCL6 and translocated c-MYC. 3′-protruding ends would direct DSB repair through homologous recombination, as indicated by their exclusive presence in S/G2 and recruitment of Rad52/Rad51, leading to SHM, upon mispair by error-prone DNA polymerases modulated by cross-linking of the B cell receptor for antigen.
Introduction
SHM provides the structural substrate for the selection of higher affinity antibody mutants by antigen (affinity maturation). It introduces point mutations with rare deletions or insertions into rearranged V(D)J immunoglobulin (Ig) genes, the germline human BCL6 protooncogene and the c-MYC protooncogene translocated into the Ig locus at a rate of 10−3 to 10−4 per base per cell generation, while virtually sparing the Ig constant (C) region (Storb et al., 2001). SHM occurs mainly in germinal center (GC) B cells. It is induced by crosslinking of the B cell receptor for antigen (BCR) and contact with activated CD4+ T cells, involving CD40:CD40L and CD80/CD86:CD28 coengagement (Zan et al., 1999, 2000, 2001; Gurrieri et al., 2002). It favors transitions over transversions and extends 1.5–2.0 kb downstream of the transcription initiation site, with preference for the RGYW/RGY and its reverse complement WRCY/RCY motif (mutational RGYW/RGY hotspot) (Chang and Casali, 1994; Storb et al., 2001; Diaz and Casali, 2002; Honjo et al., 2002; Papavasiliou and Schatz, 2002a).
The mechanism that underlies SHM remains speculative. DSBs are associated with the process and AID is required for both SHM and class switch DNA recombination (CSR) (Muramatsu et al., 2000; Honjo et al., 2002; Martin et al., 2002; Okazaki et al., 2002). Like SHM, DSBs preferentially target the RGYW/RGY hotspot within the Ig V(D)J but not the C region of hypermutating B cells (Sale and Neuberger, 1998; Bross et al., 2000; Papavasiliou and Schatz, 2000). Predominant occurrence of DSBs in the S/G2 phase of the cell cycle suggests repair of these DNA lesions through homologous recombination (HR) (Papavasiliou and Schatz, 2000). During this process, the intervention of the translesion polymerase ζ, which extends DNA strand past mispair, possibly in concert with another poorly processive and error-prone polymerase, would lead to introduction of mutations through mispairing (Papavasiliou and Schatz, 2000; Zan et al., 2001; Diaz and Casali, 2002; Papavasiliou and Schatz, 2002a). AID intervention in SHM seems to follow rather than precede the formation of (blunt) DSBs (Bross et al., 2002; Papavasiliou and Schatz, 2002b). AID may directly deaminate DNA and induce transition mutations of C-G pairs (Di Noia and Neuberger, 2002; Chaudhuri et al., 2003) or edit an unknown pre-mRNA to generate mRNA encoding an endo- or exonuclease or a factor involved in the DNA repair process (Okazaki et al., 2002; Doi et al., 2003).
A model for SHM in which mutations are introduced into DNAs during DSB repair (Papavasiliou and Schatz, 2000; Diaz and Casali, 2002; Papavasiliou and Schatz, 2002a) could explain the salient feature of the process. However, the available data do not define the induction requirements, the modalities, the nature of DSBs, or their role and that of AID in SHM. Using human B cells, we found that, like SHM, DSBs specifically target germ-line Ig V, BCL6, and c-MYC DNA but not Cμ, PIM1, PAX5, or α-fetoprotein (AFP), which do not normally undergo SHM. These DSBs are blunt ended and are likely repaired through nonhomologous end-joining (NHEJ) without the insertion of mismatches, i.e., point mutations. In rearranged Ig V genes, translocated c-MYC and BCL6 of hypermutating B cells, DSBs are modified in a CD40-induced and AID-dependent fashion to yield free 5′- and 3′-protruding ends, which critically initiate DNA strand invasion and repair of these resected DNA ends in S/G2 through HR. During this repair process, mispair insertion and extension by translesion error-prone polymerases, which are regulated upon cross-linking of the BCR (Zan et al., 2001; Diaz and Casali, 2002), would lead to insertion of somatic point mutations.
Results
DSBs Occur in Ig VHDJH and BCL6 but Not in Cμ or PIM1, PAX5, and AFP DNA of Hypermutating B Cells
In human B cells, SHM targets the BCL6 transcriptional repressor gene in response to the same inducing stimuli, i.e., BCR crosslinking and contact with activated CD4+ T cells, and with the same modalities as the Ig V(D)J genes (Zan et al., 2000, 2001). SHM is negligible in pre-GC IgD+CD38− B cells, increases significantly in IgD+CD38+ centroblasts, decreases in IgD−CD38+ centrocytes, and extinguishes in post-GC memory IgD−CD38− B cells (Pascual et al., 1994; Liu et al., 1996; Wilson et al., 2000; Zan et al., 2001). Genomic DNA was purified from these cells, linker-ligated, serially 2-fold diluted into unligated genomic DNA, and used as template to analyze DSBs in the Ig H chain and BCL6 loci as well as in PAX5, PIM1, and AFP using specific ligation-mediated (LM)-PCRs, which rely on the availability of 5′-phosphorylated blunt ends for DNA amplification. LM-PCR using reverse primers specific for “down-stream” DNA ends in rearranged VHDJH and BCL6, and forward primers specific for “upstream” DNA ends in BCL6, Cμ, PAX5, PIM1, and AFP (see Supplemental Table S1 and Figure S2 at http://www.immunity.com/cgi/content/full/18/6/727/DC1) revealed DSBs in Ig VHDJH and BCL6 DNA, but not in Cμ, PAX5, PIM1, or AFP of pre-GC IgD+CD38− and post-GC IgD−CD38− B cells (Figure 1, T4−). In IgD+CD38+ and IgD−CD38+ B cells undergoing SHM, such blunt DSBs were increased in frequency. The failure to amplify DNA ends in Cμ, PAX5, PIM1, or AFP was not due to the lack of specificity of the Cμ, PAX5, PIM1, or AFP primers, as these effectively amplified the respective intact genomic DNA and “detected” free blunt ends artificially inserted in these genes in a dose-dependent fashion (see Supplemental Figures S2B, S2C, and S3), thereby emphasizing the in vivo origin of the amplified free DNA ends in VHDJH and BCL6. DNA amplification was not influenced by “apo-ptotic” DSBs, as the proportion of apoptotic cells was negligible in all the purified pre-GC, GC, and post-GC B fractions (see Supplemental Figure S4A) and high levels (more than 70%) of apoptotic cells had no impact on the specific detection of VHDJH DSBs (Figure S4B), or cell division, as different rates of B cell proliferation did not affect the levels of detected, specific BCL6 DSBs (Figure S4C).
Figure 1. DSBs Occur in Ig VHDJH and BCL6, but not Cμ, PAX5, PIM1, or AFP, and Include a High Proportion of Resected DNA Ends in Hypermutating GC B Cells.
