Abstract
Background
Esophageal cancer (EC) is one of the most aggressive malignant gastrointestinal tumors; however the traditional therapies for EC are not effective enough. Great improvements are needed to explore new and valid treatments for EC. We aimed to screen the differentially expressed miRNAs (DEMs) in esophageal cancer and explore the pathogenesis of esophageal cancer along with functions and pathways of the target genes.
Material/Methods
miRNA high-throughput sequencing data were downloaded from The Cancer Genome Atlas (TCGA), then the DEMs underwent principal component analysis (PCA) based on their expression value. Following that, TargetScan software was used to predict the target genes, and enrichment analysis and pathway annotation of these target genes were done by DAVID and KEGG, respectively. Finally, survival analysis between the DEMs and patient survival time was done, and the miRNAs with prediction potential were identified.
Results
A total of 140 DEMs were obtained, 113 miRNAs were up-regulated including hsa-mir-153-2, hsa-mir-92a-1 and hsa-mir-182; while 27 miRNAs were down-regulated including hsa-mir comprising 29a, hsa-mir-100 and hsa-mir-139 and so on. Five miRNAs (hsa-mir-103-1, hsa-mir-18a, hsa-mir-324, hsa-mir-369 and hsa-mir-320b-2) with diagnostic and preventive potential were significantly correlated with survival time.
Conclusions
The crucial molecular targets such as p53 may provide great clinical value in treatment, as well to provide new ideas for esophageal cancer therapy. The target genes of miRNA were found to play key roles in protein phosphorylation, and the functions of the target genes during protein phosphorylation should be further studied to explore novel treatment of EC.
MeSH Keywords: Esophageal Neoplasms; Genes, vif; Survival Analysis
Background
Esophageal cancer (EC) is the sixth leading cause of cancer-related death worldwide [1] and it is also one of the most aggressive malignant tumors in the gastrointestinal tract, affecting more males than females. It is reported the incidence of EC has increased rapidly in recent years, especially in the United States and other developed countries [2]. In China, the EC mortality rate is second only to gastric cancer [3]. The commonly used treatment method for EC is surgery combined with chemotherapy or radiotherapy; however, as most patients with EC are diagnosed at an advanced stage, this method is not very effective, with a 5-year survival rate of 5–10% [4]. To study the detailed molecular mechanism of EC and develop effective therapies, it is of great importance to explore and identify new biological markers.
MicroRNAs (miRNAs) consist of an abundant class of naturally occurring endogenous, small non-coding RNAs at a length of 18–25 nucleotides, which can repress protein translation by binding to the target mRNAs [5]. Numerous studies have shown that miRNAs play important regulatory roles in many physiological processes, such as cell proliferation, differentiation, apoptosis, and cell death [5,6]. In addition, miRNAs can target up to several hundred mRNAs, which makes them very powerful regulators, and aberrant miRNA expression can disturb a multitude of cell signaling pathways and profoundly influence cancer onset and progression. Changes in the expression of miRNAs have been observed in a variety of human tumors, including EC [7–9]. Although the expression differences do not necessarily represent causal events in carcinogenesis, these changes may regulate some genes that are very important during the process of tumor pathogenesis and may contribute to the classification and prognosis of tumors. For example, microRNA-21 is reported to be over-expressed in esophageal squamous cell carcinoma, it can target PDCD4 at the posttranscriptional level, and it regulates cell proliferation and invasion, which may serve as a novel therapeutic target in EC [10]. The molecular mechanisms by which miRNAs contribute to the development of esophageal cancer have been reported by Tian et al., who found that KLF4 mRNA is a direct target of miR-10b and that KLF4 can partly inhibit ESCC cell migration and invasion initiated by miR-10b. Although previous studies have proved that miRNAs contribute to the initiation, development, invasion, and metastasis of tumors, their underlying molecular mechanisms remain unclear. Therefore, it is urgent to identify and study sensitive and specific molecular markers to understand the potential pathogenesis and to improve early diagnosis of EC.
This study screened differentially expressed miRNAs (DEMs) in EC and analyzed the target genes of these DEMs by DAVID. Through performing survival analysis between the DEMs and patient survival time to identify miRNAs with prediction potentials, we hoped to explore the pathogenesis of EC and to determine molecular markers for early diagnosis and treatment of EC.
