Abstract
Recent studies suggest that actin filaments are essential in how a cell controls its nuclear shape. However, little is known about the relative importance of membrane tension in determining nuclear morphology. In this study, we used adhesive micropatterned substrates to alter the cellular geometry (aspect ratio, size, and shape) that allowed direct membrane tension or without membrane lateral contact with the nucleus and investigate nuclear shape remodeling and orientation on a series of rectangular shapes. Here we showed that at low cell aspect ratios the orientation of the nucleus was regulated by actin filaments while cells with high aspect ratios can maintain nuclear shape and orientation even when actin polymerization was blocked. A model adenocarcinoma cell showed similar behavior in the regulation of nuclear shape in response to changes in cell shape but actin filaments were essential in maintaining cell shape. Our results highlight the two distinct mechanisms to regulate nuclear shape through cell shape control and the difference between fibroblasts and a model cancerous cell in cell adhesion and cell shape control.
Keywords: Micropatterned materials, Nuclear morphology, Actin filaments, Cell shape
Introduction
Normal physiological aging [1, 2] and pathological situations [3, 4] alter the size and shape of the nucleus. The geometry of the nucleus is known to have a significant influence on cell proliferation [5], gene expression [6], and protein synthesis [7] but the mechanisms through which nuclear morphology alters cell function remains unclear. The nucleus is enclosed by the nuclear envelope that isolates the chromosomes from the cytoplasm and typically has either an oval or round shape. The nuclear envelope is a double membrane with the outer membrane connected externally to the endoplasmic reticulum (ER), and to the inner nuclear membrane at each nuclear pore complex. Under the inner membrane lies a meshwork of nuclear lamina proteins composed of intermediate filament lamins that provide mechanical support and anchor the nuclear pore complexes [8, 9]. The nuclear size is not directly related to the DNA content as the nuclei of cells in different tissue differ in size, but have identical DNA content. The nuclear size is influenced by transport between the nucleus and cytoplasm through nuclear pores [10], other cellular structures such as ER [11], lamin [12, 13], nuclear envelop associated proteins [14], and lipid biosynthesis [15, 16]. In different species, the nuclear size is found to be proportional to the overall size of the cell [17, 18]. Previous studies of nuclear morphology have focused on the structural role of the nuclear lamina and nuclear envelope [2, 19]. It is known for example, that alterations in lamina proteins in mutated cells [20, 21] and cancer cells [22] are associated with abnormally shaped nuclei. However, the role of physical factors transduced through the cell membrane and cytoplasm in altering the shape of the nucleus have been the subject of only more recent investigations [6].
Recent studies have shown that perinuclear actin networks, consisting of actin filament bundles called transmembrane actin-associated nuclear lines [23] or actin cap [24], together with their regulatory proteins [25] exert significant control on nuclear shape. Molecules referred to as linkers of nucleus to cytoskeleton (LINC) anchor actin cap fibers directly to the nucleus [26–29]. Micromanipulation techniques have demonstrated further that compressive forces, driven by cytoskeletal tension in endothelial cells on elongated adhesive islands, alter the morphology of the nucleus [5] and offer insights to the spatial coordination between cell and nuclear shape. However, questions remain on the role of membrane tension. For example, in previous investigation confining cells to rectangular shapes, the membranes are locally physical compressing the nucleus as well as the global compressive forces driven by actomyosin tension, it is unknown the individual effect of each.
Changes in the nuclear size and shape have been associated with cancer and enlarged nuclei typically represent more metastatic [22, 30–32]. However, it remains unknown if the coordination between cell and nuclear shape is different in cancer cells. In this study, we investigate nuclear shape remodeling on a series of rectangular shapes that allow direct membrane tension or without membrane lateral contact with the nucleus. The roles of cytoskeleton and tension in these patterned cells on nuclear shapes were investigated by various pharmacological agents while maintaining the cell shape independently by culturing cells on single adhesive islands of different rectangular shapes. Finally, the nuclear shape remodeling was investigated in a model adenocarcinoma cell to investigate the regulation of nuclear shape in response to changes in cell shape in cancer cells.
Materials and Methods
Materials
Tissue culture dishes were purchased from Fisher Scientific (catalog no. 430166) and used as received. Polydimethylsiloxane (PDMS; Sylgard 184) was obtained from Dow Corning (Midland, MI) and used at 10:1 (w: w) base: curing agent. Cytochalasin D and nocodazole were obtained from Sigma Co. Y27632 was purchased from Calbiochem. Alexa 488 or 594-phalloidin, 4′, 6-diamidino-2-phenylindole (DAPI), and Alexa Fluor® 488 goat anti-mouse IgG1 (γ1) (secondary antibody) were purchased from Molecular Probes (Eugene, OR). Phosphate buffered saline (PBS), Iscove's modified Dulbecco's medium (IMDM) or Dulbecco's Modified Eagle Medium (DMEM), and Fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA).
Preparation of poly (OEGMA-co-MA)
Random copolymers of Oligo(ethylene glycol) methacrylate (OEGMA) and methacrylic acid (MA) (Scientific Polymer Products, NY) were prepared by free radical polymerization of 10 wt % methanolic solutions of the two monomers (80:20 OEGMA to MA mass ratio) at 60 °C using an electric-heated thermostatic water bath. Polymerizations were initiated with 1 wt % (with respect to monomer) 2,2′-azobis(2-amidinopropane) dihydrochloride (Wako, VA) and allowed to react for 24 hours.
Preparation of patterned tissue culture dishes
Micropatterns consisting of two different areas (900 and 2500 µm2) of various geometric forms (square, rectangle and parallelogram) were fabricated on silicon wafers using standard photolithographic techniques. From this silicon master, complementary PDMS replicas were prepared following the soft lithography procedure developed by Whitesides and co-workers [33] and used as stamps in subsequent microcontact printing steps to form background of poly (OEGMA-co-MA) copolymer directly on cell culture dishes. Patterned dishes were sterilized under UV for at least 1 hour before plating cells.
Cell culture
NIH 3T3 fibroblasts and A549 cells (Human lung carcinoma-derived alveolar epithelial cell line) were purchased from ATCC and cultured in IMDM and DMEM respectively, supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml) at 37 °C in a humidified atmosphere containing 5% CO2. Sub-confluent monolayers were dissociated with 0.25% trypsin-EDTA solution, re-suspended in IMDM or DMEM with 10% FBS, and then plated on micropatterned culture dishes.
