Abstract
Low temperature is a major abiotic stress that impedes plant growth and development. Brassica juncea is an economically important oil seed crop and is sensitive to freezing stress during pod filling subsequently leading to abortion of seeds. To understand the cold stress mediated global perturbations in gene expression, whole transcriptome of B. juncea siliques that were exposed to sub-optimal temperature was sequenced. Manually self-pollinated siliques at different stages of development were subjected to either short (6 h) or long (12 h) durations of chilling stress followed by construction of RNA-seq libraries and deep sequencing using Illumina's NGS platform. De-novo assembly of B. juncea transcriptome resulted in 133,641 transcripts, whose combined length was 117 Mb and N50 value was 1428 bp. We identified 13,342 differentially regulated transcripts by pair-wise comparison of 18 transcriptome libraries. Hierarchical clustering along with Spearman correlation analysis identified that the differentially expressed genes segregated in two major clusters representing early (5–15 DAP) and late stages (20–30 DAP) of silique development. Further analysis led to the discovery of sub-clusters having similar patterns of gene expression. Two of the sub-clusters (one each from the early and late stages) comprised of genes that were inducible by both the durations of cold stress. Comparison of transcripts from these clusters led to identification of 283 transcripts that were commonly induced by cold stress, and were referred to as “core cold-inducible” transcripts. Additionally, we found that 689 and 100 transcripts were specifically up-regulated by cold stress in early and late stages, respectively. We further explored the expression patterns of gene families encoding for transcription factors (TFs), transcription regulators (TRs) and kinases, and found that cold stress induced protein kinases only during early silique development. We validated the digital gene expression profiles of selected transcripts by qPCR and found a high degree of concordance between the two analyses. To our knowledge this is the first report of transcriptome sequencing of cold-stressed B. juncea siliques. The data generated in this study would be a valuable resource for not only understanding the cold stress signaling pathway but also for introducing cold hardiness in B. juncea.
Keywords: Brassica juncea, transcriptome, cold stress, low temperature, silique, RNA-seq
Introduction
The Brassicaceae family, which includes nearly 3500 species and 350 genera is one of the 10 most economically important plant families (Warwick and Black, 1991). Within the family, species of the genus Brassica comprise multiple vegetables (cabbage, broccoli, brussels sprout, cauliflower, turnip—B. oleracea), oilseeds (B. rapa, B. juncea, B. napus), and condiments (B. nigra, B. carinata, B. juncea; Branca and Cartea, 2011). B. juncea (n = 18) is an amphidiploid species derived from interspecific crosses between two diploid progenitor parents, B. nigra (n = 8) and B. rapa (n = 10) (Prakash and Hinata, 1980). It is grown as an oilseed crop in India (brown or Indian mustard), as a leaf vegetable in China, and as a condiment in western countries (Rakow, 2004). India is the third largest producer of rapeseed-mustard in the world after China and Canada. This crop accounts for nearly one-third of the edible oil produced in India, making it the country's key edible oilseed crop. The major impediments in harnessing the true yield potential of mustard are biotic stresses such as blight, aphids, white rust and abiotic stresses such as frost, high temperature, salinity, and drought.
Low temperature is one of the most intimidating abiotic stresses that affect plant growth and development, thereby limiting the distribution of crop species. Based on its intensity, cold stress can be broadly classified into chilling and freezing stresses. Exposure to temperatures below 0°C results in freezing stress, whereas chilling stress occurs at temperatures ranging from 0 to 20°C. Plants such as rice, maize and tomato that grow in tropical and subtropical regions are chilling sensitive whereas the plants from temperate region are chilling tolerant (Solanke and Sharma, 2008; Chinnusamy et al., 2007). Plants have the ability to acquire tolerance to chilling and freezing conditions if they are pre-exposed to non-freezing temperatures, through a process known as cold acclimation (Levitt, 1980). Cold acclimation helps plants to fine tune their metabolism and improve freezing tolerance by initiating signaling cascades that leads to several biochemical and physiological changes, including modification of membrane lipid composition and changes in gene expression (Shinozaki and Yamaguchi-Shinozaki, 1996; Thomashow, 1998; Gilmour et al., 2000; Chinnusamy et al., 2003). The altered gene expression leads to accumulation of several protective proteins such as antifreeze proteins (Griffith et al., 1997), late embryogenesis abundant (LEA) proteins (Antikainen and Griffith, 1997), heat shock proteins (HSP) (Wisniewski et al., 1996), cold-regulated (COR) proteins and various metabolites such as amino acids, soluble sugars, organic acids, pigments (Krause et al., 1999), polyamines (Bouchereau et al., 1999), and antioxidants (Hausman et al., 2000). These metabolites and proteins help in protecting plant membranes and prevent cell disruption during cold stress by stabilizing membrane lipids, proteins, maintaining hydrophobic interactions, ion homeostasis and scavenging the reactive oxygen species (ROS) (Hare et al., 1998; Gusta et al., 2004; Chen and Murata, 2008; Janská et al., 2010).
Spatial and temporal gene expression changes have traditionally been studied by comparing levels of steady state transcripts. With advancements of molecular techniques, it is now possible to generate information on alterations in transcripts levels at whole genome level. Using cDNA microarrays or whole genome arrays, the expression pattern of genes in response to chilling stress has been analyzed in Arabidopsis, rice, sunflower and several other plants (Seki et al., 2002; Rabbani et al., 2003; Fernandez et al., 2008). Seki et al. (2001) used a full-length cDNA microarray of 1300 Arabidopsis genes, and identified 19 COR (cold-regulated) genes, among which the newly identified genes were ferritin, a nodulin-like protein, LEA protein and glyoxalase. In another study, Fowler and Thomashow (2002) reported 306 COR genes using microarray for 8000 genes. Of these 306 genes, 218 were up-regulated and 88 were down-regulated and 45 of these COR genes were found to be expressing under the control of CBF1. Different ecotypes of Arabidopsis exposed to non-freezing cold stress exhibited different transcriptome level signatures (Barah et al., 2013). Transcriptome profiling of cold stress subjected 3-week-old B. rapa plants resulted in identification of genes encoding CBF/DREB like transcription factor, ERD10, RD29A/COR78, COR47/RD17 (Lee et al., 2008).
Low temperature has a negative impact on different stages of plant reproductive development leading to premature abortion of seeds and fruits (Thakur et al., 2010). Likewise pod-filling stages in B. juncea are highly susceptible to frost stress injury thereby resulting in significant reduction in crop yield. Rajasthan, being the largest producer of mustard in India, contributes to 47.2% of the total production. Injury caused by episodic incidents of frost in semi-arid plains of Rajasthan has caused huge financial losses to the farmers. Cold waves and frost stress affects mustard cultivation in Rajasthan where night temperature in winters fall below −4.4°C. The loss in crop production due to frost stress in 2006 in Rajasthan alone was 344,400 tons resulting in total economic loss of Rs 5906 million (Prasada Rao et al., 2010). Since low temperature stress is a major limiting factor in B. juncea crops, it is significant to study the response of plants to stress and the underlying mechanism of tolerance. The first step in achieving this goal is to understand the plant response to non-freezing cold stress at the molecular level. To gain an in-depth knowledge of the global gene expression changes in developing siliques of B. juncea that were exposed to chilling stress, RNA-seq was employed to generate the transcriptome profile. Manually, self-pollinated siliques (5 DAP-30 DAP) of B. juncea were subjected to either short (6 h) or longer (12 h) durations of cold stress followed by RNA extraction, library construction and sequencing using Illumina's next generation sequencing platform. This is the first genome wide report of transcriptional response in B. juncea siliques that were exposed to cold stress. Deciphering the global gene expression changes in cold-stressed developing siliques, would be useful in understanding of the molecular pathway of cold stress and devising future strategies for enhancing low temperature tolerance in Indian mustard.
Materials and methods
Plant material and growth conditions
B. juncea var. Varuna seeds were obtained from National Seed Centre, IARI, New Delhi. Seeds were sown in the field at University of Delhi, South Campus during the growing season (October-March) of mustard. Plants were grown under field conditions and self-pollination was initiated at the time of flowering.
Controlled self-pollination and stress treatment of B. juncea var. Varuna
B. juncea var. Varuna was manually self-pollinated with pollen derived from the same cultivar. All the open flowers and siliques were removed from the flowering twigs followed by emasculation of unopened buds. Fresh pollen was dusted on the exposed stigma followed by bagging to avoid cross-pollination. The manually self-pollinated siliques were subjected to cold stress at 5, 10, 15, 20, 25, and 30 days after pollination (DAP). Self-pollinated branches were excised and placed in a beaker filled with water and subjected to cold treatment at 4°C for 6 h and 12 h followed by immediate harvesting. Light conditions were maintained during cold stress to simulate natural conditions. For control samples, self-pollinated branches were excised and placed in water under field conditions. Self-pollinated siliques of different stages of development (5–30 days) were frozen in liquid nitrogen and stored at −80°C until further use. The temperature range in the field on the day of stress and harvesting was between 17°C (minimum) and −24°C (maximum). The entire experiment was designed such that control samples corresponding to different DAPs, were harvested on the same day. To study the anatomical stages of embryo development in B. juncea var. Varuna, resin sectioning was performed. The detailed procedure for staging and the results obtained is given in Supplementary Datasheet 1.
Library preparation and data analysis
Total RNA was extracted from stressed as well as control siliques using Qiagen's plant RNA extraction kit as per the manufacturer's protocol. RNA-seq libraries were prepared utilizing TruSeq RNA sample preparation kit v2 (Illumina Inc., USA) as per the manufacturer's protocol and sequenced on HiSeq 2000 (Illumina, USA). The raw reads from sequencing run of all stages was subjected to a quality filtering utilizing “NGS QC toolkit” (Patel and Jain, 2012). Low quality reads whose 70% of the read length had phred score less than 20 (Q ≤ 20) were discarded followed by trimming of adapter sequences. Orphan reads remaining after applying these filtering criteria were also discarded. The QC filtered paired reads of all the samples were merged together into two files containing left and right reads, respectively. These merged files were used to reconstruct the transcripts using Trinity assembler with default parameters (Haas et al., 2013). The statistics of the assembled transcripts were measured utilizing TrinityStats.pl script of Trinity. QC filtered reads of each of the samples were aligned to the assembly using Bowtie2 aligner (Langmead and Salzberg, 2012) and quantification of each transcript was carried out with RSEM tool (Li and Dewey, 2011). Transcripts having less than 1 read count in all the samples were considered as misassembled or artifacts and hence discarded from the assembly. The statistics of the filtered assembly were again extracted using TrinityStats.pl script. Samples were normalized with the TMM normalization method (Robinson and Oshlack, 2010) of trinity and differentially expressed transcripts (with p < 0.001) were isolated using trinity differential expression module by applying a minimum cutoff of 2-fold change on log2 scale. Generation of heat map, Spearman correlation matrix, and cluster analysis of differentially expressed transcripts was performed using R-based tools. The reconstructed transcripts were annotated using FastAnnotator online server tool with default parameters (Chen et al., 2012a).
