Abstract
Purpose
Recent studies have shown that 7-12% of endometrial cancers (ECs) are ultramutated due to somatic mutation in the proofreading exonuclease domain of the DNA replicase POLE. Interestingly, these tumors have an excellent prognosis. In view of the emerging data linking mutation burden, immune response and clinical outcome in cancer, we investigated whether POLE-mutant ECs showed evidence of increased immunogenicity.
Experimental design
We examined immune infiltration and activation according to tumor POLE proofreading mutation in a molecularly defined EC cohort including 47 POLE-mutant tumors. We sought to confirm our results by analysis of RNAseq data from the TCGA EC series and used the same series to examine whether differences in immune infiltration could be explained by an enrichment of immunogenic neoepitopes in POLE-mutant ECs.
Results
Compared to other ECs, POLE-mutants displayed an enhanced cytotoxic T cell response, evidenced by increased numbers of CD8+ tumor infiltrating lymphocytes and CD8A expression, enrichment for a tumor-infiltrating T cell gene signature, and strong upregulation of the T cell cytotoxic differentiation and effector markers T-bet, Eomes, IFNG, PRF and granzyme B. This was accompanied by upregulation of T cell exhaustion markers, consistent with chronic antigen exposure. In-silico analysis confirmed that POLE-mutant cancers are predicted to display more antigenic neo-epitopes than other ECs, providing a potential explanation for our findings.
Conclusions
Ultramutated POLE proofreading-mutant ECs are characterized by a robust intratumoral T cell response, which correlates with, and may be caused by an enrichment of antigenic neo-peptides. Our study provides a plausible mechanism for the excellent prognosis of these cancers.
Keywords: POLE, ultramutation, endometrial cancer, tumor immunology, biomarker
INTRODUCTION
Endometrial cancer (EC) is the commonest gynecological malignancy in the Western world, and affects approximately 150,000 women each year in Europe and the US combined (1). ECs have traditionally been classified into endometrioid (EEC) and non-endometrioid (NEEC) tumors according to clinical and histopathological criteria. However, recent work has shown that this dualistic model can be improved upon by a molecular classification into subgroups that more accurately reflect underlying tumor biology and clinical outcome (2, 3).
One interesting subgroup is the 7-12% of ECs with somatic mutation in the proofreading exonuclease domain of the DNA replicase POLE (2, 4-6). Polymerase proofreading is essential for ensuring fidelity of DNA replication (7), and in keeping with its dysfunction, POLE proofreading-mutant cancers have a frequency of base substitution mutation among the highest in human cancer (2, 8, 9). POLE-mutant ECs display other distinctive features, including a characteristic mutation signature, with a preponderance of C>A transversions and bias for particular amino acid substitutions, and strong associations with endometrioid histology, high grade, and microsatellite stability (MSS) (2, 4-6, 10). We and others have recently shown that, despite the association with high grade, POLE-mutant ECs have an excellent prognosis (5, 6, 11). However, the reasons for this were unclear.
Although the ability of the immune system to suppress malignant disease has long been recognized (12), the last few years have seen a remarkable increase in our understanding of the complex and dynamic interplay between cancers and the host immune response. For example, preclinical and translational studies have confirmed that tumor missense mutations can lead to presentation of antigenic neo-epitopes by MHC class I molecules, resulting in activation of T cell-mediated cytotoxicity (13-16). Consequently, mutations that cause strongly antigenic epitopes are likely to undergo negative selection in developing tumors (13, 14). Cancers also demonstrate multiple alternative mechanisms of immune escape, including downregulation of HLA class I expression, and upregulation of immunosuppressive molecules including PD1/PD-L1, TIM3, LAG3 and TIGIT in a phenomenon referred to as adaptive immune resistance (17, 18). Despite this, it is clear that in many cancers the immune system retains a degree of control over tumor growth – illustrated by the association between increased density of tumor infiltrating lymphocytes (TILs), particularly CD8+ cytotoxic T cells, and favorable outcome in multiple cancer types, including EC (19-22). Interestingly, a recent study has shown that CD8+ cell infiltration correlates strongly with the number of predicted antigenic mutations in tumors (23), suggesting that the immunogenicity of cancers is determined at least partly by their mutational burden (24). These data are consistent with the observations that hypermutated microsatellite unstable (MSI) endometrial and colorectal cancers typically display greater TIL density than other tumors (25, 26).
During our previous studies (4-6), we noted that POLE-mutant ECs frequently displayed strikingly high TIL density, often accompanied by a Crohn’s-like reaction. Similar observations have recently been reported following pathological review of POLE-mutant TCGA ECs (27). We hypothesized that this may represent infiltration by cytotoxic T lymphocytes, which could in turn contribute to the favorable prognosis of these tumors. We also speculated that this might relate to an increase in antigenic neo-epitopes in POLE-mutant ECs secondary to ultramutation. We tested this using a molecularly defined cohort of ECs, including 47 POLE-mutant tumors, and the recently published TCGA series (2).
