SUMMARY
Mitochondrial dysfunction is a common feature in neurodegeneration and aging. We identify mitochondrial dysfunction in xeroderma pigmentosum group A (XPA), a nucleotide excision DNA repair disorder with severe neurodegeneration, in silico and in vivo. XPA deficient cells show defective mitophagy with excessive cleavage of PINK1 and increased mitochondrial membrane potential. The mitochondrial abnormalities appear to be caused by decreased activation of the NAD+-SIRT1-PGC-1α axis triggered by hyperactivation of the DNA damage sensor PARP1. This phenotype is rescued by PARP1 inhibition or by supplementation with NAD+ precursors that also rescue the lifespan defect in xpa-1 nematodes. Importantly, this pathogenesis appears common to ataxia-telangiectasia and Cockayne syndrome, two other DNA repair disorders with neurodegeneration, but absent in XPC, a DNA repair disorder without neurodegeneration. Our findings reveal a novel nuclear-mitochondrial cross-talk that is critical for the maintenance of mitochondrial health.
INTRODUCTION
While significant advances have been made in our understanding of many DNA repair disorders, it remains an ongoing mystery in the field of aging and DNA metabolism why some patients with DNA repair deficiency develop neurodegeneration while others are spared. Accumulation of nuclear DNA damage has been associated with accelerated aging disorders and normal aging, however, the connection between DNA damage and neurodegeneration is less clear (Hoeijmakers, 2009; Rass et al., 2007). Recently, mitochondrial involvement has been proposed in two major DNA repair disorders ataxia-telangiectasia (AT) and Cockayne syndrome (CS) (Scheibye-Knudsen et al., 2012; Valentin-Vega et al., 2012). As the power center of cells, mitochondria are involved in a plethora of metabolic networks and play a significant role in sustaining the life and health of humans (Green et al., 2011). Neurons may be particularly vulnerable to mitochondrial alterations because of their high energy demands. Accordingly, mitochondrial dysfunction has been proposed to underlie numerous neurodegenerative diseases. Mitochondrial alterations may therefore represent an attractive explanation for the neurological phenotype in some DNA repair disorders. However, in the case of CS and AT, the mechanisms leading from a DNA repair defect to mitochondrial dysfunction have been unclear. Because mitochondria are involved in a large number of processes, defects in a variety of pathways could lead to secondary mitochondrial alteration. To address this we recently developed an in silico method to screen potential diseases for mitochondrial involvement using a clinical database, www.mitodb.com (Scheibye-Knudsen et al., 2013). We have continuously expanded this database and screened for known DNA repair disorders with neurodegeneration. An unexpected finding was that patients with xeroderma pigmentosum (XP) group A (XPA) had a significant clinical mitochondrial phenotype.
XP is a rare autosomal recessive disorder characterized by severe sun sensitivity leading to a greatly increased risk of UV induced skin cancers. XPA was the first disorder shown to be caused by defective DNA repair laying the foundation for numerous discoveries and creating a field of research. In addition to the dermatological ailments XPA patients often suffer from various degrees of neurodegeneration with progressive cerebral and cerebellar atrophy, neuropathy and sensorineural hearing loss (DiGiovanna and Kraemer, 2012). The neurological features in XPA share substantial similarities to what is often observed in mitochondrial diseases where progressive cerebellar degeneration, peripheral neuropathy and sensorineural hearing loss are highly prevalent. Notably, the neurological deficits seen in AT and CS show extensive clinical overlap with XPA perhaps indicating a common pathogenesis in these three disorders (Table S1).
From an aging perspective mitochondria are particularly important because it has been proposed that progressive mitochondrial dysfunction leads to declining cellular function, tissue decay and finally death. Indeed, a number of pathways known to extend lifespan also appear to preserve mitochondrial function. SIRT1 is a NAD+ dependent deacetylase that regulates longevity possibly through maintenance of mitochondrial homeostasis (Mouchiroud et al., 2013). Mitochondrial health can, however, be moderated in several ways. Regulation and elimination of reactive oxygen species (ROS) through antioxidants, augmentation of mitochondrial DNA repair and mitochondrial autophagy (henceforth mitophagy) have all been suggested to be important mediators of longevity. SIRT1 has been proposed to increase mitochondrial health through the transcription factor PGC-1α that appears to be important for the regulation of ROS and mitochondrial biogenesis particularly in muscle (Lagouge et al., 2006). In the brain, however, mitophagy has been proposed to be central in maintaining neuronal health (Youle and van der Bliek, 2012).
We herein report that XPA deficiency leads to mitochondrial alterations in nematodes, mice, rats and humans through an inhibition of SIRT1. This novel mechanism appears in the neurodegenerative DNA repair disorders XPA, CS and AT, but not in a related DNA repair disorder, XP group C (XPC), where neurodegeneration is rarely seen. In XPA, CS and AT, SIRT1 attenuation leads to decreased mitophagy through the depression of PGC-1α and UCP2. Importantly SIRT1 appears to be inhibited by the activation of the DNA damage sensor poly-ADP-ribose polymerase-1 (PARP1). Indeed, we find that the mitophagic defect in XPA, CS and AT can be rescued pharmacologically using PARP inhibitors or by overexpressing UCP2. Further, NAD+ precursors are able to rescue the mitochondrial defects in XPA cells and extend lifespan in xpa-1 worms, indicating a possible therapeutic strategy for this disease. We present a model explaining the mitochondrial defect seen in XPA, CS and AT through a previously unknown nuclear-mitochondrial cross talk. We also propose a key role for the longevity gene SIRT1 in selective mitophagy underscoring the importance of mitochondrial maintenance in brain health and aging.
RESULTS
XPA deficiency leads to mitochondrial pathology
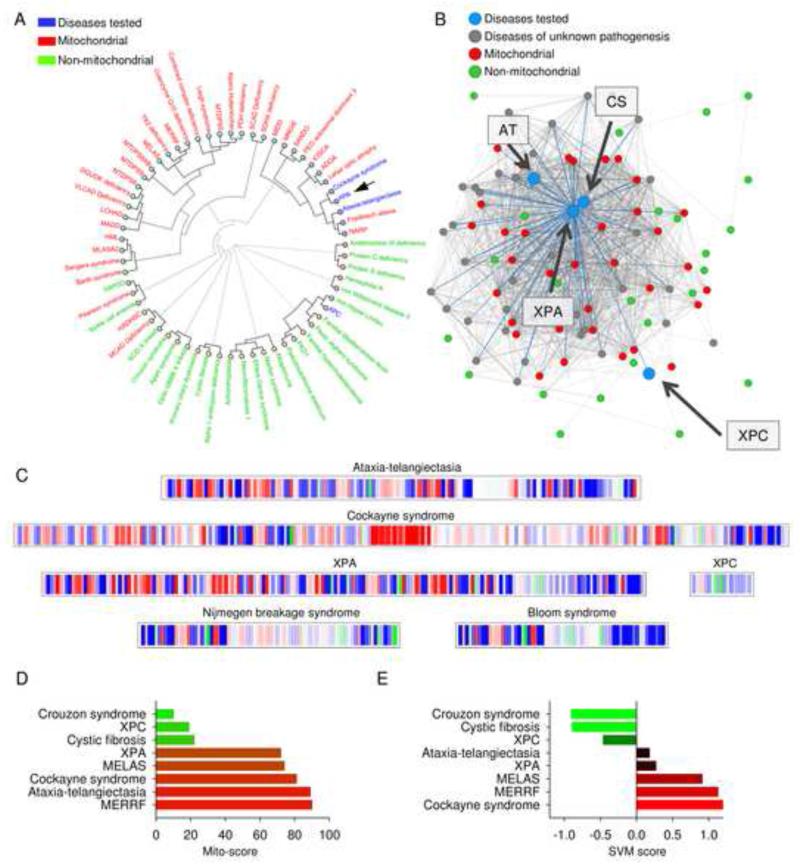
Using our bioinformatics tool, www.mitodb.com, we reported that CS and AT clustered with known mitochondrial disease (Scheibye-Knudsen et al., 2013). In pursuing the goal of understanding how DNA repair leads to neurodegeneration we further screened the database for DNA repair defective disorders with prominent clinical mitochondrial defects. Using the bioinformatics tools we found that XPA was predicted to have a significant mitochondrial involvement as evidenced by the clinical phenotypical similarity with other recognized mitochondrial diseases (Figure 1; Table S1 for the clinical features of XPA, CS and AT). Hierarchical clustering based on the prevalence of signs and symptoms revealed that XPA associates with the two abovementioned diseases, CS and AT. XPA, CS and AT in turn associated with a number of known mitochondrial diseases (Figure 1A). Interestingly, XPC, which does not have neurological features, did not associate with the mitochondrial diseases (Figure 1A). The network in figure 1B shows how the diseases associate with each other based on overlapping signs and symptoms. Each dot represents a disease and the connecting lines between the diseases represent shared traits. The more traits that are shared the closer two diseases will associate with each other. As can be seen XPA, CS, and AT are closely linked with known mitochondrial diseases while XPC does not link with the mitochondrial disorders (Figure 1B). Furthermore, a mitochondrial barcode for XPA was generated that revealed a dominating red impression which indicates substantial mitochondrial involvement, while other DNA repair disorders (XPC, Nijmegen breakage syndrome and Bloom syndrome) without neurodegenerative phenotypes show much less red in the barcode (Figure 1C). In addition, two quantitative scores were calculated. A mito-score of more than 50 indicates overlap with more mitochondrial than non-mitochondrial diseases while a positive score from the support vector machine (SVM) indicates a likely mitochondrial involvement. In support of a possible mitochondrial dysfunction, XPA had a mito-score of 72 and a SVM score of 0.27 (Figure 1D-E; and further on www.mitodb.com).