Real time PCR validation
Total RNA was extracted from three independent biological replicates from stressed as well as control siliques (5–30 DAP) using Qiagen's plant RNA extraction kit as per the manufacturer's protocol. 10 μg of total RNA was subjected to DNaseI treatment with 1 U DNaseI (NEB, USA). The reaction was carried out at 37°C for 10 min followed by heat inactivation at 65°C for 10 min. 2.5 μg of DNase-treated RNA was used for cDNA synthesis with iScript reverse transcriptase (BioRad, USA) according to the manufacturer's protocol. The expression of actin gene (ACT7) was found to be stable in our transcriptome database and hence was used as the normalization control in real time PCR. The primers used for ACT7 were forward primer: 5′ TGGGTTTGCTGGTGACGAT 3′ and reverse primer: 5′ TGCCTAGGACGACCAACAATACT 3′. Primers were designed for selected transcripts from transcriptome database and real time PCR was performed using SYBR green I master mix (Roche, GmBH) on CFX-Connect™ Real time system (BioRad, USA). Details of the primers are presented in Supplementary Table 1. Relative expression of the transcripts was calculated using ΔΔCt method (Livak and Schmittgen, 2001). Scatter plot and Pearson correlation coefficient was calculated in MS-excel using log2 values of qPCR and digital expression data.
The RNA-seq data discussed in this publication has been deposited in NCBI‘s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE73201 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73201).
Results
De novo assembly of B. juncea silique transcriptome data
B. juncea var. Varuna plants were controlled self-pollinated at the time of flowering. Controlled self-pollinated siliques of B. juncea at different stages of development (5 DAP-30 DAP) were subjected to cold stress at 4°C for capturing the changes in gene expression. The siliques from control plants were also used to study the anatomical details of embryo development in B. juncea var. Varuna. Longitudinal sections of fertilized ovule showed first division of zygote at 5 DAP, first longitudinal division of embryo at 10 DAP, globular stage of embryo at 15 DAP, heart shape embryo at 20 DAP, torpedo stage of embryo at 25 DAP and cotyledonary immature embryo at 30 DAP (Supplementary Datasheet 1).
RNA-seq libraries were constructed from 18 different samples that encompassed six development stages (5, 10, 15, 20, 25, and 30 DAP) of B. juncea siliques, obtained by controlled self-pollination, and three different time points (0 h, also referred as control, 6 h and 12 h) of cold stress. Details of the experimental design and analysis pipeline are presented in Figure 1. As it was difficult to harvest enough embryos after controlled pollination, entire siliques were harvested for tissue collection. Sequencing of 18 libraries generated a composite of approximately 820 million reads, which were subjected to quality filtering using “NGS QC toolkit” (Patel and Jain, 2012). The adapter contaminated reads as well as reads whose 70% bases had Phred score less than 20 (Q ≤ 20) as well as orphan reads were discarded. This resulted in approximately 700 million high quality paired reads (Table 1).
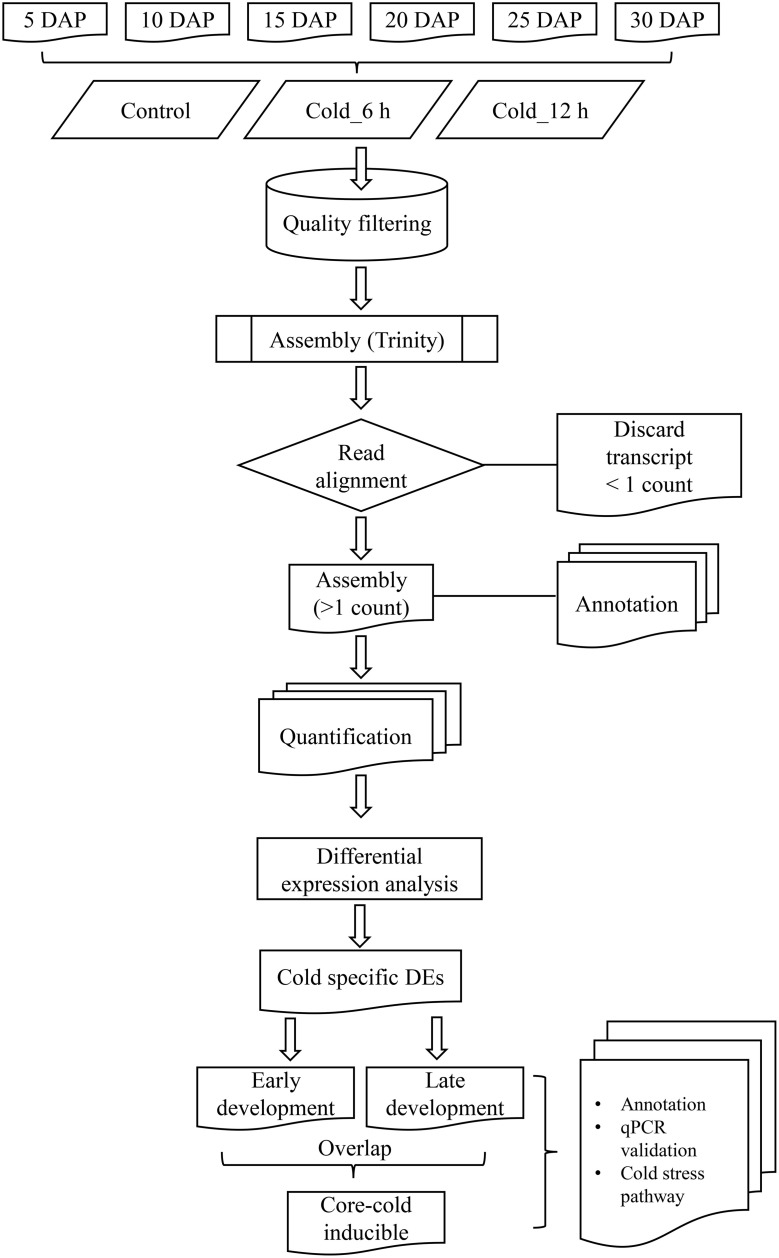
Figure 1.
Flowchart representing computational pipeline employed for analysis of transcriptome sequenced from B. juncea siliques.
Table 1.
Quality filtering and statistics of raw reads obtained in transcriptome libraries of B. juncea siliques exposed to low temperature for different durations.
| S. No. | Sample | No. of raw reads | No. of HQ reads | Percentage (%) | Mapped reads | Percentage mapping (%) |
|---|---|---|---|---|---|---|
| 1 | 5 DAP_C | 34,996,934 | 33,098,872 | 94.58 | 23,672,240 | 71.52 |
| 2 | 5 DAP_6 h | 46,283,995 | 43,352,879 | 93.67 | 31,094,528 | 71.72 |
| 3 | 5 DAP_12 h | 27,407,889 | 21,593,463 | 78.79 | 15,463,603 | 71.61 |
| 4 | 10 DAP_C | 31,916,211 | 30,033,245 | 94.10 | 21,664,687 | 72.14 |
| 5 | 10 DAP_6 h | 38,077,842 | 35,335,917 | 92.80 | 25,477,680 | 72.10 |
| 6 | 10 DAP_12 h | 20,851,323 | 16,264,475 | 78.00 | 11,803,985 | 72.58 |
| 7 | 15 DAP_C | 36,410,242 | 34,111,247 | 93.69 | 24,585,074 | 72.07 |
| 8 | 15 DAP_6 h | 40,661,329 | 37,646,457 | 92.59 | 27,322,541 | 72.58 |
| 9 | 15 DAP_12 h | 44,470,147 | 36,420,384 | 81.90 | 26,589,923 | 73.01 |
| 10 | 20 DAP_C | 54,387,436 | 51,174,780 | 94.09 | 37,227,245 | 72.75 |
| 11 | 20 DAP_6 h | 55,097,716 | 51,238,446 | 93.00 | 37,445,913 | 73.08 |
| 12 | 20 DAP_12 h | 48,736,907 | 38,808,054 | 79.63 | 28,375,618 | 73.12 |
| 13 | 25 DAP_C | 34,719,115 | 28,553,791 | 82.24 | 20,805,257 | 72.86 |
| 14 | 25 DAP_6 h | 46,760,019 | 37,279,174 | 79.72 | 26,906,383 | 72.18 |
| 15 | 25 DAP_12 h | 92,720,961 | 78,646,125 | 84.82 | 58,031,931 | 73.79 |
| 16 | 30 DAP_C | 45,741,906 | 37,042,168 | 80.98 | 26,990,691 | 72.86 |
| 17 | 30 DAP_6 h | 63,185,861 | 50,328,733 | 79.65 | 37,024,361 | 73.57 |
| 18 | 30 DAP_12 h | 58,774,016 | 50,083,797 | 85.21 | 36,735,973 | 73.35 |
Raw reads were filtered to remove adapter sequences and low quality reads using NGS QC tool kit. The percentage of reads retained after QC filtering is depicted in column 5.