MATERIALS AND METHODS
Patients and tumors
Tumors were selected from the PORTEC-1 and PORTEC-2 studies (n=57) (28, 29) and EC series from the Leiden University Medical Centre (LUMC) (n=67) and the University Medical Centre Groningen (UMCG) (n=26), to provide similar numbers of low (grade 1/2) and high grade (grade 3) tumors of the common molecular subtypes: POLE wild-type, microsatellite stable (MSS); POLE wild-type, microsatellite unstable (MSI); and POLE (proofreading) mutant (Table S1). All cases were of endometrioid histology (EEC), and all POLE-mutant tumors were MSS. Of 373 cases reported in the TCGA series, 245 had paired whole exome and RNAseq data and were informative for this analysis (2). Of these 197 were EECs, four mixed EEC/NEECs, and 44 serous (NEEC) cancers. Ethical approval for tumor molecular analysis was granted at LUMC, UMCG and by Oxfordshire Research Ethics Committee B (Approval No. 05\Q1605\66).
POLE mutation and microsatellite instability status
DNA was extracted from FFPE blocks and sequencing of POLE hotspot exons 9 and 13 (which contain around 90% of pathogenic proofreading mutations) was performed in tumors from the PORTEC, LUMC and UMCG series as previously reported (4, 5). All mutations were confirmed in at least duplicate PCR reactions. Details of whole exome sequencing performed by the TCGA have been previously reported (2). Pathogenic POLE proofreading mutations were defined as somatic variants within the exonuclease domain associated with ultramutation and a frequency of C>A transversions of ≥20% (10).
MSI status was determined in the PORTEC, LUMC and UMCG cases by five-marker panel of microsatellites as reported previously (30), with exception of four cases in which this failed, where status was determined by MLH1, MSH2, MSH6 and PMS2 immunohistochemistry (6). Determination of MSI in the TCGA series was by seven-marker panel, with MSI-H defined as alteration at ≥3 markers (2). Classification of POLE mutant TCGA ECs was also informed by results of recent analysis of mononucleotide repeats in 48 genes (10). In our analysis, only MSI-H cases were classified as MSI – we excluded one TCGA case for which MSI status could not be determined. The single tumor that displayed both POLE proofreading mutation and MSI by these criteria was assigned to the POLE mutant group on the basis of its characteristic mutation signature, in accordance with a recent report (10).
Histological assessment
Tumors were evaluated for the presence/absence of TILs and for Crohn’s like reaction by a gynecologic pathologist (TB) blinded to other clinicopathological data.
Immunohistochemistry and cell quantification
Following de-paraffinization, antigen retrieval and blocking of peroxidase activity, whole slides were incubated overnight at 4°C (CD8, CD3) or room temperature (TIA-1, HLA, FoxP3) with primary antibodies against CD8 (1:50, clone C8/144B, DAKO, Agilent technologies, Glostrup, Denmark), CD3 (1:25, clone F7.2.38, DAKO), HCA2 and HC10 (both 1:800, kindly provided by Prof. Dr. J. Neefjes, the Netherlands Cancer Institute), FoxP3 (1:100, mAbcam 450, Abcam, Cambridge, UK), and TIA-1 (1:400, clone 2G9A10F5, Beckman Coulter, Miami FL, USA). Sections were subsequently incubated with either anti-mouse Envision+ reagent (K4000, DAKO) for 30 minutes (CD8 primary), RAMpo (1:100) and GARpo (1:100) secondary and tertiary antibodies, (CD3 primary), or BrightVision-Poly/HRP (Poly-HRP-GAM/R/R; DPV0110HRP; ImmunoLogic) (HCA2, HC10, FoxP3 and TIA-1) before 3,3-diaminobenzidine (DAB) treatment and haematoxylin counterstaining. Slides were then dehydrated and mounted before digitalization (ScanScope, Aperio Technologies, USA or Ultra Fast Scanner 1.6 RA. Philips), and analysis.
CD8+, CD3+, FOXP3+ and TIA-1+ cell numbers were quantified in intraepithelial and intrastromal regions in the center of the tumor (CT) and the invasive margin (IM) as previously reported (19, 22). For each region, the mean number of positive cells in eight high power fields (200μm × 200μm) was calculated. For analysis of HLA expression, the percentage of tumor cells with membranous HCA2 and HC10 staining was quantified as previously described (31). In each case, scoring was performed independently by two observers, blinded to other clinicopathological data.