Material and Methods
miRNA microarray data and patient information
The miRNA expression data and the corresponding medical information of the patients were downloaded from the TCGA database, and there were a total of 200 samples (187 esophageal cancer samples and 13 normal samples) and the medical information was for 169 patients. These miRNA expression data were sequenced by Illumina HiSeq system and the standardized miRNA data were level 3 data in the TCGA database.
Firstly, these miRNA data were standardized, and then the data with expression values zero were removed. The miRNA data in level 3 were downloaded, which included a total of 1046 comments for the miRNA expression values. These miRNA data in level 3 had already been standardized within the sample and the standardization between samples was performed using generalized linear model in an R language Limma package to eliminate batch effects between samples.
Screening of differentially expressed miRNA
SAMR [11] package in R software was used to screen the differentially expressed miRNA between normal tissue samples and esophageal cancer tissue samples. The differential expression degree was shown by logFC and p, while Log2FC was used to indicate the differential expression degree of miRNA between differentially expressed tumor samples and normal samples. Down-regulated and up-regulated miRNAs were expressed as logFC <−1 and logFC >1, respectively, both with FDR <0.05. Principal component analysis method was used to efficiently distinguish the differentially expressed miRNAs between esophageal cancer tissue samples and normal tissue samples.
Survival analysis
All the medical information for patients was summarized and subjected to statistical analysis so as to obtain the cutoff value of medical information. Then Kaplan-Meier method was used to study the distribution of survival time in different disease states, while log-rank test was used to determine the differences of survival ability under various disease states. In addition, univariate Cox regression model was applied to study the relationship between differentially expressed miRNAs and survival time of patients. The original data were arranged sequentially by survival days, then displayed the survival state (status, death was 1, while survival was 2), cumulative survival, standard error of the survival rate, cumulative event, and number remaining of each sample. Finally, the survival rates of patients were obtained according to the results.
Screening and verification of the molecular markers for miRNAs
The data were classified into 2 cohorts (training group and test group), and the expression profiling of miRNAs in training group was analyzed with Cox regression model. Then the miRNAs associated with survival (p<0.01) and 5 miRNAs significantly associated with survival (p<0.005) were found obtained. A multivariate Cox regression model was constructed based on the 5 miRNAs to predict patient survival and the parameters of each miRNA had a Cox regression correlation coefficient; therefore, we could give a risk factor to each patient. According to this model, the patients with higher risk scores had weaker viability compared to those with lower risk scores. Then we calculated the median risk scores of all the patients, and considered this value as the boundary. It was defined as high risk when the risk score was above the boundary, while it was low risk below the boundary. Kaplan-Meier model was utilized to observe the survival time distribution of every variable and log-rank detection method was used to determine the significances among various classifications under the same variables.
Analysis of the target genes of miRNAs
Firstly, the target genes of the differentially expressed miRNAs in Mirtarbase [12] database were extracted, then GO functional annotation and KEGG pathways of these target genes were analyzed by DAVID [13]. A P value of function enrichment and a corrected P value after multiple testing (Benjamini correction) were calculated using DAVID software according to each GO value.
Results
Expression differences of miRNAs among various samples
There were a total of 187 esophageal cancer samples and 13 normal samples in 200 samples, which included 1046 of miRNA expression values. The differences between normal and cancer samples were observed by principal component analysis and cluster analysis. As shown in Figure 1, the normal samples and tumor samples could be separated, indicating that the data could be analyzed in further study.
Figure 1.
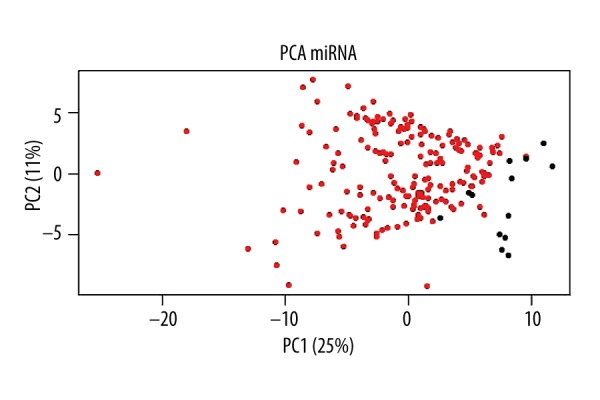
Principal component analysis of the miRNA expression values. Principal component analysis was complicated according to the miRNA expression differences between normal and cancer samples. The horizontal axis shows that the first principal component accounted for 25%, while the vertical axis shows that the second principal component accounted for 11%. The red dots and black spots indicate cancer samples and normal samples, respectively. The figure clearly shows that normal samples gathered on the right, while cancer samples gathered on the left, indicating that there were significant differences between the 2 samples.