Figure 1. Patients suffering from XPA are phenotypically similar to patients suffering from mitochondrial diseases.
(A) Hierarchical clustering of diseases based on the prevalence of clinical parameters using mitodb.com. (B) A representation of how XPA, XPC, CS and AT associate within the disease network. Each dot represents a disease and the connecting lines represent one or more shared sign and/or symptom. The shorter and thicker the line the greater the prevalence of the shared symptoms. (C) The mitochondrial barcode of XPA, XPC, CS, AT, Nijmegen breakage syndrome and Bloom syndrome. Each vertical bare represent a sign or symptom that is shared with another disease in the database. Mitochondrial (red), non-mitochondrial (green) and diseases of unknown pathogenesis (blue). A predominantly red barcode will indicate similarities with mitochondrial diseases while green will indicate non-mitochondrial involvement. (D) and (E) The mito-score (D) and the support vector machine (SVM) score (E) of XPA, XPC, CS and AT as well as two non-mitochondrial diseases Crouzon syndrome and cystic fibrosis and two bona fide mitochondrial disorders mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) and myoclonic epilepsy associated with ragged-red fibers (MERRF). See also Table S1.
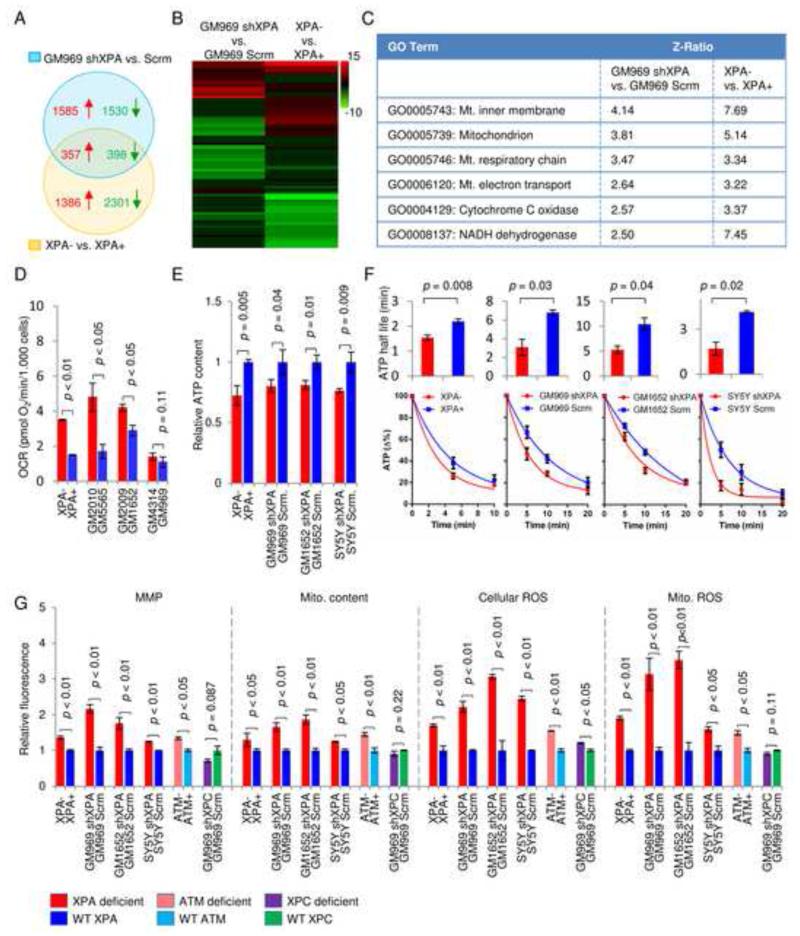
To gain additional information regarding the pathogenesis in XPA we performed gene expression microarray in two XPA deficient cell lines and their controls: (1) an immortalized XPA patient cell line reconstituted with WT XPA (XPA+) or an empty vector (XPA-), and (2) a primary human fibroblast GM969 with stable shRNA knockdown of XPA or with scrambled shRNA. We then identified common alterations in gene expressions between these cell lines as XPA dependent. A Venn diagram shows an overlap between the two abovementioned groups of 755 significantly changed genes in which 357 were upregulated and 398 were downregulated (Figure 2A). Gene ontology pathway analysis detected a number of significantly changed pathways (Figure 2B). Among the common 16 upregulated pathways between the cell lines (Figure S1A), 6 pathways were associated with mitochondrial metabolism as evidenced by the significant increase in Z-ratios. The array analysis thus supports the findings from the clinical database (Figure 2C).
Figure 2. XPA deficiency increases mitochondrial metabolism, membrane potential and ROS formation.
(A) Venn diagrams illustrating the overlap of induced (red, z-score > 1.5) and repressed (green, z-score < −1.5) genes between two groups of XPA deficient cells by microarray (n=3). (B) Heat map of microarray data showing significantly changed GO-pathways. (C) Common statistically significantly upregulated mitochondrial pathways in XPA deficient cells compared with controls. Also see Figure S1A. (D) Oxygen consumption rates in primary fibroblasts from XPA patients and their age and sex matched controls (means ± SD, n=3). (E) and (F) ATP levels and ATP consumption in XPA deficient and control cells were detected using a luciferase based assay after inhibition of ATP production with oligomycin and 2-deoxyglucose at time=0 (means ± SD, n=3). Data in (F) was fitted to an exponential decay curve and half-lives were calculated (means ± SD, n=3). (G) Flow cytometry was used to measure mitochondrial membrane potential (MMP) using TMRM staining, mitochondrial content using MitoTracker Green, cellular reactive oxygen species (ROS) using dihydroethidium and mitochondrial ROS using Mitosox (means ± S.D., n=3). See also Figure S1 and Figure S2.
We next performed a number of in vivo tests to determine how XPA might affect mitochondrial function. We first measured overall oxygen consumption rates (OCR). In addition to the XPA− cells and their complemented control XPA+ cells, three primary human fibroblasts from XPA patients and their sex and age matched controls were also used for OCR investigation. All XPA patient cell lines showed increased OCR compared to controls although for one pair (GM4314 vs GM969) the difference did not reach significance (Figure 2D). To gain further insight into the potential regulatory role of XPA in mitochondrial function additional XPA deficient cell lines were generated by stable shRNA knockdown: two primary human fibroblasts (GM969 and GM1652) and the SH-SY5Y human neuroblastoma cells reflecting the significant neurological involvement in XPA patients. Increased oxygen consumption may stem from the increased ATP consumption. By measuring ATP levels at various time points after inhibition of glycolysis using 2-deoxyglucose and oxidative phosphorylation using oligomycin, we found that basal ATP levels were decreased and ATP consumption was increased (Figure 2E-F). XPA deficiency also increased mitochondrial membrane potential (MMP), mitochondrial content, and both total as well as mitochondrial ROS (Figure 2G). To control for potential nonspecific effects of the shRNA which targets the 3′ UTR of XPA, a second shRNA which matches the coding region of XPA was used in the experiment and the same results were acquired (Figure S1B). Both shRNAs show over 90% knockdown efficiency (Figure S1C). Interestingly, this mitochondrial phenotype is also seen in AT− (Figure 2G) and CS− cells (Scheibye-Knudsen et al., 2012). In contrast, we did not observe this mitochondrial phenotype in XPC knockdown cells (Figure 2G and Figure S1D). Immunoblots of two mitochondrial markers COX-4 and VDAC as well as immunofluorescence of cells probed with COX-4 confirmed the finding of an increased mitochondrial content in XPA− cells (Figure S2A-B). Since it has been proposed that CSB and ATM act within the mitochondrial matrix to regulate mitochondrial function (Aamann et al., 2010; Valentin-Vega et al., 2012), we isolated mitochondria from cells to investigate whether XPA might be present in this compartment. Although we found trace amounts of XPA in mitochondria particularly after oxidative damage using menadione these traces were removed by treating the mitochondrial suspension with proteinase K (Figure S2C). This indicates that under our conditions XPA is not significantly present in the mitochondrial matrix, and that the mitochondrial alterations may be secondary to a nuclear defect.