Immunofluorescence
Following de-paraffinization, antigen retrieval and blocking of peroxidase activity, whole slides were stained overnight at 4°C with primary antibody against TIA-1 (1:50, ab2712, Abcam). Sections were subsequently incubated with anti-mouse Envision+ reagent (K4000, DAKO) for 30 minutes and HRP visualized using cyanine 5 tyramide signal amplification (TSA) according to the manufacturer’s instructions (PerkinElmer). Next, whole slides were stained overnight with primary antibody against CD8 (1:25, clone C8/144B, DAKO) and a biotinylated antibody against fibronectin (1:50, ab6584, Abcam). Slides were incubated with GaM-AF555 (1:150 Life Technologies) and streptavidin-dylight488 (1:150, Thermo Scientific), counterstained with DAPI (Life Technologies) and mounted in prolong gold mounting medium (Life Technologies). Immunofluorescent slides were scanned using a TissueFaxs imaging system (TissueGnostics, Austria). Processed channels were merged using Adobe Photoshop CS5 (Adobe).
Leukocyte methylation scores
Leukocyte methylation scores (syn1809223) (32, 33) were downloaded from Synapse (https://www.synapse.org/) and annotated according to MSI and POLE status.
TCGA RNAseq data
Details of the TCGA RNAseq analysis have been previously reported (2). RSEM normalized (34) and raw RNAseq count data were downloaded from FireBrowse (http://firebrowse.org/?cohort=UCEC&download_dialog=true) accessed 11/11/14. After removal of normal tissue controls and technical duplicates, 245 samples with RSEM normalized and 231 samples with raw count data were informative for analysis.
Gene set enrichment analysis
TCGA raw counts were annotated by molecular subtype prior to normalization and ranking of genes differentially expressed between POLE-mutant (n=16) and other (n=205) ECs using DESeq (35). Gene set enrichment analysis (GSEA) (36) was then performed with the PreRanking setting, using GO Biological Processes and C7 Immunologic Signatures sets from the Molecular Signatures Database (MSigDB) http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=BP, and a published 200-gene T cell tumor infiltration gene signature (18).
Prediction of antigenic neo-epitopes
We created an algorithm to estimate the immunogenicity of individual tumors taking into account the following considerations: i) to generate a functional neo-epitope a missense mutation must be expressed; ii) most functional neo-epitopes identified to date are predicted to bind MHC class I molecules (IC50 < 500nM) by NetMHCPan (23, 37, 38); iii) the likelihood that a neo-epitope is antigenic is reduced if the corresponding wild-type peptide also binds the MHC with similar affinity as T cells to the epitope may be centrally deleted or tolerized (39). Our strategy was similar to others reported recently (15, 23, 38, 40). For each tumor we calculated all possible 9mers for every missense mutation in expressed genes (defined as non-zero reads from RNAseq) and calculated the binding affinity of the mutant and corresponding wild-type peptide for HLA-A*02:01 (as a single model example HLA allele) using NetMHCPan 2.8 (37). In the event that several peptides had an IC50 < 500nM, the strongest binder was used for analysis. We defined antigenic mutations as neo-epitopes predicted to bind MHC molecules (IC50 < 500nM) for which the corresponding wild-type peptide was not predicted to bind MHC (IC50 > 500nM).
Statistical analysis
We used the non-parametric Mann-Whitney test for all comparisons of continuous data and Spearman’s rho to analyze correlation between variables. Categorical variables were compared using Fisher’s exact test. All statistical tests were two-sided, with a P value of <0.05 taken to indicate significance. Except where indicated, statistical tests were unadjusted. Statistical analyses were performed using STATA (Texas), and Prism 6.0 (GraphPad, LaJolla).
RESULTS
POLE proofreading-mutant ECs show increased lymphocytic infiltrate
Preliminary analysis of H&E-stained sections suggested that POLE proofreading-mutant ECs frequently displayed a prominent lymphocytic infiltrate and Crohn’s-like lymphocytic reaction (Figure S1A,B). Formal quantification of this in a set of 150 ECs comprising approximately equal numbers of POLE proofreading-mutant/microsatellite stable (POLE-mutant), POLE wild-type/microsatellite-unstable (MSI) and POLE wild-type/microsatellite-stable (MSS) subtypes of low and high grade (Table S1), confirmed that TILs were more frequent in POLE-mutant (22/47) than in both MSS (8/54; P=0.0009, Fisher’s exact test), and MSI (10/49; P=0.009) subtypes (Figure S1C). Crohn’s-like reaction was also significantly more common in POLE-mutant than other tumors (P<0.001 both comparisons; Figure S1D).