Genomic DNA from freshly isolated human tonsil IgD+CD38−, IgD+CD38+, IgD−CD38+, and IgD−CD38− B cells was treated with T4 DNA polymerase (T4+) or nil (T4−) before being linker-ligated. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially 2-fold diluted into unligated homologous genomic DNA and used as templates in LM-PCR (lanes 1–8 in each panel). The amplified DNA was blotted and then probed with [γ-32P]-ATP labeled gene-specific oligonucleotide probes. Linker-ligated genomic DNA diluted in water was used as template to amplify β-actin DNA.
To extend our analysis to the detection of 3′- and 5′-protruding ends, genomic DNA was treated, before linker ligation, with T4 DNA polymerase, which trims back 3′ -overhangs while “filling in” 3′-recessed ends, thereby yielding blunt DNA ends (see Supplemental Figure S3 at http://www.immunity.com/cgi/content/full/18/6/727/DC1). Pretreatment with T4 polymerase significantly increased the levels of detected DSBs in Ig VHDJH and BCL6 of the hypermutating GC IgD+CD38+ and IgD−CD38+ B cells, but not in the nonhypermutating pre-GC IgD+CD38− and post-GC IgD−CD38− B cells (Figure 1, T4+). It also failed to reveal DSB levels in Cμ, PAX5, PIM1, or AFP in all four B cell fractions. In hypermutating IgD+CD38+ and IgD−CD38+ B cells, VHDJH and BCL6 DSBs comprised both 3′- and 5′-protruding ends, which accounted for the majority of the total DSBs (see Supplemental Figure S5; not shown). Hence, resected but not blunt DNA ends are characteristic of hypermutating Ig V genes and BCL6.
DSBs Occur in Germline VH and c-MYC but They Increase in Frequency and Are Modified to Yield Resected DNA Ends after Rearrangement or Translocation into the Ig H Chain Locus
The occurrence of resected DSBs in IgD+CD38+ and IgD−CD38+ B cells together with the occurrence of blunt DSBs in IgD+CD38− or IgD−CD38− B cells led us to hypothesize that genes that hypermutate only after rearrangement or translocation into the Ig locus, such as V genes and the c-MYC protooncogene, respectively, would accumulate blunt DSBs when in germline configuration, but would incur resected DSBs after rearrangement or translocation. By LM-PCR amplification of downstream DNA ends of germline VH genes using reverse primers targeting the recombination signal sequence (RSS) heptamer and nonamer of V3 gene family members (see Supplemental Table S1 at http://www.immunity.com/cgi/content/full/18/6/727/DC1), as well as LM-PCR amplification of the c-MYC upstream DNA ends using forward primers targeting a sequence 5′ of the c-MYC exon 1 initiation of transcription, we demonstrated the presence of DSBs in germline V3 genes and c-MYC in both hypermutating and nonhypermutating normal human B cells (Figure 2). These DSBs were blunt, occurred in the absence of AID expression and targeted (75.6%) the RGYW/RGY hotspot (see Supplemental Table S6). The differential amplification of germline and translocated c-MYC DNA in hypermutating Ramos B cells confirmed the presence of blunt DSBs in the germ-line allele and showed high levels of resected DSBs in the c-MYC allele translocated into the Ig H chain locus (Figure 2B). Such resected DSBs were concomitant with high levels of somatic point-mutations. Hence, the VH and c-MYC genes display an inherent propensity to “breakage” to yield blunt DSBs, even in germline configuration and in the absence of AID. Rearrangement or translocation of these genes into the Ig locus together with AID expression results in significant levels of resected DSBs and SHM.
Figure 2. DSBs Occur in Germline VH Genes and Are Blunt Ended; Resected DSBs Occur Only in the Translocated and Actively Hypermutating c-MYC Allele, as They Do in Rearranged and Hypermutating Ig VHDJH Gene Segments.
(A) Genomic DNA (8 ng) from 1280 IgD+CD38−, IgD+CD38+, IgD−CD38+, and IgD−CD38−B cells (same as experiment of Figure 1) was treated with nil (T4−) or T4 DNA polymerase (T4+), linker-ligated, and then serially 2-fold diluted into unligated homologous genomic DNA and used as template in LM-PCR for analysis of germline V3 gene and c-MYC DSBs. AID and β-actin transcripts were detected by RT-PCR using serially 2-fold diluted cDNA as a template. The top drawing schematizes the strategy utilized to detect free ends in germline V3 and c-MYC DNA. Arrows depict primers, thick bars depict [γ-32P]-ATP labeled gene-specific oligonucleotide probes, bars with ticks depict distances, and simple bars depict the expected maximal length of the amplified product. For the downstream end, the expected maximal length of the amplified product corresponds to the distance intervening between the 5′-end of the second round LM-PCR reverse primer and the transcription initiation site (the usual 5′ boundary of the SHM target); for the upstream end, the expected maximal length of the amplified product corresponds to the distance intervening between the 5′-end of the second round LM-PCR forward primer and an area approximately 2.0 Kb downstream of the initiation of transcription site (the usual 3′ boundary of the SHM target). (B) Differential amplification of germline c-MYC DNA and c-MYC t(8;14) translocated in human Ramos B cells. Linker-ligated genomic DNA (8 ng from 1280 cells) was serially 2-fold diluted into unligated genomic DNA and used as template in LM-PCR. Identical amounts of linker-ligated genomic DNA diluted in water was used as template to amplify germline and translocated c-MYC sequences. Numbers at the bottom are mutation frequencies in the germline and translocated c-MYC gene after a 6-week culture. The top drawing schematizes the strategy utilized used to detect DSBs in the germline and translocated c-MYC.
Germline Ig VH and c-MYC DSBs Occur in Pro-B, Pre-B, T Lymphocytes, and Nonlymphoid Cells but Are Blunt Ended Only
We hypothesized that the higher density of DSB upstream ends found in Ig V genes (Bross et al., 2000; Papavasiliou and Schatz, 2000) (see Supplemental Figure S7) reflects the amplification of multiple copies of different germline VH genes of the same family by the upstream end-specific primer. This cannot occur in BCL6, which exists as one copy per aplotype and, indeed, shows comparable intensity of amplified free upstream and downstream DNA ends (Figures 1 and S5). The inherent propensity to breakage of germline VH, BCL6, and c-MYC DNA (Figures 2A and S7) is further supported by the finding that these genes, but not Cμ, incur DSBs at a high frequency also in nonhypermutating RS4;11 pro-B cells, Nalm-6 pre-B cells, U266 plasmacytoma cells, Hs27 fibroblasts, HuVec endothelial cells, HeLa cervix cells, and normal human CD4+ T cells (BCL6 not shown). These DSBs occur in the absence of AID expression, are blunt ended only (Figure 3), and preferentially target the mutational RGYW/RGY hotspot (not shown).
Figure 3. Blunt DSBs Occur in Germline Ig V3 and c-MYC DNA of Pro-B, Pre-B Cells, T Lymphocytes, and Nonlymphoid Cells.