Differential expression analysis of miRNA
We obtained 140 differentially expressed miRNAs between normal and EC samples after analysis by SAMR [11] in R package, including 113 up-regulated and 27 down-regulated (Figure 2). The up-regulated miRNAs accounted for 77.1% of the total differentially expressed miRNAs, including hsa-mir-153-2, hsa-mir-92a-1, and hsa-mir-182, while the down-regulated miRNAs accounted for 22.9%, including hsa-mir-29a, hsa-mir-100, and hsa-mir-139 (Table 1).
Figure 2.
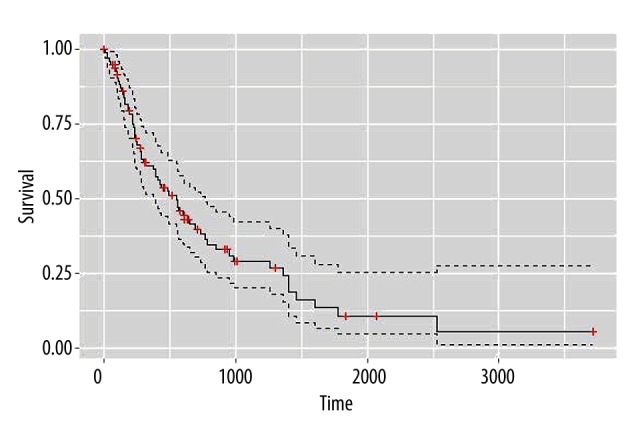
Overall survival status changes of patients. The survival curves were drawn according to patient survival time. The vertical axis shows survival time and the horizontal axis shows survival rates, the red crosses on the curve are cut-off values. As seen from the figure, patient viability decreased gradually over time.
Table 1.
Six significantly differentially expressed miRNAs.
| Type | miRNA ID | logFC | P value | FDR |
|---|---|---|---|---|
| Up-regulated miRNA | hsa-mir-21 | 1.470062 | 18.29724 | 1.16E-16 |
| hsa-mir-93 | 1.937027 | 13.3289 | 1.70E-09 | |
| hsa-mir-106b | 1.717675 | 9.561354 | 4.45E-08 | |
| Down-regulated miRNA | hsa-mir-490 | −3.50374 | 2.269419 | 5.43E-14 |
| hsa-mir-204 | −3.40881 | 3.141546 | 3.08E-14 | |
| hsa-mir-490 | −3.50374 | 2.269419 | 5.43E-14 |
Survival analysis
The censoring rate 61.54% was relatively high judged from the medical information of patients in Table 2, indicating that there were still many patients alive at the end of the study or they may directly depart from the study. A survival curve was drawn according to patient survival time, as shown in Figure 2, which shows that although there was a high cutoff value, the medical information about esophageal cancer is sufficient and suitable to predict the biological targets of miRNAs in the following study.
Table 2.
Summary of the medical information for esophageal cancer patients.
