ABSTRACT
Bacterial resistance to antibiotics is precipitating a medical crisis, and new antibacterial strategies are being sought. Hypothesizing that a growth-restricting strategy could be used to enhance the efficacy of antibiotics, we determined the effect of FDA-approved iron chelators and various antibiotic combinations on invasive and multidrug-resistant extraintestinal pathogenic Escherichia coli (ExPEC), the Gram-negative bacterium most frequently isolated from the bloodstreams of hospitalized patients. We report that certain antibiotics used at sublethal concentrations display enhanced growth inhibition and/or killing when combined with the iron chelator deferiprone (DFP). Inductively coupled plasma optical emission spectrometry reveals abnormally high levels of cell-associated iron under these conditions, a response that correlates with an iron starvation response and supraphysiologic levels of reactive oxygen species (ROS). The high ROS level is reversed upon the addition of antioxidants, which restores bacterial growth, suggesting that the cells are inhibited or killed by excessive free radicals. A model is proposed in which peptidoglycan-targeting antibiotics facilitate the entry of lethal levels of iron-complexed DFP into the bacterial cytoplasm, a process that drives the generation of ROS. This new finding suggests that, in addition to restriction of access to iron as a general growth-restricting strategy, targeting of cellular pathways or networks that selectively disrupt normal iron homeostasis can have potent bactericidal outcomes.
IMPORTANCE The prospect that common bacteria will become resistant to all antibiotics is challenging the medical community. In addition to the development of next-generation antibiotics, new bacterial targets that display cytotoxic properties when altered need to be identified. Data presented here demonstrate that combining subinhibitory levels of both iron chelators and certain antibiotics kills pathogenic Escherichia coli. The mechanism of this effect is the production of supraphysiologic levels of reactive oxygen species, likely powered by the excessive import of iron. These findings were consistent for both clinically relevant and no longer clinically used antibiotics and may extend to Staphylococcus aureus as well.
INTRODUCTION
Antibiotics are compounds that inhibit or kill bacteria and may have saved more lives than any other medical intervention, aside from vaccination (1). However, the development of strains resistant to antibiotics is precipitating a medical crisis. It is estimated that each year in the United States there are 900,000 cases of antibiotic-resistant infections, with an estimated cost of over 20 billion U.S. dollars (2). Several factors contribute to resistance, including the over- and misuse of these drugs (which generates evolutionary pressure that selects for resistant strains), horizontal gene transfer (which allows elements that confer resistance to be transferred among species or genera), and the high level of genetic diversity generated from de novo mutation (which creates more fit members when they are faced with antibiotics [3, 4]). Numerous strategies have been employed to combat the resistance problem, including the reduced use of antibiotics in livestock, the development of next-generation antibiotics with little established resistance, the use of biologics such as phage to kill bacteria or probiotics to stimulate the host immune system, and the combination of different antibiotics into a type of killing cocktail (3, 5). Most antibiotics function by disrupting one of three critical cellular functions, including the inhibition of DNA replication (e.g., quinolones), the inhibition of protein biosynthesis (e.g., aminoglycosides), and the inhibition of cell wall biosynthesis (e.g., β-lactams) (5). In addition to finding new compounds, there is also a great need to discover new targets and mechanisms to kill bacterial cells that differ from traditional approaches.
Nutritional immunity is the term used to describe the host's sequestration of critical nutrients to prevent the growth and replication of bacteria during an active infection. A component of nutritional immunity is the sequestration of metals, especially iron. Bacterial replication is absolutely dependent on the acquisition of iron from host sources. The disruption of bacterial iron metabolism has dramatic negative consequences on virulence in vitro and in vivo (6–9). Because an estimated 30% of all enzymes require metals as a cofactor and iron is critical for such cellular events as DNA biosynthesis, the trichloroacetic acid cycle, oxidative stress defense, and energy transduction (7, 9, 10), targeting of iron-dependent processes represents a viable strategy for antimicrobial development. Indeed, there are a growing number of studies evaluating the use of iron chelators as antibacterials, with efficacy demonstrated in some cases (11–13) but not others (14, 15). Inspired by the way mammals restrict bacterial growth to prevent infection (16, 17), we report here that the combined use of iron chelators and sublethal concentrations of some antibiotics generates a potent response that kills the cells of a model Gram-negative blood pathogen (extraintestinal pathogenic Escherichia coli [ExPEC]). Investigation of the mechanism behind this response links it to a supraphysiologic elevation in the levels of cellular iron content coupled to an iron starvation response. This state, in turn, promotes the development of high cellular levels of reactive oxygen species (ROS) that ultimately kill the cell.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
ExPEC CP9 (18) and methicillin (MET)-resistant Staphylococcus aureus TCH1516 (19) were isolated from hospitalized patients and were kind gifts of James Johnson (University of Minnesota) and Sarah Highlander (J. Craig Venter Institute). A chloramphenicol (CHL) resistance gene was inserted into CP9 for selection (20). Clinical E. coli isolates (ELZ4013, ELZ4045, ELZ4046, ELZ4234, ELZ4251, and ELZ4486) (21) were generously provided by Lynn Zechiedrich (Baylor College of Medicine, Houston, TX). All experiments were performed with brain heart infusion (BHI) medium or cation-adjusted Mueller-Hinton broth (CAMHB; both from Teknova). The following antibiotics and reagents were used at the concentrations indicated in the figure legends: ampicillin (AMP; from Amresco), cefotaxime (CTX; from Chem-Impex), CHL (from EMD), ciprofloxacin (CIP; from Cellgro), daptomycin (from SellechChem), gentamicin (GEN; from Chem-Impex), levofloxacin (LVX; from TCI), meropenem (MEM; from TCI) MET (from USP), spectinomycin (SPT; from MP), trimethoprim (TMP; from Chem-Impex), vancomycin (VAN; from RPI), deferiprone (from Sigma-Aldrich), deferoxamine (from Sigma-Aldrich), 2,2′-dipyridyl (from Alfa Aesar), ortho-nitrophenyl-β-d-galactopyranoside (ONPG; from Thermo Fisher Scientific), 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA; from Calbiochem), glutathione (from Calbiochem), l-ascorbic acid (from G-Biosciences), propidium iodide (PI; from Fisher Scientific), Tween 20 (from Sigma-Aldrich), and Triton X-100 (from Sigma-Aldrich).
Growth of bacteria. (i) Endpoint assays using OD to measure growth.
All growth assays were performed with 96-well plates and 200 μl of BHI medium at a starting optical density at 600 nm (OD600) of 0.01. Antibiotics (or detergents), chelators, iron or other metals, DCFH-DA, and antioxidants were added to wells at the times and concentrations indicated in the figure legends for each particular experiment. The cultures were then maintained under continuous shaking for 18 h at 37°C, and the OD600 was recorded with a plate reader (BioTek synergy HT) at 30-min intervals. Bacterial growth was quantified by determining the difference in OD between the start and end of the experiment, calculated as OD600-18 h − OD600-0 h.
(ii) Time-kill assays using CFU enumeration on LB agar.
E. coli CP9 or S. aureus TCH1516 was grown in 5 ml of BHI medium overnight at 37°C with vigorous shaking. The next day, bacteria were subcultured at a starting OD600 of 0.01 in 5 ml of CAMHB supplemented with an antibiotic, DFP, or an antibiotic with DFP or not supplemented at all. The concentrations of the antibiotics and DFP were as follows: DFP, 150 μg/ml for CP9 and 900 μg/ml for TCH1516; AMP, 2.5 μg/ml for CP9 and 500 μg/ml for TCH1516; CTX, 62.5 ng/ml for CP9; CHL, 250 μg/ml for CP9; LVX, 0.1 μg/ml for TCH1516; MET, 1 mg/ml for CP9; VAN, 250 μg/ml for CP9. At t = 0, 2, 5, 8, and 24 h (for CTX, additional measurements were made at 16, 18, 20, and 22 h), the CFU count of each sample was determined by 10-fold serial dilutions and drip plating of 20 μl of each diluent. Synergy was defined as a 2-log10 difference in the number of CFU per milliliter between the combination and its most active constituent after 24 h, as well as a 2-log10 difference between the combination and the starting inoculum, as defined by National Committee for Clinical Laboratory Standards (NCCLS) (22).