Treatment with Pharmacological agents
Cells plated on the patterned culture dish were allowed to attach and spread to the adhesive island for 15–18 hours prior to incubation with various pharmacological agents. Cells were incubated with 1.0 µM cytochalasin D for 35 minutes, 20 µM Y27632 for 30 minutes and 5.0 µM nocodazole for 1 hour at 37 °C prior to being fixed and stained.
Immunofluorescence staining
Cells were plated at a density of approximately 8,000-cells/cm2 and allowed to reach confluence within the cell-adhesive patterns on the tissue culture dishes. After drug treatment for some cases, cells were fixed with 3.7% paraformaldehyde for 15 minutes, washed in phosphate buffered saline, permeabilized with 0.2% Triton X-100 for 5 minutes and then blocked with 3% BSA (bovine serum albumin) for 30 minutes. Samples were then rinsed with PBS and incubated with Alexa 594-phalloidin and DAPI to stain for F-actin and nuclei respectively. Phase-contrast and fluorescence micrographs were collected using a CCD camera (SPOT CAM, Diagnostic Instruments Inc.) mounted on a Nikon TE-2000 inverted microscope with 40× objective lens. Confocal images were obtained from Zeiss LSM 710 NLO Confocal Microscope under 63× or 40× plan apochromat water objective lens. Images were processed with Metamorph software (Ver 6.0r4, Universal Imaging, Westchester, PA) or Zeiss Meta software version 3.5. Multiple Z-section images (0.2 µm thick sections) were analyzed and processed using a combination of Zeiss LSM Image Browser, Metamporph, and ImageJ (http://rsbweb.nih.gov/ij/). The nuclear height on the micropatterns for different aspect ratios were measured by staining the nuclei with DAPI and imaged confocally with a z-resolution of 0.15 um. Only a single cell confined within each pattern was considered for analysis.
Assessment of cell and nuclear morphology
Cells were fixed and stained with DAPI and phalloidin to show nucleus and F-actin. Cells with spreading area within ±15% of the adhesive island area were included in the quantification of cell and nuclear morphology. Projected cell and nuclei length and width was calculated by interactive tracing the maximum intensity projection image of a cell [F-actin or nuclei (DAPI)], thresholding, and applying the Integrated Morphometry Analysis (IMA) routine. The length was calculated by IMA as the span of the longest chord through the object; the width is the dimension perpendicular to the length. Quantification of cell nuclear elongation and determination of orientation were performed on an image analysis routine in Metamorph or by using NIH ImageJ software.
Statistical analysis