XPA deficiency up-regulates autophagy
Since previous reports showed altered macro autophagy (hereafter referred to as autophagy) in AT and CS (Scheibye-Knudsen et al., 2012; Valentin-Vega et al., 2012), we speculated that an autophagic defect in XPA could explain the increased mitochondrial content. To address this hypothesis, we examined the changes in autophagosomes, autolysosomes, and several autophagy-related proteins. Surprisingly, we detected increased levels of the LC3-II signal which indicates increased autophagy in all four XPA deficient human cells as well as in rat neurons subjected to shRNA knockdown of XPA (Figure S3A-B). Increased LC3-II was also detected in CSB− and ATM− cells (Figure S3A right panel). Treatment with the lysosome inhibitors bafilomycin A1 and chloroquine revealed increased LC3-II in XPA− cells compared with XPA+ cells, indicating that XPA-deficiency may induce autophagosome synthesis (Figure S3A, first panel). Accordingly, quantification of autophagy using an mRFP-GFP tandem fluorescent-tagged LC3 plasmid (Kimura et al., 2007) revealed increased numbers of both autophagosomes and autolysosomes compared to controls (Figure S3C-E). Further p62 levels were decreases in XPA− compared with controls (Figure S3F) indicating increased autophagy (Klionsky et al., 2012). This was supported by increases in Beclin-1 levels and Atg12-Atg5 conjugation in XPA− cells compared with WT (Figure S3F) reflecting activation of autophagy (Klionsky et al., 2012). Notably, these changes were also seen in CSB and ATM deficient cells (Figure S3F). Furthermore, activity of the mechanistic target of rapamycin complex 1 (mTORC1), which down-regulates autophagy, was inhibited as evidenced by decreased level of phosphorylated p70S6, a downstream target of mTORC1 (Figure S3F).
Since we observed increased ATP consumption and increased ROS formation in XPA deficient cells we speculated that AMPK activation might be responsible for the increased autophagy. Indeed, increased phosphorylation of the AMPKα subunit at Thr172 was observed in XPA deficient cells compared with XPA+ cells indicating activation of AMPK (Figure S3G). Furthermore, XPAcells treated with either a AMPK phosphorylation inhibitor dorsomorphin (DM) or a ROS scavenger N-acetyl-L-cysteine (NAC) had lower levels of phosphor-AMPKα (Figure S3G). This led to increased mTOR activation as shown by increased p70S6 phosphorylation. Collectively, these results suggest that autophagy induction in XPA deficiency is at least partially attributed to activation of AMPK and inhibition of mTOR.
XPA deficiency impairs mitophagy and drives apoptosis
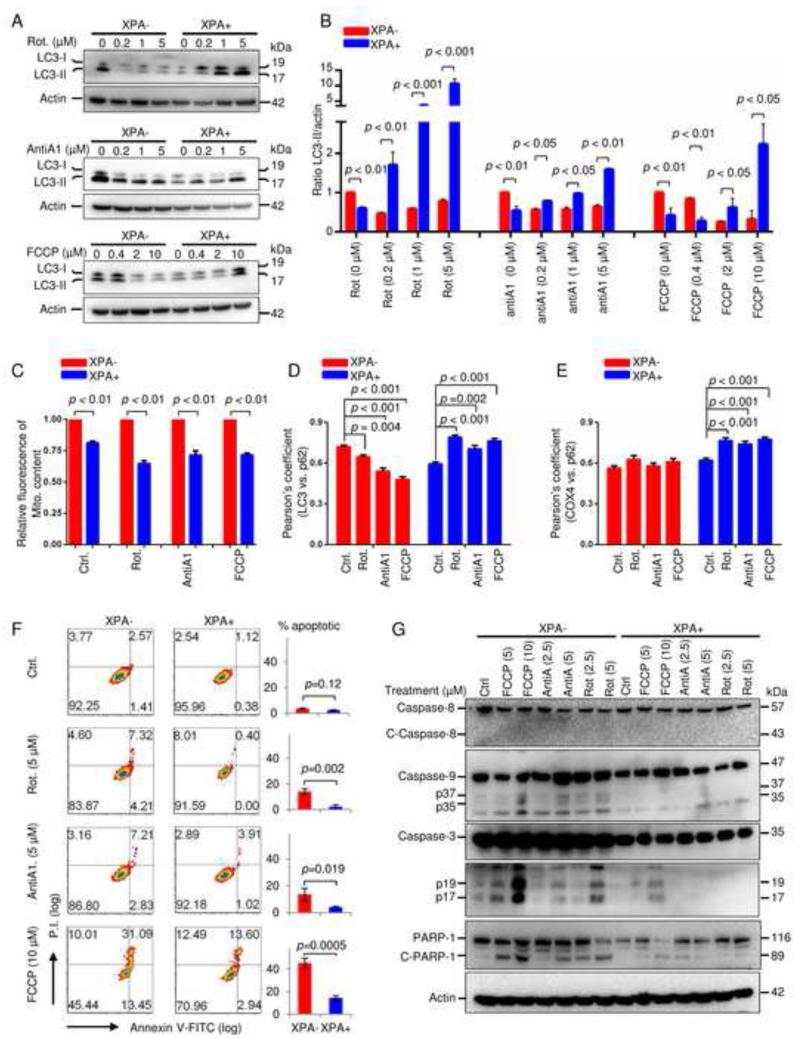
Increased autophagy could, however, not explain the increased mitochondrial content. We therefore asked whether the mitochondrial alterations could be caused by specific defects in mitophagy. Three mitochondrial toxins were used to induce mitophagy: rotenone (a mitochondrial complex I inhibitor), antimycin A1 (a complex III inhibitor), and the mitochondrial uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP). Immunoblots revealed that the toxins induced a significant increase in LC3-II levels in WT cells while LC3-II levels decreased in XPA− cells, indicating an impaired mitophagic process in the absence of XPA protein (Figure 3A-B). These findings were substantiated by increased loss of mitochondria in control cells compared to XPA− cells after treatment with various mitochondrial toxins that induce mitophagy (Figure 3C). We then investigated changes in protein colocalization among autophagy/mitophagy-associated proteins, including LC3 vs. p62, and COX4 vs. p62. As expected the mitochondrial toxins enhanced the colocalization of both LC3 vs. p62 and COX4 vs. p62 in XPA+ cells as measured by the Pearson’s coefficient, while XPA− cells showed loss of colocalization of LC3 vs. p62 and COX4 vs. p62 (Figure 3D-E and Figure S4A-B), suggesting deficient relocalization of p62 to damaged mitochondria.
Figure 3. XPA deficiency impairs cellular mitophagy and sensitizes cells to caspase-9 regulated apoptosis under multiple mitochondrial stresses.
(A) Cells were treated with mitochondrial toxins for 24 h, and LC3 was detected by immunoblot in total cell lysates. Rotenone: a mitochondrial complex I inhibitor, antimycin A: a mitochondrial complex III inhibitor, FCCP: a mitochondrial uncoupler. (B) Quantitative values of relative LC3-II levels normalized to Actin (means ± S.D., n=3). (C) Relative values of mitochondrial content in XPA− and XPA+ cells after different treatments using flow cytometry (means ± S.D., n=3). (D, E) Quantification of colocalization of LC3 and p62 (D) or COX4 and p62 (E) after treatment with mitochondrial toxins as determined by Pearson’s correlation coefficient using confocal microscopy (means ± S.E.M., n>50). See also Figure S4A and S4B. (F) Detection of apoptosis using flow cytometry in XPA− and XPA+ cells with Annexin-V/PI staining after mitochondrial stressors. Quantification is shown on the right (means ± S.D., n=3). (G) Immunoblot of the expression of proteins involved in caspase-8 regulated and caspase-9 regulated apoptotic pathways in XPA− and XPA+ cells exposed to different mitochondrial toxins. See also Figure S4.