(iii) Growth of E. coli clinical strains.
All growth assays were performed as described above for the “endpoint assays” in the presence of VAN, DFP, or both. The concentrations of the reagents used were as follows: ELZ4013, 100 μg/ml VAN and 125 μg/ml DFP; ELZ4045, 250 μg/ml VAN and 300 μg/ml DFP; ELZ4046, 500 μg/ml VAN and 300 μg/ml DFP; ELZ4234, 400 μg/ml VAN and 300 μg/ml DFP; ELZ4251, 2 mg/ml VAN and 125 μg/ml DFP; ELZ4486, 500 μg/ml VAN and 300 μg/ml DFP.
MIC90 determinations.
MIC90s were determined by adapting the NCCLS standard broth microdilution method (23) using BHI medium with a starting OD600 of 0.01 and an inoculum volume of 2 μl.
Fur reporter assay.
The PryhB-lacZ reporter plasmid (pAML23) (24) and a vector-only control (pQF50) (25) (both generously provided by Shelley Payne, University of Texas at Austin) were transformed into E. coli CP9 by the heat shock method (26). E. coli carrying each plasmid was grown with aeration at 37°C until mid-log phase (8 h for CTX plus DFP and VAN plus DFP) was reached. Supernatant were separated from cell pellets, and both fractions were analyzed for β-galactosidase activity as described by Miller (27) with ONPG as the substrate; 1 Miller unit = 1,000 × (OD420 − 1.75 × OD550)/(t × v × OD600), where t = 15 min and v = 0.1 ml.
Measurement of total cellular iron content.
E. coli CP9 was grown in the presence of antibiotic, DFP, or antibiotic with DFP as described above, except that the experiment was performed with 5 ml of culture (150 ml for antibiotic plus DFP) to obtain enough cells to measure the iron content. After 18 h, cells were collected, washed three times with phosphate-buffered saline (PBS), serially diluted, and plated on LB agar to determine the CFU count of each sample. About 5 × 109 cells from each sample were lysed in concentrated HNO3 and analyzed by inductively coupled plasma optical emission spectrometry (ICP-OES; Agilent 725; Department of Earth and Atmospheric Sciences at the University of Houston) (28). Supernatants (1-ml aliquots) of the cells grown in BHI medium were also collected, treated with concentrated HNO3, and analyzed for iron concentrations.
Measurement of intracellular ROS levels.
E. coli CP9 was seeded at a starting OD600 of 0.01 under various conditions (see the legend to Fig. 6) in the presence or absence of the antioxidants glutathione and ascorbate. Cultures were then grown with shaking at 37°C for 18 h, and then DCFH-DA (500 μM) was added. Bacteria continued to grow for another 2 h with shaking at 37°C, after which both the OD600 and fluorescence (excitation wavelength, 485 ± 20 nm; emission wavelength, 528 ± 20 nm) were measured every 15 min (readings at 2 h after the addition of DCFH-DA are reported) (29). Data were recorded as the amount of fluorescence per cell.
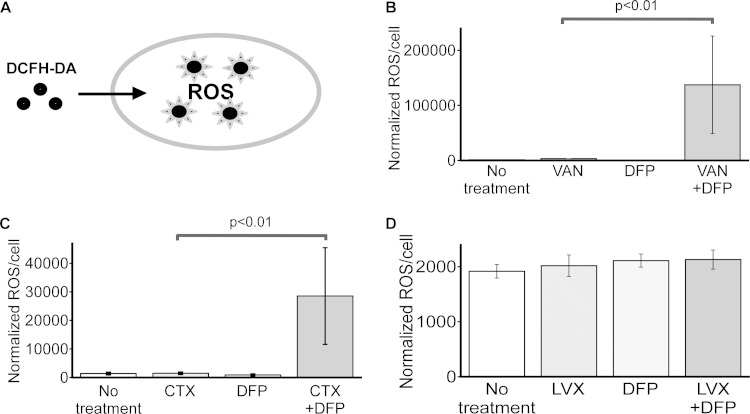
FIG 6.
Antibiotics and iron restriction promote increased intracellular ROS levels in E. coli. (A) Diagram depicting the reaction of DCFH-DA with ROS to generate fluorescence. (B to D) E. coli CP9 was cultured in BHI medium in the presence or absence of an antibiotic (VAN at 250 μg/ml, CTX at 62.5 ng/ml, or SPT at 10 μg/ml), DFP (150 μg/ml), or both for 18 h, and intracellular ROS levels were measured by using DCFH-DA as described in Materials and Methods. Data are the mean values and standard deviations from three independent experiments. P values were determined by Student's t test.
Membrane permeability assay.
Bacteria grown overnight were subcultured 1:50 in fresh medium until the OD600 reached 0.4. Bacteria were treated with nothing, an antibiotic (VAN at 250 μg/ml or CTX at 62.5 ng/ml), DFP (150 μg/ml), or both. At different time points (t = 0, 0.5, 1, 2, 4, 6, and 24 h), 1 ml of culture was resuspended in PBS, mixed with PI (final concentration, 10 μM; Fisher), and incubated at room temperature for 10 min. Alternatively, cells were washed three times after PI addition with no change in the experimental outcome. After PI addition, cells were subjected to OD600 and fluorescence (excitation wavelength, 530 ± 20 nm; emission wavelength, 590 ± 35 nm) measurements (30). Data were normalized to cell numbers.
Statistics.
All measurements represent the mean and standard deviation of at least three independent experiments. Significance (P value) was determined by Student's t test.
RESULTS
Iron restriction enhances the efficacy of some antibiotics.