Increased density of intratumoral CD8+ lymphocytes in POLE-mutant ECs
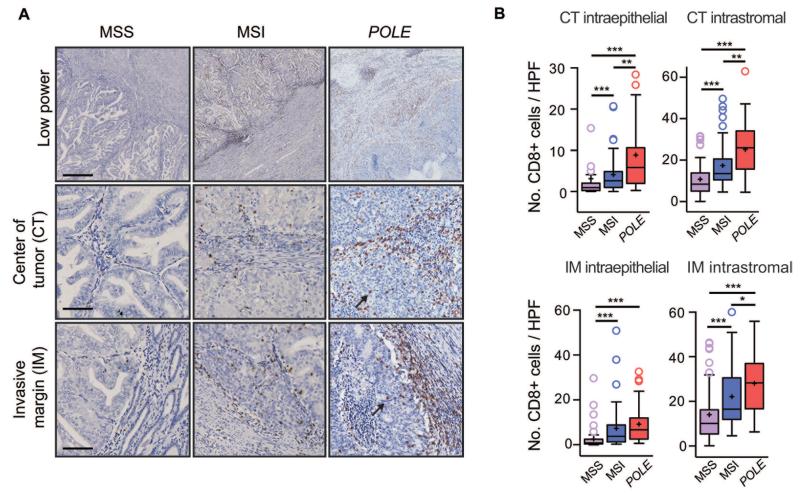
Mindful of the relationship between cytotoxic T cell infiltrate and favorable cancer outcome (19-22), and the excellent prognosis of POLE-mutant ECs (5, 6, 11), we next examined whether POLE-mutants showed evidence of increased T cell infiltrate in our EC cohort. While as anticipated (25), CD8+ cell numbers in intraepithelial and intrastromal compartments in the tumor center (CT) and the invasive margin (IM) were higher in MSI than MSS ECs (P<0.0001, all comparisons, Mann-Whitney test), in POLE-mutant tumors the density of CD8+ infiltrate was frequently striking (Figure 1A), and significantly exceeded that of both MSS (P<0.0001 for all four regions) and MSI cancers in the CT (median 5.9 vs. 2.6 intraepithelial CD8+ cells per high power field [HPF], P=0.001; 26.0 vs. 13.5 intrastromal CD8+ cells, P=0.002) (Figure 1B). Furthermore, the proportion of tumors with numbers of CD8+ cells exceeding the median in all four regions was substantially higher in POLE-mutant (60.0%) than MSI (31.3%, P=0.007, Fisher’s exact test) and MSS tumors (7.2%, P<0.0001). Staining for CD3 and the cytolytic marker TIA-1 in a subset of cases confirmed increased T cell density in POLE-mutant tumors (Figure S2A,B) and suggested that the infiltrate contained lymphocytes capable of cytotoxic activity (Figure S3A,B). Co-immunofluorescence confirmed co-expression of TIA-1 in the CD8+ lymphocytes comprising the POLE-mutant tumor infiltrate (Figure 2A-F), further supporting the conclusion that these cells were capable of mediating an anti-tumor effect.
Figure 1. Increased CD8+ lymphocyte infiltration in POLE-mutant endometrial cancers.
(A) Results of CD8 immunohistochemistry by EC molecular subtype shown at low magnification (upper panels) and high power views of the center of the tumor (CT) and invasive margin (IM). Arrows highlight intraepithelial CD8+ cells in POLE-mutant tumor. Scale bars correspond to 500 μm in upper panel and 100 μm in the middle and lower panels. (B) Quantification of CD8+ cell number in intraepithelial and intrastromal compartments in the CT and IM by EC molecular subtype. Boxes represent the interquartile range (IQR), with upper whisker indicating the 75th percentile plus 1.5 × IQR, and the lower whisker the 25th percentile minus 1.5 × IQR. The median and mean values are indicated by a horizontal line and cross respectively. Statistical comparison between groups was made by unadjusted two-sided Mann-Whitney test, *, **, and *** correspond to P<0.05, P<0.01, and P<0.001 respectively.