All the data are shown as the mean ± the standard deviation (SD). Pairwise comparisons between means of two different groups were performed using a Student t-test. The differences between two groups of data are considered statistically significant or statistically highly significant if the Student t-test gives a significance level P (P value) smaller than 0.05 or 0.001, respectively. Multiple comparisons were performed using analysis of variance (ANOVA), followed by Tukey's post test to determine differences.
Results and Discussion
Changes in cell morphology influence nuclear shape
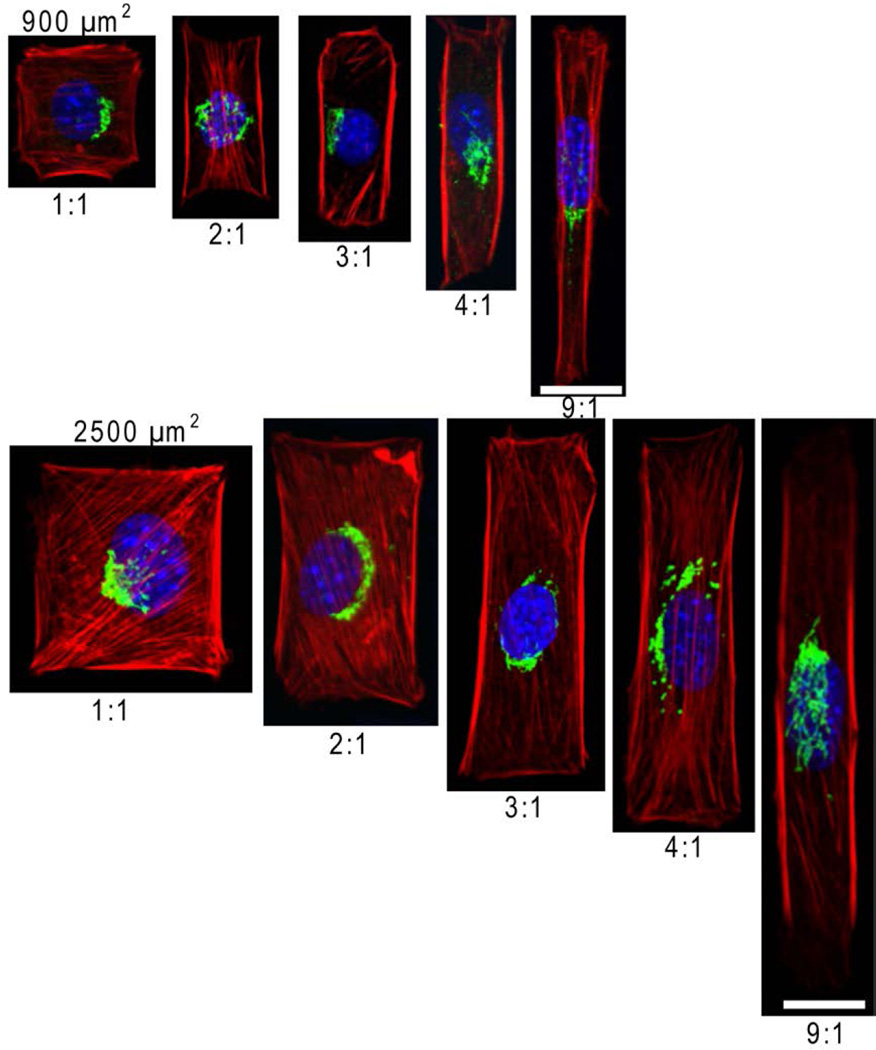
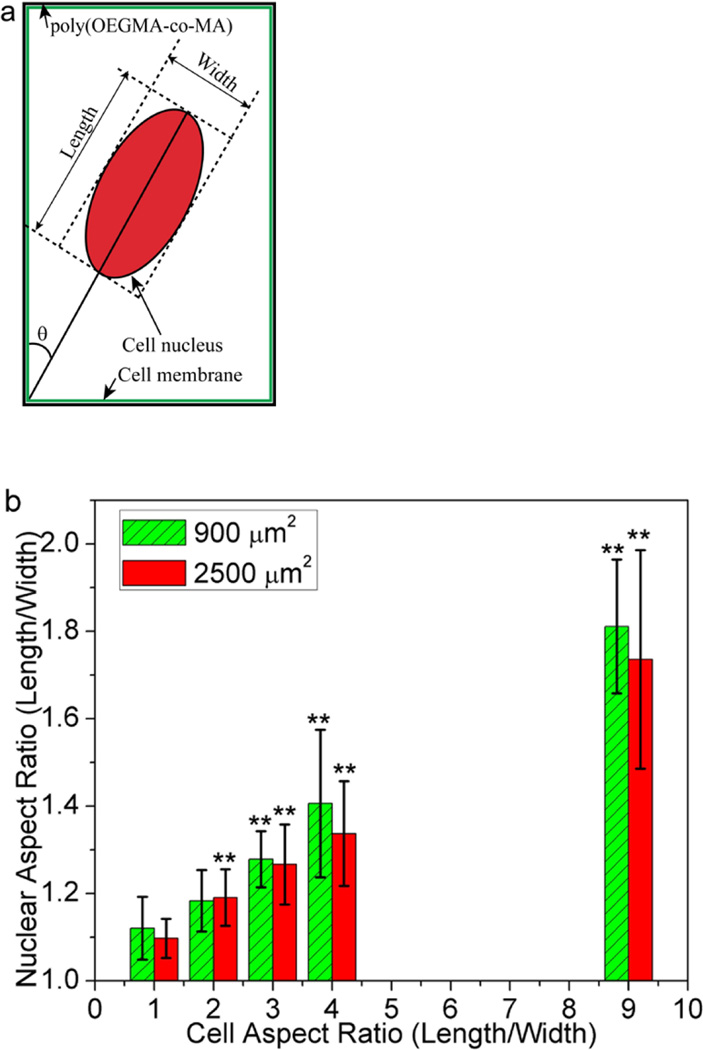
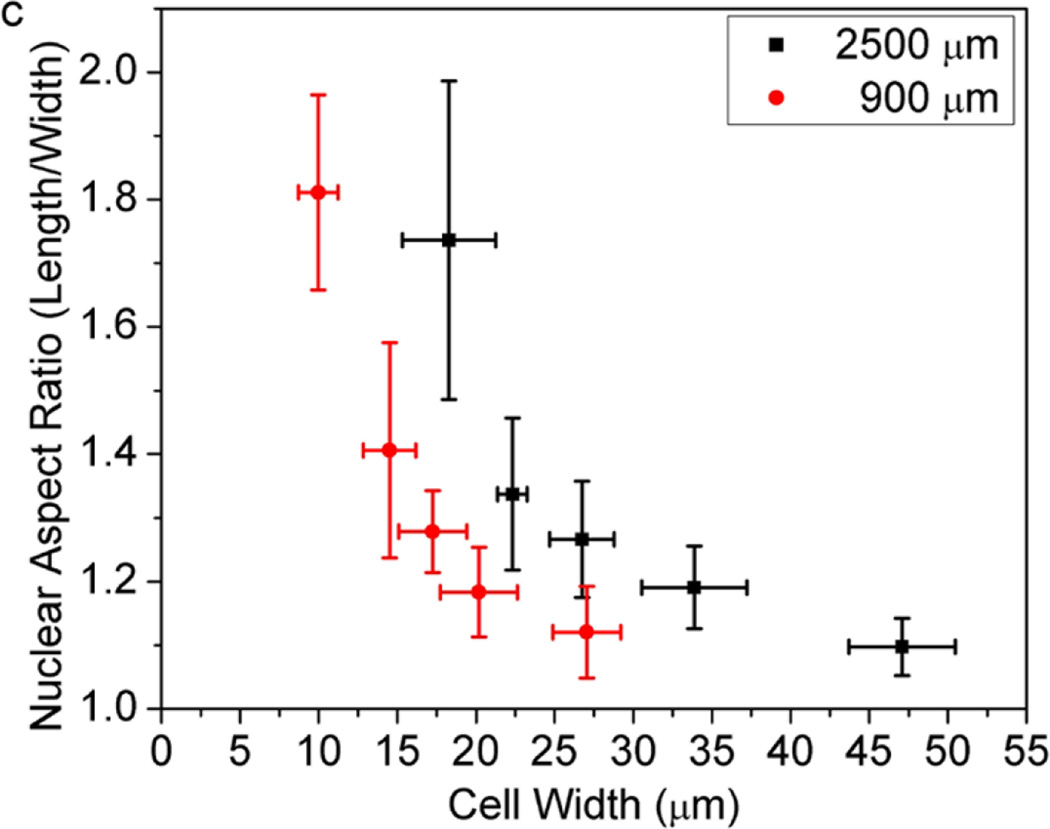
We cultured cells on adhesive islands of equal areas (900 µm2 and 2500 µm2) with different aspect ratios to examine if and how nuclear shapes adapt in relation to constraints on cell shape. The rectangular islands on which cells attach are areas of bare tissue culture dish defined by a background of cell resistant poly(OEGMA-MA). As cells spread to adopt the elongated shapes of their confining islands, their nucleus also elongate along the longer axis of their cell body (Figure 1). Assigning the longest dimension of cell nuclei as the length, and the width as the direction perpendicular to the length (Figure 2a), we measured the aspect ratio of the nuclei for individual cells constrained within rectangular islands of varying length to width ratio. As Figure 2b shows, cells on square adhesive islands exhibit rounded nuclei with aspect ratios close to 1. Elongation of the overall cell shape up to length to width ratio of 9:1 results in further elongation of the cell nuclei that reaches a length to width ratio of approximately 1.75. The extent of nuclear elongation that accompanies elongation of the cell body is more pronounced for cells confined in the smaller 900 µm2 islands. Figure 2c, which plots the nuclear aspect ratio for fixed areas and varying cell width, shows that for a given cell width, the nucleus is consistently more elongated for the more spread 2500 µm2 cells. This implies that nuclear deformation is controlled not just by local constraints on cell shape, but also by the overall cell geometry. Earlier studies suggest that cytoskeletal tension increases with cell spreading [34]. A larger cytoskeletal tension on cells spread to larger areas would explain the greater nuclear elongation for the more spread 2500 µm2 cells. This is supported by consistent observations shown representatively in Figure 1 wherein actin stress fibers are more visible and oriented parallel to the length of more spread 2500 µm2 cells.