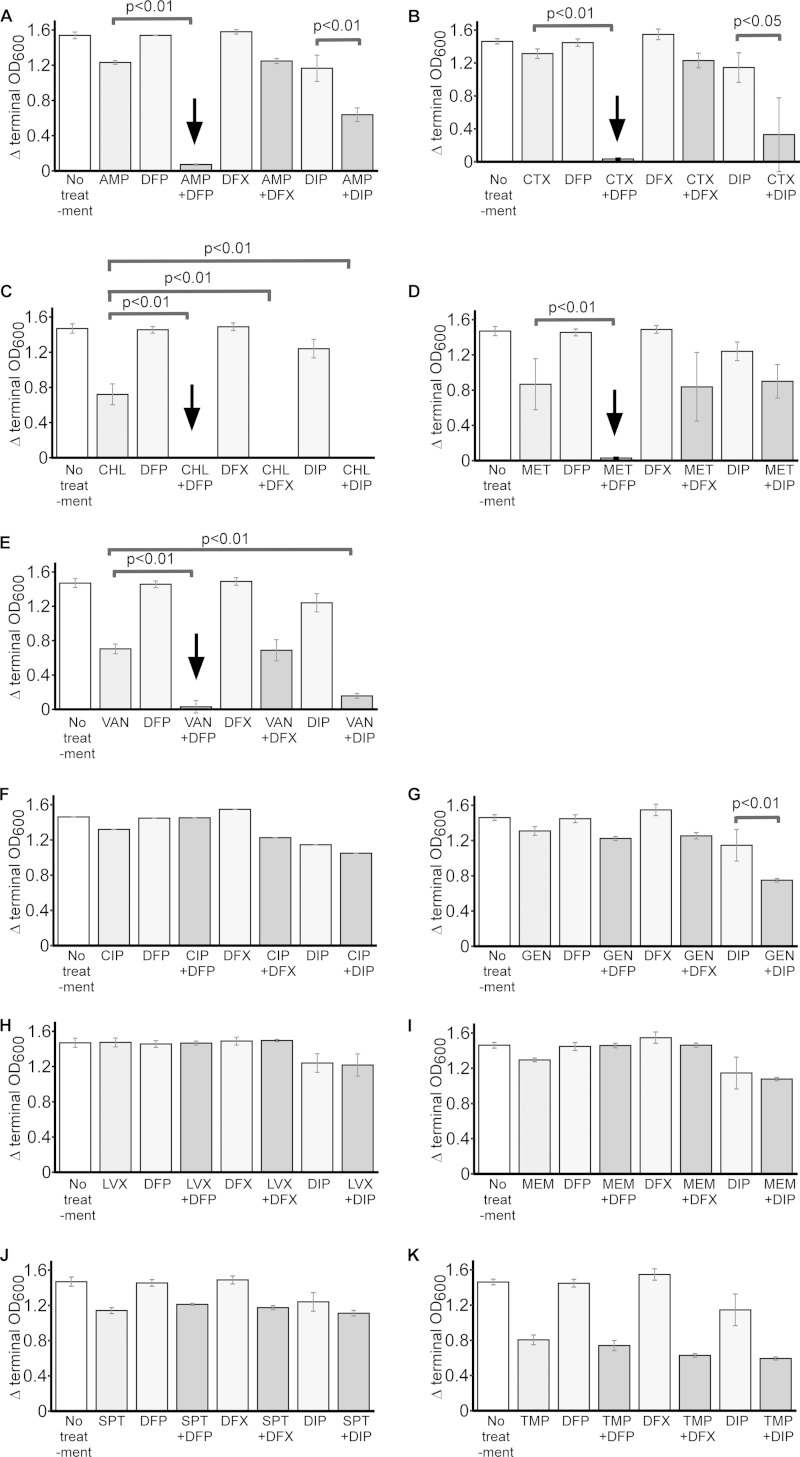
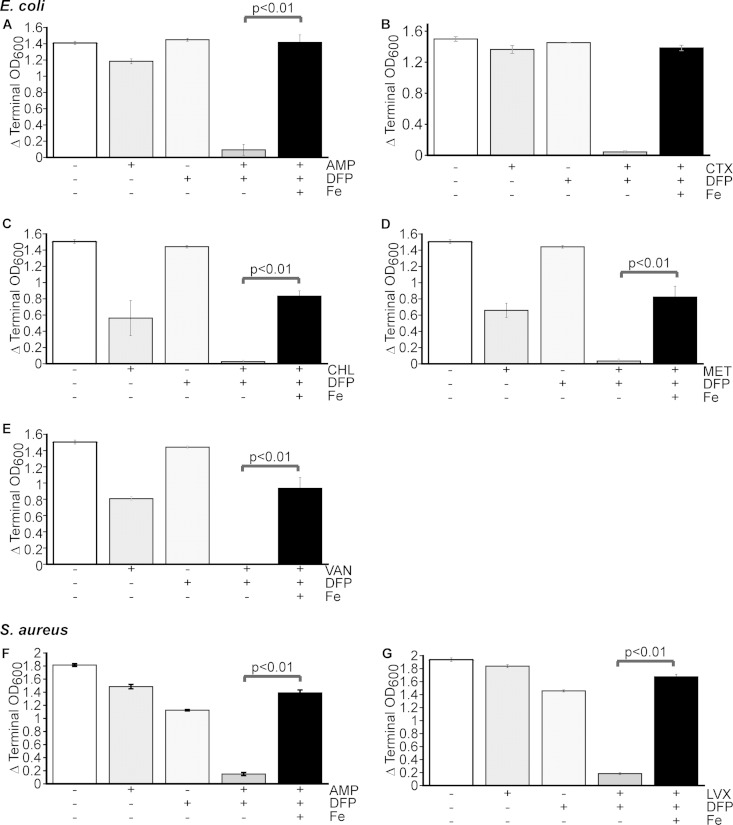
We sought to determine if restricting access to iron would enhance the effectiveness of antibiotics. Therefore, we determined the MIC90s of 11 different antibiotics representing all of the major antibiotic classes against ExPEC strain CP9, a common cause of bacteremia that harbors multidrug resistance (31) (see Table S1 in the supplemental material). Seven of the antibiotics were also tested against S. aureus, another serious multidrug-resistant species (see Table S1). ExPEC and S. aureus are the Gram-negative and Gram-positive species, respectively, most frequently isolated from the blood of infected patients (32). These bacteria were also tested for sensitivity to the FDA-approved iron chelators DFP and DFX and an iron chelator commonly used in the research laboratory, DIP. DFP and DFX are compounds that are used to treat iron overload in humans and also demonstrate the ability to restrict the growth of bacteria in vitro (12). The MIC90s of all of the antibiotics and chelators tested could be reliably determined (see Table S1). We next evaluated the growth of each species when it was exposed to various combinations of antibiotics and chelators. Five of the 11 antibiotics, when combined with an iron chelator at sublethal levels, demonstrated a negative effect on the growth of ExPEC strain CP9 that was greater than that of each individual compound or the untreated control (Fig. 1A to E). This included the clinically used antibiotic CTX, as well as AMP, CHL, MET, and VAN. Another, SPT, also demonstrated this effect but only at concentrations that were close to the MIC90 (data not shown). Interestingly, although DFX and DIP showed some effect when combined with antibiotics, DFP demonstrated significant efficacy in each of the five cases and thus was chosen for additional studies to examine the mechanism of this effect. Of note, a similar effect on S. aureus was observed but only with two of the seven antibiotics tested (AMP and LVX) (see Fig. S1 in the supplemental material). The enhanced inhibitory effect observed when antibiotics were combined with DFP seems to be related to the iron status of the cell or medium, because the addition of iron restored bacterial growth under these conditions (Fig. 2). Taken together, these data suggest that DFP and certain antibiotics have potent growth inhibition activity when combined, although they are not inhibitory when used individually. The data also suggest that this effect is dependent on E. coli's access to iron.
FIG 1.
Iron chelators and antibiotics restrict the growth of pathogenic E. coli. ExPEC strain CP9 was cultured in BHI medium in the presence or absence of antibiotics, iron chelators, or both, and the OD600 was recorded for 18 h (OD600-18 h − OD600-0 h). The iron chelators used were DFP (150 μg/ml), DFX (1 mg/ml), and DIP (200 μM). The antibiotics used were AMP (2.5 μg/ml) (A), CTX (62.5 ng/ml) (B), CHL (250 μg/ml) (C), MET (1 mg/ml) (D), VAN (250 μg/ml) (E), CIP (15 ng/ml) (F), GEN (6.25 μg/ml) (G), LVX (0.01 μg/ml) (H), MEM (16 ng/ml) (I), SPT (10 μg/ml) (J), and TMP (0.4 μg/ml) (K). Data are the mean values and standard deviations from at least six separate replicates. P values were determined by Student's t test. Panels A to E indicate the antibiotics that had negative effects on bacterial growth when combined with an iron chelator, and the arrows show the effect of DFP.
FIG 2.
Antibiotic-DFP-mediated growth inhibition is iron specific. E. coli CP9 (A to E) and S. aureus TCH1516 (F, G) were cultured in BHI medium in the presence or absence of an antibiotic, DFP, or both and an antibiotic plus DFP with added iron (800 μM FeSO4). DFP was added at a final concentration of 150 μg/ml for CP9 and 900 μg/ml for TCH1516. Data are the mean values and standard deviations from three independent experiments. P values were determined by Student's t test. (A) AMP (2.5 μg/ml); (B) CTX (62.5 ng/ml); (C) CHL (250 μg/ml); (D) MET (1 mg/ml); (E) VAN (250 μg/ml); (F) AMP (500 μg/ml); (G) LVX (0.1 μg/ml).
The combination of DFP and an antibiotic is cytotoxic.