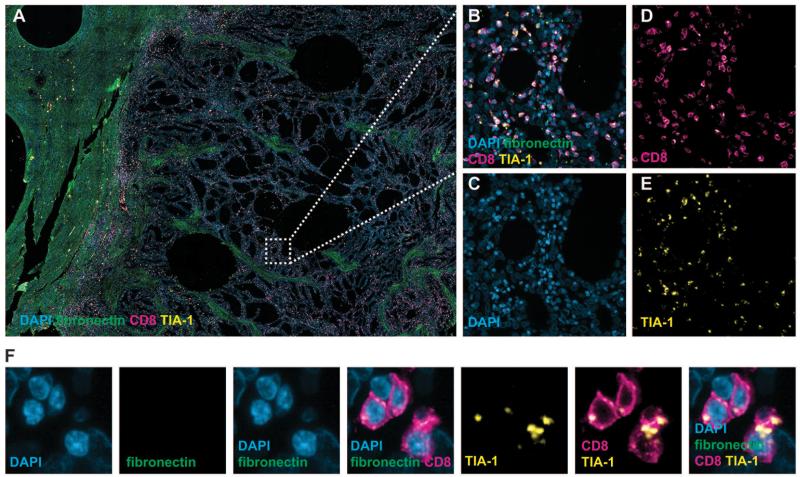
Figure 2. CD8+ infiltrating lymphocytes in POLE-mutant tumors show cytolytic potential.
(A) Low-magnification image of POLE-mutant tumor following co-immunofluorescence (Co-IF) staining for DAPI (nuclei), fibronectin (extracellular matrix), CD8 and the cytolytic marker TIA-1. (B-E) Co-IF images of the tumor center following staining for all markers (B), DAPI (C), CD8 (D) and TIA-1 (E) confirming co-expression of TIA-1 by tumor-infiltrating CD8+ lymphocytes, also shown at high magnification (F).
Interestingly, in light of the correlation previously reported between B and T cell subsets at the IM (41), we found that dense CD20 stromal infiltrate in this region was more common in POLE-mutants (Figure S4A), while a tendency to increased numbers of FOXP3+ cells in both MSI and POLE-mutant tumors (Figure S4B) was also notable, given that this has been associated with favorable cancer prognosis in some studies (41).
Cytotoxic T cell infiltration and activation in POLE mutant ECs in TCGA series
We sought to confirm our results using the TCGA EC series (2), in which the improved clinical outcome of POLE-mutant tumors was first suggested (Figure 3A). Of 244 informative tumors in this study, 157 were MSS, 69 MSI and 18 POLE-proofreading-mutant (the single tumor with both POLE proofreading mutation and MSI was categorized as POLE-mutant according to its mutation spectrum, in keeping with a recent report (10)). 43 of the MSS tumors were NEECs, while all MSI and POLE-mutant ECs cases were EECs (Table S1).
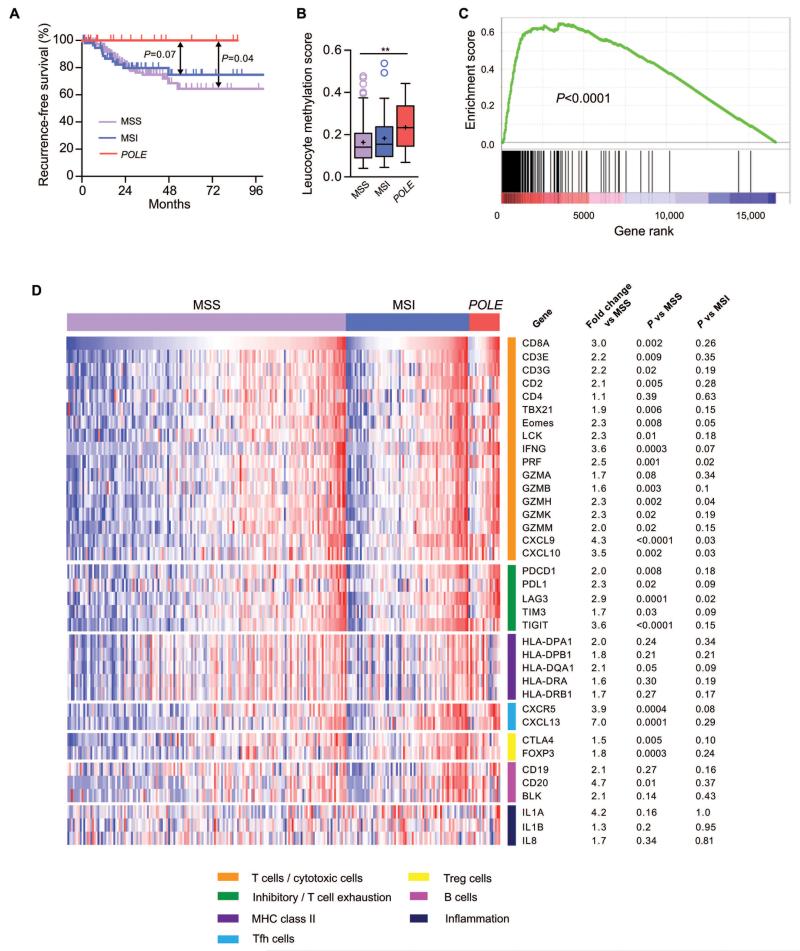
Figure 3. Clinical outcome and T cell response according to tumor molecular subtype in TCGA endometrial cancers.