Since studies on the crosstalk between autophagy/mitophagy and apoptosis indicate possible opposite roles on cell viability (Green et al., 2011), we wanted to investigate whether XPA deficient cells were more susceptible to apoptosis induced by mitochondrial toxins than controls. After treatment with the abovementioned mitochondrial toxins, XPA deficient and control cells were stained with Annexin V/PI and sorted by flow cytometry. The results revealed that XPA− cells were more susceptible to mitochondrial stress-induced apoptosis as evidenced by more early and late apoptotic cells compared to control cells (Figure 3F). For example, 5 μM rotenone did not significantly induce apoptosis in control cells, while it triggered nearly 15% apoptotic cell death in XPA− cells.
Furthermore, when exposed to 10 μM FCCP for 24 h, the fraction of apoptotic cells increased from 16.5% in XPA+ cells to over 40% in XPA− cells. Consistently, protein markers of apoptosis including cleaved caspase-3 (p17 and p19 fragments) and the caspase substrate PARP1 were both increased in mitochondrial stress-exposed XPA− cells (Fig. 3G). This appeared to be due to increased mitochondrial apoptosis signaling since we only detected cleaved Caspase-9 (p35 and p37 fragments), and not cleaved Caspase-8 in XPA− cells exposed to mitochondrial stress (Figure 3G). Taken together, these results indicate that XPA deficiency leads to a shift from mitophagy to apoptosis under mitochondrial stress.
The mitophagy defect in XPA deficient cells involves mitochondrial hypertrophy and a voltage-dependent increase in PINK1 proteolysis
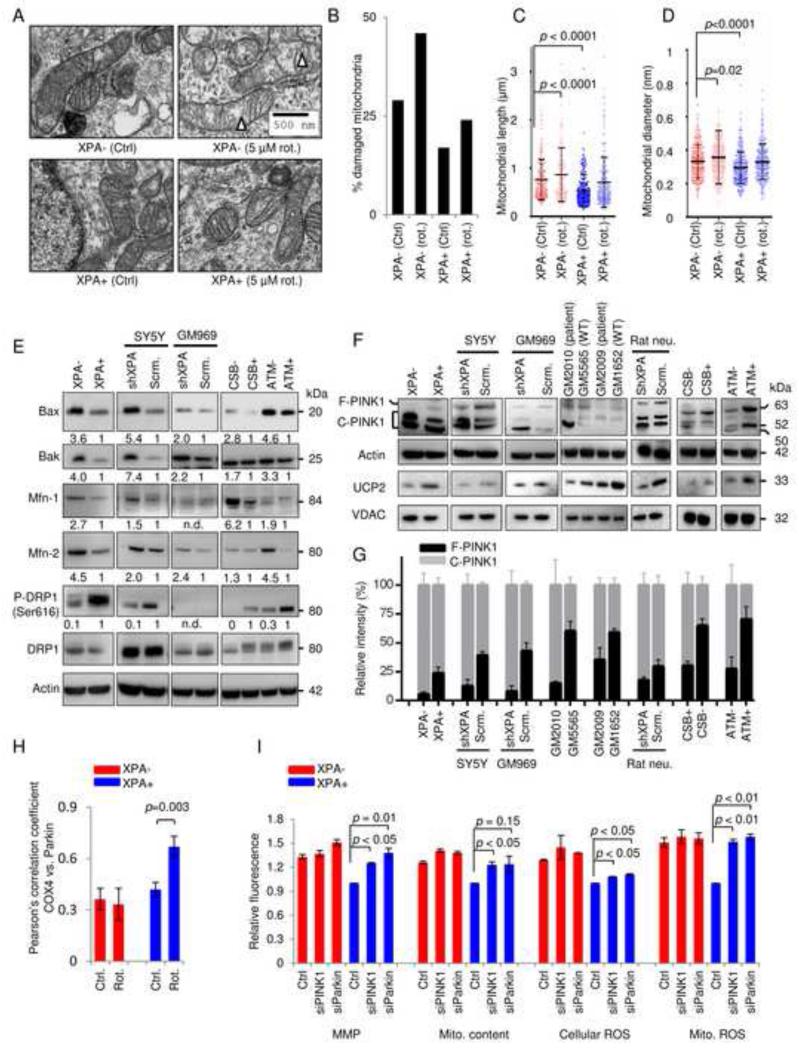
It is generally believed that small fragmented mitochondria with depolarized MMP are more easily degraded by mitophagy in a PINK1/Parkin-dependent pathway. Reduction in MMP facilitates the accumulation of full length PINK1 on the outer mitochondrial membrane of damaged mitochondria leading to Parkin recruitment and initiation of selective mitophagy (Youle and van der Bliek, 2012). To investigate if mitochondrial size might be altered we performed electron microscopy on XPA+ and XPA− that revealed increased amounts of damaged mitochondria, increased mitochondrial diameter and increased mitochondrial length in XPA− cells compared to WT cells (Figure 4A-D). Since mitochondrial size and fragmentation are regulated via a balance between fusion and fission we next investigated proteins involved in these processes. Specifically, we detected the mitofusion proteins, mitofusin-1 (Mfn-1), Mfn-2, Bax and Bak, as well as the mitofission protein dynamin related protein 1 (DRP1). Indeed, both XPA− and shXPA SY5Y cells showed an upregulation of the above-mentioned mitofusion proteins, and a decrease of phosphorylated DRP1 (Ser616) indicating inhibited mitochondrial fission (Figure 4E). This pattern was also seen in CSB and ATM deficient cells (Figure 4E).
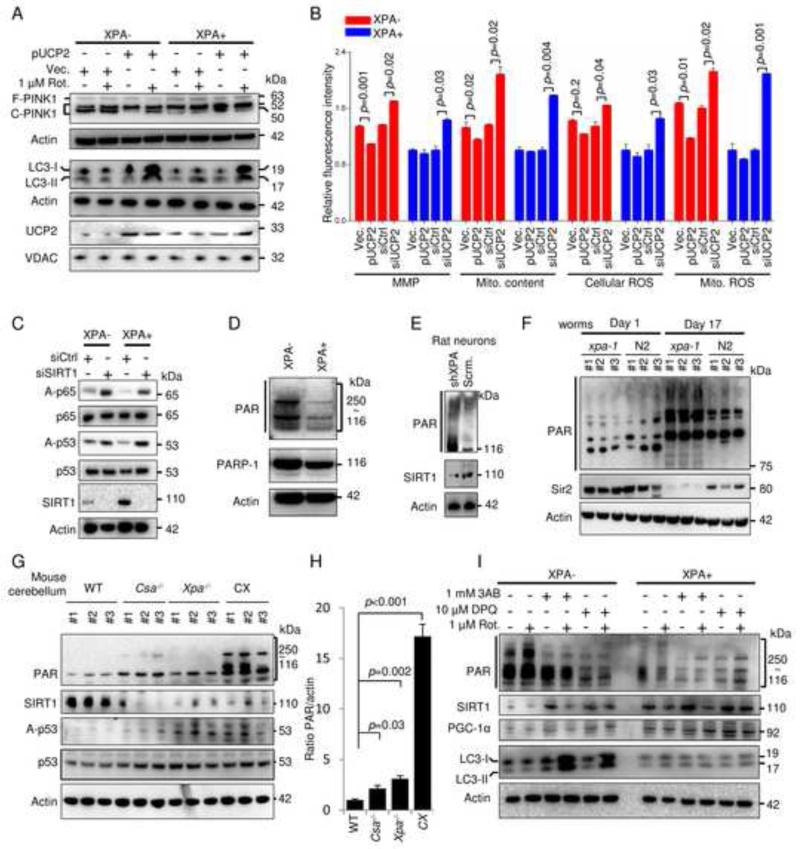
Figure 4. XPA deficiency suppresses mitophagy by up-regulation of mitochondrial fusion and cleavage of PINK1.