| Covariates | Category | Total |
|---|---|---|
| Age, years, no (%) | <60 | 82 |
| ≥60 | 87 | |
| Gender, no (%) | Male | 23 |
| Female | 146 | |
| Vital status | Alive | 104 |
| Dead | 65 | |
| Disease stage, no (%) | Stage I | 20 |
| Stage II | 68 | |
| Stage III | 55 | |
| Stage IV | 6 | |
| Lymph node involvement, no (%) | N0 | 20 |
| N1 | 63 | |
| N2 | 12 | |
| N3 | 7 | |
| NX | 2 | |
| M stage, no (%) | M0 | 126 |
| M1 | 2 | |
| M1a | 4 | |
| MX | 17 | |
| T stage, no (%) | T0 | 1 |
| T1 | 30 | |
| T2 | 35 | |
| T3 | 83 | |
| T4 | 5 | |
| R stage, no (%) | R0 | 126 |
| R1 | 12 | |
| R2 | 1 | |
| RX | 7 |
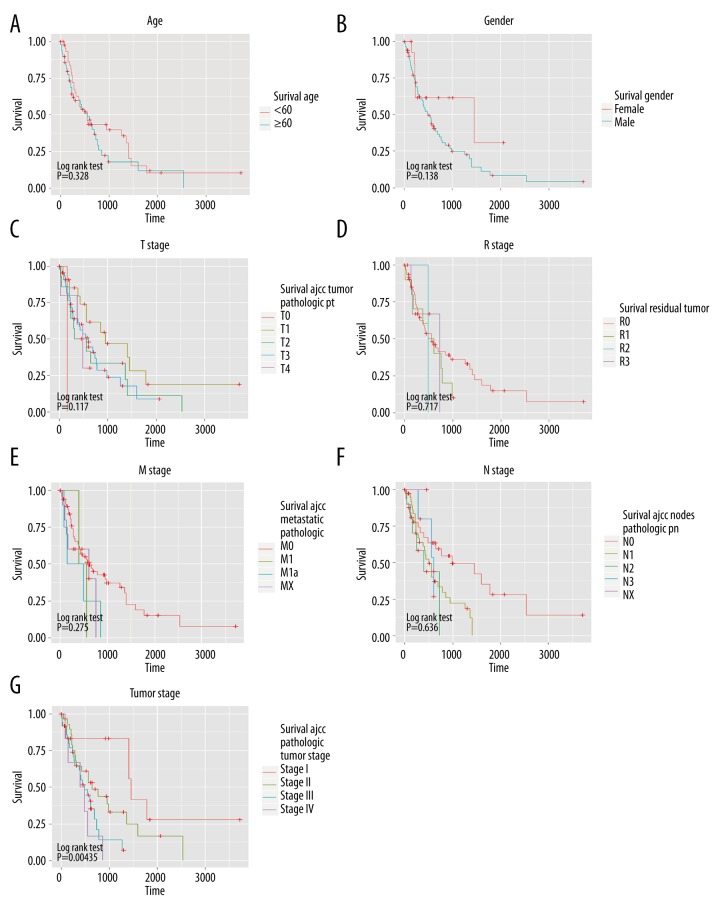
The relationship between the survival time and various variables (age, sex, T stage, R stage, M stage, N stage, and tumor stage) was verified by Kaplan and log-rank methods (Figure 3). We found there were significant differences between different tumor statuses and the overall survival time. According to the results in Figure 3, univariate Cox regression model was applied to analyze the differences between miRNA and survival time of patients under each state. Correlation coefficients in results are demonstrated in the heat map in Figure 4. The molecular markers of miRNAs were selected when P<0.1 and there were significant results in at least 2 separate categories.
Figure 3.
Changes in patient survival status in different classifications. (A–G) Represent the relationships between age, gender, T stage, R stage, M stage, N stage, tumor stage, and survival time, respectively. P-value of log rank test is showed in the lower left corner of each figure.
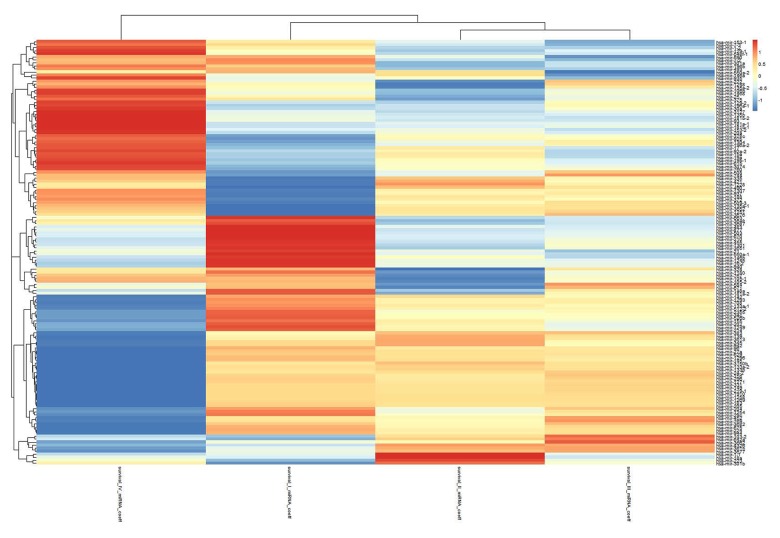
Figure 4.
Predictive capabilities of miRNAs under different tumor statuses. The abscissa and the ordinate show different tumor statuses and different miRNAs, respectively. This heat map shows that the capacity of the 140 differentially expressed miRNAs of tumors affected the survival under various statuses. Red represents strong positive correlations between miRNA expression and survival time, while green represents strong negative correlations.