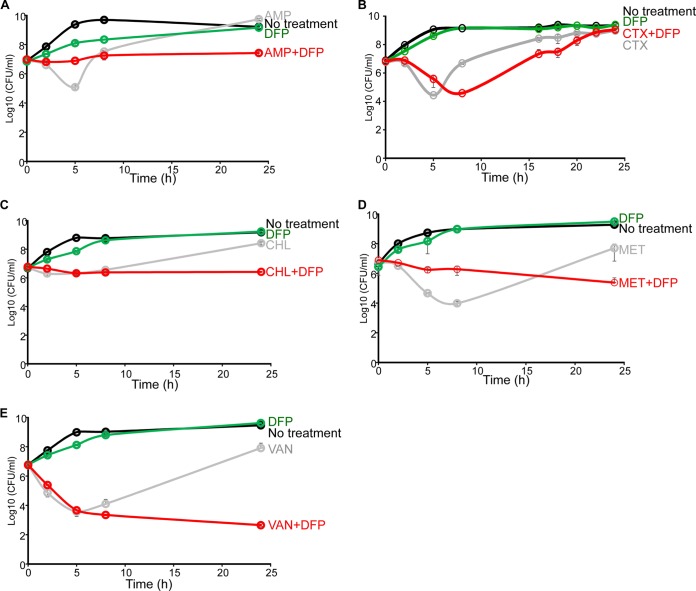
We next investigated the mechanism by which this combination of compounds with different modes of action achieved its negative effect on bacterial cells. Time-kill assays, as defined by the National Committee for Clinical Laboratory Standards, are a favored methodology for distinguishing between bacteriostatic and bactericidal activities, as well as if an effect of this type is synergistic or additive (33). When examined in this fashion, all of the combinatorial antibiotics except CTX for E. coli (four for E. coli and two for S. aureus), when in the presence of DFP, reduced the cell number below that of the starting inoculum over time and proved more potent at reducing the overall density of the population relative to each individual drug at 24 h (Fig. 3; see Fig. S2 in the supplemental material; P < 0.01, except for CTX-DFP). Of note, the CTX-DFP combination demonstrated antibacterial activity for most of the growth phase but returned to individually treated levels at 24 h (P = 0.56). The VAN-DFP combination could technically be defined as synergy (see Materials and Methods). In fact, when we assessed the effects of VAN plus DFP on six ExPEC isolates from the blood of patients at Texas Children's Hospital (all of which demonstrated diverse antibiotic resistance profiles [21]), in each case, the combination treatment was more effective than each compound given alone (see Fig. S3 in the supplemental material; P < 0.01). These data indicate that the combination of an antibiotic and a chelator, especially DFP, which are not as effective when used individually, has a powerful cytotoxic effect. ExPEC was chosen to investigate the significance of this mechanism throughout this study. For a summary of the effects of all of the antibiotics that were tested in combination with DFP on the growth of these bacteria, see Table S2 in the supplemental material.
FIG 3.
Time-kill assays with antibiotics and DFP that demonstrate enhanced efficacy against E. coli. E. coli CP9 was cultured in 5 ml of BHI medium overnight at 37°C with vigorous shaking. After 18 h, bacteria were subcultured at a starting OD600 of 0.01 in 5 ml of CAMHB supplemented with an antibiotic (panel A, AMP at 2.5 μg/ml; panel B, CTX at 62.5 ng/ml; panel C, CHL at 250 μg/ml; panel D, MET at 1 mg/ml; panel E, VAN at 250 μg/ml) or with an antibiotic with DFP at 150 μg/ml or not supplemented at all. The CFU counts of the cultures were determined by 10-fold serial dilution and drip plating of 20 μl of each diluent at the times indicated. Data are the mean values and standard deviations from three independent experiments.
Iron chelation and an antibiotic generate an iron starvation response.
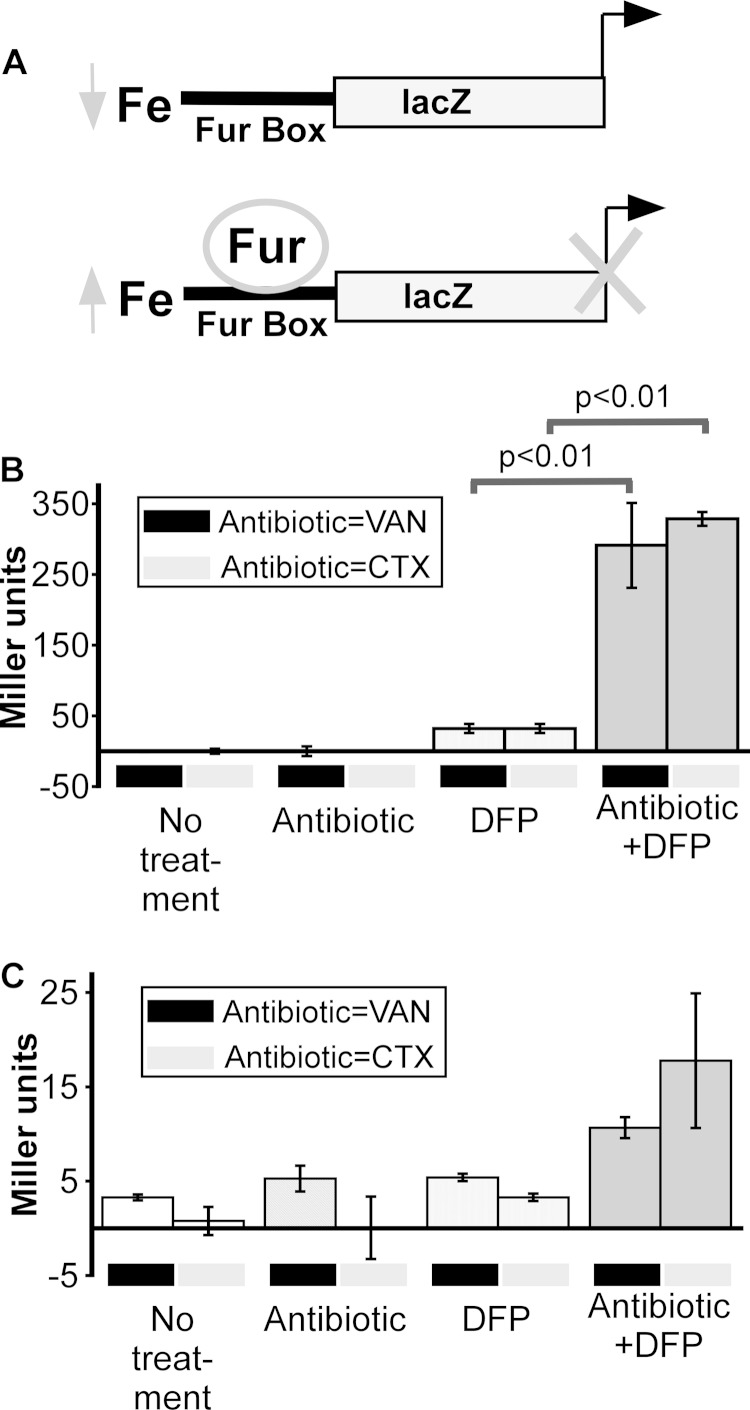
We reasoned that the observed combined effect of an antibiotic and a chelator on bacterial cells was related to an inability of the cells to acquire enough iron and thus measured the transcriptional activity of Fur. Fur is a master regulator of iron-responsive genes and is active as a transcriptional repressor when intracellular ferrous iron (Fe2+) levels are high but inactive (not repressive) when ferrous iron levels are low (34, 35). The reporter plasmid pAML23 (PryhB-lacZ), which contains a canonical Fur DNA binding element (Fur box) from ryhB cloned upstream of a promoterless lacZ gene, is well accepted as a reporter of Fur activity (Fig. 4A) (24, 36). The expression of lacZ, with the readout being β-galactosidase activity, is inversely correlated with the Fur activity in that when high levels of β-galactosidase activity are observed, Fur does not act as a repressor (24, 36). Because of the number of experiments that would need to be performed to examine the mechanism of this combined effect, we restricted additional efforts to two antibiotics: CTX, because of its current use as a β-lactam antibiotic in the hospital setting, and VAN, a former first-line antibiotic used to treat Gram-positive infections and to which there is now widespread resistance (37, 38). ExPEC harboring PryhB-lacZ was incubated in the presence or absence of each antibiotic either alone or in combination with DFP, and the levels of β-galactosidase activity were measured in both the cells and culture supernatants (Fig. 4B and C). Cells harboring the reporter plasmid had detectable β-galactosidase activity in the presence of DFP (suggestive of removal of Fur repression and a low level of access to intracellular ferrous iron). Surprisingly, ExPEC demonstrated significantly high β-galactosidase activity (5- to 10-fold, suggestive of even lower levels of access to ferrous iron) when these cells were cultured in the presence of both DFP and either CTX or VAN (Fig. 4B, far right panels). Very little signal, relative to that in the cells, was associated with the culture supernatants (Fig. 4C), which suggests that an intracellular response was being examined. This finding suggests that ferrous iron levels in E. coli treated with both an antibiotic and DFP are either extremely low or that Fur is unable to bind iron because it is sequestered in another form. In both cases, the net effect is that Fur is not active as a repressor of the reporter under combinatorial conditions.