(A-D) XPA− and XPA+ cells were treated with 5 μM rotenone or vehicle for 24 h, and electron microscopy was performed (A). Triangles indicate damaged mitochondria. Quantification of damaged mitochondria (B), mitochondrial length (C) and mitochondrial diameter (D), (means ± S.D., n=14, with >150 mitochondria counted per group). (E) Immunoblot of proteins involved in mitochondrial size regulation. Bax, Bak, mitofusin-1 (Mfn-1), and Mfn-2 participate in mitochondrial fusion whereas phosphorylation of DRP1 at Ser616 is involved in mitochondrial fission. (F) Protein expression of PINK1 and UCP2 levels in XPA deficient cells and tissues. (G) Quantification of (F) showing full length and cleaved PINK1 normalized to total PINK1 (means ± S.D., n=3). (H) The colocalization between COX-4 and Parkin in XPA− and XPA+ cells treated with 5 μM rotenone for 24 h and quantified using the Pearson’s correlation coefficient (means ± S.D., n=3 and Figure S4D). (I) Various mitochondrial parameters in XPA+ and XPA− cells after siRNA knockdown of PINK1 and Parkin (means ± S.D., n=3 and Figure S4F-G for knockdown efficiency).
Since PINK1 is retained at the outer mitochondrial membrane of depolarized mitochondria and XPA deficiency led to hyperpolarization of the membrane, we next investigated the stability of full length PINK1 (Wang et al., 2011). As expected, we found increased cleaved PINK1 as well as less full-length PINK1 in XPA deficient cells compared with controls (Figure 4F-G). Since the MMP may be regulated by mitochondrial uncoupling proteins (UCPs) we next investigated the protein levels of UCP1, 2, 3 and 4 (Figure S4C). Immunoblots with purified mitochondrial fractions from XPA− and XPA+ cells revealed that UCP2 was significantly decreased in XPA− cells (Figure 4F). Further, the same results were also found in two primary fibroblasts from XPA patients as well as in primary rat neurons and human SH-SY5Y cells subjected to XPA shRNA knockdown (Figure 4F-G). Interestingly, CSB and ATM deficient cells also displayed increased cleavage of PINK1 as well as lower levels of UCP2 (Figure 4F-G). Based on this we predicted that XPA deficient cells would be less able to recruit Parkin to damaged mitochondria after stress. Indeed, mitochondrial stress with rotenone failed to increase the colocalization of Parkin and COX-4 in XPA− cells as indicated by no significant change in the Pearson’s correlation coefficient while it was increased in control cells (Figure 4H and Figure S4D). To investigate if the defect in PINK1 stabilization and Parkin recruitment was contributing to the mitochondrial phenotype we performed siRNA knockdown of these two proteins in XPA− and XPA+ cells. This led to an increase in mitochondrial content, MMP and ROS production in WT cells only indicating that XPA, PINK1 and Parkin are epistatic in the pathogenesis of the mitochondrial dysfunction (Figure 4I and figure S4F-G). These novel results collectively indicate that XPA, CSB or ATM deficiency trigger mitochondrial fusion and hyperpolarization of the mitochondrial membrane through a down-regulation of UCP2 leading to cleavage of PINK1 and an inability to clear damaged mitochondria.
The NAD+-SIRT1-PGC-1α axis regulates the expression of UCP2 which can rescue the mitophagy defect in XPA− deficient cells
UCP2 upregulation is associated with attenuation of mitochondrial ROS production and neuroprotection (Islam et al., 2012). To investigate whether UCP2 augmentation could rescue the mitochondrial phenotype in XPA− cells, we transiently transfected UCP2 plasmid into cells followed by rotenone treatment. UCP2 was expressed in XPA− cells, leading to less cleaved PINK1, and more LC3-II after rotenone treatment (lane 2 vs. lane 4, Figure 5A). UCP2 transfection in XPA− cells also lowered MMP, and decreased mitochondrial content and mitochondrial ROS (Figure 5B). This effect was also observed in ATM deficient cells where ectopic UCP2 expression led to a near complete rescue of the mitochondrial phenotype (Figure S4E). Accordingly, siRNA knockdown of UCP2 in XPA− and XPA+ cells led to an exacerbation of the mitochondrial phenotype, particularly in the WT cells (Figure 5B and S5A). Considering that UCP2 is regulated by the upstream transcription factor PGC-1α and that this factor in turn is regulated by SIRT1 (Bai et al., 2011; Lagouge et al., 2006), we measured the expression level and activity of these two proteins. In XPA− cells, SIRT1 was less expressed corresponding with an increased acetylation of its downstream targets NF-κB (p65) and p53 as compared to controls (Figure 5C). In agreement with this, knockdown of SIRT1 by siRNA down-regulated PGC-1α and UCP2 (lanes 5 and 7 in Figure S5B) led to deficient mitophagy as demonstrated by lower levels of LC3 signal from a relative value of 1.5 to 0.8 of control after rotenone exposure (Figure S5B, lanes 6 and 8). SIRT1 uses NAD+ as a substrate and is regulated by NAD+ levels (Mouchiroud et al., 2013). Indeed, NAD+ levels were decreased (see below). NAD+ metabolism is intimately tied to the DNA damage responses through PARP1 (Bai et al., 2011). PARP1 acts as an early responder to DNA damage, where it creates a PAR-polymer using NAD+ as a substrate. Since XPA is caused by a defect in DNA repair we speculated that the decrease in NAD+ may be due to increased PARP1 activity. Indeed, this appeared to be the case (PAR, Figure 5D). Increased PARP activation and decreased SIRT1 levels were also observed in primary rat neurons after XPA knockdown (Figure 5E). It has recently been suggested that age associated PAR accumulation could be conserved from nematodes to mammals (Mouchiroud et al., 2013) and we thus investigated the consequence of XPA− deficiency by examining Sir2 (the nematode homolog of mammalian SIRT1) and PAR levels in C. elegans. Immunoblots were performed on whole nematode extracts of synchronized adult 1-day and 17-day old xpa-1(ok698) mutants (xpa-1, the nematode homologue of human XPA) and WT (N2) worms. Consistent with previous reports (Mouchiroud et al., 2013), PARylation increased with age in both types of worms. Interestingly, aged xpa-1 nematodes had significantly higher levels of PAR and decreased Sir2 compared with age-matched WT controls (Figure 5F). Since the neurodegeneration in Xpa−/− mice is very mild, we further investigated the correlation between PARylation and neurodegeneration in a Xpa−/−/Csa−/− (CX) mice that display early onset cerebellar ataxia (Brace et al., 2013) similar to the Xpa−/−/Csb−/−mice with increased apoptosis of cerebellar external granular layer neurons (Murai et al 2001). Here, a dramatic increase in PAR levels was observed (Figure 5G-H). The age associated increase in PAR levels in XPA deficiency thus appears to be conserved from worms to mammals and importantly was also found in CSB and ATM cells but not in XPC knockdown cells (Figure S5C-D). To test whether PARP activation might be driving the mitochondrial phenotype we utilized PARP inhibitors on the XPA− cells. Both XPA− and control cells were pre-treated for 12 h with two PARP inhibitors, 3-aminobenzamide (3AB) and 3,4-Dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone (DPQ), and then exposed to rotenone. 3AB and DPQ significantly inhibited PARylation in XPA− cells tested by immunoblot and confocal microscopy (Figure 5I and S5E). Accompanying the downregulation of PAR was an upregulation of SIRT1 and PGC-1α, and a stimulation of rotenone-induced mitophagy in XPA− cells shown by an increased LC3-II signal (lane 2 vs. lanes 4 and 6 in Figure 5I). These data support the notion that the DNA repair deficiency in XPA leads to PARP1 activation and an attenuated NAD+-SIRT1-PGC-1α axis resulting in defective mitophagy.
Figure 5. The NAD+-SIRT1-PGC1-α axis regulates the expression of UCP2 which can rescue the mitophagy defect in XPA-cells.
(A) XPA− and XPA+ cells were transfected with pUCP2 or an empty vector for two days, and then treated with 1 μM rotenone or vehicle for 24 h, followed by immunoblot for proteins as indicated. (B) XPA− and XPA+ cells were transfected with pUCP2, vector, control siRNA or siRNA targeting UCP2 for two days, subsequently indicated parameters were analysed by flow cytometry (means ± S.E.M., n=3). (C) Immunoblot of XPA− and XPA+ cells after transfection with control siRNA or siRNA targeting SIRT1 (siSIRT1). (D) Representative immunoblot of PAR and PARP-1 in XPA− and XPA+ cells. (E) Immunoblot showing expression of PAR and SIRT1 in primary rat neurons with shRNA XPA knockdown or control shRNA. (F) Immunoblot of whole cell extracts from young (day 1) and old (day 17) xpa-1 mutant and WT (N2) nematodes. (G) Immunoblot of cerebellar protein levels in 2-week old WT, Csa−/−, Xpa−/− or Csa−/−/Xpa−/− (CX) mice and quantification in (H) (means ± S.D., n=3). (I) Immunoblot of XPA− and XPA+ cells treated with two PARP inhibitors, 3AB (1 mM) and DPQ (10 μM), for 12 h, followed an additional 24 h treatment in the presence of 1 μM rotenone or vehicle. See also Figure S5.