Through univariate survival analysis, the miRNAs associated with patient overall survival in different tumor statuses were screened, as shown in Figure 4. Five miRNAs that may be considered as biomarkers were selected: including hsa-mir-103-1, hsa-mir-18a, hsa-mir-324, hsa-mir-369, and hsa-mir-320b-2.
Construction of predictive disease model
A mathematical model for miRNA and survival time was constructed by Cox multivariate regression model using data in the training group, and the prognosis formula was as follows: Prognostic score=(0.054 × expression level of hsa-mir-103-1) + (0.36 × expression level of hsa-mir-18a) + (0.14 × expression level of hsa-mir-324) + (0.37 × expression level of hsa-mir-369) + (−0.46 × expression level of hsa-mir-320b-2). Among which, 4 miRNAs belonged to risky type, 1 belonged to protective type.
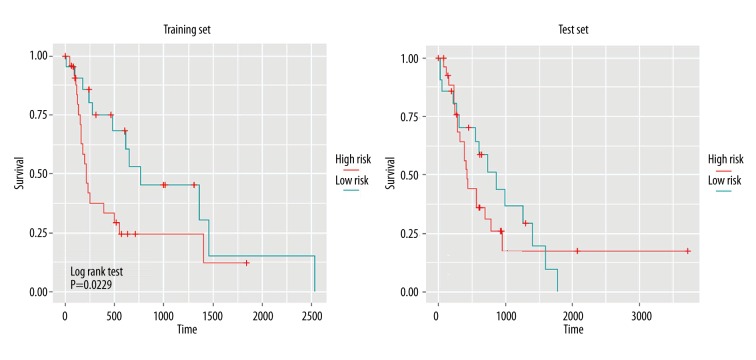
The patients were divided into high-risk and low-risk groups, and the differences between the 2 groups were analyzed by the log rank test (Figure 5). Significant differences were displayed between high-risk and low-risk patients in training group and test group, demonstrating that this model was feasible in actual prediction. The prediction results of miRNA in patients are displayed in Figure 6, which shows the expression differences of miRNA between high-risk and low-risk patients. The results show that lower risk coefficient is associated with longer survival time.
Figure 5.
Determination of patient survival risk by Kaplan-Meier analysis based on expression levels of 5 miRNAs. The abscissa shows the survival time, while the ordinate shows the survival rate. Red and blue represent high-risk and low-risk patients, respectively. P-value at the lower left corner indicates differences between the 2 risk groups.
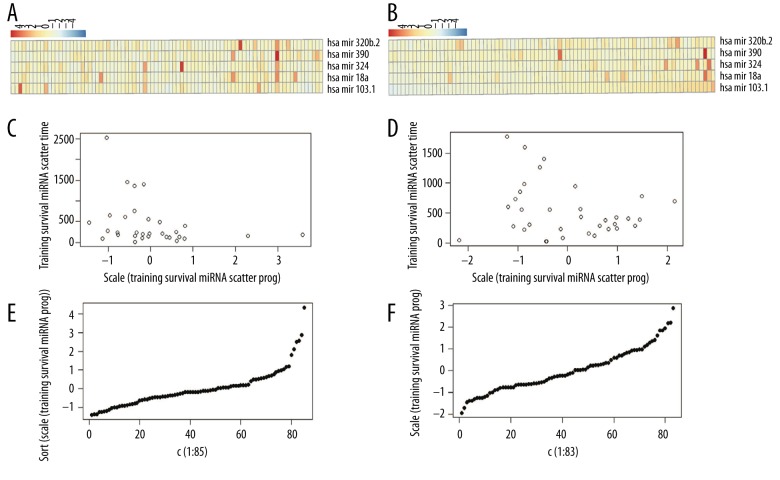
Figure 6.
Analysis of the risk coefficients predicted by the 5 miRNAs. (A) Risk coefficients analysis of training set data; (B) Heat map for expression levels of 5 miRNAs in training set; (C, D) Relationship between patient survival time and risk coefficients; (E, F) Distribution of risk coefficients.