Blunt DSBs were detected at a high frequency in the unmutating unrearranged germline V3 (downstream end) and c-MYC (upstream end) genes of normal GC IgD−CD38+B cells, Ramos B cells, Nalm-6 pre-B cells, RS4;11 pro-B cells, U266 plasmacytoma, Hs27 fibroblasts, HuVec, HeLa cells, and normal human CD4+T cells. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially 2-fold diluted into unligated genomic DNA and used as template in LM-PCR. AID and β-actin transcripts were detected by RT-PCR using serially 2-fold diluted cDNAs as templates. The 4-fold difference (two dilutions) in Ramos c-MYC T4− and T4+ DNA amplification was due to the presence of some protruding ends in the translocated and hypermutating c-MYC allele, as shown in Figure 2B.
The Generation of Resected DSBs Is Induced by CD40-Dependent AID Expression
To define the stimuli responsible for DSB induction, we treated IgM+IgD+ CL-01 B cells, our human monoclonal model of inducible GC differentiation (Zan et al., 1998; Cerutti et al., 1998a; Schaffer et al., 1999), with anti-BCR Ab, cocultured them with activated CD4+ T cells (Zan et al., 1999, 2000, 2001), and then determined the extent of genomic DSBs. These experimental conditions effectively induce Ig VHDJH and BCL6 SHM in human B cells in vitro (Zan et al., 1999, 2000, 2001; Gurrieri et al., 2002). Coculture of CL-01 B cells with activated CD4+ T cells with or without BCR crosslinking significantly increased (by up to 32-fold) the frequency of DSBs. Most of these newly induced DSBs involved resected DNA ends and were concomitant with significantly increased AID expression and insertions of somatic point mutations in the expressed VHDJH genes (Figure 4A, T4− and T4+). These findings were confirmed in freshly isolated normal human peripheral blood CD19+ cells cultured in the presence of human trimeric (ht)CD40L and/or IL-4 and/or anti-BCR Ab. In these B cells, CD40 signaling induced AID expression and significantly enhanced the minimal levels of spontaneously occurring DSBs and IL-4- and BCR-induced AID expression (Figure 4B). DSB induction was htCD40L dose dependent, as increasing amounts of htCD40L progressively increased the levels of blunt (by approximately 4-fold at 100–500 ng/ml of htCD40L) and resected (by approximately 16-fold at 2500 ng/ml) DSBs (Figure 4C). It was also time dependent, as the frequency of blunt DSBs increased by more than 16-fold at 24 hr and that of resected DSBs by more than 32-fold at 48 hr (Figure 4D).
Figure 4. The Generation of Resected DSBs Is Induced by CD40-Dependent AID Expression.
(A) Activated CD4+ T cells critically enhance DSBs to yield resected DNA ends. IgM+IgD+ CL-01 B cells were cultured for 48 hr in the presence or absence of activated CD4+ T cells upon pretreatment with anti-BCR Ab or nil. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially 2-fold diluted into unligated genomic DNA and used as template in LM-PCR. Linker-ligated genomic DNA serially diluted in water was used as template to amplify β-actin DNA. AID and β-actin transcripts were detected by RT-PCR using serially 2-fold diluted cDNAs as templates. Mutation rates in VHDJH cDNA were measured after a 2-week culture.
(B) CD40 signaling induces AID expression and DSB resection. Freshly isolated human peripheral blood CD19+ B cells were cultured with nil, IL-4, anti-BCR Ab, or anti-BCR Ab plus IL-4 in the presence or absence of htCD40L (500 ng/ml). Downstream blunt and resected VHDJH DSBs were detected by specific LM-PCR after 48 hr of culture. AID and β-actin transcripts were analyzed by RT-PCR using serially 2-fold diluted cDNAs as a template.
(C) Dose-dependent enhancement of VHDJH DSBs by htCD40L. IgM+IgD+ CL-01 B cells were cultured in the presence of increasing amounts of ht-CD40L for 48 hr before measuring blunt and resected DSBs. Linker-ligated genomic DNA diluted in water was used as template to amplify β-actin DNA.
(D) Time-course analysis of DSB enhancement by htCD40L. IgM+IgD+ CL-01 B cells were cultured in the presence of htCD40L (500 ng/ml) and VHDJH DSBs were measured after 0, 2, 12, 24, and 48 hr. Linker-ligated genomic DNA diluted in water was used as template to amplify β-actin DNA.
5′-Protruding End DSBs Occur in G1, S, and G2/M while 3′-Protruding End DSBs Occur Only in S and G2/M
To investigate the relationship of DSBs with the cell cycle, we sorted propidium iodide-treated CD40L-stimulated IgM+IgD+ CL-01 B cells into G1, S, and G2/M fractions and characterized the VHDJH DSBs for the involvement of 3′- and 5′-protruding ends. LM-PCR after DNA pretreatment with nil, T4 DNA polymerase, 3′→5′ dsDNA-specific exonuclease (Exo) III, Exo III followed by 3′→5′ ssDNA-specific Exo T, shrimp alkaline phosphatase (SAP), which catalyzes the release of 5′-phosphate groups from DNA, alone or SAP followed by 3′→5′ and 5′→3′ ssDNA-specific Exo VII (Figures 5, S3, and S5) showed that both blunt and resected DSBs were more frequent (4- to 32-fold) in S and G2/M than in G1 (Figure 5). 5′-protruding DNA ends were found in G1, S, and G2/M, but 3′-protruding DNA ends only in S and G2/M. Comparable findings were derived from the analysis of hypermutating Ramos B cells (Figure S8), further emphasizing that DSBs yielding 3′-protruding VHDJH ends occur at a stage of the cell cycle when HR is operational.
Figure 5. Cell Cycle-Associated Generation of DSBs.
DSBs that include 5′-protruding ends accumulated in G1, S, and G2/M, while 3′-protruding ends accumulate predominantly in S and G2/M. IgM+IgD+ CL-01 B cells stimulated by htCD40L for 48 hr were stained with propidium iodide and then sorted into G1, S, and G2/M fractions. Genomic DNA was purified and submitted to different treatments before being linker-ligated. Linker-ligated DNA (8 ng from 1280 B cells) was serially 2-fold diluted and used as a template in LM-PCR to amplify downstream ends of VHDJH DSBs. Dashed frames enclose the LM-PCR amplifiable pretreated DNA ends. DNA treatments included: nil (a), T4 DNA polymerase, which trims back 3′-overhangs while filling in 3′-recessed ends, thereby yielding blunt DNA ends (b), 3′→5′ dsDNA-specific exonuclease Exo III (c), Exo III followed by 3′→5′ ssDNA-specific Exo T (d), SAP, which catalyzes the release of 5′-phosphate groups from DNA, only (e) or SAP followed by 3′→5′ and 5′→3′ ssDNA-specific Exo VII (f). Shown is β-actin DNA amplified using the linker-ligated untreated (nil) genomic DNA as a template. Comparable β-actin DNA amplification was obtained from linker-ligated genomic DNA pretreated with T4 DNA polymerase, Exo III, Exo III, and then Exo T, SAP only, or SAP and then Exo VII.