PARP inhibition and NAD+ precursor treatment rescues the mitochondrial phenotype in vivo
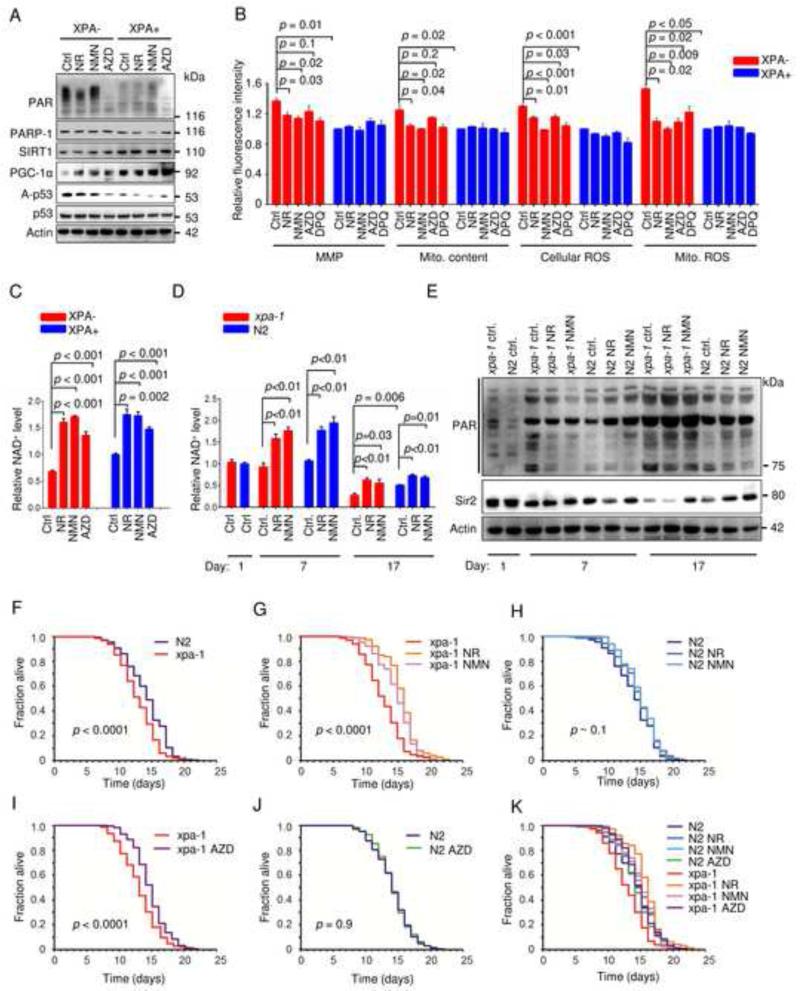
Recent work has suggested that supplementation with the NAD+ precursors nicotinamide riboside (NR) and nicotinamide mono-nucleotide (NMN) may attenuate the effect of the age-associated activation of PARP (Canto et al., 2012; Mouchiroud et al., 2013). We therefore treated the cells with NR and NMN as well as the PARP inhibitor AZD2281 (olaparib). This treatment appeared to increase PGC-1α levels and led to SIRT1 activation as shown by decreased acetylation of p53 (Figure 6A). These treatments also appeared to attenuate the mitochondrial phenotype as evident by a decrease in MMP, mitochondrial content, cellular and mitochondrial ROS production (Figure 6B). Indeed, NAD+ levels increased substantially in both XPA deficient and WT cells (Figure 6C). To further address the effect of these compounds on the XPA phenotype we did lifespan studies on WT (N2) and xpa-1 worms. There was no difference in the NAD+ levels in young adult worms when comparing WT and xpa-1 extracts (Figure 6D). After 7 days in culture the NR and NMN treatment significantly increased NAD+ in both genotypes and this corresponded with a decrease in PARylation levels (Figure 6D-E). Notably, very old worms (day 17) showed significantly decreased levels of NAD+, particularly in xpa-1, and this decrease was rescued to some extent by NR and NMN treatment. These findings corresponded with increased PAR levels and decreased Sir2 expression (Figure 7E). Since increased PAR and decreased NAD+ levels are associated with decreased longevity we next explored the lifespan of xpa-1 worms. As previously described (Arczewska et al., 2013), xpa-1 nematodes had a shorter mean lifespan than WT N2 worms (12.7 days vs 14.1 days, Figure 6F,K). Treatment with the PARP inhibitor AZD2281, NR and NMN extended the mean lifespan of xpa-1 (mean life-span: xpa-1 AZD2281 = 14.6 days, xpa-1 NR = 15.7 days and xpa-1 NMN= 14.9 days, Figure 6G-I), and increased the lifespan of WT worms although this latter increase did not reach significance (mean life-span: N2 AZD2281=14.2 days, N2 NR=14.6 days, N2 NMN = 14.9 days, Figure 6H-K).
Figure 6. NAD+ augmentation rescues the mitochondrial and aging phenotype in XPA− cells and xpa-1 worms.
(A) Immunoblot of XPA− and XPA+ cells treated for 24 h with the NAD+ precursors nicotinamide riboside (NR), nicotinamide mononucleotide (NMN) or the PARP inhibitor AZD2281. (B) Mitochondrial parameters of XPA− and XPA+ cells treated for 24 hours with NR, NMN, AZD2281 or DPQ (means ± S.E.M., n=3). (C) NAD+ levels of XPA− and XPA+ cells treated for 24 hours with NR, NMN or AZD2281 (means ± S.E.M., n=3). (D) NAD+ levels of WT (N2) or xpa-1 mutant worms treated with NR or NMN throughout their lifespan (means ± S.E.M., n=3). (E) Immunoblot of WT (N2) or xpa-1 mutants treated with NR or NMN throughout their lifespan. (F-K) Lifespan curves of WT (N2) or xpa-1 mutants treated with NR, NMN or AZD2281 throughout their lifespan (Kaplan Meyer survival curves were calculated from populations of 199 to 491 animals in each group, significance was calculated by the log-rank test).
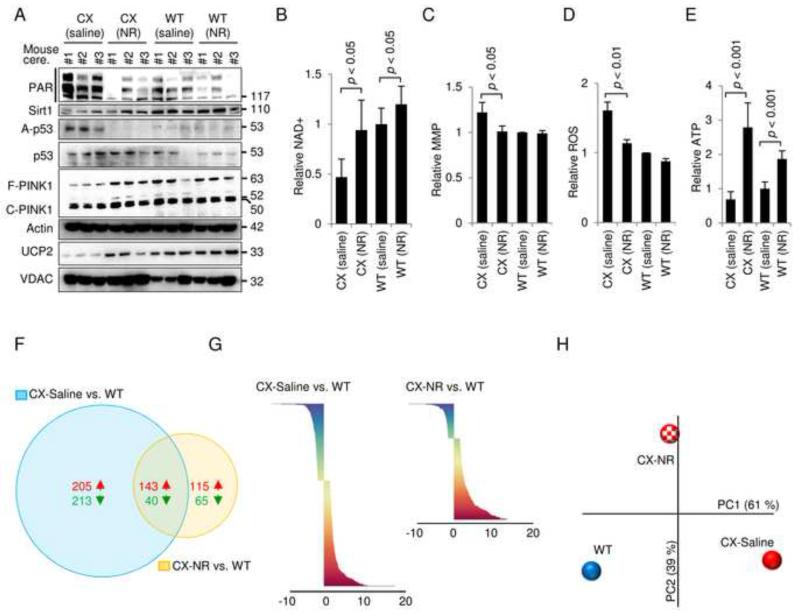
Figure 7. The NAD+ precursor nicotinamide riboside can rescue the mitochondrial phenotype of CX mice in vivo.