FIG 4.
Antibiotics stimulate an iron starvation response. (A) Diagram depicting the function of PryhB-lacZ as an intracellular reporter of the response of bacteria to iron depletion. When the intracellular free iron levels are low, Fur will not repress the transcription of lacZ. (B, C) E. coli CP9 was cultured in BHI medium in the presence or absence of an antibiotic (VAN at 250 μg/ml or CTX at 62.5 ng/ml), DFP (150 μg/ml), or both, and β-galactosidase activity was measured at 8 h in both the cell pellet fraction (B) and the culture supernatant fraction (C). Data are the mean values and standard deviations from three independent experiments. P values were determined by Student's t test.
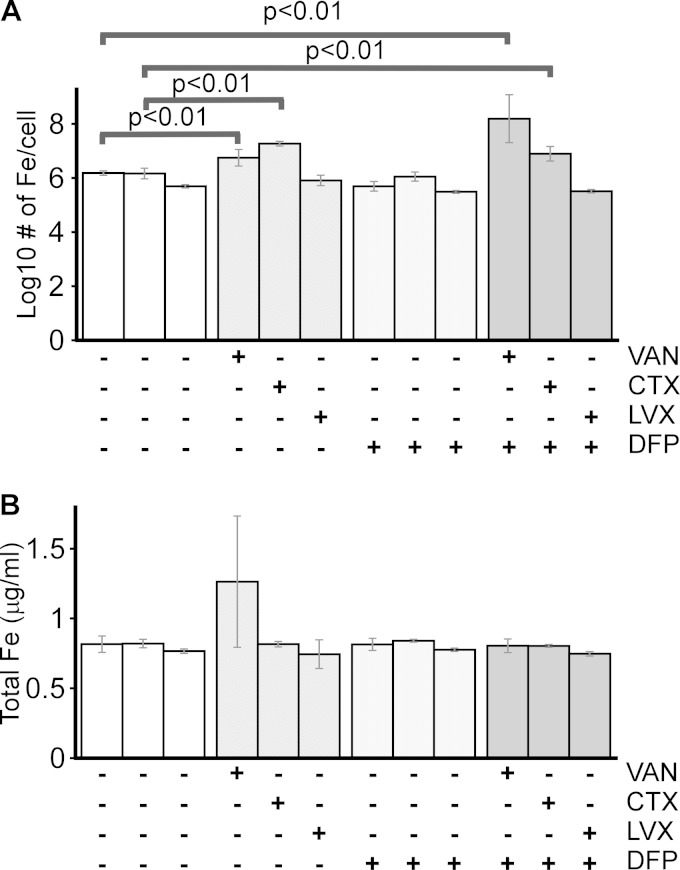
Iron chelation and antibiotics promote high levels of cell-associated iron.
Surprised by the iron starvation response observed when E. coli is treated with DFP and antibiotics, we sought a better understanding of the state of cellular iron under these conditions. Therefore, we used ICP-OES to measure the levels of iron in ExPEC treated with only DFP, an antibiotic, or both DFP and an antibiotic. Surprisingly, cells treated with VAN or CTX in combination with DFP had nearly 100 and 10 times, respectively, the levels of intracellular iron in cells treated with only the antibiotic or the chelator or not treated at all (Fig. 5A). This was not true of cells treated with LVX, an antibiotic that did not show a negative effect on ExPEC growth when combined with DFP (Fig. 1H and 5A, far right column). The latter control links not only the cytotoxic effect of a chelator and an antibiotic with high iron levels, it also suggests that the increase in iron upon combined treatment is a specific response to certain antibiotics and chelators. Of note, the total iron levels present in cell and supernatant samples were unchanged between treatments, indicating that the changes observed were due to alterations in cellular iron instead of some strange complication from the medium (Fig. 5B). When considered in the context of the results from Fig. 1 to 5, these data suggest that starvation of ExPEC of extracellular free iron (by chelation) in the context of certain antibiotics promotes the build-up of intracellular iron.
FIG 5.
Antibiotics and iron chelation induce excessive iron accumulation. E. coli CP9 was cultured in BHI medium in the presence or absence of an antibiotic (VAN at 250 μg/ml, CTX at 62.5 ng/ml, or LVX at 0.01 μg/ml), DFP (150 μg/ml), or both. After 18 h, the amount of iron present in the cell pellet (A), calculated as the number of iron atoms per cell, or the total iron present in the sample (B) (Fesupernatant plus Fecell pellet), was determined by ICP-OES. The data in panel A were normalized to the level of iron present in untreated cells (left columns). Data are the mean values and standard deviations from three independent experiments. P values were determined by Student's t test.
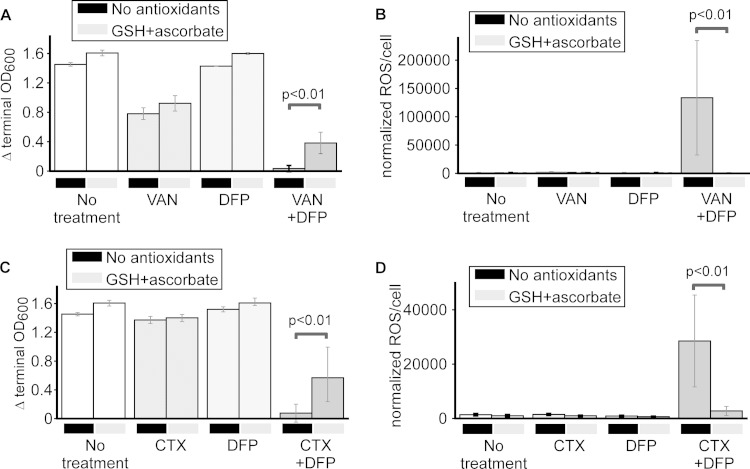
Iron chelation and antibiotics drive the production of reactive oxygen species.
We hypothesized that the accumulation of iron in chelator-antibiotic-treated E. coli may interact with H2O2 (via Fenton chemistry to generate hydroxyl radicals) or O2 (via a perferryl ion to yield superoxide radicals) to generate toxic levels of ROS (39). To determine whether ExPEC subjected to both an antibiotic and DFP is under oxidative stress, the levels of intracellular ROS were measured with the membrane-permeating fluorescent dye DCFH-DA, which fluoresces upon association with oxygen radicals (Fig. 6A). Upon the addition of VAN, CTX, or DFP to ExPEC, there was no difference in the level of intracellular ROS from that in the untreated control (Fig. 6B and C). Interestingly, when both VAN and CTX were combined with DFP, two scenarios that promoted cell death (VAN-DFP) or growth inhibition (CTX-DFP) and increased cellular iron, there was also a marked upswing in the steady-state level of cell-associated ROS compared to that in untreated or singly treated E. coli (Fig. 6B and C). Furthermore, cells treated with DFP and LVX, a combination that did not show enhanced antibacterial activity, did not demonstrate a concomitant increase in cellular ROS (Fig. 6D). These findings indicate that exposure of ExPEC to a combination of an antibiotic and iron chelation produces a highly oxidative cellular environment, a state not observed when each compound is used alone.
Antioxidants rescue E. coli from antibiotic-DFP cytotoxicity and lower ROS levels.