Resected VHDJH Gene DSBs Recruit Rad52 and Rad51 and Blunt VH Gene DSBs Recruit Ku70/Ku86
DSBs can be repaired through two different and competing pathways. Opposing broken DNA ends can be rejoined with no or little regard for sequence homology by NHEJ or through HR, a homologous template-directed recombination process. Both pathways share the phosphorylated H2A histone family member X (γ-H2AX), which is activated by and recruited to DSBs, and the Rad50/Mre11/Nbs1 protein complex, but also require additional components that are unique to each process (van Gent et al., 2001; D’Amours and Jackson, 2002). Namely, NHEJ requires DNA-PKcs, Ku70, Ku86, XRCC4, and DNA ligase IV, while HR uses homologs of the yeast Rad52 epistasis protein group, including Rad51 and Rad52, which together form a strong complex (Shinohara and Ogawa, 1998; Song and Sung, 2000). The two pathways can compete for DSB repair. For instance, HR is elevated in mutant mice defective in Ku70 (Pierce et al., 2001). On the other hand, chicken DT40 cell mutants defective in HR become more sensitive to ionizing radiation when carrying an additional mutation in the gene encoding the Ku70 homolog (Takata et al., 1998) and scid/RAD54−/− double mutant mice are much more sensitive to radiation than RAD54−/− single mutants (Essers et al., 2000).
We adapted a modified chromatin-immunoprecipitation (ChIP) assay to “pull-down” γ-H2AX, Nbs1, Mre11, Ku70/Ku86, Rad51, or Rad52 and analyzed by specific LM-PCR the coprecipitated DNA, i.e., the DNA to which these proteins were bound. We reasoned that if a 3′-protruding end modification were critical in SHM to direct the repair process toward HR for template-directed strand synthesis by an error-prone DNA polymerase, then the precipitation of Rad52/Rad51 would lead to the identification of free 3′-protruding DNA ends, and these would be found only in rearranged VHDJH genes. On the other hand, if blunt DSBs were repaired through NHEJ, then the precipitation of Ku70/Ku86 would lead to the identification of blunt DNA ends in germline VH genes, which, as we clearly show (Figures 2A, 3, and 6), effectively break but do not hypermutate. Specific Abs to γ-H2AX, Mre11, Nbs1, and Ku70/Ku86 revealed these proteins as bound to germline VH and rearranged VHDJH free DNA ends in hypermutating Ramos cells (Figure 6). T4 polymerase pretreatment showed these DNA ends to be blunt. In the same hypermutating B cells, Abs to Rad51 or Rad52 revealed the association of these proteins only with rearranged VHDJH DNA, which comprised resected but not blunt DNA ends, as indicated by the successful LM-PCR amplification of DNA pretreated with T4 polymerase. The failure to precipitate any amplifiable Cμ DNA strongly reinforces the specificity of our experimental approach and further confirms the lack of DSBs in Cμ exons. Analysis of the DNA precipitated by specific Abs to Rad52 or Rad51 using a fill-in approach to amplify 3′-recessed termini and a 3′-anchored PCR modified for dUTP insertion to exclude the amplification of 5′-protruding and blunt ends demonstrated the presence of both 5′- and 3′-protruding DNA ends at a high frequency (not shown). The breakpoints of these 5′-and 3′-protruding ends preferentially (more than 88%) targeted the RGYW/RGY hotspot (see Supplmental Table S6). Hence, resected DSBs are characteristic of hypermutating genes and recruit trans-factors dedicated to the HR pathway, while blunt DSBs occur in nonhypermutating genes and recruit trans-factors dedicated to the NHEJ pathway.
Figure 6. Resected VHDJH DNA Ends Recruit Rad52 and Rad51 but VH Blunt DNA Ends Do Not.
Genomic DNA was precipitated from hypermutating Ramos B cells by Abs specific for human γ-H2AX, Mre1, Nbs1, Ku70/Ku86, Rad52, or Rad51. The precipitated DNA was treated with either T4 polymerase (T4+) or nil (T4−) and then linker-ligated before being serially 2-fold diluted (from 1.0 × 104, 5.0 × 103, 2.5 × 103, 1.25 × 103, 625, 313, 156, and 78 Ramos B cells, lanes 1–8) for the detection of VHDJH and germline V3 free downstream ends or Cμ DSBs free upstream end. Control Abs are nonintentionally immunized normal rabbit IgG, nonintentionally immunized normal goat IgG, and MOPC-21 IgG1 mAb with irrelevant binding activity. Shown are results from one of four ChIP assays yielding comparable results.
AID Is Critical for the Generation of Resected DNA Ends
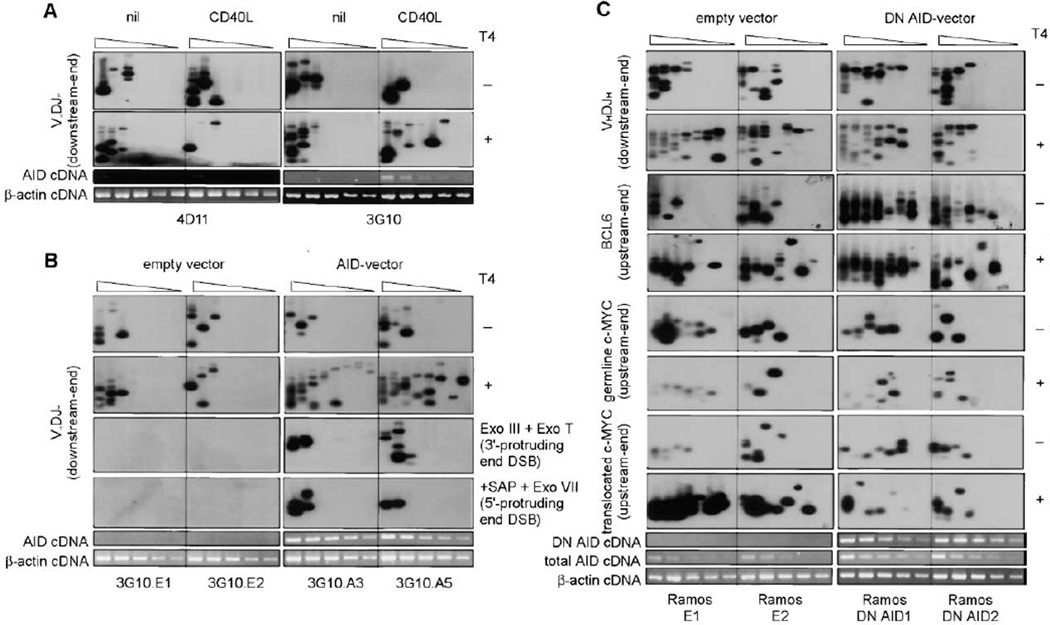
We hypothesized that AID’s role in SHM critically involves the generation of resected DNA ends, particularly 3′-protruding ends, which are essential to direct the DNA repair process toward HR. Indeed, AID expression was upregulated in the hypermutating IgD+CD38+ and IgD−CD38+ B cells, in which the VHDJH and BCL6 DNA DSBs were characteristically resected, but not in the nonmutating IgD+CD38− and IgD−CD38− B cells, in which DSBs were blunt (Figures 1 and 2A). Further, the CD40-dependent induction of AID expression and SHM was associated with generation of resected DSBs in B cells (Figures 4A and 4B). Accordingly, the CD40-dependent induction of AID expression in CL-01 subclone 3G10 B cells resulted in the generation of resected DSBs, while in subclone 4D11, in which CD40 signaling failed to increase AID, no resected ends were generated (Figure 7A). To test the hypothesis that AID is critical for the generation of resected DSBs, we expressed AID in the SHM-inducible IgM+IgD+ CL01 subclone 3G10 B cells by transfection with a plasmid vector containing human AID cDNA, driven by the CMV promoter, or nil, and then analyzed the VHDJH DSB downstream ends. Expression of AID did not increase the level of blunt DSBs, but induced significantly higher levels of both 5′-and 3′-protruding DNA ends, which accounted for more than a 32-fold increase in total DSBs (Figure 7B). Conversely, expression of a dominant negative (DN) AID cDNA (Papavasiliou and Schatz, 2002b) in spontaneously hypermutating Ramos B cells abrogated the generation of resected ends and concomitant SHM in VHDJH, BCL6, and translocated c-MYC, but did not affect the occurrence of blunt DSBs in germline c-MYC (Figure 7C; not shown). In most cases, as exemplified by the E1/DN AID1 pair, DN AID overexpression reverted the level of blunt DSBs to the level of the total DSBs detected in B cells transfected with empty vector, indicating that resected DNA ends arise from the modification of blunt DSBs. In some cases, however, as exemplified by downstream end VHDJH and upstream end BCL6 DSBs in the E2/DN AID2 pair, DN AID overexpression reduced the level of resected but not that of DSBs, raising the possibility that some resected DNA ends emerge as such. Hence, AID, which is induced by CD40 signaling, is critical for the generation of resected DNA ends.