(A) Immunoblot of cerebellar proteins in 3-month old WT and CX mice treated with saline or nicotinamide riboside (NR) subcutaneous injections for 2 weeks (500 mg NR/kg body weight). Each lane is a separate mouse. (B) NAD+ levels in the cerebellum of the mice described in (A) (means ± S.D., n=4). (C) The mitochondrial membrane potential of isolated cerebellar mitochondria from mice described in (A) (means ± S.D., n=4). (D) ROS production in isolated cerebellar mitochondria from mice described in (A) (means ± S.D., n=4). (E) ATP levels in the cerebellum of mice as described in (A) (means ± S.D., n=4). (F) Venn diagram of significantly changed gene in the cerebellum when comparing CX-saline with WT-saline and CX-NR treated with WT-saline treated (n=3). (G) An overview of the significantly changed genes when comparing CX-saline vs WT-saline and CX-NR vs WT-saline (n=3). (H) Principal component analysis of the average Z-scores of all the genes in each group (n=3).
Recent work has shown that short term treatment with NAD+ precursors rescues the aging phenotype in mouse skeletal muscle (Gomes et al., 2013). A phenotype of the CX mice is death at weaning if the mice are fed a hard chow (Brace et al., 2013). We therefore tested if NR supplementation in the drinking water of the mothers could rescue the death at weaning of the CX pups. This did not appear to be the case (data not shown) indicating that either the death at weaning phenotype was unaffected by NR, or that the method of NR supplementation in the water was inadequate. Instead we investigated the cerebellum, a target organ in XPA, in 3 months old CX mice that were fed a soft palatable chow which increases survival at weaning (Brace et al., 2013). We treated the CX mice with 2 weeks of daily subcutaneous NR injections (500 mg NR/kg body weight). Strikingly this short term treatment decreased PAR levels, increased SIRT1 activation (decreased p53 acetylation), led to retention of full length PINK1 and increased the expression of UCP2 in the cerebellum from CX double knockout mice (Figure 7A). Further NAD+ levels, MMP and mitochondrial ROS production were normalized by this treatment (Figure 7B-D). ATP levels increased significantly in both genotypes after NR treatment (Figure 7E). Since alterations in NAD+ levels appeared to affect the activity of central transcription factors such as SIRT1 we hypothesized that NR treatment would rescue the transcriptional differences between WT and CX mice. Indeed, gene microarray data showed that the cerebellar transcriptome of the CX mice was partially normalized by the NR treatment. For CX-saline compared with WT-saline there were 601 significantly changed genes while CX-NR compared with WT-saline only showed 363 significantly changes genes (Figure 7F-G). Principal component analysis of the entire unselected gene set revealed great separation of the CX-saline and WT group on principal component 1 (PC1, x-axis, Figure 7H). There was no difference in CX-saline and WT groups on PC2 (y-axis) indicating that PC1 explained the main differences in the transcriptome between the two genotypes. A potential normalizing effect of NR treatment on the transcriptome should therefore be an effect on PC1. Indeed, NR treatment appeared to drive the entire transcriptome of CX mice towards WT (Figure 7H). PC2 appeared to be an effect of NR that led to changes independent of the genotype since there was no difference in this parameter between WT-saline and CX-saline. Taken together these data suggests that PARP activation drives an accelerated aging phenotype in XPA and that pharmacological intervention with PARP inhibitors or compounds that increase NAD+ can partially normalize this phenotype.
DISCUSSION
In this study we present evidence for a mitochondrial involvement in the XPA phenotype through in silico modeling, microarray analysis and a number of functional metabolic assays on human cells, rat neurons, mice and worms. Importantly, the mitochondrial phenotype in XPA appears similar to what is observed by us and others in AT and CS. This phenotype may be caused by defective mitophagy due to excessive cleavage of PINK1 by hyperpolarized mitochondria. These effects may dependent upon depression of the NAD+-SIRT1-PGC-1α-UCP2 axis that is triggered by over-activation of the DNA damage sensor PARP1. Our findings highlight the importance of mitophagy in neuronal health and provide insight into the neurodegenerative phenotype in XPA, ATM and CS patients which is not explained merely by their classic DNA repair functions.
Is XPA a mitochondrial disease?
XPA deficiency appears to lead to mitochondrial dysfunction across species and in a variety of cell types. This indicates that a DNA repair defect may yield a similar mitochondrial phenotype in different tissues. In the context of mitochondrial diseases, defects are often observed in fibroblast cultures from patients although the clinical phenotype is most commonly observed in brain or muscle tissue. The general consensus regarding this phenomenon is that brain and muscle are particularly prone to changes in ATP production and defective mitochondria would therefore predominantly manifest as neurodegeneration or myopathies even though the mitochondrial defect may be present in all tissues. Similarly, the DNA repair deficiency in XPA, CS and ATM is presumably present in all tissues. However, in these disorders neurodegeneration could be caused by an increased ATP consumption leading to an inability to meet the energetic demand of neurons.
NER, in which XPA is a central player, does not take place in mitochondria and the XPA protein itself was not found in this compartment. Thus, the mitochondrial dysfunction we observe appears to be secondary to the nuclear function of XPA. This may be in contrast to what has been reported for AT and CS where ATM and CSB were present in mitochondria (Aamann et al., 2010; Valentin-Vega et al., 2012). Interestingly, the neurodegeneration seen in XPA is often less severe than what is seen in AT and CS. If ATM and CSA/B act within the mitochondria, a combined nuclear and mitochondrial defect could lead to a more severe mitochondrial phenotype than what is seen in XPA. However, a possible role of these proteins in the mitochondria may be separate from their effect on mitophagy since we observe similar mitophagic defects in XPA, CSB and ATM deficient cell lines.
Roles of XPA outside nucleotide excision DNA repair
Though the XPA protein is primarily involved in NER, evidence suggests that it has additional roles in the repair of oxidative DNA lesions. For example, increased ROS levels have been found in xpa-1 nematodes and Xpa−/− mice, and oxidative nucleotide damage has been shown in the brains of XPA patients (Arczewska et al., 2013; Hayashi et al., 2005; Maria Berra et al., 2013). Similarly, increased ROS production also exists in CS and AT patient cells (Scheibye-Knudsen et al., 2012; Valentin-Vega et al., 2012). Accordingly, it seems that defects in different pathways of DNA repair may induce the same oxidative stress phenomenon. In light of our current findings increased ROS induced DNA damage could be a secondary effect of an increased MMP and decreased UCP2 in these disorders.
XPA has also been implicated in several other cellular processes. XPA deficient cells have decreased levels of retinoic acid and treatment of XPA deficient cells with retinoic acid rescued the UV-sensitivity of these cells through an unknown mechanism (Ding et al., 2001). Interestingly, it has been shown that retinoic acid can alter the mitochondrial function through increased expression of UCP1 and UCP2 (Rial et al., 1999). In light of our findings increasing UCPs may be particularly important and could potentially explain the beneficial effect of retinoic acid in XPA. In addition, XPA has been proposed to be a transcription factor and is recruited to promoters of genes even in the absence of exogenous damage (Le May et al., 2010). Since several studies have linked CSB to transcription it is possible that the neurological alterations in CS and XPA may be exacerbated by a transcriptional deficiency. It is, however, still not clear what molecular function XPA, CSB and ATM would have in transcription and more research is warranted.
Defective mitophagy due to perturbation of the NAD+-SIRT1-PGC-1α axis
Excessive ROS and increased ATP consumption may trigger higher basal autophagy in DNA repair deficient cells through AMPK driven inhibition of mTOR independent of a defect in mitophagy. AMPK activation could be an adaptive mechanism relative to PARP activation and PAR recycling that would increase ATP consumption. In this regard SIRT1 and UCP2 depression could be a response to meet the increased ATP demand after DNA damage since increasing MMP will lead to greater mitochondrial ATP output. Under normal circumstances DNA repair removes the damage, shuts down PARP and allows MMP to return to normal. In XPA, CS and AT persistent DNA damage may lead to continuous activation of PARP, increased MMP and consequently defective mitophagy. Simultaneously the increased ROS production and AMPK activation appear to drive mTOR inhibition and increase basal autophagy. Importantly, XPC deficient cells do not appear to have PARP activation perhaps relating to why these patients rarely develop neurodegeneration.