Figure 1.
Cell shape and spreading area affect nucleur morphology. for typical organization of actin filaments (red), Golgi (green) and DAPI-stained nuclei (blue).. NIH 3T3 cells were confined in individual adhesive island with area of 900 or 2500 µm2 with aspect ratios varying from 1 to 9. The scale bar respresents 20 µm.
Figure 2.
Correlation between nuclear shape and cell morphology. (a) Schematic characterization of the length, width and orientation angle of the cell nucleus; (b) Plot of the nuclear aspect ratio as a function of the NIH 3T3 cell aspect ratio. Data are shown as Mean ± SD with 8 ≤ n ≤ 12. ANOVA followed by Tukey's post tests yield the following p values p=0.052 (ratio 2), p=6.2×10−5 (ratio 3), p=9.0×10−5 (ratio 4), p=6.6×10−11 (ratio 9) for 900 µm2 and p=5.4×10−4 (ratio 2), p=4.2×10−7 (ratio 3), p=7.7×10−6 (ratio 4), p=5.9×10−7 (ratio 9) for 2500 µm2, respectively. Statistical significance is calculated with respect to cell aspect ratio 1, *p<0.05, **p<0.005; (c) Plot of the nuclear aspect ratio as a function of the NIH 3T3 cell width. Data are shown as Mean ± SD with 8 ≤ n ≤ 12.
Cell morphology mediates Golgi apparatus polarity
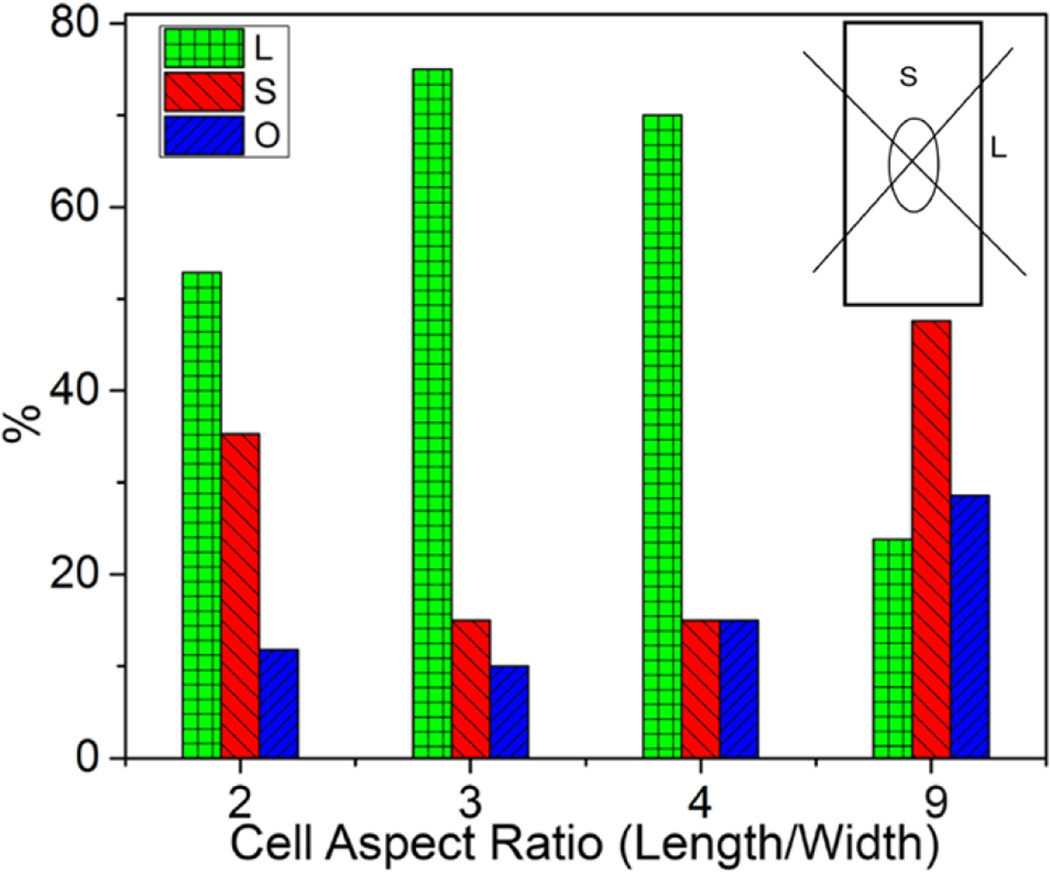
The localization of Golgi apparatus relative to the cell nucleus is widely considered to be an indicator of cell polarity with the Golgi oriented typically towards the leading edge [35]. In migrating cells, the Golgi position relative to the nucleus depends on the geometry imposed by the substrate [36] with the cellular shape and the leading edge redirecting continuously during migration. However, for immobile cells, the influence of cell shape on the orientation of Golgi complex remains unclear. We observed that, in the absence of gradients in soluble factors, Golgi complex do not exhibit random juxtanuclear position, but tend to polarize as cells adapt to elongated geometries (Figure 1). To quantity the role of cell shape on Golgi orientation, we fixed and stained cells on 900 µm2 patterns of varying aspect ratios. The area around the nucleus was divided by two intersecting perpendicular lines centered at the centroid of the nucleus. The orientation of Golgi apparatus was defined as S when the majority of Golgi orient toward either short edge, L when the majority of Golgi orient toward either long edge, and O when the majority of the Golgi overlaps with the nucleus (Figure 3). At a high aspect ratio of 9, the Golgi is located predominantly toward the short edges, but is oriented preferentially toward the long edges of the rectangular shape at lower aspect ratios of 2, 3 and 4. This result shows that the position of the Golgi depends on the cell shape even when cells are not migrating, and is consistent with earlier observations [37]. However, the increasing preferential orientation of the Golgi toward the long edge with increasing cell aspect ratio from 2 to 3 has not been observed earlier, and is unexpected since the cytoplasmic area available for this orientation decreases with decreasing width. This suggest spatial polarization of Golgi complex is not based solely on spatial constraints, which would have led to preferential orientation toward the short edge with increasing cell elongation due to increasing of the cytoplasm toward the short edge. The localization of Golgi towards the short edge only occurs at the highest aspect ratio, when there is practically no cytoplasmic space available for Golgi orientation towards the long edges.