Figure 7. The Generation of Resected DNA Ends Requires AID.
(A) CD40 signaling upregulates AID expression and induces associated resected DSBs in AID/SHM-inducible IgM+IgD+3G10 B cells but not in AID/SHM-noninducible IgM+IgD+4D11 B cells (cultured for 48 hr in the presence of htCD40L). Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially 2-fold diluted into unligated genomic DNA and used a template in LM-PCR. AID and β-actin transcripts were analyzed by RT-PCR using serially 2-fold diluted cDNA as template.
(B) AID overexpression induces the generation of resected DNA ends. AID/SHM-inducible 3G10 IgM+IgD+ B cells were transfected with either a vector expressing human AID (subcultures 3G10.A3 and 3G10.A5) or an empty vector control (subcultures 3G10.E1 and 3G10.E2). Blunt and resected VHDJH DSB downstream ends were then detected by LM-PCR using serially 2-fold diluted DNA as template. AID and β-actin transcripts were analyzed by RT-PCR using serially 2-fold diluted cDNA as template.
(C) DN AID expression abolishes the generation of resected DNA ends. Paired samples of Ramos B cells transfected with an empty pcDNA vector (E1, E2) or a vector containing a human DN AID construct (DN AID1, DN AID2) were analyzed for blunt or resected VHDJH DSBs (downstream ends), BCL6 DSBs (upstream ends), and DSBs in germline and translocated c-MYC (upstream ends) by specific LM-PCR. Total AID, DN AID, and β-actin transcripts were analyzed by RT-PCR using serially 2-fold diluted cDNA as a template. Depicted are the results from two of four experimental pairs yielding comparable results.
Discussion
In this study, we showed that, consistent with the targeting specificity of the SHM machinery, DSBs target specifically the Ig, BCL6, and c-MYC genes while sparing the Cμ region and the PIM1, PAX5, and AFP genes. We also showed that DSBs occur at high frequency not only in hypermutating rearranged Ig VHDJH, BCL6, and translocated c-MYC genes, but also in the nonhypermutating germline forms of these genes. These DSBs are blunt ended and are repaired through NHEJ, as indicated by the recruitment of Ku70/Ku86, Mre11, and Nsb1 but not Rad52/Rad51 to these broken DNA ends. In VHDJH genes, BCL6, and translocated c-MYC of hypermutating GC B cells, DSBs increase in frequency to include abundant resected DNA ends, the generation of which requires AID expression, as induced by engagement of B cell CD40 by CD40L on activated CD4+ T cells. Like blunt DNA ends, such resected ends target the RGYW/RGY hotspot. They include 3′-protruding ends, which recruit Rad52 and Rad51 to direct DNA repair through the HR pathway, as further indicated by the prevalence of resected DSBs in the S and G2 phases of the cell cycle, when this DNA repair pathway is dominant.
Ig V, BCL6 and c-MYC genes break in nonhypermutating B cells, Pro-B cells, Pre-B cells, plasmacytoma cells, T cells, and nonlymphoid cells, such as fibroblasts, endothelial cells, and cervix cells, at a frequency comparable to that of hypermutating B cells (Figure 3). This breakage is AID independent and yields blunt DNA ends only. CD40-induced and AID-dependent resection of blunt ends to generate 3′- and 5′-protruding ends is necessary for SHM to unfold (Figures 4 and 7). The DSBs detected by us and other investigators (Bross et al., 2000, 2002; Papavasiliou and Schatz, 2000; Papavasiliou and Schatz, 2002b) could be generated in vivo, thereby reflecting an important feature of certain genes and a critical role in SHM, or introduced “nonspecifically” in vitro during DNA purification. By showing that DNA breakage is a specific and exquisite function inherent only to the genes that can undergo SHM, i.e., Ig V (but not CH), BCL6, and c-MYC, our findings provide strong evidence against the contention that DSBs are nonspecific, i.e., introduced during DNA purification. That DSBs occur in vivo and are present prior to DNA purification is further emphasized by the pull-down of broken dsDNA ends in our ChIP assays utilizing specific Abs to Ku70/Ku86, Mre11, Nsb1, and Rad52/Rad51 (Figure 6, Table S6).
The upstream end of the blunt DSBs in the Ig VHDJH DNA of Ramos and CL-01 B cells was detected at a significantly higher frequency than its downstream counterpart (Bross et al., 2000; Papavasiliou and Schatz, 2000). It has been suggested to reflect a “protection” of the upstream end that remains blunt while the downstream end is not protected but is further processed in preparation for repair or “modification” that causes it to be unreactive with the blunt end linker, thereby yielding a much less intense signal in LM-PCR (Papavasiliou and Schatz, 2000). The comparable efficiency of amplification of upstream and downstream ends of both blunt and resected DSBs in BCL6 of normal human B cells ex vivo (Figures 1 and S5) and CL-01 B cells stimulated by activated CD4+ T cells (not shown), together with the demonstration of blunt DSBs occurring at a high frequency in unrearranged VH genes (Figures 2, 3, and S7) indicates that the high frequency of amplification of upstream DNA ends of VHDJH DSBs is due to the high frequency of DSBs occurring in multiple germline members of the same VH gene family as detected by the V3 and V4 upstream (forward) primers used for the LM-PCR (Figure S7).