We find that SIRT1 may have a central role in mitophagy through regulation of UCP2 via PGC-1α. PGC-1α is particularly interesting because loss of this central transcription factor leads to neurodegeneration (Lagouge et al., 2006; St-Pierre et al., 2006). In support of a role in neuronal health UCP2 suppresses mitochondrial ROS production and leads to neuroprotection in a Parkinson’s disease model (Andrews et al., 2005). This is pertinent since evidence suggests that Parkinson’s disease may be caused by defective mitophagy. Accordingly the loss of UCP2, as we observe, leads to mitochondrial hyperpolarization and increased import, cleavage and removal of the central mitophagic kinase PINK1. A recent study in nematodes showed increased PARP activation and lower NAD+ levels in aged WT nematodes, as well as Sir2 dependent life span extension by PARP inhibition, thus supporting the link between PARP1and SIRT1 with aging (Mouchiroud et al., 2013). In agreement with this, we also detected higher PARylation in old than young nematodes. More importantly, we found that the lifespan defect in xpa-1 worms could be rescued by treatments with both the NAD+ precursors NR and NMN and through PARP inhibition using AZD2281 (olaparib). Quite strikingly, only two weeks of treatment with NR rescued the mitochondrial phenotype in the cerebellum of the CX mice as well as attenuated the transcriptional changes in this organ. This study thereby adds to the growing evidence of a highly coordinated nuclear-mitochondrial cross talk in the regulation of life− and health-span.
Conclusion
In closing we present herein the first evidence of a mitochondrial dysfunction in XPA based on in silico algorithms, transcriptomics analysis and cellular biology. In addition we describe a novel unifying mechanism leading from the causal endogenous DNA repair defect to mitochondrial and mitophagic dysfunction in XPA, CS and AT. Further we present a new pharmacological avenue for therapeutic intervention in XPA using NAD+ precursors.
EXPERIMENTAL PROCEDURES
Database
Prediction of mitochondrial pathology of XPA was generated using the on-line database www.mitodb.com (Scheibye-Knudsen et al., 2013).
Detection of Autophagy and Mitophagy
Monitoring autophagy/mitophagy was carried out by detecting LC3-II/actin ratio, protein levels of different autophagy-associated proteins, and numbers of autophagic elements (Klionsky et al., 2012). Autophagic flux was measuring using two autophagy inhibitors bafilomycin A1 (100 nM) and chloroquine (10 μM). Autophagic elements were detected by transient transfection of cells with ptfLC3 plasmid (Addgene, ID 21074, deposited by Tamotsu Yoshimori) and images by confocal microscopy (Eclipse TE-2000e, Nikon). Mitophagy was induced with three mitochondrial toxins: rotenone, Antimycin A1, and FCCP. After treatment with indicated doses for 24 h, cells were harvested and prepared for western blotting.
Mitochondrial Parameters
Mitochondrial membrane potential (MMP), mitochondrial content and ROS at both cellular and mitochondrial levels were measured by a BD Accuri™ C6 flow cytometer as previously described (Scheibye-Knudsen et al., 2012).
ATP and NAD+/NADH Quantitation
ATP concentration was detected using an ATPlite™ Luminescence Assay System (PerkinElmer) as per manufacturer’s instruction. Measurement of NAD+/NADH was performed by using a commercial NAD/NADH assay kit (#ab65348) by following the provided protocol.
Worm studies
Lifespan analysis was performed at 20 °C for Bristol N2 and xpa-1 (opk698) mutants grown on classical NGM plates, or NGM plates supplemented with 500 μM NR, 500 μM NMN, or 100 nM Olaparib (AZD2281) seeded with E. coli OP50 as food source (Fensgard et al., 2010). We calculated mean, standard deviation of the mean, and P value using the log-rank test, from pooled population of animals.
Nicotinamide riboside supplementation on CX (Csa−/−/Xpa−/−) mice
CX and WT (C57BL/6) mice of 3 months of age were given subcutaneous interscapular injections of 500 mg NR/kg body weight/day or the equivalent volume of saline for a consecutive of 14 days at 4:00 pm (Gomes et al., 2013). On day 15, mice were sacrificed and half of a cerebellum was harvested for purification of mitochondria, with the left half snap-frozen, homogenized, and aliquoted for western blotting, microarray, and detection of ATP and NAD+. Freshly purified mitochondria were used for detection of MMP and mito ROS with FACS.
See supplementary information for detailed methods.
Supplementary Material
Highlights.
Xeroderma pigmentosum group A (XPA) phenocopies mitochondrial diseases in silico.
XPA deficiency leads to mitochondrial and mitophagic dysfunction across species.
Dysfunctional mitophagy is caused by PARP1 activation and attenuation of SIRT1.
This mechanism is also seen in ataxia-telangiectasia and Cockayne syndrome.
Acknowledgements
We thank Drs. Kevin Becker, Yongqing Zhang and Elin Lehrmann for their help in performing microarray and data analysis, and Dr. Magdalena Misiak for preparing rat neurons. We thank Drs. Mark Mattson and Peter Sykora for reading this manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute on Aging. H.N. was supported by a grant from the research council of Norway.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, 3rd, Stevnsner T, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews ZB, Horvath B, Barnstable CJ, Elsworth J, Yang L, Beal MF, Roth RH, Matthews RT, Horvath TL. Uncoupling protein-2 is critical for nigral dopamine cell survival in a mouse model of Parkinson’s disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:184–191. doi: 10.1523/JNEUROSCI.4269-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arczewska KD, Tomazella GG, Lindvall JM, Kassahun H, Maglioni S, Torgovnick A, Henriksson J, Matilainen O, Marquis BJ, Nelson BC, et al. Active transcriptomic and proteomic reprogramming in the C. elegans nucleotide excision repair mutant xpa-1. Nucleic acids research. 2013;41:5368–5381. doi: 10.1093/nar/gkt225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell metabolism. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace LE, Vose SC, Vargas DF, Zhao S, Wang XP, Mitchell JR. Lifespan extension by dietary intervention in a mouse model of Cockayne Syndrome uncouples early postnatal development from segmental progeria. Aging cell. 2013;12:1144–1147. doi: 10.1111/acel.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell metabolism. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. The Journal of investigative dermatology. 2012;132:785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Ichikawa M, Furukawa A, Tomita S, Tanaka K, Ichikawa Y. Low synthesis of retinoic acid due to impaired cytochrome P450 1a1 expression in mouse xeroderma pigmentosum fibroblasts. The international journal of biochemistry & cell biology. 2001;33:603–612. doi: 10.1016/s1357-2725(01)00037-1. [DOI] [PubMed] [Google Scholar]
- Fensgard O, Kassahun H, Bombik I, Rognes T, Lindvall JM, Nilsen H. A two-tiered compensatory response to loss of DNA repair modulates aging and stress response pathways. Aging. 2010;2:133–159. doi: 10.18632/aging.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) Induces a Pseudohypoxic State Disrupting Nuclear-Mitochondrial Communication during Aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Araki S, Kohyama J, Shioda K, Fukatsu R. Oxidative nucleotide damage and superoxide dismutase expression in the brains of xeroderma pigmentosum group A and Cockayne syndrome. Brain & development. 2005;27:34–38. doi: 10.1016/j.braindev.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. The New England journal of medicine. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Islam R, Yang L, Sah M, Kannan K, Anamani D, Vijayan C, Kwok J, Cantino ME, Beal MF, Fridell YW. A neuroprotective role of the human uncoupling protein 2 (hUCP2) in a Drosophila Parkinson’s disease model. Neurobiology of disease. 2012;46:137–146. doi: 10.1016/j.nbd.2011.12.055. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Molecular cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Maria Berra C, de Oliveira CS, Machado Garcia CC, Reily Rocha CR, Koch Lerner L, de Andrade Lima LC, da Silva Baptista M, Martins Menck CF. Nucleotide excision repair activity on DNA damage induced by photoactivated methylene blue. Free radical biology & medicine. 2013;61C:343–356. doi: 10.1016/j.freeradbiomed.2013.03.026. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo Y, Viswanathan M, Schoonjans K, et al. The NAD+/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- Rial E, Gonzalez-Barroso M, Fleury C, Iturrizaga S, Sanchis D, Jimenez-Jimenez J, Ricquier D, Goubern M, Bouillaud F. Retinoids activate proton transport by the uncoupling proteins UCP1 and UCP2. The EMBO journal. 1999;18:5827–5833. doi: 10.1093/emboj/18.21.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, 3rd, Cooper M, Spencer R, de Cabo R, et al. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. The Journal of experimental medicine. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Scheibye-Alsing K, Canugovi C, Croteau DL, Bohr VA. A novel diagnostic tool reveals mitochondrial pathology in human diseases and aging. Aging. 2013;5:192–208. doi: 10.18632/aging.100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Maclean KH, Tait-Mulder J, Milasta S, Steeves M, Dorsey FC, Cleveland JL, Green DR, Kastan MB. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.