(A) Kaplan-Meier curves demonstrating recurrence-free survival of POLE wild-type, microsatellite stable (MSS, n=147), microsatellite unstable (MSI, n=63) and POLE proofreading mutant (POLE, n=18) ECs in the TCGA series (note that survival data were not available for all cases). Comparison between subgroups was made by two-sided log-rank test. (B) Leucocyte methylation scores according to EC molecular subtype. Unadjusted comparison between groups was made by two-sided Mann-Whitney test. ** indicates P<0.01. (C) Gene set enrichment analysis (GSEA) of 200-gene tumor-associated T cell signature (Ref. 18) in POLE-mutant ECs compared to other tumors. Raw RNAseq counts were normalized and ranked using DESeq prior to GSEA analysis with pre-ranking. (D) Heatmap showing expression of immunological genes according to EC molecular subtype. RSEM-normalised RNAseq expression data were log2 transformed, zero centred and assigned unit variance. For each gene, the mean fold change in expression in POLE mutants relative to MSS ECs was calculated and expression compared between POLE mutants, MSS and MSI ECs by unadjusted, two-sided Mann-Whitney test.
We first examined leucocyte methylation scores, which estimate the proportion of a heterogeneous tumor sample that consists of leucocytes (32, 33). After confirming that scores correlated strongly with CD8A expression (ρ=0.65, P<0.0001), we noted that, following exclusion of MSI and POLE-mutant tumors, both leucocyte methylation scores and CD8A expression did not differ between EECs and NEECs (P=0.52 and P=0.16 respectively, Mann-Whitney test). We therefore included tumors of both histologies in the MSS cohort in all subsequent analyses. Leucocyte methylation scores were similar in MSI and MSS tumors (median 15.5% vs 14.2%, P=0.2), in contrast to a significant increase in POLE mutants (median 23.3%; P=0.006 vs. MSS, P=0.07 vs MSI) (Figure 3B), concordant with our previous results. Given the biological differences between NEECs and EECs, we formally confirmed that these results were essentially unaltered following exclusion of the former from the MSS group (median 14.7%, P=0.008 vs POLE-mutant ECs).
We proceeded to explore whether infiltration of POLE-mutant ECs by cytotoxic T cells was manifest as immune expression signatures and/or increased expression of key immunological genes. Agnostic pathway analysis of TCGA RNAseq data by gene set enrichment analysis (GSEA) demonstrated significant enrichment of immune-related pathways in POLE-mutant ECs compared to other tumors, including Immune Response (normalized enrichment score [NES] 4.12 FDR q<0.0001) and Immune System Process (NES 3.77, q<0.0001). GSEA also confirmed that POLE-mutant cancers showed striking enrichment of a recently reported, highly specific 200 gene signature corresponding to tumor T cell infiltration (18) (Figure 3C).
Focused analysis of genes involved in T cell-mediated cytotoxicity confirmed that, compared to MSS tumors, MSI ECs had higher expression of CD8A (2.1 fold, P=0.0005) and interferon γ (IFNG) (2.1 fold, P=0.0006) though expression of the cytotoxic differentiation and activation markers T-bet (TBX21), Eomes, perforin and granzymes B,H,K and M was either essentially unchanged (≤1.1fold) or not significantly increased. In contrast, and once again consistent with our previous results, POLE-mutants demonstrated substantial upregulation of CD8A (3.0 fold vs all MSS tumors, P=0.002; 3.2 fold vs MSS EECs only, P=0.004), accompanied by significant increases in T-bet (1.9 fold, P=0.006), Eomes (2.3 fold, P=0.008), IFNG (3.6 fold, P=0.0003), PRF (2.5 fold, P=0.001), granzymes B,H,K and M (1.6 to 2.3 fold, P=0.002 to 0.02), and the IFNG-induced cytokines CXCL9 (4.3 fold, P<0.0001) and CXCL10 (3.5 fold, P=0.002) (Figures 3D, S5). Upregulation of most of these genes in tumors has been shown to predict good prognosis (19, 41). POLE-mutants also demonstrated striking upregulation of the T follicular helper genes CXCL13 (7.0-fold, P=0.0001) and CXCR5 (3.9-fold, P=0.0004), which have recently been shown to strongly predict favorable outcome in colorectal cancer (41). Notably, despite limited numbers, in several cases expression of cytotoxic markers and cytokines exemplifying effector status in POLE mutants significantly exceeded that in MSI tumors (e.g. PRF, P=0.02; GZMH P=0.04; CXCL9/10 both P=0.03) (Figures 3D, S5). Collectively, these data not only corroborated our previous finding that POLE mutant tumors had greater T lymphocyte infiltration than other ECs, but also strengthened the conclusion that these lymphocytes were capable of exerting anti-tumor activity.