During the preparation of this manuscript, it has been reported that AID is dispensable in the generation of Ig V gene (blunt) DSBs (Bross et al., 2002; Papavasiliou and Schatz, 2002b). Here we show that generation of blunt DSBs, which even at high frequency do not give rise to somatic mutations, does not require AID. Comparably high levels of blunt DSBs occur in germline VH, c-MYC, and BCL6 (not shown) of not only hypermutating B cells, which express AID, but also of (nonhypermutating) lymphoid and nonlymphoid cells, which do not express AID (Figure 3), further indicating that DNA breakage is a highly specific function that is inherent only to genes that have the potential to undergo SHM. The requirement for transcription in the generation of DSBs (Papavasiliou and Schatz, 2000) may be fulfilled here, as germline Ig and/or TCR V genes can be transcribed in B and T lymphocytes (Cayre et al., 1981; Picard and Schaffner, 1984; Yancopoulos and Alt, 1985; Baer et al., 1988; Berman et al., 1991; Koenig et al., 1997), and SHM can occur in rearranged VHDJH genes transcribed at a low level (Fukita et al., 1998). The finding that the vast majority of blunt and resected DSBs target the RGYW/RGY hotspot (Table S6) further suggests a specialized function inherent to this motif and indicates that these lesions can be the precursors of resected DNA ends, which are necessary for SHM.
Our findings show that DSB resection is both critical for SHM and dependent on AID expression, as induced by CD40 signaling. We show here that AID is not necessary for the emergence of blunt DSBs but it is critical for the generation of resected DSBs, which are intimately associated with SHM. Most resected DSBs are generated through modification of blunt DSBs while some may emerge as such (Figures 1, 2B, S5, and data not shown). DSB resection occurs at both upstream- and downstream ends and is directly or indirectly AID dependent. AID could directly deaminate DNA, thereby converting C to U and giving rise to U-G mispairs (Di Noia and Neuberger, 2002; Chaudhuri et al., 2003). This could yield an abasic site through the intervention of uracil-DNA-glycosylase (UDG), followed by incision of the abasic DNA strand by (apurinic/apyrimidic) AP endonuclease. Such a process could occur on a strand of a blunt DSB or on opposite strands of intact DNA to give rise to resected DSBs. Alternatively, AID may edit mRNA for an exonuclease or endonuclease involved DSB resection (Doi et al., 2003). These two possibilities are not mutually exclusive, as AID can function both as a DNA deaminator and an mRNA editor. Regardless of the precise role of AID, the contention that DSB resection is critical for SHM is strongly supported by our demonstration that in hypermutating B cells, anti-Rad52 and anti-Rad51 Abs pulled-down exclusively rearranged Ig V(D)J DNA with 5′- and 3′-protruding ends, which in most cases identified the RGYW/RGY hotspot, but not germ-line V DNA with free blunt ends (Figure 6, Table S6, and not shown).
Consistent with the demonstrated staging of the SHM process (Liu et al., 1996; Wilson et al., 2000; Zan et al., 2001), the frequency of in vivo resected DSBs is virtually nil in pre-GC B cells, increases significantly in GC centroblasts/centrocytes, and extinguishes in post-GC memory B cells. Consistent with the target specificity of SHM, resected DSBs occur only in actively hypermutating Ig VHDJH genes, BCL6, and the translocated c-MYC allele but not in their nonhypermutating germline counterparts (Figure 2B), further emphasizing the SHM-related specificity of the DNA resection process. The abundance of resected DSBs in the translocated c-MYC of Ramos cells, in which c-MYC translocation led to the deletion of the Ig H intronic μ enhancer, suggests that this element is either not involved in the generation of resected DNA ends or is dispensable in this function and can be substituted by other DNA elements. This possibility is further suggested by the occurrence of resected DSBs in BCL6, which undergoes SHM without translocation in the Ig locus (Zan et al., 2000, 2001).
We propose here that SHM can begin with the introduction DSBs in the RGYW/RGY hotspot. The free DNA ends at the break sites would then be subjected to repair according to a stringent mechanism governing pathway choice (Figure S11) (Wu et al., 2003). Blunt DNA ends would bind the Ku70/Ku86 heterodimer, which is recruited together with Mre11, Nbs1, and γ-H2AX to direct repair through NHEJ without insertion of point mutations in non-B cells and nonhypermutating B cells. In hypermutating B cells, DSBs are resected in a CD40-induced and AID-dependent fashion. 5′-protruding ends (3′-recessed termini) can be filled in G1 (Figures 5 and S8), thereby allowing for ligation of the two free DNA ends by NHEJ. 3′-protruding ends can recruit Rad52, which in turn mediates Rad51 binding and directs the repair process toward HR. This would critically underlie SHM, as suggested by our findings in the human and those in DNA-PKcs-deficient mice. These mice are defective in the NHEJ but not the HR pathway and show a complete block in CSR but intact SHM (Bemark et al., 2000). Mismatches, i.e., point mutations, are inserted during the process of strand repair by HR in S/G2. Point mutations may also be inserted during filling-in of 3′-recessed termini in G1 (Faili et al., 2002). In either case, mismatch insertion is dependent on BCR signaling that critically modulates the expression of selected translesion DNA polymerases, including polymerases ζ and perhaps ι. DNA polymerase ζ is central to DNA damage-induced mutagenesis (Johnson et al., 2000; Zan et al., 2001; Diaz and Casali, 2002) and effectively extends DNA past mispairs, as inserted by polymerase ι (Zan et al., 2001; Diaz and Casali, 2002) or other translesion polymerases. Defining these DNA repair steps and the precise role of AID in DSB resection will further our overall understanding of the SHM process.
Experimental Procedures
Human B Lymphocytes
The phenotypic and functional features of the human monoclonal IgM+IgD+ CL-01 B cell line were reported (Cerutti et al., 1998a, 1998b; Schaffer et al., 1999; Zan et al., 1999, 2000, 2001; Schaffer et al., 2003). The AID/SHM-noninducible 4D11 and AID/SHM-inducible 3G10 monoclonal IgM+IgD+ B cell lines were subclones of CL-01 IgM+IgD+ B cells. The monoclonal human Ramos B cells used here were selected for high rate of spontaneous SHM in the rearranged and expressed Ig V4-34DXP1JH6b allele and the BCL6 locus. The highly preferential targeting of RGYW/RGY by both DSBs and somatic mutations (see Figures S9 and S10 at) indicated that the modalities of the two processes in these B cells are consistent with those reported in both the human and the mouse (Sale and Neuberger, 1998; Bross et al., 2000; Papavasiliou and Schatz, 2000, 2002b). IgM+IgD+ B cells were prepared from human peripheral blood by T cell depletion and subsequent positive selection using a mAb to human IgD (Southern Biotechnology Associates Inc., Birmingham, AL), magnetic Microbeads conjugated with goat anti-mouse IgG, and a MACS magnetic sorter (Miltenyi Biotec Inc., Auburn, CA) (Zan et al., 2001). Tonsil IgD+CD38−, IgD+CD38+, IgD−CD38+, and IgD−CD38− B cells were selected upon incubation of purified IgD+ B cells with MultiSort Release Reagent (Miltenyi Biotec Inc.), FITC-conjugated mouse mAb to CD38 (PharMingen, BD Biosciences, San Diego, CA), and anti-FITC MicroBeads (Miltenyi Biotec Inc.) (Zan et al., 2001). The homogeneity of the separated cell fractions was assessed using a FACS calibur (Becton Dickinson, Immunocytometry Systems, BD Biosciences). The human activated CD4+ T cells used for the SHM induction experiments were prepared as reported (Zan et al., 1999, 2000).