We next determined if the elevated ROS level was responsible for the cytotoxicity and growth inhibition of ExPEC treated with both an antibiotic and DFP. To test this hypothesis, we grew cells with or without an antibiotic or DFP or an antibiotic plus DFP in the presence or absence of two known antioxidants, glutathione (a small peptide) and ascorbate (a vitamin) (40, 41). As shown in Fig. 7A and C (far right column), the addition of antioxidants improved the growth of E. coli in the presence of either VAN or CTX with DFP, an effect not observed in cells left untreated or treated with these compounds singly. This enhancement of growth in the presence of antioxidants was matched by a concomitant decrease in the levels of antibiotic-DFP-induced ROS in combination-treated but not singly treated cells (Fig. 7B and D). Taken as a whole, these findings suggest that the elevated ROS level is responsible for the cytotoxicity and/or growth inhibition observed in combination-treated cells, a process that is likely catalyzed by the high levels of cell-associated iron.
FIG 7.
Antioxidants decrease antibiotic/chelation-induced ROS levels and rescue growth. E. coli CP9 was cultured in BHI medium in the presence or absence of an antibiotic (VAN at 250 μg/ml or CTX at 62.5 ng/ml), DFP (150 μg/ml), or both, and growth (A and C) and ROS levels (B and D) were measured in the presence or absence of antioxidants (5 mM glutathione [GSH] and 5 mM ascorbate). Data are the mean values and standard deviations from at least six separate replicates. P values were determined by Student's t test.
Antibiotic-mediated bacterial membrane integrity disruption and chemical and physical properties of the chelators contribute to antibiotic-DFP cytotoxicity.
Cell wall-targeting and aminoglycoside antibiotics, the antibiotics that showed enhanced efficacy when combined with DFP in this study, have been reported to affect bacterial membrane permeability, whereas the antibiotics that were not effective with DFP do not demonstrate this property (42). We tested this hypothesis by using the entry of PI into cells to measure the integrity of E. coli exposed to VAN or CTX. Indeed, both antibiotics increased membrane permeability to PI, an effect that lingered throughout the life of the culture (see Fig. S4 in the supplemental material) (P < 0.01 when t = 1, 2, 4, or 6 h for VAN-DFP; P < 0.01 when t = 6 h for CTX-DFP). A curious finding, and one that is consistent with the main interpretation of this work, was that the addition of DFP further increased membrane permeability. The molecular reason underlying this observation is not currently known, but we did note that DFP and DIP are similar in size, cell-permeating ability, and affinity for iron; they are also the chelators that generally demonstrated an inhibition or killing effect when combined with antibiotics (for a summary, see Table S3 in the supplemental material). These properties are different for DFX, which is larger, has less cell-permeating ability, and has a higher affinity for iron. Indeed, DFX did not readily combine with antibiotics to produce antibacterial activity. Since a permeable membrane correlates with antibacterial activity, it suggests that a compromised membrane may facilitate the entry of DFP-Fe3+ complexes into the bacterial cell. In fact, the addition of two different detergents (Tween 20 and Triton X-100) with DFP had a greater antibacterial effect than either alone (see Fig. S5 in the supplemental material).
DISCUSSION
Hypothesizing that altering bacterial iron homeostasis could form a viable entry point for the development of new antimicrobial strategies, we demonstrate in this report that the number one cause of Gram-negative bacteremia, E. coli (43), a species that harbors extensive resistance to many classes of drugs, undergoes (i) a cytotoxic response to sublethal doses of clinically current and no longer used antibiotics when combined with the iron chelator DFP, a process that is (ii) dependent on the presence of iron and associated with (iii) an iron starvation response, as measured by the derepression of the ferric uptake regulator Fur, (iv) a dramatic increase in the levels of cell-associated iron, and (v) a rise in the level of intracellular oxygen radicals, the alleviation of which by antioxidants increases the survival of these cells. Other noteworthy findings of this work include that this response was also observed in other clinical strains of E. coli (suggesting that it may be a universal property of E. coli in general), that it is correlated with an antibiotic-dependent induction of the permeability of the outer membrane, and that the cytotoxic or growth-inhibitory properties of a chelator and an antibiotic in some, but not all, cases also hold true for S. aureus. In addition, the strain of E. coli used in this study was engineered to contain the gene conferring resistance to CHL (20) but can be killed by combining CHL with DFP. The latter finding suggests that iron chelators can enhance the effectiveness of antibiotics even when the strain is already resistant. The findings reported here strongly suggest that the novel combination of seemingly unrelated drugs may have dramatic and negative effects on bacterial cells. The investigation of these effects may reveal previously unknown mechanisms by which bacteria can be killed, including the molecular players involved and the properties they modulate. Considering the need for not only new drugs but also new antimicrobial targets to combat the growing resistance problem, and the rise of the concept of antibiotic “adjuvants” (44), this work highlights the modulation of microbial iron homeostasis as a potential therapeutic entry point.
The eukaryotic hosts restrict the access of bacteria to key metals such as iron by sequestering the iron in proteins in cells or through high-affinity chelation by small molecules (45). This process can be mimicked in culture through the use of natural and synthetic compounds, such as DFP, DFX, or DIP, that preferentially bind iron. Under these conditions, the expected outcome is inhibition of growth; i.e., with insufficient iron, bacteria will simply slow their metabolism and not replicate. This was, in fact, observed in our time-kill studies when E. coli was cultured in the presence of only DFP. However, what emerged from these experiments is that if two divergent stresses are applied, one related to restriction of access to nutrients (a chelator) and the other to inhibition of a critical cellular process (an antibiotic), the cells undergo a three-hit effect that severely restricts their ability to adequately recover. The first hit, induced by the antibiotic, likely relates to the disruption of physiology and leads to a reduction in the initial number of viable cells seeded into a culture. The second hit, induced by the chelator, likely affects the ability of the cells to attain or make nutrients that would allow them to ramp up their metabolism to compensate for the first insult. The final hit, which likely keeps persister cells (which are what was assayed throughout this study) in a state of poor health and thus unable to replicate, relates to the buildup of iron and iron-catalyzed ROS. The latter state resembles, in some fashion, a recently described process observed in eukaryotic cells termed “ferroptosis,” i.e., cell death due to dysregulation of iron homeostasis (46).
Kohanski et al. have noted that antibiotics, particularly those that are bactericidal, induce the formation of ROS in bacterial cells, albeit in the absence of iron chelation (47). These findings were subsequently challenged by Keren et al. and Liu et al. (48, 49), who questioned whether antibiotics kill without the requirement of oxygen, which would be needed to generate ROS. The mechanistic details of these studies have not been worked out, but data presented here may help shed light on what is likely to grow into an important topic. There are numerous conditions and variables that must be considered before we entertain the idea that a ROS-based killing model is an explanation for these results. For example, data reported here demonstrate that ROS generation can be a mechanism that induces cell death in bacteria, but only when DFP is present. In addition, not all of the antibiotics tested displayed a combinatorial effect when combined with an iron chelator and not all combinatorial effects were observed in another species (the Gram-positive bacterium S. aureus). Of all of the major antibiotic classes tested, five of them, and only when combined with an iron chelator, inhibited the growth of or killed E. coli at 24 h postinoculation. Only two of them showed the same effect on S. aureus. Interestingly, one antibiotic (AMP) showed the effect on both E. coli and S. aureus; incidentally, it is one of the three suggested by Kohanski and coworkers to induce ROS-mediated cell death. Taken as a whole, the data suggest that some antibiotics do have the ability, under the right conditions, to induce ROS (in support of Kohanski et al.) but that this mechanism does not hold true for all antibiotics (in support of Keren et al. and Liu et al.), and iron chelation by DFP is a requirement, at least under the conditions used in this study. In the study of Kohanski et al., it was shown that DIP rescued the killing effect of bactericidal antibiotics (a positive effect), whereas in our study, we showed that DIP synergized with antibiotics (a negative effect). Kohanski et al. analyzed the first 3 h after the addition of reagents, whereas we monitored bacterial growth for up to 24 h. As shown in the time-kill assay (Fig. 3; see Fig. S2 in the supplemental material), for the first few hours assayed, antibiotic-DFP-treated cells show greater growth than those treated with the antibiotic alone, which would be considered a “rescue” phenotype by Kohanski et al. Thus, we reasoned that this discrepancy could be explained by differences in the methods used in the two studies, including the times assayed, the concentrations used, and even the strains tested (we assessed the responses of pathogens). In contrast, Luo et al. (VAN plus deferasirox) and Zhu et al. (CHL plus hexadentate 3-hydroxypyridin-4-one) both also showed that iron chelation can synergize with antibiotics in killing S. aureus (50, 51), which is consistent with our data. More work is needed to define the exact nature of these interactions.