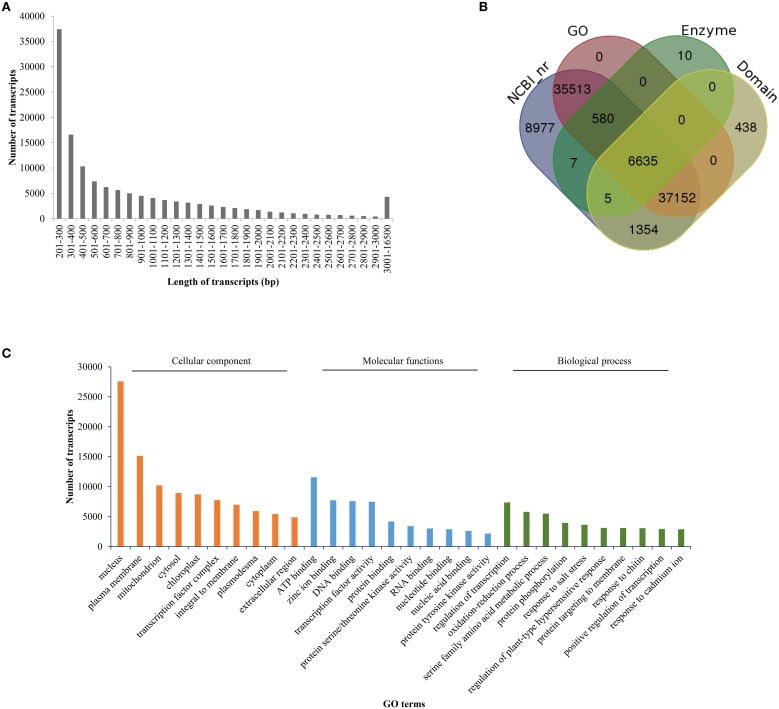
The filtered HQ reads were subjected to assembly using the de novo assembler “Trinity” (Haas et al., 2013), which employs “de-Bruijn” graph approach (at a k-mer of 25 nucleotides) to assemble the reads into contigs. The preliminary assembly generated 212,124 contigs (hereafter referred to as “transcripts”). Combined length of these transcripts was 156 Mb and the N50 value of the assembly was 1202 bp. The obtained length of transcripts ranged between 201 and 14,199 bp and the GC content of assembled transcripts was found to be 41.92%. These transcripts also included isoforms and therefore represented 143,414 unigenes. The combined length of the unigenes was 83 Mb and had a N50 value of 822 bp. Average length of transcripts and unigenes (from the preliminary assembly) was found to be 737 and 577 bp, respectively. The preliminary assembly was subjected to assembly correction by aligning the HQ paired reads to the assembled transcripts with the help of Bowtie2 module of RSEM. More than 70% of the paired reads mapped to the transcripts of the preliminary assembly (Table 1). Quantification of each transcript was carried out using RSEM tool. Transcripts having expression value of less than 1 read count in all the 18 samples were removed from the assembled data. The final filtered assembly was reanalyzed for various parameters, including N50 value, transcript number, unigene number and average transcript length. Following assembly correction and filtering, 133,641 transcripts were retained, the combined length of which was 117 Mb. The average length of transcripts was 875 bp and the N50 value of the assembly was 1428 bp. While the maximum length of the transcript was found to be 16 kb, the minimum length was 200 bp. The distribution of transcript length is presented in Figure 2A. The 133,641 filtered transcripts represented 83,899 unigenes. The average length of the unigenes was 692 bp with a N50 value of 1225 bp (Table 2).
Figure 2.
Size distribution and gene ontology analysis of the assembled transcripts. (A) Assembled transcripts were evaluated for their size distribution and the number of transcripts in each size range is presented. (B) Overlap in the distribution of annotated transcripts based on their databases hits. The LAST search was used to identify transcripts in NCBI nr database whose output was further annotated with Blast2GO for assigning gene ontology terms. PRIAM database and RPS blast were employed for identification of enzymes and conserved domains, respectively. (C) Gene ontology categorization of annotated B. juncea transcripts on the basis of cellular components, molecular functions and biological processes. The numbers of transcripts in the ten most enriched GO terms for each of the categories are shown.
Table 2.
Assembly statistics of B. juncea transcriptome.
| Parameters | Uncorrected | Corrected |
|---|---|---|
| Number of transcripts | 212,124 | 133,641 |
| Number of unigenes | 143,414 | 83,899 |
| Assembly length (Mb) | 156 | 117 |
| Minimum transcript length (bp) | 201 | 201 |
| Average transcript length (bp) | 737 | 875 |
| N50 (bp) | 1202 | 1428 |
Annotation of assembled B. juncea siliques transcripts
To gain an insight on probable functions of each transcript, assembly was subjected to annotation analysis utilizing web-based tool “FastAnnotator”, which uses four different annotation protocols: (1) LAST (Local Alignment Search Tool) search to identify best hits in NCBI non-redundant (nr) database, (2) Blast2GO (https://www.blast2go.com/) to assign gene ontological (GO) terms, (3) PRIAM (http://priam.prabi.fr/) for identification of enzymes along with their EC numbers, and (4) RPS BLAST to predict functional domains in transcript sequences by searching the domain models (Pfamv26) from Conserved Domain Database (CDD; http://www.ncbi.nlm.nih.gov/cdd; Chen et al., 2012a). The LAST search against NCBI nr database resulted in annotation of approximately 67% transcripts (90,223 out of 133,640 transcripts) with a minimum e-value of 10−5 (Supplementary Datasheet 2). In addition to the NCBI-based annotation, 438 transcripts were annotated by domain search and 10 were annotated with enzyme database. Out of the 90,671 annotated transcripts, 79,880 transcripts could be associated with at least one GO term. Nearly, 7237 transcripts had at least one enzyme hit and 45,584 transcripts had >50% identity with previously reported protein domains (Figure 2B). Similarly, 8977, 438, and 10 transcripts exhibited exclusive hits to NCBI nr, domain and enzyme databases, respectively. The top 10 GO terms (associated with the annotated transcripts) in various biological processes, molecular functions and cellular components are presented in Figure 2C. The top GO terms in biological processes were regulation of transcription, oxidation-reduction, protein phosphorylation and stress responses. On the basis of localization in cellular components, maximum number of transcripts localized to nucleus followed by plasma membrane and mitochondria. ATP binding, zinc ion binding and DNA binding were the most enriched GO terms in the molecular function category.
Identification of differentially expressed transcripts (DETs) in developing siliques
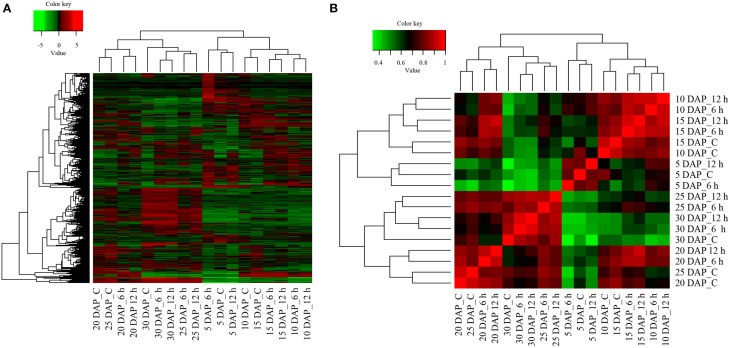
Approximately, 13,342 transcripts exhibiting significant differential expression were identified by pair-wise comparisons (Supplementary Datasheet 3) in all possible combinations of the 18 samples. On the basis of expression profile, DETs were clustered to generate a heat map (Figure 3A) as well as Spearman correlation matrix (Figure 3B). Broadly, differentially expressed transcripts of 5, 10, and 15 DAP clustered in one clade whereas those of 20, 25, and 30 DAP clustered together in another clade, thereby enabling us to categorize and identify gene expression changes occurring either during the early (occurring in 5–15 DAP) or late (occurring in 20–30 DAP) stages of silique development. Transcripts of controls corresponding to 10 and 15 DAP clustered together. Similarly, transcripts present in controls of 20 and 25 DAP clustered together. Nonetheless, in most of the cases 6 h and 12 h cold stressed differentially expressed transcripts clustered together. The only exception was at 5 DAP whose control samples clustered with the 12 h cold stress samples, whilst 6 h samples was placed in a different sub-clade (Figures 3A,B). Following clustering, transcripts displaying similar expression pattern in response to cold stress were cataloged for each developmental stages separately and their expression pattern is depicted in Supplementary Figures 1–6.
Figure 3.
Analysis of differentially expressed transcripts and their inter-relationships in B. juncea siliques. (A) Heat map of differentially expressed transcripts in control and cold stressed B. juncea siliques at different stages of development. Differentially expressed transcripts identified by pair-wise comparisons of all the 18 samples were clustered together on the basis of their expression values and heat map was plotted with TMM-normalized FPKM values. (B) Spearman correlation matrix of all the samples indicating extent of similarity between each possible pair. All the 13,342 transcripts exhibiting differential expression were clustered together and Spearman correlation matrix was plotted on TMM-normalized FPKM values. The key for sample labels should be interpreted as x DAP_y, where x denotes number of days; DAP represents days after pollination and y denotes either unstressed siliques –“C” or duration of cold stress in hours –“h”.