Figure 3.
Cell shape leads to polarized organization of Golgi complex. Golgi orientation for NIH 3T3 fibroblast on 900 µm2 rectangular patterns with different aspect ratios. On the nucleus (O), short (S) and long (L) edges represent the position of Golgi apparatus relative to the nuclei.
The role of actomyosin on nuclear deformation depends on cell shape
To investigate the role of myosin-mediated contractility on nuclear shape remodeling, Y-27632 a Rho associated protein kinase (ROCK) inhibitor and regulator of myosin mediated contractility [38], was added to cells on rectangular micropatterns with varying aspect ratio. At the highest aspect ratio of 9, when the cell width is reduced to less than the width of nuclei of unconstrained cells, the nuclei remain elongated along the direction of the long cell axis suggesting that factors other than myosin are involved in maintaining nuclear elongation (Figure 4).
Figure 4.
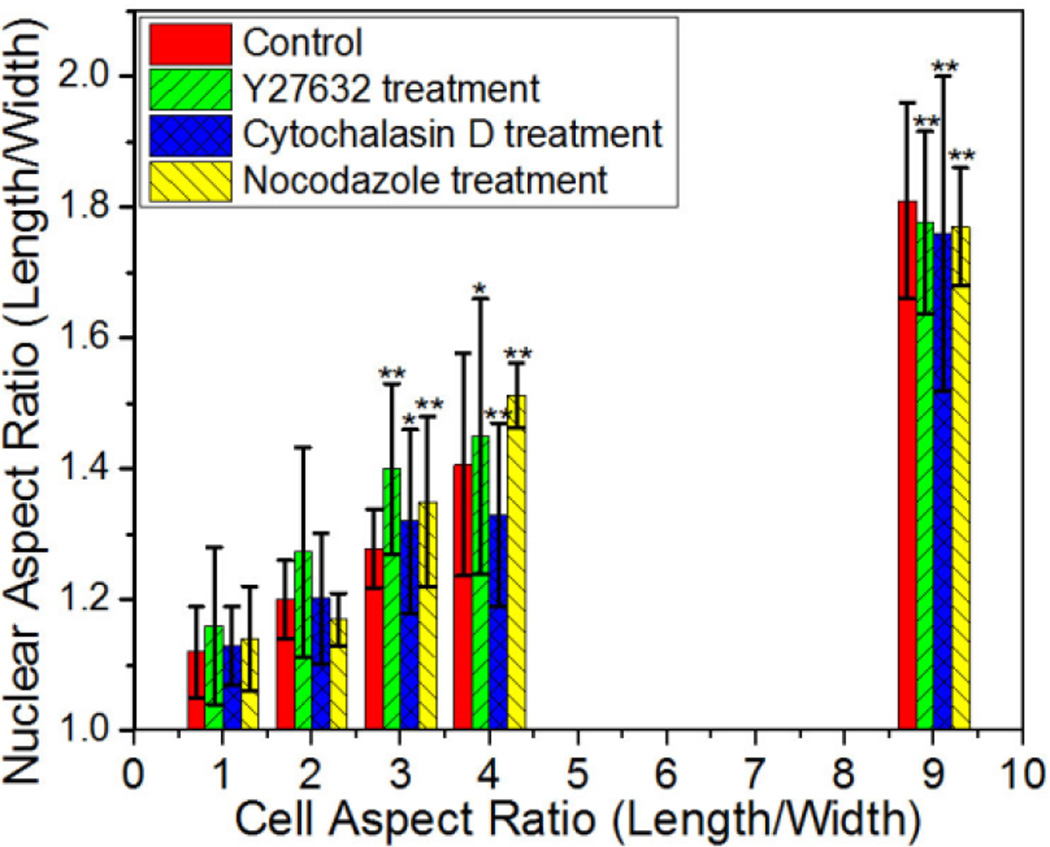
Quantification of nuclear aspect ratio for NIH 3T3 cells treated with the indicated pharmacological agents on 900 µm2 rectangular patterns. Data are shown as Mean ± SD with 8 ≤ n ≤ 11. ANOVA followed by Tukey's post tests yield the following p values: p=0.07 (ratio 2), p=3.89×10−4 (ratio 3), p= 2.56×10−4 (ratio 4), p=2.89×10−7 (ratio 9) for cytochalasin D; p=0.15 (ratio 2), p=9.64×10−4 (ratio 3), p= 0.018 (ratio 4), p=4.3×10−6 (ratio 9) for Y27632; p=0.43 (ratio 2), p=0.0038 (ratio 3), p= 1.4×10−4 (ratio 4), p=2.21×10−6 (ratio 9) for nocodazole. Statistical significance is calculated with respect to shape ratio 1 treated for cytochalasin D, Y27632, or nocodazole respectively. *p<0.05, **p<0.005.