Mechanisms of immune escape in POLE-mutant ECs
POLE-mutant cancers have significantly better prognosis than other ECs (5, 6, 11), as evidenced by the absence of recurrences in the TCGA series (2). However, the presentation of all patients in this study with clinically detectable tumors, in some cases with lymphatic spread, indicates that any immune response was, at best, only partially successful in suppressing POLE-mutant EC growth. We therefore explored potential mechanisms of immune escape in these tumors.
We first considered the possibility that POLE-mutant ECs may escape from immune surveillance by loss or inactivation of components required for antigen presentation. While 31.8% of POLE mutant ECs in our study set of 150 ECs showed loss of HLA class I protein expression by IHC, this was not significantly different to that observed in MSI (28.6%, P=0.82, Fisher’s exact test) or MSS (20.0%, P=0.24) tumors and was not reflected in increased CD8+ cell numbers. Similarly, although we found a tendency to over-representation of POLE mutants among TCGA ECs with HLA class I gene expression in the lowest quartile, this was also not significantly different from other molecular subtypes (P=0.07 vs MSS).
Interestingly, while mutations in MHC pathway components were common in POLE-mutant tumors in the TCGA series, most were of unlikely pathogenicity, with exception of two tumors with potentially functional variants. The first, a beta 2-microglobulin R117* mutation also detected in a POLE-mutant CRC (9), had a variant allele fraction of 0.59 and is likely to affect stability of the MHC complex by disruption of the interaction with the HLA heavy chain (42). The second, an HLA-B S112R substitution with variant allele fraction 0.71, lies near the F pocket essential for peptide display, and is predicted to be deleterious by both Mutation Assessor and SIFT (Table S2). However, the effect of each variant on antigen presentation is, at present, uncertain.
We proceeded to examine whether the cytotoxic T cell response in POLE-mutant ECs may be attenuated by upregulation of immunosuppressive mediators – a phenomenon termed adaptive immune resistance (17). We found that the T cell exhaustion markers LAG3, TIM-3 and TIGIT, and the T cell inhibitors PD1 and CTLA4 were strongly correlated with CD8A expression across all ECs of all molecular subtypes (ρ=0.65 to 0.87; P<0.0001, all cases), though the correlation with PD-L1 was more modest (ρ=0.34, P<0.0001) (Figure S6A). Interestingly, while expression of these markers in MSI compared to MSS ECs was either unchanged/minimally altered (TIM-3, CTLA4, PD-L1) or moderately increased (LAG3 1.9 fold, P<0.002; TIGIT 2.2 fold, P<0.0001), in POLE mutants all were significantly, and substantially upregulated (e.g. LAG3 2.9 fold, P<0.0001 vs. MSS, P<0.02 vs. MSI; TIGIT 3.6 fold, P<0.0001 vs. MSS, P<0.15 vs. MSI) (Figures 3D, S6B), consistent with prolonged antigen stimulation. However, as noted above, the overall increase in expression of cytotoxic effector markers suggested that this upregulation was insufficient to fully suppress the T cell response in POLE mutants.
POLE proofreading-mutant ECs are likely to display increased numbers of antigenic neopeptides
We hypothesized that the T cell response in POLE-mutant ECs might be due to an excess of antigenic neo-epitopes as a consequence of ultramutation. To quantify this, we analyzed the TCGA EC series using a methodology similar to several recent studies (23, 40). Our algorithm was based on three assumptions – first, that a mutation must be in an expressed gene to exert an effect, second, for a neopeptide to act as an antigen it must bind MHC class I molecules (IC50 <500nM), and third, that neopeptides for which the corresponding wild-type peptide also binds MHC molecules are less likely to be immunogenic due to central deletion or tolerization (39).