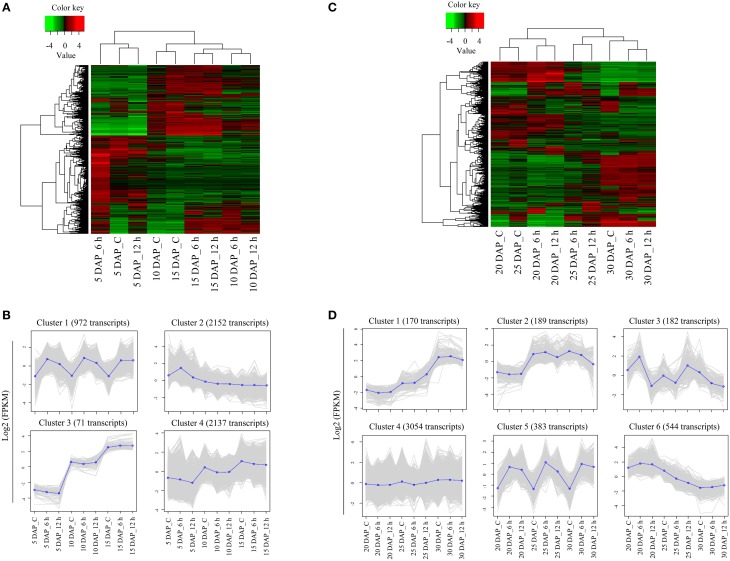
The samples (control and cold stress treated) from 5, 10, and 15 DAP were considered as early stages of silique development. Detailed analysis of early stage samples identified 5332 transcripts exhibiting ≥2-fold difference in expression levels (on a log2 scale), which were subsequently used for generation of a heat map. As evident in Figure 4A, the hierarchical clustering of samples resulted in two parent clades where 5 DAP samples clustered under one clade whereas 10 and 15 DAP samples were part of another clade. Heat map and the associated clusters of expression profile (Figures 4A,B) showed that expression of a large number of transcripts is modulated significantly in response to cold stress. Based on the expression patterns of transcripts, four major categories comprising of 972, 2152, 71, and 2137 transcripts in clusters 1, 2, 3, and 4, respectively were identified (Figure 4B). Clusters 1 and 2 contained transcripts that were up-regulated by cold stress, whereas clusters 3 and 4 had transcripts that were developmentally regulated, as their expression remained unchanged under cold stress. Expression of the transcripts belonging to cluster 1 increased in response to both 6 h and 12 h of cold stress. More importantly, this trend was observed in all the initial stages (i.e., 5, 10, and 15 DAP) of silique development. Transcripts belonging to cluster 2 were also up-regulated by cold stress. However, the expression of these transcripts increased only at 6 h of cold stress in 5 DAP samples. Their expression was largely stable in all the other stages and time points.
Figure 4.
Detailed analysis of differentially expressed genes in early and late stages of siliques exposed to cold stress. Heat maps of differentially expressed transcripts in control and cold stressed B. juncea siliques at early (A) and late (C) stages of development. Differentially expressed transcripts identified by pair-wise comparisons of 9 samples of each stage were clustered together on the basis of their expression values and heat map was plotted with TMM-normalized FPKM values. Clusters of transcripts showing similar expression profile in early (B) and late (D) stages of silique development under control and stress conditions. The average expression of each cluster was plotted as dark blue line whereas gray lines represent expression of individual transcripts. The key for sample labels should be interpreted as x DAP_y, where x denotes number of days; DAP represents days after pollination and y denotes either unstressed siliques –“C” or duration of cold stress in hours –“h”.
The stages of silique development at 20 DAP, 25 DAP, and 30 DAP were categorized as late stages and these stages compositely had 4522 differentially expressed transcripts. Hierarchical clustering revealed that control samples of 20 DAP and 25 DAP clustered together. However, cold stress samples of 6 h and 12 h (at all the stages) were placed in the same cluster (Figure 4C). From the hierarchical clustering, we identified six major patterns of gene expression in the late stages of silique development (Figure 4D). Out of the six clusters, only two clusters (clusters 3 and 5) had transcripts that were induced by cold stress. The expression of transcripts in cluster 3 increased (w.r.t. to the corresponding controls) after 6 h of cold stress in 20 DAP and after 12 h of cold stress in 25 DAP. Noticeably, expression of transcripts in cluster 5 had a similar pattern as cluster 1 of early stage as these transcripts were induced by both 6 h and 12 h of cold stress in all the later stages of silique development.
Identification and characterization of transcripts inducible by cold stress in B. juncea siliques
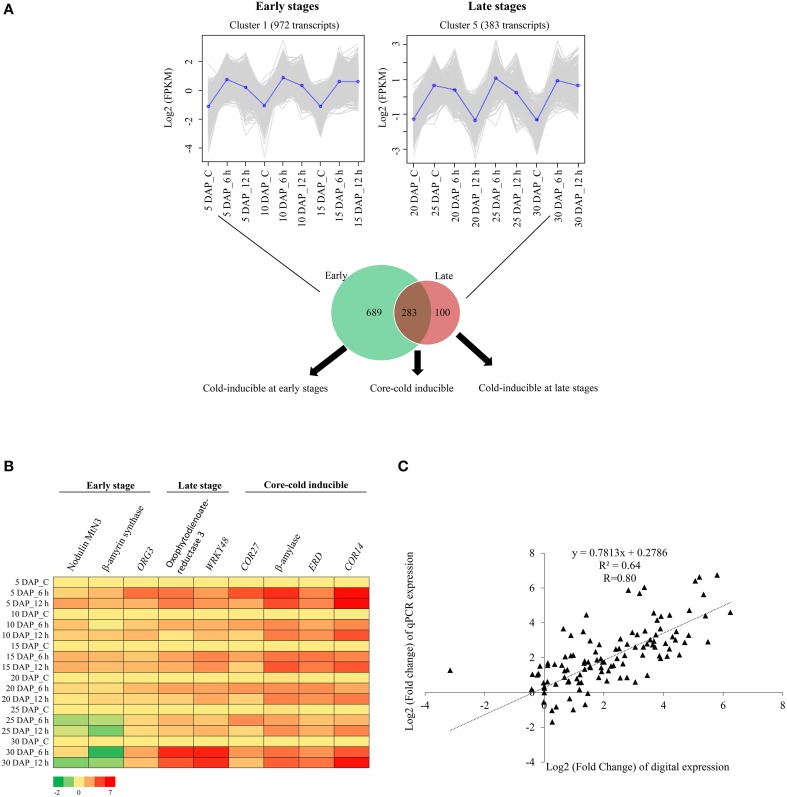
To identify the core group of genes that are induced in both the early and late stages of silique development, the transcripts from cluster 1 of the early stage (Figure 4B) was compared with those from cluster 5 of the late stage (Figure 4D). Comparison of 972 transcripts of cluster 1 with 383 transcripts of cluster 5 led to identification of 283 transcripts that were induced by cold stress in all the stages of embryo development. The 283 transcripts thereby constituted what is being referred to as the “core cold-inducible” transcripts. Additionally, this analysis revealed that 689 transcripts were specifically induced by cold stress in the early stages of silique development, whereas expression of 100 transcripts increased under cold stress, in the late stages of silique development. Results of this analysis are presented in Figure 5A and the list of the transcripts is provided in Supplementary Datasheets 4–6. To further assign relevance, transcripts from each of these subsets were annotated and their gene ontology terms in the biological processes category were identified (Supplementary Figure 7). Details of this analysis are presented below.
Figure 5.
Identification and validation of cold-inducible transcripts from early and late stages of silique development in B. juncea. (A) Transcripts of cluster 1 (early stage) and cluster 5 (late stage) were compared and candidates that were either specific or common to both the stages of silique development were identified. (B) Heat map showing relative abundance of selected cold-inducible transcripts belonging to the early, late and core subsets as determined by qPCR. (C) Scatter plot showing correlation between digital and qPCR expression profiles. Pearson correlation coefficient (R) was calculated between log2 fold change values of qPCR and digital expression. The key for sample labels should be interpreted as x DAP_y, where x denotes number of days; DAP represents days after pollination and y denotes either unstressed siliques –“C” or duration of cold stress in hours –“h”.
Annotation of subsets
Functional annotations of the three subsets were fetched from the annotated assembly. More than 70% of the transcripts from each of the subsets were annotated with different databases. Out of 689 transcripts in early inducible subset, 492 were annotated with different databases. Similarly, annotations were possible for 231 and 72 transcripts of core-cold inducible and cold-inducible subsets at late stages of silique development, respectively (Supplementary Figure 7). Majority of the transcripts in all the three categories were annotated by GO search as well as by the domain structure databases. However, none of the transcripts were annotated specifically from enzyme database alone.
Considering only the top 20 GO terms, gene ontology analysis revealed that some of the biological processes were exclusively associated with specific categories. For example GO terms, “MAPKK” and “protein targeting to membrane” associated exclusively with cold-inducible subset of early silique development. Similarly, the other subset, containing cold-inducible transcripts in the later stages of silique development, associated specifically with GO terms like “oxidation-reduction process,” “response to UV-B,” and “stomatal complex morphogenesis.” The GO terms “photoperiodic response,” “photo-morphogenesis,” “two-component signal transduction,” and “myoinositol hexaphosphate processes” were linked to the core-group transcripts. GO terms representing transcripts involved in response to various abiotic and biotic stresses like cold, wounding, fungal infection etc., were present in all the three subsets.
Identification of TFs, TRs, and kinases inducible by cold stress in B. juncea siliques
Transcription factors (TFs) are proteins, which directly bind to the DNA sequence and modulate transcription of a gene, whereas transcriptional regulators (TRs) are proteins, which interact with other proteins to regulate gene transcription. Apart from these proteins, kinases are known for their role in relaying stress signals to the downstream signaling components. Therefore, an in-depth analysis of the transcriptome data was performed to identify TFs, TRs, and kinases that are inducible by cold stress in the three subsets of cold-inducible transcripts.
Transcription factors (TFs)
We identified 120 transcripts belonging to 22 TF families from the three subsets. These included members of various stress-responsive TFs such as WRKY, HSFs, MYBs, AP2-EREBs, NACs etc. Nineteen of the 22 families were detected in the early cold-inducible category and interestingly 12 of these were induced by cold exclusively during the early stages of silique development. Members belonging to 5 families of TFs were present in the later stages of development, however only 1 of these was exclusive to later stages. Maximum members (25) were detected for the Myb family; out of which 15 were cold-inducible exclusively in the early stages subset while another 10 were part of the core-group of transcripts. None of the Myb TF family member was inducible by low temperature in the later stages of silique development. The next major category of TF was AP2-EREBP type, which had 10 cold-inducible transcripts in the early group, 1 in late group and 4 in the core group (Table 3).
Table 3.