To determine whether the microtubule and actin filaments are involved in the regulation of nuclear shape, cells confined to different rectangular shapes were treated with nocodazole to depolymerize microtubules and cytochalasin D to inhibit actin polymerization [39]. For varying cell aspect ratios, the nuclear aspect ratio of cells with depolymerized microtubules are statistically equal to cells not treated with nocodazole, suggesting that microtubules do not contribute significantly to nuclear elongation. In contrast, when actin polymerization was blocked by cytochalasin D, nuclear aspect ratios decreased for cell aspect ratio of 4. Nonetheless, the nuclei remains highly elongated at the highest cell aspect ratio of 9 when the cell width is less than the width of nuclei in unconstrained cells. This suggests that actin filaments play an important role in controlling the nuclear shape, with other factors becoming increasingly dominant when the cell width is reduced to less than the natural width of nuclei. Khatau et al. [24] have elucidated how the shape of nuclei is controlled by cell shape through the perinuclear actin cap composed of actomyosin filaments bundles that are dynamically connected to the nuclear envelope. They found that treatment with latrunculin B, a F-actin depolymerizing drug, leads to almost complete abrogation of the regulation of nuclear shape by cell shape. In their study, fibronectin-coated micropatterned stripes were used, which constrained the width of cells, but not their spreading area. The widths examined in this earlier study, 30 to 50 µm were also wider than the natural width of nuclei in unconstrained cells. As shown in Figure 2c, cells constrained within 2500 µm2 rectangular islands exhibit relatively small changes in nuclear shape until the width of islands is comparable to the width of the nuclei. These observations of cell and nuclear elongation in the presence or absence of cytoskeleton modifying agents suggest that nuclear shape is regulated primarily by the actin caps, but at the narrowest cell widths, equal to or less than the natural width of nuclei, the lateral membrane tension and/or intermediate filament network become more dominant in driving the elongation of the nucleus. This is consistent with earlier studies using micropipettes to systematically distort cells, the results of which showed that actin microfilaments mediate nuclear shape at low distortion, while at high distortion, intermediate filament alone can mediate force transfer to the nucleus [40].
Orientation of nuclei along the long axis of the cell requires actin filaments
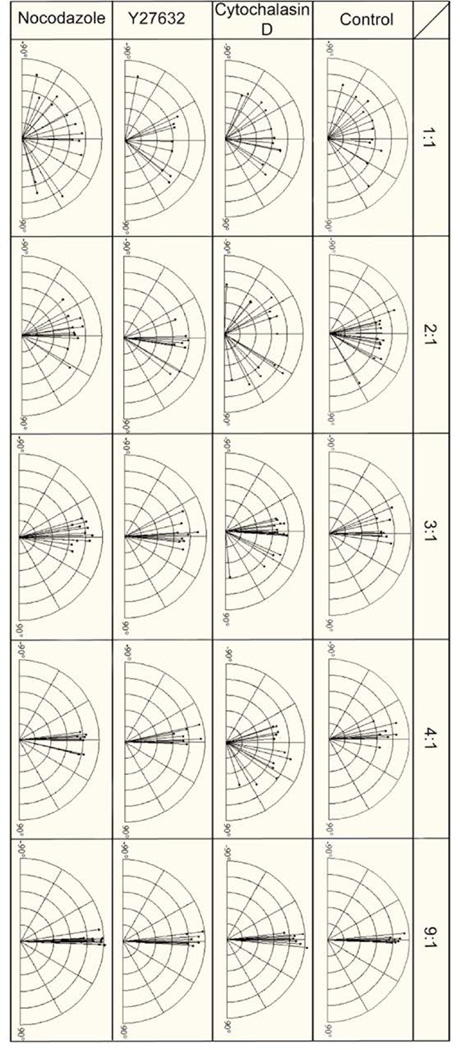
To examine how the elongated morphology of the cell influences the orientation of cell nuclei, we measured the angle between the length of cell body and the longest dimension of the cell nucleus. A 0° orientation angle represents alignment of the nucleus parallel to the long axis of cells within the rectangular islands. Figure 5 summarizes the angular orientation of nuclei for NIH 3T3 cells in rectangular islands of varying aspect ratios. The nuclei showed no preferential alignment for cells adopting square shapes, but became progressively more aligned to the length of the rectangular shapes as the aspect ratio increases. To investigate the roles of myosin mediated contractility, actin filaments, and microtubules on mediating nuclei alignment, cells of varying aspect ratios were exposed to drugs inhibiting ROCK (Y27632), actin polymerization (cytochalasin D) and microtubule polymerization (nocodazole). When treated with Y27632 and nocodazole, the nucleus remains oriented along the long axis of the cell. This implies that cellular tension and microtubules are not required for nuclear orientation. In contrast, disruption of microfilaments with cytochalasin D decreases the alignment of the nucleus significantly at aspect ratios from 2 to 4. At the highest aspect ratio of 9, cytochalasin D treated cells did not significantly alter the nuclear alignment suggesting other factors such as lateral membrane tension or intermediate filament are sufficient to orient the nucleus.
Figure 5.
Effects of different cell shape on the length and orientation of nuclei for NIH 3T3 cells under different treatments of cytochalasin D, nocodazole and Y27632 compared to standard culture conditions.
Correlation between cell and nuclear morphology for model cancer cells
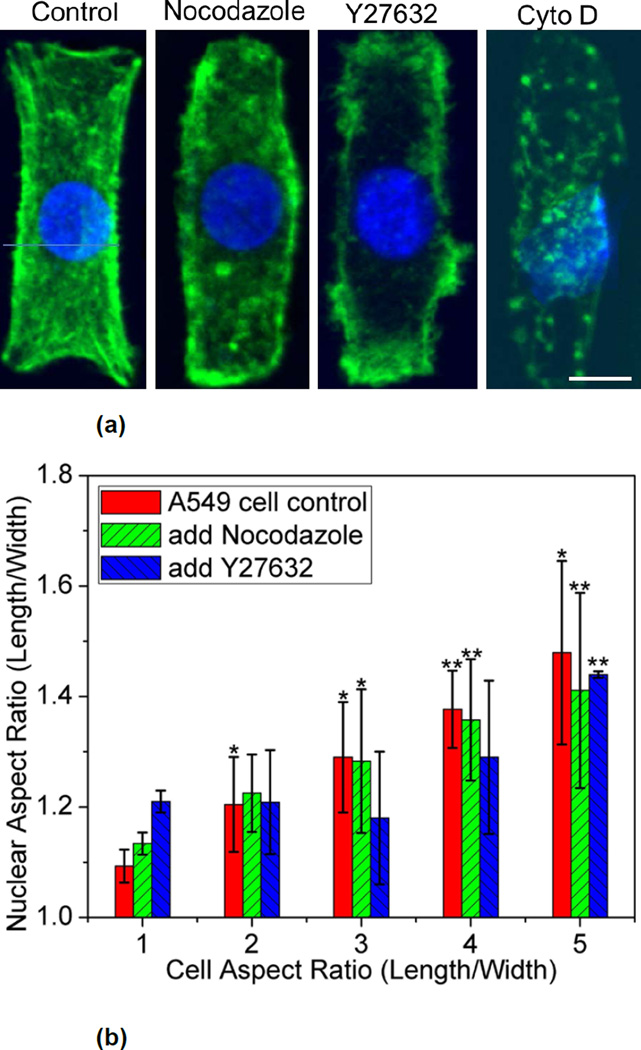
To examine how spatial coordination between cell and nuclear shape differs in cancer cells, adenocarcinoma human alveolar basal epithelial cells, (A549) was selected as a model system. A549 cells spread to adopt the rectangular islands of varying aspect ratios after 18 hours and were then fixed and stained to visualize their nuclei and actin filaments. The nuclei of most A549 cells on non-patterned tissue culture dishes exhibit abnormal non-ellipsoidal shapes that are displaced significantly from the geometric center of the cell. Upon confinement in rectangular patterns, the nucleus of individual A549 cells adopted elliptical shapes that elongate with the aspect ratio of the confining patterns. Elongation is observed at a lower cell aspect ratio of 2 (Figure 6), suggesting that A549 cell nuclei are more sensitive to changes in cell shape compared to fibroblasts, which exhibit statistically significant elongation at aspect ratios of 3 and higher (Figure 2b).