Analysis of the target genes for 5 miRNAs
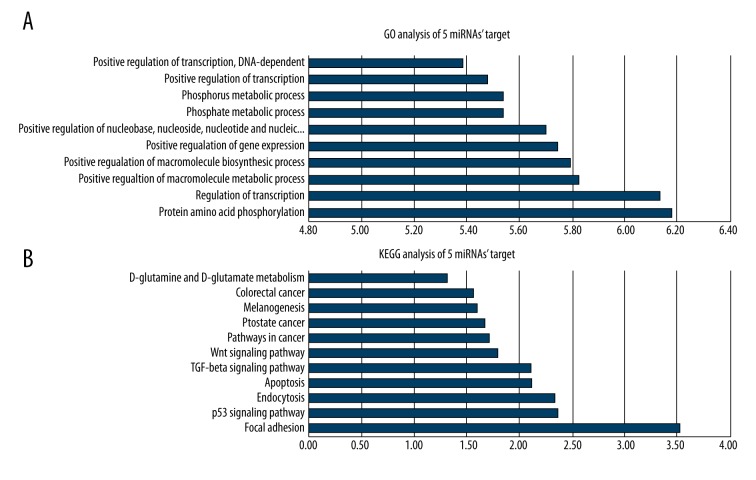
Target genes of 5 miRNAs were predicted by TargetScan. A total of 2147 target genes were predicted. These target genes were then subjected to GO annotation and KEGG pathway analysis by DAVID (Figure 7). The results of GO annotation demonstrated the main functions of these target genes were associated with protein phosphorylation and transcriptional regulation, while the results of KEGG analysis showed they were mainly related to cell adhesion and P53 signaling pathway.
Figure 7.
GO and KEGG analysis of the target genes of differentially expressed miRNAs. (A) GO analysis of the target gene; (B) KEGG analysis of the target gene. The abscissa represents degree of significance, while the vertical axis represents functional annotation; the greater the degree of significance, the greater the correlation between the target gene and annotation.
Discussion
Early diagnosis of cancer, especially the non-invasive tumors, has gradually gained much attention. Through detection of nucleic acid or protein in blood, early diagnosis of susceptible populations has also turned into a focus in cancer screening studies. However, several conventional serum tumor markers, such as carcinoembryonic antigen (CEA) [14], lacked enough sensitivity and specificity to facilitate early diagnosis of cancer, so the significance of non-invasive biomarkers as a detection method should be highlighted [15]. At the same time, non-invasive methods also helps tumor typing, individualized treatment, and improving real-time tracking of tumor treatment. miRNA as a focus in anti-cancer research will gain growing attention.
In this study, we screened a total of 140 differentially expressed miRNAs between normal and EC samples. Using medical information of patients, we predicted the miRNAs that could be considered as molecular targets, and 5 miRNAs (hsa-mir-103-1, hsa-mir-18a, hsa-mir-324, hsa-mir-369, and hsa-mir-320b-2) were finally selected. Guo et al. [16] for the first time reported that high expression of miR-103/107 showed a negative prognostic effect on the survival of EC patients. Moreover, it was found that low expression levels of miR-103/107 demonstrated a close correlation with a long overall and disease-free survival for EC patients. Therefore, miR-103/107 may serve as a useful diagnostic tool or a candidate drug target for EC management. mir-18a also had been reported in previous studies to be significantly over-expressed in several carcinomas, such as gastric cancer [17], hepatocellular carcinoma, colorectal carcinomas, and pancreatic carcinoma [18]. In addition, miR-18a may have the potential to function as a potential tumor suppressor by targeting K-Ras so as to regulate proliferation and anchorage-independent growth of human cancer cells [19]. Ras is involved in the performance of critical steps in tumorigenesis, and dysregulation of Ras is frequently observed in human cancers. K-Ras is the most frequently mutated isoform of Ras in cancer and the percentage of human tumors harboring oncogenic Ras mutations is high; therefore, interrupting the Ras-signaling pathway may become a major focus of new drug development. Generally speaking, miR-18a could be considered as a potential target or therapeutic agent for cancer therapy. Disregulation of miR-324 was found to be associated with macrophage dysfunction and colon cancer [20]. miR-369 and miR-320b-2 have also been studied in other cancers, but little research has been conducted on their roles in EC.