Major TF, TR and kinases family induced by low temperature belonging to three subsets (cold-inducible at early stage, cold-inducible at late stage, and core-cold inducible) of siliques development.
| Type | Early stage specific | Late stage specific | Core-cold specific |
|---|---|---|---|
| TRANSCRIPTION FACTORS | |||
| ABI3VP1 | 1 | ND | ND |
| AP2-EREBP | 10 | 1 | 4 |
| bHLH | 8 | ND | ND |
| bZIP | 2 | ND | ND |
| C2C2-CO-like | ND | ND | 12 |
| C2C2-Dof | 5 | 1 | ND |
| C2C2-GATA | 1 | ND | ND |
| C2H2 | 5 | ND | ND |
| C3H | 5 | 2 | 5 |
| FHA | 1 | ND | ND |
| HB | 4 | ND | 1 |
| HSF | ND | 1 | ND |
| MADS | 1 | ND | ND |
| MYB | 15 | ND | 10 |
| NAC | 7 | ND | ND |
| PLATZ | 2 | ND | 1 |
| Sigma70-like | ND | ND | 2 |
| SBP | 1 | ND | ND |
| TCP | 1 | ND | ND |
| Tify | 2 | ND | ND |
| Trihelix | 2 | ND | ND |
| WRKY | 6 | 1 | ND |
| TRANSCRIPTIONAL REGULATORS | |||
| ARID | 1 | ND | ND |
| GNAT | 1 | ND | ND |
| Orphans | 7 | ND | 31 |
| Pseudo ARR-B | ND | ND | 6 |
| TRAF | ND | ND | 1 |
| KINASES | |||
| Calcium dependent protein kinase | 1 | ||
| Domain of unknown function 26 (DUF26) kinase | 1 | ||
| Legume lectin domain kinase | 1 | ||
| Leucine rich repeat kinase I | 1 | ||
| Leucine rich repeat kinase XI & XII | 1 | ||
| Other protein kinase | 3 | ||
| SNF1-related protein kinase (SnRK) | 7 | ||
| Unknown function kinase | 1 | ||
ND, not detected.
The core-cold inducible subset consisted of well-known positive regulators of cold stress pathway like CBFs, AP2-EREB, constans-like 1, DREB2-2, and members of Myb TF family whereas late stage cold-inducible subset included HSFA6b and AP2-EREB members. Transcription factors were maximally represented in early stages of silique development. Members of WRKY family have been implicated in abiotic as well as biotic stress (Kayum et al., 2015). WRKY23 and WRKY46 were expressed under cold stress in early stages of silique development. SPL family of TFs is a functionally diverse set of proteins and is known to be involved in regulating plant growth and development (Preston and Hileman, 2013). In early stages of development, SPL13 was found to be cold-inducible. Similarly, zeaxanthin epoxidase the first enzyme in ABA synthesis pathway (Schwartz et al., 1997) was inducible by cold stress in the early stages of siliques development. ABA is a plant hormone well known for its involvement in seed development as well as abiotic stress response (Nakashima and Yamaguchi-Shinozaki, 2013). There is a direct evidence of light regulated expression of CBFs in cold stress. Expression of CBFs is regulated by circadian-rhythm and depends upon red to far-red ratio of light (Fowler et al., 2005). In our data sets, it was found that phytochrome interacting transcription factor was inducible by cold stress during early developmental stages whereas circadian-rhythm regulating factor was up-regulated in core-cold inducible datasets.
Transcriptional regulators (TRs)
Transcriptional regulators are group of proteins, which interact with other proteins and affect transcription of genes. We were able to identify five families of cold-inducible TRs in developing siliques. Two families each were detected exclusively in early and core group of transcripts. None of the cold-inducible TR members were detected exclusively during late stages of silique development (Table 3). Members belonging to Orphan TRs, were abundantly represented in the core group of cold-inducible transcripts.
Kinases
Kinases play an important role in signal transduction by relaying the signal through protein phosphorylation. In our dataset, cold-inducible kinases were found only during the early phases of silique development. None of the kinases were up-regulated by cold stress either in later stages of development or were part of the core-cold inducible subsets. Families of kinases that are up-regulated by cold stress in the early development are presented in Table 3. A total of 16 transcripts encoding for kinases were identified out of which 7 belonged to the SNF1-related protein kinases.
Expression profiling of differentially regulated transcripts by quantitative PCR (qPCR)
Nine transcripts representing all the three subsets (cold-inducible at early stage, core-cold inducible, and cold-inducible at late stage) were selected for validation of digital expression profile. Total RNA extracted from B. juncea siliques (5 DAP-30 DAP) that were subjected to 6 h and 12 h of cold stress, was used for quantification of the relative expression by qPCR. The heat map of relative expression for the 9 transcripts is depicted in Figure 5B and the corresponding bar graphs are given in Supplementary Figures 8–10. The expression pattern obtained by qPCR was in concordance with the expression pattern inferred by the RNA-seq data as shown by high Pearson correlation coefficient of 0.8 (Figure 5C). Three of the transcripts exclusive to early stages include β-amyrin synthase, ORG3-like transcription factor, and Nodulin MtN3 (SWEET13). The expression of Nodulin MtN3 (SWEET13) was up-regulated by cold stress in both 5 and 15 DAP of silique development, whereas β-amyrin synthase was inducible in 5, 10, and 15 DAP. ORG3-like transcription factor was inducible in cold stress subjected 5 DAP. These transcripts were not inducible or had a reduced expression in response to cold stress during the late stages (20, 25, and 30 DAP) of silique development. The cold-induced expression of Oxophytodienoate reductase 3 and WRKY transcription factor 48 (genes whose digital expression were high only in the late stages) was found in early stages also, however, their induction levels at 30 DAP were much higher than early stages. Four core cold-inducible transcripts (COR27, COR14, putative β-amylase, and early response to dehydration) were found to exhibit high expression at all the stages of silique development in response to cold stress.
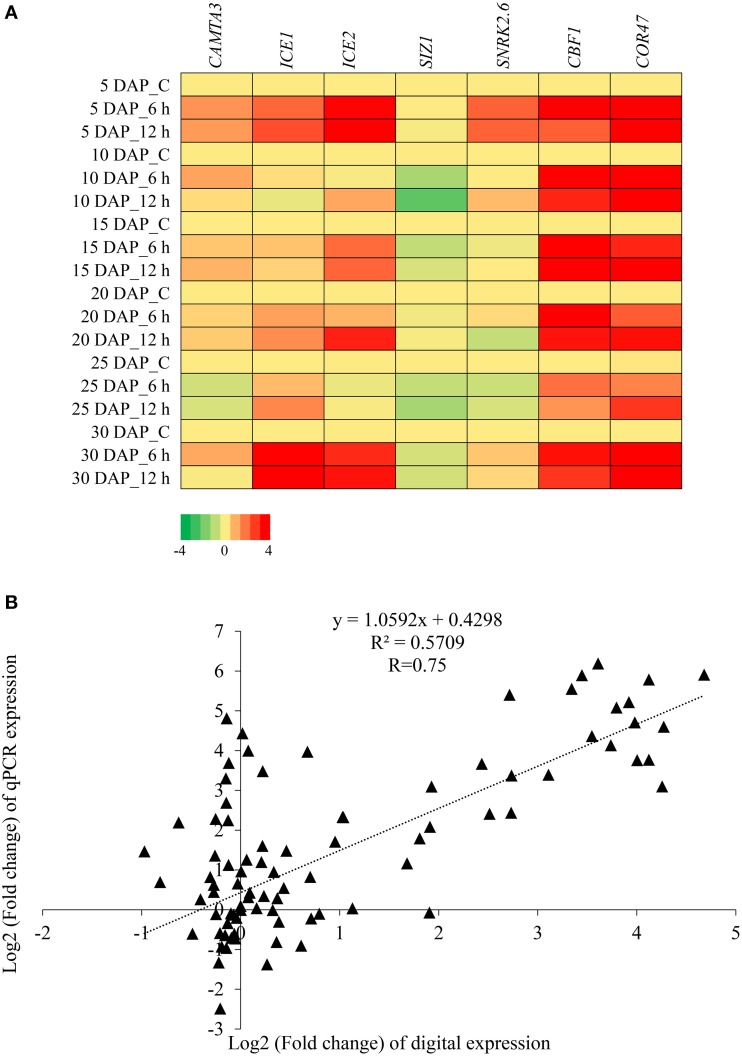
An additional set of seven transcripts, comprising known cold signaling pathway genes, were selected for expression analysis by qPCR. The heat map of relative expression profiles of the transcripts are depicted in Figure 6A and the bar graph of these genes are given in Supplementary Figure 11. CBF1 and COR47 were found to be inducible at all stages of silique development under cold stress, whereas SnRK2.6 and CAMTA3 were inducible at specific stages of silique development. The expression profiles of ICE1 and ICE2 were largely similar, and were induced after cold stress at 5 DAP and 30 DAP. However, increase in transcript level of ICE2 was also observed in 15 and 25 DAP after cold stress. We failed to observe any significant increase in the expression of SIZ1 in any of the developing stage on exposure to 6 h and 12 h of cold stress. The expression pattern obtained by qPCR was in agreement with the expression pattern inferred by the RNA-seq data. Pearson correlation coefficient (R) was 0.75 (Figure 6B).
Figure 6.
Expression profiling of B. juncea transcripts homologous to cold-stress signaling pathway components. (A) Heat map showing relative abundance of genes belonging to cold signaling pathway as determined by qPCR. (B) Scatter plot showing correlation between digital and qPCR expression profile. Pearson correlation coefficient (R) was calculated between log2 fold change values of qPCR and digital expression.
Discussion
The sowing period of mustard in India ranges from mid-October to first week of November (Shekhawat et al., 2012) and pod filling occurs at approximately 75–80 days after sowing (http://www.nuziveeduseeds.com/mustard-how-to-grow/). The minimum mean temperature during the pod filling stages ranges from 10 to 14°C (http://www.imd.gov.in/doc/Winter2010.pdf). As the early pod filling stages are sensitive to frost injury, it is important to understand the molecular response of developing siliques to low temperatures so that the gathered knowledge could be used for generating frost resilient B. juncea plants. To fully comprehend cold stress-mediated transcriptional response chilling stress was imposed to manually self-pollinated B. juncea siliques followed by RNA-seq. In several previous studies only vegetative parts of plants were exposed to cold stress and although it is likely that cold stress during the vegetative phase of plant growth affects the reproductive phases, we imposed cold stress to the developing siliques of B. juncea as cold and frost stress largely affects post-fertilization stages. Our study allowed us to focus on identification of specific changes in gene expression in post-fertilization stages, which define the yield under stress conditions. Nonetheless, it would be worthwhile to identify genes that impact developmental programming in plants exposed to cold stress during the vegetative phase. In the present study excised self-pollinated branches of field-grown mustard plants were subjected to low temperature in a cold chamber. To normalize the wounding response, self-pollinated branches were excised and placed in a beaker containing water under field conditions and this tissue was used as control. Cold stress was imposed for either 6 h or 12 h to identify gene expression changes in transcription factors and kinases, which are normally induced early as well as their downstream target genes that are up-regulated after a longer period of stress. With the help of transcriptome sequencing we were able to identify subsets of cold responsive genes that are expressed in early and/or late stages of silique development. As reference genome for mapping the sequencing reads was not available, paired-end sequencing was performed to obtain an improved de novo assembly. In addition to the sequencing length, it is important that libraries should be sequenced with adequate depth so as to capture low abundance transcripts. As per the estimates published in a recent report, sequencing of >200 million paired-end reads can optimally discover rare transcripts and their isoforms in human genome (Sims et al., 2014). In the current study, approximately 700 million purity-filtered reads were generated, collectively from all the libraries in B. juncea (whose genome size is estimated to be one third of the human genome), and we believe that this depth is sufficient for discovering even the rare isoforms. Assembly, of transcripts was performed using trinity assembler, which resulted in 133,641 transcripts and composed 117 Mb of sequences.