Figure 6.
(a) Representative of nuclei (blue) and actin (green) in A549 cells treated with different pharmacological agents. The scale bar respresents 10 µm. (b) Average the nuclear aspect ratio as a function of the cell aspect ratio in control and nocodazole or Y27632 treated A549 cells on rectangular micropatterns with different aspect ratios. Data are shown as Mean ± SD with 8 ≤ n ≤ 15. ANOVA followed by Tukey's post tests yield the following p values: p=0.13 (ratio 2), p=0.029 (ratio 3), p= 0.0021 (ratio 4), p=0.0033 (ratio 5) for nocodazole; p=0.85 (ratio 2), p=0.75 (ratio 3), p= 0.56 (ratio 4), p=1.01×10−5 (ratio 5) for Y27632. Statistical significance is calculated with respect to shape ratio 1 in control, nocodazole, or Y27632 treated cells respectively. *p<0.05, **p<0.005.
Similar to fibroblast cells, an insignificant change in nuclear morphology was observed with the microtubule depolymerizing agent nocodazole. At aspect ratio of 3 and 4, rounding of the nucleus was observed when myosin mediated contractility was inhibited by Y27632, implying that ROCK is necessary for nuclear remodeling for A549 cells (Figure 6). In contrasts to fibroblasts, A549 cells do not spread to fully occupy the rectangular patterns and reduce their spreading area substantially when exposed to cytochalasin D. Moreover, the nucleus, which is located typically at the geometric center of their cell body in rectangular islands, became offset from the center upon exposure to cytochalasin D. Unlike 3T3 fibroblasts, actin filaments have a critical role in maintaining the shape of A549 cells in these patterns.
Conclusions
We have utilized micropatterned adhesive islands to control the size and aspect ratio of cells and determined how cell shape influences the shape of the nucleus. The ability to define the cell morphology while keeping the cell spreading constant is achieved by organizing cells with the cell resistant ink poly(OEGMA-co-MA). This ink, developed in our laboratory [41], can be printed directly on cell culture dishes to mask the background region. As demonstrated here, this simple and versatile approach for defining the spreading area and shape of individual or groups of cells can be used to investigate fundamental questions in cell biology and tissue engineering.
We observed a direct correlation between the aspect ratio of the cell and the nuclei. The onset of this coupling occurs at lower aspect ratios for cells with larger spreading areas. We characterized the orientation of the elongated nuclei relative to the cell shape and found that the nuclei of 3T3 cells exhibited preferential alignment along the length of rectangular patterns that was most pronounced at higher cell aspect ratios. In addition, we examined the role of microtubules, actin filament and myosin-mediated contractility through nocodazole, cytochalasin D and Y27632 treatment respectively to cells with different aspect ratios. The results show that actin filaments play a critical role in regulating nuclear shape and orientation consistent with earlier studies [5, 6, 24].
Since cells with the absence of actin filaments at high cell aspect ratios can maintain the nuclear elongation, this suggests two regimes of cell-nuclear shape control. At the larger cell widths, nuclear shape and orientation are influenced significantly by actin filaments, through the perinuclear actin cap [24]. The physical connection with the ECM from the focal adhesions at the cell membrane can transmit mechanical forces to nuclear LINC complexes and lamina through the cytoskeleton [42]. Recent studies also demonstrated that the physical properties of the ECM can modulate the nuclear shape [42]. Fibroblasts grown on soft substrates produced cells with round nuclei while stiff substrate lead to flattened nuclei [43]. When cell widths are constrained to less than the natural nuclear width of unconstrained cells, elongation and alignment of the nucleus occurs even after disruption of actin filaments. A possible mechanism that regulates nuclear shape at high aspect ratios is the plasma membrane tension from the surface tension of the lipid bilayer contributed from the adhesion to the extracellular matrix on the substrate. Plasma membrane tension has been identified as a regulator of cell spreading and motility [44,45] and could play a role in maintaining the nuclear shape.
The micropatterning approach used here enables control of cell morphology in two dimensions. We observed that the maximum height of the cells at their nucleus does not change significantly. For 900 um2 micropatterns, this averages from 3.4 to 3.9 µm as the aspect ratio is elongated from one to nine. However, the variability in cell height for a given aspect ratio is large and we are unable to make a statistically robust inference on whether the aspect ratio indeed regulates the height of the nucleus. In vivo, cells are surrounded in a three dimensional environment. To address the question of how third dimensional constraints influence nuclear shape, other techniques, possibly using dielectrophoresis in conjunction with in situ photopolymerization [46] or microfluidic devices [47] would be more suitable. Studies have also shown that proliferating and quiescent cells differ remarkably in their nuclear architecture and spatial arrangement of chromosomes [48], thus suggesting the importance of monitoring cell cycle progression in future studies.