Cell Cultures
All cultures were performed in RPMI-1640 medium supplemented with 10% heat-inactivated FBS (Sigma-Aldrich, Inc., St. Louis, MO), 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. Human IL-4 (Schering-Plough Corp., Kenilworth, NJ) and htCD40L (Immunex Corp., Seattle, WA) were added to cultures as indicated, at concentrations of 200 U/ml and 20–2500 ng/ml, respectively. CL-01 B cells were induced to undergo SHM by co-culture with activated CD4+ T cells and upon BCR cross-linking by IMMUNO-BEAD®-conjugated rabbit Ab to human Ig μ chain and rabbit Ab to human Ig (H + L chain) (Irvine Scientific, Irvine, CA) (anti-BCR Ab) (Zan et al., 2001).
Analysis of DNA DSBs by LM-PCR
For LM-PCR, genomic DNA was prepared from 5 × 105 cells using an anion-exchange resin (QIAGEN Genomic-tips, Qiagen Sciences, Germantown, MD) and then ligated in a 25 µl reaction volume with a double-strand anchor linker (Schlissel et al., 1993). For quantification of broken DNA ends, linker-ligated DNA was serially 2-fold diluted (64, 8, 4, 2, 1, 0.5, 0.25, 0.125, and 0.0625 ng of linker-ligated DNA derived from 10240, 1280, 640, 320, 160, 80, 40, 20, and 10 cells, respectively) into unligated genomic DNA and used as template in LM-PCR or serially diluted in water and used as template for β-actin DNA amplification. A first set of primers, consisting of the linker primer BW1 and the specific forward primer (priming the upstream DNA end) or reverse primer (priming the downstream DNA end) was used for the first round of PCR (12 cycles with a 50°C annealing step). A second set of primers, consisting of the same BW1 oligonucleotide and a nested specific forward or reverse primer was used for the second round of PCR (25 cycles with a 60°C annealing step; 1 µl of the 25 µl first round reaction was used to prime the 25 µl of second round reaction) (Table S1). Amplified DNA was fractionated through 1.5% agarose, blotted onto Hybond-N+ membranes (Amersham Biosciences, Inc., Piscataway, NJ), and hybridized to [γ-32P]-ATP labeled gene-specific oligonucleotide probes. The homology of the Ig VH primers and probes with germline VH sequences (VBASE, MRC Centre for Protein Engineering, Cambridge, UK, http://www.mrc-cpe.cam.ac.uk/vbase-ok.php?menu=901) was determined using MacVector 6.5.3 (Accelrys, Burlington, MA). The procedure used to extract genomic DNA does not introduce detectable amounts of DSBs, as demonstrated by the analysis of Cμ DNA, and compares favorably to the agarose embedding or “Agarose plug” (Schlissel et al., 1993; Papavasiliou and Schatz, 2000) and the “whole cell lysate” or “Direct” (Bross et al., 2000) methods (Figures 1, 3, 4C, 6, S2, S3, and not shown). Genomic DNA was treated with different enzymes before being used as a template for the amplification of DSB upstream or downstream DNA ends. Total DSBs were detected by treating DNA with T4 DNA polymerase in the presence of dNTPs (200 µM); DSBs involving 3′-protruding ends were detected after sequential pretreatment with Exo III and Exo T; DSBs involving 5′-protruding ends were detected after sequential pretreatment with SAP and Exo VII (Figures 5, S3, S5, and S8).
AID and DN AID Construction and Overexpression
The full-length human AID cDNA was amplified using the primers AID-F2 (5′-GAGGCAAGAAGACACTCTGG-3′) and AID-R2 (5′-GTGA-CATTCCTGGAAGTTGC′3′). The full-length human DN AID cDNA containing the H56R/E58Q mutations (Papavasiliou and Schatz, 2002b) was generated by PCR-targeted mutagenesis. AID and DN AID cDNAs were cloned directly into the pcDNA3.1 expression vector (pcDNA3.1/V5-His TOPO TA Expression Kit, Invitrogen Life Technologies). The construct and empty pcDNA vectors were used to transfect CL-01 subclone 3G10 and Ramos cells, which were then cultured in the presence of G418. After 21 days, the selected cells were harvested to prepare mRNA and genomic DNA to analyze the expression of AID or DN AID by specific RT-PCR using the appropriate oligonucleotide primers (see below) and determine the frequency and nature of the DSBs.
RT-PCR Amplification of AID, DN AID, and β-Actin Transcripts
RNA was extracted from 2 × 106 cells using the RNeasy Mini Kit (Qiagen). mRNA was reverse transcribed using the SuperScript Pre-amplification System (Invitrogen Life Technologies). cDNA was used as a template for the amplification of AID transcripts (primers: forward AID-F 5′-TGCTCTTCCTCCGCTACATCTC-3′ and reverse AID-R 5′-AACCTCATACAGGGGCAAAAGG-3′) and β-actin transcripts (Zan et al., 2001). The AID or DN AID transcript encoded by the expression vectors were amplified use a vector forward primer located downstream of the transcriptional start site (5′-ACGACT CACTATAGGGAGACC-3′) together with the AID reverse primer AID-R. RT-PCRs were made semiquantitative by varying the number of amplification cycles and performing dilution analysis to ensure a linear relationship between the amount of cDNA used and the intensity of the PCR product.
Cell Cycle Study
Ramos B cells or IgM+IgD+ B CL-01 cells that had been cultured with htCD40L were fixed in 70% ethanol by overnight incubation at 4°C and then stained for 2 hr at room temperature in a solution containing 100 µg/ml propidium iodide (Sigma-Aldrich Co.), 100 U/ml RNase A and 0.1% NP-40 before being submitted to flow cytometry.
ChIP Assays
Ramos cells (2.5 × 107) were treated with 1% formaldehyde for 10 min at room temperature to crosslink chromatin. After washing with cold PBS containing protease inhibitors, crude chromatin was separated by addition of nuclei-lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 0.5 M NaCl, 1% Trition-X-100, 0.5% sodium deoxycholate, and 0.5% sarkosyl [pH 8.0]), resuspended in IP-1 buffer (20 mM Tris-HCl, 200 mM NaCl, 2 mM EDTA, 0.1% sodium deoxycholate, 0.1% SDS, and protease inhibitors) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and sonicated to yield 200–1000 bp DNA fragments. The fragmented chromatin was precleared with agarose beads bearing protein A or G (Santa Cruz Biotechnology, Inc.) and then incubated with purified rabbit or goat IgG to human Rad51 (Novus Biologicals, Littleton, CO), Rad52 (Santa Cruz Biotechnology, Inc.), γ-H2AX (Upstate Biotech, Waltham, MA), Nsb1 (Santa Cruz Biotechnology, Inc.), Mre11 (Santa Cruz Biotechnology, Inc.), or mouse mAb to human Ku70/Ku86 (Lab Vision/NeoMarkers, Fremont, CA), overnight at 4°C. The immune complexes were isolated using beads bearing protein A or G. After washing, they were eluted with the 50 mM Tris-HCl, 0.5% SDS, 200 mM NaCl, 100 µg/ml Proteinase K (pH 8.0), and “elution buffer.” The eluates were heated at 65°C overnight to reverse crosslinks. DNA was recovered by phenol extraction and ethanol precipitation and was finally solubilized in TE buffer. The recovered DNA was treated with T4 DNA polymerase (T4+) or nil (T4−), linker-ligated, and then serially 2-fold diluted in untreated homologous DNA. The immunoprecipitated DNA was specified by LM-PCR using the Ramos reverse VH primers, the germline reverse V3 primers, and the forward Cμ primers.