Approximately 13,000 differentially regulated transcripts were identified by pair-wise comparison of all the 18 samples. Based on the expression profile of DETs, two distinct clades were observed- one comprising of 5, 10, and 15 DAP samples and another of 20, 25, and 30 DAP samples. Because of the clustering pattern, we categorized gene expression changes according to the early and late phases of silique development. It was also observed that cold stress subjected samples (i.e., 6 h and 12 h) clustered together and were distinct from control samples. We identified multiple transcripts that were regulated by cold stress. Notably genes that code for proteins like dehydrin, DREB1B, early response to dehydration, low-temperature induced (Lti), glycine rich RNA binding protein, calcineurin B-like (CBLs), CBL-interacting protein kinases (CIPKs) etc. were up-regulated by low temperature stress. In several studies these proteins were shown to be involved in cellular response to cold stress (Gilmour et al., 1998; Fowler and Thomashow, 2002; Maruyama et al., 2004).
To identify the core components of cold stress signaling in developing siliques, we compared the transcripts that are cold-inducible of the early stages (5, 10, and 15 DAP) with those of late stages (20, 25, and 30 DAP). A total of 1072 transcripts were inducible by cold stress out of which 283 transcripts were detected from both early and late stages of silique development. This subset was therefore considered as the core group of cold-inducible transcripts. In addition, 689 transcripts and 100 transcripts were induced specifically in early and late stages of silique development respectively. These transcripts were therefore categorized as either early or late components of the cellular response to cold stress in developing siliques of B. juncea. To gain a better understanding about the role of these genes, we annotated them on the basis of their homology and more than 70% of the transcripts were annotated using existing databases. Multiple GO categories whose genes were involved in various abiotic and biotic stresses were highlighted in all the three subsets. Transcripts belonging to GO category “Circadian Rhythm” constituted the second largest number in the core group of transcripts. It is now well-known that circadian rhythmicity and cold stress mediated gene expression are interconnected (Espinoza et al., 2008; Dong et al., 2011; James et al., 2012; Maibam et al., 2013). Our results also show that transcripts belonging to circadian rhythmicity are cold-inducible throughout the silique development phases. Additionally, some of the transcripts of circadian rhythmicity were inducible by cold specifically in the later phases of silique development.
Another major GO category whose transcripts were inducible by low temperature was “long-day photoperiodism.” Regulation of cold-inducible genes and freezing tolerance by light quality and duration have been previously reported in Arabidopsis (Franklin and Whitelam, 2007; Lee and Thomashow, 2012) and therefore, cold-induction of transcripts involved in photoperiodism is an additional evidence that these processes are intimately linked. A GO category whose transcripts were specifically up-regulated in the later stages of cold stress was “oxidation-reduction” process. One of the harmful effects that cold stress imposes is generation of ROS. Some of the genes that counteract the effects of ROS are cold-inducible (Lee et al., 2002; Zhu et al., 2007; Shi et al., 2014). Though generation of ROS is a function of stress, the enrichment of cold-inducible transcripts involved in oxidation-reduction process only in the later phases (which are exposed to similar cold stress as the early stages) is quite intriguing and requires further investigations.
A total of 120 transcripts belonging to 22 different TF (transcription factor) families were identified from the 1072 cold-induced transcripts. A large number of the TF transcripts (79) were cold-inducible specifically during early stages of silique development. These stages are vulnerable to the low temperature stress and therefore cold induction of kinases in these stages is possibly a mechanism to produce protective proteins by phosphorylation-mediated activation of the upstream TFs. During later stages of embryo development, LEA proteins start accumulating (Battaglia et al., 2008), and provide protection to the cells from ensuing desiccation. The core-cold-inducible subset has 4 members of AP2/EREBP family, which included one CBF (DREB1), two DREB2 members, and a single ERF. The core group also has 4 and 8 members of C2C2-constans like and C2C2-dof like TF family members, respectively, which are known to play regulatory roles in Arabidopsis in response to cold (Mikkelsen and Thomashow, 2009). The major family of TF enriched in core-cold (10 transcripts) as well as early stage cold-inducible (15 transcripts) subset is MYB TF family. The role of MYB genes in regulating genes of cold stress signaling pathway and in governing tolerance to cold stress is well documented (Vannini et al., 2004; Agarwal et al., 2006; Dai et al., 2007). CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), also belongs to MYB TF family and positively regulates expression of CBFs in Arabidopsis (Dong et al., 2011). Various transcripts coding for other TFs like NAC and WRKY were also identified from early stage cold-inducible subset. Transcript having homology to AtHSFA6b was induced by cold stress in later stages. These results are similar to the unpublished work from our lab where we found that Arabidopsis HSFA6b was inducible by low temperature.
Interestingly, it was found that a significant number of transcripts coding for transcriptional regulators (TRs) were inducible by low temperature. Some of the transcripts displayed inducibility in all the stages, whereas others were inducible only in the early stages. None of the TRs were cold-inducible exclusively in the later stages. One of the cold-inducible TR in early stages belonged to the GNAT (Gcn5-related N-acetyltransferase) family. Though it has been shown that CBF1 interacts with GCN5 (Stockinger et al., 2001; Mao et al., 2006), a direct role of GCN5 in promoting acetylation at COR promoters was not observed (Pavangadkar et al., 2010). Nonetheless, identification of a highly cold-inducible homolog of GNAT in B. juncea indicates that histone acetylation plays a critical role in modulating gene expression during cold stress. Involvement of histone modifications in cold stress is further supported by identification of a cold-inducible transcript of ARID (AT-rich interaction domain) family, members of which function as chromatin remodelers and histone demethylases (Tu et al., 2008; Lu and Tobin, 2011; Lin et al., 2014). Apart from TRs and TFs, multiple families of kinases induced by cold stress specifically during the early stages of embryo development were identified. The largest group of cold-inducible transcripts belonged to SnRK family. Members of SnRK families are involved in plant response to abiotic stresses (Ma et al., 2009; Umezawa et al., 2009; Vlad et al., 2009). Though, SnRK3 family members are routinely linked with abiotic stress response (Kim et al., 2003; Fujii and Zhu, 2009; Fujii et al., 2011), a recent study showed that the SnRK2 family member OST1 kinase phosphorylates ICE1, thereby increasing its stability and freezing tolerance in A. thaliana (Ding et al., 2015).
Quantitative PCR was employed to validate the expression pattern congregated from RNA-seq data. Nine transcripts representing all the three stages (cold-inducible at early stage, core-cold inducible and cold-inducible at late stage) were selected for qPCR analysis. The 4 transcripts from core-cold inducible category included COR27, COR14, putative β-amylase, and early responsive to dehydration. Similar to the digital expression data, these genes were found to be inducible by cold stress in all the stages of silique development. Mikkelsen and Thomashow (2009) identified COR27 along with COL1 to be cold-inducible and as well as regulated by circadian clock. COR14 is a cold stress regulated gene isolated in barley (Crosatti et al., 1995). Higher levels of COR14 were observed in cold-tolerant variety during cold acclimation as compared to susceptible variety (Cattivelli et al., 1995). β-amylase hydrolyzes α-1,4 glycosidic linkages of polyglucan chains at the non-reducing end to produce maltose which in turn protects membranes, proteins, and photosynthetic electron transport chain during severe temperature stress (Kaplan and Guy, 2004). Cold stresses and dehydration result in the up-regulation of stress-induced genes such as RD (responsive to dehydration), ERD (early responsive to dehydration), COR (cold regulated), LTI (low-temperature induced), and KIN (cold-inducible). During cold acclimation, DREBs bind to the promoter region of the downstream target genes such as RD29A, ERD10, COR15A, RD17, and Kin2, subsequently resulting in low temperature tolerance (Seki et al., 2002). Enhanced expression of COR, RD, and ERD have been observed in transgenic Arabidopsis plants overexpressing DREB1A when exposed to low temperature (Liu et al., 1998; Kasuga et al., 1999). Transcripts selected from “cold-inducible at early stage” subset include Nodulin MtN3 family protein, β-amyrin synthase (bAS), and Transcription factor ORG3-like. Members of nodulin MtN3 family are polytopic membrane proteins having MtN3/saliva domain and some of the members of these family like AtSWEET15 are cold-inducible, however their functional role in cold stress has not yet been characterized (Seo et al., 2011; Yuan and Wang, 2013). β-amyrin synthase is involved in biosynthesis of the secondary metabolite β-amyrin, which serves as an intermediate in the synthesis of triterpene glycosides associated with plant defense (Kemen et al., 2014). We found that bAS gene is mildly inducible by low temperature stress, which is in agreement with its previously reported stress-inducibility in Bruguiera gymnorrhiza and Glycirrhiza glabra (Basyuni et al., 2009; Nasrollahi et al., 2014).