What is the evidence of ROS generation in E. coli under conditions of iron chelation plus antibiotics? First, incubation of bacteria with an antibiotic and a chelator yielded high fluorescence when DCFH-DA, a broad-spectrum, ROS-activatable, fluorescent dye (52), was included. Whereas DCFH-DA casts a wide net that can capture many types of ROS, a cited disadvantage is that its high reactivity also means that it can be nonspecifically oxidized by Fe2+ in the presence of O2 or H2O2 (53). Although this is generally considered a limitation of this dye, for the purpose and hypothesis of our study, it is an advantage because it can detect two types of ferrous iron-mediated ROS species, those generated through reaction with O2 (via a perferryl ion to yield superoxide radicals) and those generated through reaction with H2O2 (via Fenton chemistry to generate hydroxyl radicals) (39). Thus, DCFH-DA represents a good reporter dye for iron-mediated ROS because it more sensitively detects both hydroxyl and superoxide radicals, in support of our model in which exposure of bacterial cells to both an antibiotic and a chelator leads to high intracellular iron levels. Indeed, we found that bacterial cells treated with both a chelator and an antibiotic had an ∼1- to 2-log higher total amount of iron, which is consistent with excessive iron accumulation. Further support for the idea of ROS generation under these conditions comes from the finding that the fluorescence of DCFH-DA was reduced by the addition of antioxidants (vitamin C and glutathione), an effect that was cell dependent and not caused by either the quenching of dye fluorescence or the use of the chemicals as a growth-promoting nutrient (data not shown).
What is the evidence that iron is involved in this mechanism? First, the combined effect is dependent on the use of the iron chelator DFP, which suggests either that removal of free iron from the medium in the presence of an antibiotic is detrimental to the cells or that a DFP-iron complex is particularly toxic when combined with an antibiotic. It is interesting that different iron chelators demonstrated various antibacterial activities when combined with an antibiotic and that, generally, if one worked with one antibiotic, the others did as well. Such a result provides compelling evidence that the chelation of iron drives this effect but only with certain antibiotics. That different chelators demonstrate different efficacies when combined with antibiotics may be explained by the differences in their affinity for iron and their membrane-permeating ability. Second, antibiotic-DFP inhibition can be overcome by the addition of exogenous iron, further indicating that inhibition is an iron-dependent process. Third, cells treated with an antibiotic and DFP have high levels of cell-associated iron, 10 to 100 times the normal level, as determined by ICP-OES. In addition, a Δfur mutant of E. coli, when combined with the antibiotics that showed a combinatorial effect with DFP, was inhibited in the ability to grow compared to that of a wild-type strain under identical conditions (unpublished results). Finally, cells exposed to both drugs have high levels of ROS; iron is a well-known catalyst of free radical formation (39). Thus, our data strongly support a model in which alterations in the balance of healthy or harmful levels of iron are critical to the survival of bacterial cells and certain antibiotics can somehow shift this balance when iron chelators are present. This novel finding gives support to the notion that targeting of iron homeostasis may be an effective antibacterial strategy. It stops short of declaring, however, that antibiotics and DFP should be used as cotherapies in infected patients. We simply do not have sufficient data to show that this strategy will work in vivo. Therefore, the results reported here should only be taken as evidence that new “drug” formulations reveal novel and interesting ways in which bacterial cells can be killed or inhibited. As we consider new options for new antibiotics, such ways may be exploited in the future. Combination of antibiotics with iron chelation, as reported here, may be an example of this.
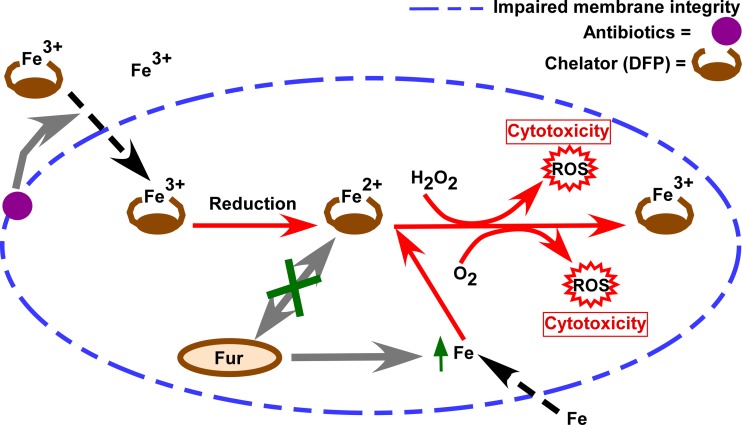
Why are antibiotics required for a combinatorial effect? Our current model (Fig. 8), one that is supported by the data presented here, as well as the published work of others, is that a sublethal concentration of DFP complexes with ferric iron in the medium (DFP-Fe3+), which is the preferred form of iron that binds DFP (54). We believe that the presence of antibiotics may lead to disruption of the bacterial membrane, which then facilitates DFP-Fe3+ entry into the cytoplasm. If iron enters the cell, the Fe3+ can then be reduced to Fe2+ because of the highly reducing environment of the cytoplasm (55). The observed inactivity of the repressor Fur under these conditions strongly suggests that this Fe2+ is not free but remains complexed with DFP, thereby preventing it from being recognized by Fur, possibly through a steric hindrance mechanism. DFP complexed with Fe2+ may interact with the much smaller H2O2 or O2 molecule, which leads to toxic levels of ROS (hydroxyl radicals and superoxide radicals, respectively) (39, 54). The sensed iron starvation of the bacterial cell may also lead to excessive iron uptake through transporters, contributing to even more intracellular ROS (although we have not tested this directly). This model is consistent with all of the available data and provides a working hypothesis for additional studies aimed at unmasking how iron homeostasis may be exploited as an antibacterial strategy.
FIG 8.
Proposed model of peptidoglycan-targeting antibiotic- and DFP-mediated toxicity to bacterial cells. DFP chelates ferric iron in the medium (DFP-Fe3+). When antibiotics are present, they destabilize the cellular membrane(s), allowing DFP-Fe3+ to readily enter the cell. DFP-Fe3+ then is reduced to Fe2+ in the highly reducing environment of the bacterial cytoplasm. Reduced DFP-Fe2+ is a known radical generator in the presence of O2 or H2O2 and leads to the production of ROS. DFP also contributes to a state of iron starvation (by shielding iron from binding to Fur), perhaps leading to excessive iron uptake, which contributes further to the generation of ROS. The supraphysiologic level of ROS generated under these conditions leads to cell death and/or inhibition of growth.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI069697 from the National Institutes of Health.
We have no conflict of interest to declare.
We thank Shelley Payne (University of Texas at Austin) for providing the pQF50 and pAML23 plasmids and Lynn Zechiedrich (Baylor College of Medicine, Houston, TX) for providing the clinical isolates of E. coli. We also thank Sarah Highlander (J. Craig Venter Institute) for contributing S. aureus strain TCH1516 and James Johnson (University of Minnesota) for ExPEC CP9.
L.M. and A.W.M. designed the experiments, analyzed the data, and wrote the paper. L.M. and Y.G. performed the experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00758-15.
REFERENCES
- 1.Waksman SA. 1947. What is an antibiotic or an antibiotic substance? Mycologia 39:565–569. doi: 10.2307/3755196. [DOI] [PubMed] [Google Scholar]
- 2.Alliance for the Prudent Use of Antibiotics. 2014. The cost of antibiotic resistance to US families and the healthcare system. http://www.tufts.edu/med/apua/consumers/personal_home_5_1451036133.pdf. Accessed 29 June 2015.