Supplementary Material
Acknowledgments
We thank Shefali Shah for her technical help. This work was supported by N.I.H. grants AI 45011, AR 40908, AG 13910, AI 07621 (to P.C.), and GM 42482 (to W.K.H).
References
- Baer R, Forster A, Lavenir I, Rabbitts TH. Immunoglobulin VH genes are transcribed by T cells in association with a new 5′ exon. J. Exp. Med. 1988;167:2011–2016. doi: 10.1084/jem.167.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Sale JE, Kim HJ, Berek C, Cosgrove RA, Neuberger MS. Somatic hypermutation in the absence of DNA-dependent protein kinase catalytic subunit (DNA-PK(cs)) or recombination-activating gene (RAG)1 activity. J. Exp. Med. 2000;192:1509–1514. doi: 10.1084/jem.192.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman JE, Humphries CG, Barth J, Alt FW, Tucker PW. Structure and expression of human germline VH transcripts. J. Exp. Med. 1991;173:1529–1535. doi: 10.1084/jem.173.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- Bross L, Muramatsu K, Kinoshita K, Honjo H, Jacobs H. DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J. Exp. Med. 2002;195:1187–1192. doi: 10.1084/jem.20011749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayre Y, Palladino MA, Marcu KB, Stavnezer J. Expression of an antigen receptor on T cells does not require recombination at the immunoglobulin JH-Cμ locus. Proc. Natl. Acad. Sci. USA. 1981;78:3814–3818. doi: 10.1073/pnas.78.6.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Zan H, Schaffer A, Bergsagel L, Harindranath N, Max EE, Casali P. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center and plasmacytoid phenotypic differentiation in a human monoclonal IgM+ IgD+ B cell line. J. Immunol. 1998a;160:2145–2157. [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Schaffer A, Shah S, Zan H, Liou HC, Goodwin RG, Casali P. CD30 is a CD40-inducible molecule that negatively regulates CD40-mediated immunoglobulin class switching in non-antigen-selected human B cells. Immunity. 1998b;9:247–256. doi: 10.1016/s1074-7613(00)80607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol. Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Diaz M, Casali P. Somatic Ig hypermutation. Curr. Opin. Immunol. 2002;14:235–240. doi: 10.1016/s0952-7915(02)00327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Doi T, Kinoshita K, Ikegawa M, Muramatsu M, Honjo T. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proc. Natl. Acad. Sci. USA. 2003;100:2634–2638. doi: 10.1073/pnas.0437710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO. J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill J-C, Reynaud CA. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Gurrieri C, McGuire P, Zan H, Yan H-J, Cerutti A, Albesiano E, Allen S, Vinciguerra V, Rai KR, Ferrarini M, et al. Chronic lymphocytic leukemia B cells undergo somatic hypermutation and intraclonal Ig VHDJH gene diversification. J. Exp. Med. 2002;196:629–639. doi: 10.1084/jem.20011693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Koenig N, Malone B, Hoch S, Schwaber J. Transcription of germline VH gene elements by normal human fetal liver. Mol. Immunol. 1997;34:333–341. doi: 10.1016/s0161-5890(97)00023-0. [DOI] [PubMed] [Google Scholar]
- Liu YJ, de Bouteiller O, Arpin C, Briere F, Galibert L, Ho S, Martinez-Valdez H, Banchereau J, Lebecque S. Normal human IgD+IgM− germinal center B cells can express up to 80 mutations in the variable region of their IgD transcripts. Immunity. 1996;4:603–613. doi: 10.1016/s1074-7613(00)80486-0. [DOI] [PubMed] [Google Scholar]
- Martin A, Bardwell PD, Woo CJ, Fan M, Shulman MJ, Scharff MD. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. The AID enzyme induces class switch recombination in fibroblasts. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Cell-cycle-regulated in DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes; merging mechanisms for genetic diversity. Cell. 2002a;109:s35–s44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hyper-mutation process. J. Exp. Med. 2002b;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Schaffner W. Unrearranged immunoglobulin lambda variable region is transcribed in kappa-producing myelomas. EMBO. J. 1984;3:3031–3035. doi: 10.1002/j.1460-2075.1984.tb02250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Neuberger MS. TdT-accessibe breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- Schaffer A, Cerutti A, Shah S, Zan H, Casali P. The evolutionary conserved sequence upstream of the human Ig Sγ3 region is an inducible promoter: synergistic activation by CD40 ligand and IL-4 via cooperative NF-κB and STAT-6 binding sites. J. Immunol. 1999;162:5327–5336. [PubMed] [Google Scholar]
- Schaffer A, Kim E, Wu X, Zan H, Testoni L, Salamon S, Cerutti A, Casali P. Selective inhibition of class switching to IgG and IgE by recruitment of the HoxC4 and Oct-1 homeodomain proteins and Ku70/Ku86 to newly identified ATTT cis-elements. J. Biol. Chem. 2003;278:23141–23150. doi: 10.1074/jbc.M212952200. [DOI] [PubMed] [Google Scholar]
- Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Song B, Sung P. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- Storb U, Shen HM, Michael N, Kim N. Somatic hypermutation of immunoglobulin and non-immunoglobulin genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:13–19. doi: 10.1098/rstb.2000.0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO. J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Wilson PC, Wilson K, Liu YJ, Banchereau J, Pascual V, Capra JD. Receptor revision of immunoglobulin heavy chain variable region genes in normal human B lymphocytes. J. Exp. Med. 2000;91:1881–1894. doi: 10.1084/jem.191.11.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zan H, Komori A, Feng J, Kim EC, Casali P. Immunoglobulin somatic hypermutation: double-strand DNA breaks, AID and error-prone DNA repair. J. Clin. Immunol. 2003;23:235–246. doi: 10.1023/a:1024571714867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- Zan H, Cerutti A, Schaffer A, Dramitinos P, Casali P. CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-β. Evidence for TGF-β but not IL-10-dependent direct SμøSα and sequential SμøSγ, SγøSα DNA recombination. J. Immunol. 1998;161:5217–5225. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ cell line in vitro: Definition of the requirements and the modalities of hypermutation. J. Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Li Z, Yamaji K, Dramitinos P, Cerutti A, Casali P. BCR engagement and T cell contact induce bcl-6 hypermutation in human B cells: association with initiation of transcription and identity with Ig hypermutation. J. Immunol. 2000;165:830–839. doi: 10.4049/jimmunol.165.2.830. [DOI] [PubMed] [Google Scholar]
- Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.