Oxophytodienoate-reductase 3 and WRKY transcription factor 48 were shortlisted from “cold-inducible at late stage” subset. Oxophytodienoate-reductase 3 catalyzes reduction of double bonds in unsaturated aldehyde and ketone leading to production of jasmonic acid, which is involved in combating various abiotic and biotic stresses (Creelman and Mullet, 1995; Schaller et al., 2000). WRKY transcription factors are involved in plant responses to various biotic and abiotic stresses (Chen et al., 2012b). Specific members of Arabidopsis WRKY TF family are regulated during early stages of cold stress (Bakshi and Oelmuller, 2014). Several WRKY family genes were found to be responsive to cold stress in soyabean, Vitis vinifera, and barley (Marè et al., 2004; Zhou et al., 2008; Wang et al., 2014).
Attempts were made to study the expression of known cold signaling pathway genes. Primers, corresponding to sequences of CBF1, CAMTA3, SIZ1, ICE1, ICE2, SnRK2.6, and COR47 were designed and expression of the above genes in cold stress subjected B. juncea siliques was quantified using real time PCR. CBFs bind to CRT/DRE cis-elements and induce the expression of downstream cold-regulated genes (Gilmour et al., 1998; Steponkus et al., 1998; Cook et al., 2004). Levels of B. juncea CBF1 increased on exposure to both 6 h and 12 h of cold stress, however CBF1 transcripts accumulated to lower levels at 12 h of cold treatment as compared to 6 h in all the development stages. This is in sync with previous reports where early induction of upstream transcription factors was observed after cold stress in A. thaliana (Chinnusamy et al., 2010). Targets of CBFs include member of Cold Regulated Genes (CORs). One of the cold-regulated genes, COR47, was validated and its transcript was up-regulated at all the stages of silique development after imposition of cold stress. Doherty et al. (2009) showed the dependence of CBFs expression on CAMTA3 as cold-induced accumulation of CBFs was substantially reduced in camta3 lines. CAMTA3 was identified as a cold-induced gene in our transcriptome datasets, which was subsequently validated by qPCR. Moreover, a direct correlation in the levels of CAMTA3 and CBF1 support the previously proposed link between the two genes. At all the stages of silique development, expression of CAMTA3 was induced by cold stress except at 25 DAP where lower levels with respect to control were observed. Similarly, cold-induced expression of CBF1 was also considerably low at 25 DAP as compared to other stages. The decrease in CAMTA3 levels at 25 DAP did not necessarily obliterate cold-inducibility of CBF1, thereby indicating that in addition to CAMTA3 other TFs might also play a role in cold stress mediated regulation of CBF1. ICE1 is the master regulator of cold stress which enhances the expression of CBFs and downstream COR genes, eventually conferring freezing tolerance (Chinnusamy et al., 2003). ICE2, a homolog of ICE1, also induces expression of CBF1/DREB1B and confers increased freezing tolerance in Arabidopsis (Fursova et al., 2009). We observed increased expression of ICE1 and ICE2 in B. juncea siliques exposed to low temperature stress. SnRK2.6 also known as open stomata 1 (OST1) is a Ser/Thr protein kinase involved in ABA signaling. Recently, OST1 was shown to phosphorylate ICE1 to increase its stability. The knockout lines of OST1 were found to be defective in freezing tolerance whereas overexpression lines exhibited enhanced tolerance (Ding et al., 2015). Though OST1 protein was not previously observed to be cold-inducible our results suggests that SnRK2.6 is up-regulated on exposure to cold stress in B. juncea siliques. It will be interesting to study whether SnRK2.6 phosphorylates ICE1 and possibly ICE2 under cold stress in B. juncea siliques. Traditionally the genes involved in cold acclimation process have been used to impart both chilling as well as freezing tolerance in plants. With the help of RNA-seq we identified genes that are up-regulated by chilling stress and potentially these genes can be utilized to introduce frost tolerance during pod filling stages in B. juncea. In addition to identifying homologs of CBF1-mediated cold stress signaling pathway genes, we also identified genes whose products have not yet been implicated in providing protection against low temperature stress. Further functional studies involving these genes will not only help us in extending our understanding on cold stress signaling but also pave way for designing frost hardy plants.
Author contributions
MA conceived, designed and supervised the research work. AJ and SK-A participated in regular discussions for designing the wet lab experiments and analysis of the sequencing data. SS and VKR contributed equally to the study. SS performed self-pollination, stress treatment, RNA isolation, prepared RNA-seq libraries, and qPCR based expression analysis. VKR performed high throughput sequencing and analyzed the transcriptome data. SS, VKR, and MA wrote the manuscript. BJ performed the anatomical staging. AJ and SK-A provided critical inputs for data presentation. AJ, SK-A, SG, and AK critically reviewed the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research work in the laboratory is supported by grants from Department of Biotechnology (DBT, BT/01/COE/08/06), India and R&D grant from University of Delhi, Delhi, India. SS was supported by DBT, India. VR is thankful for research fellowships from Council of Scientific and Industrial research (CSIR), India. We thank Dr. Kishore Gaikwad, National Research Centre of Plant Biotechnology (NRCPB), New Delhi, India; Dr. Vinod Scaria, Dr. Sridhar Sivasubbu and Shamsudheen from Institute of Genomics and Integrative Biology (IGIB), Delhi, India for helping with the sequencing runs. We also thank Profs. Deepak Pental and Akshay Pradhan, Department of Genetics, UDSC for providing space to grow B. juncea plants.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00932
References
- Agarwal M., Hao Y., Kapoor A., Dong C. H., Fujii H., Zheng X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. 10.1074/jbc.M605895200 [DOI] [PubMed] [Google Scholar]
- Antikainen M., Griffith M. (1997). Antifreeze protein accumulation in freezing tolerant cereals. Physiol. Plant. 99, 423–432. 10.1111/j.1399-3054.1997.tb00556.x [DOI] [Google Scholar]
- Bakshi M., Oelmüller R. (2014). WRKY transcription factors: jack of many trades in plants. Plant Signal. Behav. 9:e27700. 10.4161/psb.27700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P., Jayavelu N. D., Rasmussen S., Nielsen H. B., Mundy J., Bones A. M. (2013). Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics 14:722. 10.1186/1471-2164-14-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basyuni M., Baba S., Inafuku M., Iwasaki H., Kinjo K., Oku H. (2009). Expression of terpenoid synthase mRNA and terpenoid content in salt stressed mangrove. J. Plant Physiol. 166, 1786–1800. 10.1016/j.jplph.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24. 10.1104/pp.108.120725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchereau A., Aziz A., Larher F., Martin-Tangui J. (1999). Polyamines and environmental challenges: recent development. Plant Sci. 140, 103–125. 10.1016/S0168-9452(98)00218-0 [DOI] [Google Scholar]
- Branca F., Cartea E. (2011). Brassica. Chapter 2, in Wild Crop Relatives: Genomic and Breeding Resources, Oilseeds, ed Kole C. (Berlin; Heidelberg: Springer-Verlag; ), 17–36. 10.1007/978-3-642-14871-2_2 [DOI] [Google Scholar]
- Cattivelli L., Crosatti C., Rizza F. (1995). Increasing in membrane stability and COR14 accumulation associated with cold-hardening in oats. J. Gen. Breed. 49, 333–338. [Google Scholar]
- Chen T. H., Murata N. (2008). Glycinebetaine: an effective protectant against abiotic stress in plants. Trends Plant Sci. 13, 499–505. 10.1016/j.devcel.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Chen T. W., Gan R. C., Wu T. H., Huang P. J., Lee C. Y., Chen Y. Y., et al. (2012a). FastAnnotator–an efficient transcript annotation web tool. BMC Genomics 13(Suppl. 7):S9. 10.1186/1471-2164-13-S7-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012b). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. 10.1016/j.bbagrm.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., et al. (2003). ICE1:a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. 10.1101/gad.1077503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J. K., Sunkar R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639, 39–55. 10.1007/978-1-60761-702-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D., Fowler S., Fiehn O., Thomashow M. F. (2004). A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 15243–15248. 10.1073/pnas.0406069101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman R. A., Mullet J. E. (1995). Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. U.S.A. 92, 4114–4119. 10.1073/pnas.92.10.4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosatti C., Soncini C., Stanca A. M., Cattivelli L. (1995). The accumulation of a cold-regulated chloroplastic protein is light-dependent. Planta 196, 458–463. 10.1007/BF00203644 [DOI] [PubMed] [Google Scholar]
- Dai X., Xu Y., Ma Q., Xu W., Wang T., Xue Y., et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 143, 1739–1751. 10.1104/pp.106.094532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li H., Zhang X., Xie Q., Gong Z., Yang S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32, 278–289. 10.1016/j.devcel.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Doherty C. J., Van Buskirk H. A., Myers S. J., Thomashow M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. 10.1105/tpc.108.063958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M. A., Farré E. M., Thomashow M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7241–7246. 10.1073/pnas.1103741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C., Bieniawska Z., Hincha D. K., Hannah M. A. (2008). Interactions between the circadian clock and cold-response in Arabidopsis. Plant Signal. Behav. 3, 593–594. 10.4161/psb.3.8.6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder N., O'brien T. P. (1968). Plant microtechnique: some principles and new methods. Am. J. Bot. 55, 123–142. 10.2307/2440500 [DOI] [Google Scholar]
- Fernandez P., Di Rienzo J., Fernandez L., Hopp H. E., Paniego N., Heinz R. A. (2008). Transcriptomic identification of candidate genes involved in sunflower responses to chilling and salt stresses based on cDNA microarray analysis. BMC Plant Biol. 8:11. 10.1186/1471-2229-8-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S. G., Cook D., Thomashow M. F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137, 961–968. 10.1104/pp.104.058354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Thomashow M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. 10.1105/tpc.003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K. A., Whitelam G. C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39, 1410–1413. 10.1038/ng.2007.3 [DOI] [PubMed] [Google Scholar]
- Fujii H., Verslues P. E., Zhu J. K. (2011). Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 1717–1722. 10.1073/pnas.1018367108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. 10.1073/pnas.0903144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fursova O. V., Pogorelko G. V., Tarasov V. A. (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429, 98–103. 10.1016/j.gene.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Gilmour S. J., Sebolt A. M., Salazar M. P., Everard J. D., Thomashow M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. 10.1104/pp.124.4.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Zarka D. G., Stockinger E. J., Salazar M. P., Houghton J. M., Thomashow M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. 10.1046/j.1365-313x.1998.00310.x [DOI] [PubMed] [Google Scholar]