- 3.Aminov RI. 2010. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol 1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh C. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, DC. [Google Scholar]
- 6.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nairz M, Haschka D, Demetz E, Weiss G. 2014. Iron at the interface of immunity and infection. Front Pharmacol 5:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porcheron G, Garénaux A, Proulx J, Sabri M, Dozois CM. 2013. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol 3:90. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Bushmarina NA, Blanchet CE, Vernier G, Forge V. 2006. Cofactor effects on the protein folding reaction: acceleration of alpha-lactalbumin refolding by metal ions. Protein Sci 15:659–671. doi: 10.1110/ps.051904206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banin E, Brady KM, Greenberg EP. 2006. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl Environ Microbiol 72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson MG, Corey BW, Si Y, Craft DW, Zurawski DV. 2012. Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob Agents Chemother 56:5419–5421. doi: 10.1128/AAC.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes SS, Nunes A, Gomes AR, de Castro B, Hider RC, Rangel M, Appelberg R, Gomes MS. 2010. Identification of a new hexadentate iron chelator capable of restricting the intramacrophagic growth of Mycobacterium avium. Microbes Infect 12:287–294. doi: 10.1016/j.micinf.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Lesic B, Foulon J, Carniel E. 2002. Comparison of the effects of deferiprone versus deferoxamine on growth and virulence of Yersinia enterocolitica. Antimicrob Agents Chemother 46:1741–1745. doi: 10.1128/AAC.46.6.1741-1745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visca P, Bonchi C, Minandri F, Frangipani E, Imperi F. 2013. The dual personality of iron chelators: growth inhibitors or promoters? Antimicrob Agents Chemother 57:2432–2433. doi: 10.1128/AAC.02529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 17.Ganz T. 2011. Hepcidin and iron regulation, 10 years later. Blood 117:4425–4433. doi: 10.1182/blood-2011-01-258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo TA, Guenther JE, Wenderoth S, Frank MM. 1993. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol Microbiol 9:357–364. doi: 10.1111/j.1365-2958.1993.tb01696.x. [DOI] [PubMed] [Google Scholar]
- 19.Highlander SK, Hultén KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green SI, Ajami NJ, Ma L, Poole NM, Price RE, Petrosino JF, Maresso AW. 2015. A murine model of chemotherapy-induced extraintestinal pathogenic Escherichia coli translocation. Infect Immun 83:3243–3256. doi: 10.1128/IAI.00684-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson-Banning KM, Chou A, Liu Z, Hamill RJ, Song Y, Zechiedrich L. 2013. Toward repurposing ciclopirox as an antibiotic against drug-resistant Acinetobacter baumannii, Escherichia coli, and Klebsiella pneumoniae. PLoS One 8:e69646. doi: 10.1371/journal.pone.0069646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. NCCLS, Wayne, PA. [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. M07-A9, 9th ed CLSI, Wayne, PA. [Google Scholar]
- 24.Mey AR, Craig SA, Payne SM. 2005. Characterization of Vibrio cholerae ryhB: the ryhB regulon and role of ryhB in biofilm formation. Infect Immun 73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farinha MA, Kropinski AM. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol 172:3496–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergmans HEN, Van Die IM, Hoekstra WPM. 1981. Transformation in Escherichia coli: stages in the process. J Bacteriol 146:564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 28.Nhan TT. 2010. Analysis of soil extracts using the Agilent 725-OES. Agilent Technologies, Savage, MD: https://www.agilent.com/cs/library/applications/IO-034.pdf. [Google Scholar]
- 29.Eruslanov E, Kusmartsev S. 2010. Identification of ROS using oxidized DCFDA and flow-cytometry, p 57–72. In Armstrong D. (ed), Advanced protocols in oxidative stress II. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 30.Nobles CL, Green SI, Maresso AW. 2013. A product of heme catabolism modulates bacterial function and survival. PLoS Pathog 9:e1003507. doi: 10.1371/journal.ppat.1003507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson JR, Russo TA, Scheutz F, Brown JJ, Zhang L, Palin K, Rode C, Bloch C, Marrs CF, Foxman B. 1997. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapG(J96) (class I) and PrsG(J96) (class III) Gal(alpha1-4)Gal-binding adhesins. J Infect Dis 175:983–988. doi: 10.1086/514006. [DOI] [PubMed] [Google Scholar]
- 32.Gohel K, Jojera A, Soni S, Gang S, Sabnis R, Desai M. 2014. Bacteriological profile and drug resistance patterns of blood culture isolates in a tertiary care nephrourology teaching institute. Biomed Res Int 2014:153747. doi: 10.1155/2014/153747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doern CD. 2014. When does 2 plus 2 equal 5? a review of antimicrobial synergy testing. J Clin Microbiol 52:4124–4128. doi: 10.1128/JCM.01121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect Immun 77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hantke K. 2001. Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 36.Ma L, Payne SM. 2012. AhpC is required for optimal production of enterobactin by Escherichia coli. J Bacteriol 194:6748–6757. doi: 10.1128/JB.01574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Courvalin P. 2006. Vancomycin resistance in Gram-positive cocci. Clin Infect Dis 42(Suppl 1):S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 38.Suman RK, Ray IM, Mohanty NC, Mukhia RK, Deshmukh YA. 2014. Assessment of usage of antibiotic and their pattern of antibiotic sensitivity test among childhood fever. Int J Pharm Pharm Sci 6:296–299. http://innovareacademics.in/journals/index.php/ijpps/article/view/1790. [Google Scholar]
- 39.Qian SY, Buettner GR. 1999. Iron and dioxygen chemistry is an important route to initiation of biological free radical oxidations: an electron paramagnetic resonance spin trapping study. Free Radic Biol Med 26:1447–1456. doi: 10.1016/S0891-5849(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 40.Lushchak VI. 2012. Glutathione homeostasis and functions: potential targets for medical interventions. J Amino Acids 2012:736837. doi: 10.1155/2012/736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ginter E, Simko V, Panakova V. 2014. Antioxidants in health and disease. Bratisl Lek Listy 115:603–606. [DOI] [PubMed] [Google Scholar]
- 42.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network (NHSN) Team and Participating Facilities NHSN. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 44.Farha MA, Brown ED. 2013. Discovery of antibiotic adjuvants. Nat Biotechnol 31:120–122. doi: 10.1038/nbt.2500. [DOI] [PubMed] [Google Scholar]
- 45.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B, Stockwell BR. 2012. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 48.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo G, Spellberg B, Gebremariam T, Lee H, Xiong Y, French S, Bayer A, Ibrahim A. 2014. Combination therapy with iron chelation and vancomycin in treating murine staphylococcemia. Eur J Clin Microbiol Infect Dis 33:845–851. doi: 10.1007/s10096-013-2023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C-F, Qiu D-H, Kong X-L, Hider RC, Zhou T. 2013. Synthesis and in-vitro antimicrobial evaluation of a high-affinity iron chelator in combination with chloramphenicol. J Pharm Pharmacol 65:512–520. doi: 10.1111/jphp.12013. [DOI] [PubMed] [Google Scholar]
- 52.Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. 2003. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J Biol Chem 278:3170–3175. doi: 10.1074/jbc.M209264200. [DOI] [PubMed] [Google Scholar]
- 53.Kalyanaraman B, Darley-Usmar V, Davies KJA, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, Ischiropoulos H. 2012. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med 52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devanur LD, Neubert H, Hider RC. 2008. The Fenton activity of iron(III) in the presence of deferiprone. J Pharm Sci 97:1454–1467. doi: 10.1002/jps.21039. [DOI] [PubMed] [Google Scholar]
- 55.Ziegler DM, Poulsen LL. 1977. Protein disulfide bond synthesis: a possible intracellular mechanism. Trends Biochem Sci 2:79–81. doi: 10.1016/0968-0004(77)90042-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.