To further analyze the selected miRNAs and explore the possible mechanisms of the development of EC, we also predicted the target genes of these miRNAs, and 2147 target genes were obtained. Through GO annotation and KEGG analysis, we found these target genes were mainly enriched in cell adhesion and P53 signaling pathways. Focal adhesion kinase (FAK) plays a key regulatory role in survival, proliferation, migration, and invasion processes, which are all associated with the development and progression of cancer [21]. Moreover, activation of FAK can cause a number of cell biological processes, like migration, invasion, proliferation, cell attachment, and survival. All these processes are important in cell, tissue and organ structuring, functioning, and remodeling [22]. Given the important role of FAK during processes of tumor genesis and metastasis and the link to prominent oncogenes, FAK is a promising target in ongoing search for anti-cancer drugs [23]. The p53 gene can encode tumor suppressor genes, and cell division and proliferation become uncontrolled if the p53 gene is damaged by carcinogenic factors [24]. The p53 gene has important roles in various functional activities, including avoiding accumulation of damaged DNA, maintaining stability of the genome, and regulating cell differentiation and aging through induction of cell cycle arrest, promoting apoptosis, and DNA repair [25]. An early study suggested that accumulation of p53 protein possibly caused by p53 gene mutation is a very early event in human esophageal carcinogenesis, which contributed to our understanding of molecular mechanisms of EC and development of an early biomarker for early diagnosis and treatment studies [26]. In future studies, we will attempt to regulate these 5 miRNAs to adjust the normal functions of the p53 signaling pathway.
Conclusions
We screened differentially expressed miRNAs between normal and EC samples in this study and selected 5 miRNAs with potential predictive ability in EC. In addition, the target genes of these 5 miRNAs were analyzed, which may provide new insights into cancer development, as well finding new potential biomarkers and therapeutic targets. Although further studies are still needed, the results of this study expand our understanding of the pathogenesis of EC.
Footnotes
Source of support: Departmental sources
References
- 1.Kano M1, Seki N, Kikkawa N, et al. miR-145, miR-133a and miR-133b: Tumor – suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127(12):2804–14. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 2.Hu Y, Correa AM, Hoque A, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2011;128(1):132–43. doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu SG, Qin XG, Zhao BS, et al. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett. 2013;5(5):1639–42. doi: 10.3892/ol.2013.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathé EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15(19):6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27(45):5959–74. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 7.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135(2):255–60. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobryshev YV, Orekhov AN, Chistiakov AD. MicroRNAs in esophageal adenocarcinoma: Functional significance and potential for the development of new molecular disease markers. Curr Pharm Des. 2015;21(23):3402–16. doi: 10.2174/1381612821666150311124418. [DOI] [PubMed] [Google Scholar]
- 9.Tsalis K, Antoniou N, Kalfadis S, et al. Laparoscopic enucleation of a giant submucosal esophageal lipoma. Case report and literature review. Am J Case Rep. 2013;14:179–83. doi: 10.12659/AJCR.883928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang JG, Wang JJ, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clinica Chimica Acta. 2010;411(11):846–52. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 11.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA – target interactions. Nucleic Acids Res. 2011;39(Database issue):D163–69. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 14.Kosugi S, Nishimaki T, Kanda T, et al. Clinical significance of serum carcinoembryonic antigen, carbohydrate antigen 19-9, and squamous cell carcinoma antigen levels in esophageal cancer patients. World J Surg. 2004;28(7):680–85. doi: 10.1007/s00268-004-6865-y. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–18. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68(1):26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 17.Yao Y, Suo AL, Li ZF, et al. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2(6):963–70. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 18.Motoyama K, Inoue H, Takatsuno Y, et al. Over-and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34(4):1069–75. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 19.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30(6):953–59. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Wang SX, Mu R, et al. Dysregulation of the miR-324-5p-CUEDC2 axis leads to macrophage dysfunction and is associated with colon cancer. Cell Rep. 2014;7(6):1982–93. doi: 10.1016/j.celrep.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 21.van Nimwegen MJ, van de Water B. Focal adhesion kinase: A potential target in cancer therapy. Biochem Pharmacol. 2007;73(5):597–609. doi: 10.1016/j.bcp.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Ilić D, Furuta Y, Kanazawa S, et al. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377(6549):539–44. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 23.Dunty JM, Gabarra-Niecko V, King ML, et al. FERM domain interaction promotes FAK signaling. Mol Cell Biol. 2004;24(12):5353–68. doi: 10.1128/MCB.24.12.5353-5368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351(6326):453–56. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 26.Wang LD, Hong JY, Qiu SL, et al. Accumulation of p53 protein in human esophageal precancerous lesions: a possible early biomarker for carcinogenesis. Cancer Res. 1993;53(8):1783–87. [PubMed] [Google Scholar]