A549 cells exhibit similar patterns of nuclear deformation in confined geometries as 3T3 fibroblasts. However, A549 cells differ from 3T3 fibroblasts in their response to cytochalasin D. Inhibition of actin polymerization with cytochalasin D prevents A549 cells from adopting the rectangular shape of the adhesive islands. Altogether the results presented here revealed two distinct regimes in the coupling of nuclear morphology to cell shape, and also demonstrated a critical difference in the role of cytoskeleton for cell shape control for fibroblasts and a model cancerous cell. Such cell geometry and cell type dependence on actomyosin tension for cell and nuclear shape offer insights on how changes in external adhesive microenvironment modifies cell and nuclear shape, which may play a critical role in gene expression, development, and cancer progression [6].
Acknowledgment
This work was supported by the National Institutes of Health (R01EB010043) and (R01GM112017) and the National Science Foundation (CBET0928219).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brandt A, Krohne G, Großhans J. The farnesylated nuclear proteins KUGELKERN and LAMIN B promote aging-like phenotypes in Drosophila flies. Aging Cell. 2008;7:541–551. doi: 10.1111/j.1474-9726.2008.00406.x. [DOI] [PubMed] [Google Scholar]
- 2.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 5.Versaevel M, Grevesse T, Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun. 2012;3:671. doi: 10.1038/ncomms1668. doi: 610.1038/ncomms1668. [DOI] [PubMed] [Google Scholar]
- 6.Jain N, Iyer KV, Kumar A, Shivashankar G. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci USA. 2013;110:11349–11354. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2002;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerace L, Huber MD. Nuclear lamina at the crossroads of the cytoplasm and nucleus. J Struct Biol. 2012;177:24–31. doi: 10.1016/j.jsb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 11.Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl L, Cohen-Fix O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell. 2006;17:1768–1778. doi: 10.1091/mbc.E05-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy DL, Heald R. Nuclear size is regulated by importin α and Ntf2 in Xenopus . Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prüfert K, Vogel A, Krohne G. The lamin CxxM motif promotes nuclear membrane growth. J Cell Sci. 2004;117:6105–6116. doi: 10.1242/jcs.01532. [DOI] [PubMed] [Google Scholar]
- 14.Brandt A, Papagiannouli F, Wagner N, Wilsch-Bräuninger M, Braun M, Furlong EE, et al. Developmental control of nuclear size and shape by kugelkern and kurzkern . Curr Biol. 2006;16:543–552. doi: 10.1016/j.cub.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 15.Webster MT, McCaffery JM, Cohen-Fix O. Vesicle trafficking maintains nuclear shape in Saccharomyces cerevisiae during membrane proliferation. J Cell Biol. 2010;191:1079–1088. doi: 10.1083/jcb.201006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siniossoglou S. Lipins, lipids and nuclear envelope structure. Traffic. 2009;10:1181–1187. doi: 10.1111/j.1600-0854.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- 17.Gregory TR. Genome size evolution in animals. In: Gregory TR, editor. The evolution of the genome. San Diego, CA: Elsevier; 2005. pp. 3–87. [Google Scholar]
- 18.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, Horigome T, et al. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila . J Cell Sci. 2003;116:3811–3823. doi: 10.1242/jcs.00682. [DOI] [PubMed] [Google Scholar]
- 21.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 23.Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, et al. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gay O, Gilquin B, Nakamura F, Jenkins ZA, McCartney R, Krakow D, et al. RefilinB (FAM101B) targets FilaminA to organize perinuclear actin networks and regulates nuclear shape. Proc Natl Acad Sci USA. 2011;108:11464–11469. doi: 10.1073/pnas.1104211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr DA, Han M. ANChors away: an actin based mechanism of nuclear positioning. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 28.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, et al. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis . Curr Biol. 2013;23:1776–1781. doi: 10.1016/j.cub.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Edens LJ, White KH, Jevtic P, Li XY, Levy DL. Nuclear size regulation: from single cells to development and disease. Trends Cell Biol. 2013;23:151–159. doi: 10.1016/j.tcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Chow KH, Factor RE, Ullman KS. The nuclear envelope environment and its cancer connections. Nat Rev Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin D, Xia Y, Whitesides GM. Soft lithography for micro-and nanoscale patterning. Nat Protoc. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 34.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochem Biophys Res Commun. 2003;307:355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 35.Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pouthas F, Girard P, Lecaudey V, Ly TBN, Gilmour D, Boulin C, et al. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J Cell Sci. 2008;121:2406–2414. doi: 10.1242/jcs.026849. [DOI] [PubMed] [Google Scholar]
- 37.Chen B, Kumar G, Co C, Ho C. Geometric Control of Cell Migration. Sci Rep. 2013;3:2827. doi: 10.1038/srep02827. DOI:2810.1038/srep02827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishizaki T, Uehata M, Tamechika I, Keel J, Nonomura K, Maekawa M, et al. Pharmacological properties of Y-27632, a specific inhibitor of Rho-associated kinases. Mol Pharmacol. 2000;57:976–983. [PubMed] [Google Scholar]
- 39.Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar G, Wang YC, Co CC, Ho CC. Controlled Cell Engineering on Biomaterials Using Polyelectrolytes. Langmuir. 2003;19:10550–11055. [Google Scholar]
- 42.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of nuclear shape by substrate rigidity. Cell Mol Bioeng. 2013;6:230–238. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raucher D, Sheetz MP. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J Cell Biol. 2000;148:127–136. doi: 10.1083/jcb.148.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheetz MP, Dai J. Modulation of membrane dynamics and cell motility by membrane tension. Trends Cell Biol. 1996;6:85–89. doi: 10.1016/0962-8924(96)80993-7. [DOI] [PubMed] [Google Scholar]
- 46.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Probing the role of multicellular organization in three-dimensional microenvironments. Nat Methods. 2006;3:369–375. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 47.Tan W, Desai TA. Layer-by-layer microfluidics for biomimetic three dimensional structures. Biomaterials. 2004;25:1355–1364. doi: 10.1016/j.biomaterials.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 48.Bridger JM, Boyle S, Kill IR, Bickmore WA. Remodelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10:149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]