- Griffith M., Antikainen M., Hon W. C., Pihakaski-Maunsbach K., Yu X. M., Chun J. U., et al. (1997). Antifreeze proteins in winter rye. Physiol. Plant. 100, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusta L. V., Wisniewski M., Nesbitt N. T., Gusta M. L. (2004). The effect of water, sugars, and proteins on the pattern of ice nucleation and propagation in acclimated and nonacclimated canola leaves. Plant Physiol. 135, 1642–1653. 10.1104/pp.103.028308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. protoc. 8, 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare P. D., Cress W. A., Van Staden J. (1998). Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 21, 535–553. 10.1046/j.1365-3040.1998.00309.x [DOI] [Google Scholar]
- Hausman J. F., Evers D., Thiellement H., Jouve L. (2000). Compared responses of poplar cuttings and in vitro raised shoots to short-term chilling treatments. Plant Cell Rep. 19, 954–960. 10.1007/s002990000229 [DOI] [PubMed] [Google Scholar]
- James A. B., Syed N. H., Bordage S., Marshall J., Nimmo G. A., Jenkins G. I., et al. (2012).Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant cell 24, 961–981. 10.1105/tpc.111.093948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janská A., Marsík P., Zelenková S., Ovesná J. (2010). Cold stress and acclimation—what is important for metabolic adjustment? Plant Biol. 12, 395–405. 10.1111/j.1438-8677.2009.00299.x [DOI] [PubMed] [Google Scholar]
- Kaplan F., Guy C. L. (2004). β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 135, 1674–1684. 10.1104/pp.104.040808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky M. (1965). A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27, 137–138. [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Kayum M. A., Jung H. J., Park J. I., Ahmed N. U., Saha G., Yang T. J., et al. (2015). Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol. Genet. Genomics 290, 79–95. 10.1007/s00438-014-0898-1 [DOI] [PubMed] [Google Scholar]
- Kemen A. C., Honkanen S., Melton R. E., Findlay K. C., Mugford S. T., Hayashi K., et al. (2014). Investigation of triterpene synthesis and regulation in oats reveals a role for beta-amyrin in determining root epidermal cell patterning. Proc. Natl. Acad. Sci. U.S.A. 111, 8679–8684. 10.1073/pnas.1401553111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. N., Cheong Y. H., Grant J. J., Pandey G. K., Luan S. (2003). CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15, 411–423. 10.1105/tpc.006858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause E., Dathe M., Wieprecht T., Bienert M. (1999). Noncovalent immobilized artificial membrane chromatography, an improved method for describing peptide-lipid bilayer interactions. J. chromatogr. A 849, 125–133. 10.1016/S0021-9673(99)00528-2 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Lee H., Xiong L., Zhu J. K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14, 1235–1251. 10.1105/tpc.010433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Thomashow M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 15054–15059. 10.1073/pnas.1211295109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. C., Lim M. H., Kim J. A., Lee S. I., Kim J. S., Jin M., et al. (2008). Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24K oligo microarray. Mol. Cells 26, 595–605. [PubMed] [Google Scholar]
- Levitt J. (1980). Responses of Plants to Environmental Stress, 2nd Edn. New York, NY: Academic Press; Vol. 1. [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Song W., Bi X., Zhao J., Huang Z., Li Z., et al. (2014). Recent advances in the ARID family: focusing on roles in human cancer. Onco Targets Ther. 7, 315–324. 10.2147/OTT.S57023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA-binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression in Arabidopsis. Plant Cell 10, 1391–1406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu S. X., Tobin E. M. (2011). Chromatin remodeling and the circadian clock: Jumonji C-domain containing proteins. Plant Signal. Behav. 6, 810–814. 10.4161/psb.6.6.15171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Maibam P., Nawkar G. M., Park J. H., Sahi V. P., Lee S. Y., Kang C. H. (2013). The influence of light quality, circadian rhythm, and photoperiod on the CBF-mediated freezing tolerance. Int. J. Mol. Sci. 14, 11527–11543. 10.3390/ijms140611527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Pavangadkar K. A., Thomashow M. F., Triezenberg S. J. (2006). Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta 1759, 69–79. 10.1016/j.bbaexp.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Marè C., Mazzucotelli E., Crosatti C., Francia E., Stanca A. M., Cattivelli L. (2004). Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 55, 399–416. 10.1007/s11103-004-0906-7 [DOI] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., et al. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38, 982–993. 10.1111/j.1365-313X.2004.02100.x [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Thomashow M. F. (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60, 328–339. 10.1111/j.1365-313X.2009.03957.x [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. 10.1007/s00299-013-1418-1 [DOI] [PubMed] [Google Scholar]
- Nasrollahi V., Mirzaie-asl A., Piri K., Nazeri S., Mehrabi R. (2014). The effect of drought stress on the expression of key genes involved in the biosynthesis of triterpenoid saponins in liquorice (Glycyrrhiza glabra). Phytochemistry 103, 32–37. 10.1016/j.phytochem.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T. P., McCully M. E. (1981). The Study of Plant Structure: Principles and Selected Methods. Melbourne, VIC: Termarcarphi Pty. Ltd. [Google Scholar]
- Patel R. K., Jain M. (2012). NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619. 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavangadkar K., Thomashow M. F., Triezenberg S. J. (2010). Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in Arabidopsis. Plant Mol. Biol. 74, 183–200. 10.1007/s11103-010-9665-9 [DOI] [PubMed] [Google Scholar]
- Prakash S., Hinata K. (1980). Taxonomy, Cytogenetics And Origin of Crop Brassicas: A Review (Opera botanica). Vol. 55 Sweden: Lund Botanical Society. [Google Scholar]
- Prasada Rao G. S. L. H. V., Rao V. U. M., Rao G. G. S. N. (2010). Climate Change and Agriculture Over India. Delhi: P. H. I. Publisher. [Google Scholar]
- Preston J. C., Hileman L. C. (2013). Functional evolution in the plant SQUAMOSA-promoter binding protein-like (SPL) gene family. Front. Plant Sci. 4:80. 10.3389/fpls.2013.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani M. A., Maruyama K., Abe H., Khan M. A., Katsura K., Ito Y., et al. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133, 1755–1767. 10.1104/pp.103.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakow G. (2004). Species origin and economic importance of Brassica, in Brassica, eds Pua E.-C., Douglas C. (Berlin; Heidelberg: Springer; ), 3–11. [Google Scholar]
- Robinson M. D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F., Biesgen C., Müssig C., Altmann T., Weiler E. W. (2000). 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210, 979–984. 10.1007/s004250050706 [DOI] [PubMed] [Google Scholar]
- Schwartz S. H., Tan B. C., Gage D. A., Zeevaart J. A., McCarty D. R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. 10.1126/science.276.5320.1872 [DOI] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., et al. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. 10.1105/tpc.13.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. 10.1046/j.1365-313X.2002.01359.x [DOI] [PubMed] [Google Scholar]
- Seo P. J., Park J. M., Kang S. K., Kim S. G., Park C. M. (2011). An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233, 189–200. 10.1007/s00425-010-1293-8 [DOI] [PubMed] [Google Scholar]
- Shekhawat K., Rathore S. S., Premi O. P., Kandpal B. K., Chauhan J. S. (2012). Advances in agronomic management of Indian Mustard (Brassica juncea (L.) Czernj. Cosson): an overview. Int. J. Agron. 14:408284 10.1155/2012/408284 [DOI] [Google Scholar]
- Shi H., Ye T., Zhong B., Liu X., Jin R., Chan Z. (2014). AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 203, 554–567. 10.1111/nph.12812 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (1996). Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 7, 161–167. 10.1016/S0958-1669(96)80007-3 [DOI] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N. E., Heger A., Ponting C. P. (2014). Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 15, 121–132. 10.1038/nrg3642 [DOI] [PubMed] [Google Scholar]
- Solanke A. U., Sharma A. K. (2008). Signal transduction during cold stress in plants. Physiol. Mol. Biol. Plants 14, 69–79. 10.1007/s12298-008-0006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus P. L., Uemura M., Joseph R. A., Gilmour S. J., Thomashow M. F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 95, 14570–14575. 10.1073/pnas.95.24.14570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E. J., Mao Y., Regier M. K., Triezenberg S. J., Thomashow M. F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. 10.1093/nar/29.7.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur P., Kumar S., Malik J. A., Berger J. D., Nayyar H. (2010). Cold stress effects on reproductive development in grain crops: an overview. Environ. Exp. Bot. 67, 429–443. 10.1016/j.envexpbot.2009.09.004 [DOI] [Google Scholar]
- Thomashow M. F. (1998). Role of cold-responsive genes in plant freezing tolerance. Plant Phys. 118, 1–8. 10.1104/pp.118.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S., Teng Y. C., Yuan C., Wu Y. T., Chan M. Y., Cheng A. N., et al. (2008). The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 15, 419–421. 10.1038/nsmb.1400 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593. 10.1073/pnas.0907095106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C., Locatelli F., Bracale M., Magnani E., Marsoni M., Osnato M., et al. (2004). Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 37, 115–127. 10.1046/j.1365-313X.2003.01938.x [DOI] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant cell 21, 3170–3184. 10.1105/tpc.109.069179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhu W., Fang L., Sun X., Su L., Liang Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14:103. 10.1186/1471-2229-14-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick S. I., Black L. D. (1991). Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) -chloroplast genome and cytodeme congruence. TAG Theor. Appl. Genet. 82, 81–92. 10.1007/bf00231281 [DOI] [PubMed] [Google Scholar]
- Wisniewski J., Orosz A., Allada R., Wu C. (1996). The C-terminal region of Drosophila heat shock factor (HSF) contains a constitutively functional transactivation domain. Nucleic Acids Res. 24, 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wang S. (2013). Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 6, 665–674. 10.1093/mp/sst035 [DOI] [PubMed] [Google Scholar]
- Zhou Q. Y., Tian A. G., Zou H. F., Xie Z. M., Lei G., Huang J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- Zhu J., Dong C. H., Zhu J. K. (2007). Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 10, 290–295. 10.1016/j.pbi.2007.04.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.