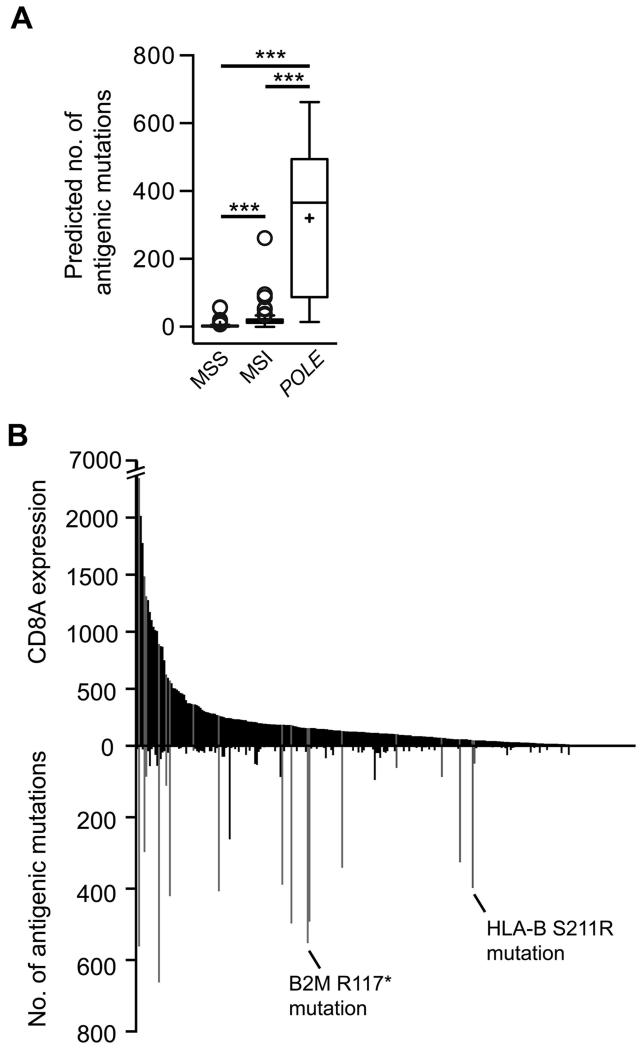
Applying these criteria, we found that 5.9% (7880/134,473) of the total number of missense mutations in TCGA ECs were predicted to be potentially antigenic. Of these, 73% (5767) occurred in POLE-mutant tumors, reflected in a significantly higher number of antigenic mutations per cancer compared to both MSI and MSS subtypes (median 365.5 vs 16 vs 2 respectively, P<0.0001 all comparisons, Mann-Whitney test) (Figure 4A), though this is likely to underestimate the number of antigenic mutations in MSI tumors as frameshift mutations were not included in our analysis. A substantial majority of antigenic mutations were in tumors with greater than median CD8A expression in both the whole series (78.4%), and the POLE-mutant subgroup (83.9%), though the strength of correlation between the two variables was modest, possibly as a result of immune escape mechanisms (Figure 4B).
Figure 4. Antigenic mutation burden and tumor CD8A expression according to POLE mutation.
(A) The number of mutations predicted to generate antigenic neo-peptides in individual TCGA tumors was calculated using exome and RNAseq data (see Methods). Box and whisker plots signify IQR ± 1.5 × IQR, mean and median as previously. Comparison between groups was made by unadjusted two-sided Mann-Whitney test, *** denotes P<0.0001. (B) Relationship between number of antigenic missense tumor mutations and tumor CD8A expression. POLE-mutant samples are highlighted in gray, together with possible mechanisms of immune escape in two cases.
DISCUSSION
By complementary analysis of two independent series totaling nearly four hundred patients, and including over sixty POLE-proofreading mutant tumors, we have shown that POLE-mutant ECs are characterized by a striking CD8+ lymphocytic infiltrate, a gene signature of T cell infiltration, and marked upregulation of cytotoxic T cell effector markers. Furthermore, we show that, as a consequence of their remarkable mutation burden, POLE proofreading-mutant cancers are predicted to display substantially more antigenic peptides than other tumors, providing a possible explanation for our findings. While our data demonstrate correlation rather than causation, the strong association between cytotoxic lymphocyte infiltration and favorable outcome in multiple cancers (19-22, 41) leads us to speculate that an enhanced T cell anti-tumor response may contribute to the excellent prognosis of POLE-mutant ECs.
During the last few years, a combination of next generation sequencing technology, improved in silico peptide-MHC binding prediction (37), and the clinical application of immune checkpoint inhibitors (43-45) have helped facilitate remarkable insights into the mechanisms of tumor immunoediting and immune escape. The intriguing observation that clinical benefit from CTLA4, PD1 and PD-L1 inhibition is greater in melanoma and cigarette smoking-associated lung cancer than most other malignancies can now be interpreted in light of the understanding that in these highly mutated tumors, adaptive immune resistance is a key enabler of disease progression (17), and its inhibition can restore the ability of T cells to respond to antigenic peptides presented by these cancers (43). In keeping with this, in melanoma response to checkpoint inhibitors has very recently been shown to correlate both with the number of predicted antigenic tumor mutations (40), and with the degree of cytotoxic T lymphocyte tumor infiltration prior to treatment (46). In light of these data, the association between the number of predicted antigenic peptides and T cell response in POLE mutant ECs in our study is noteworthy, as is the marked increase in T cell exhaustion markers, as these have recently been shown to identify tumor neo-antigen-specific CD8+ cells in cancer patients (47, 48). Despite upregulation of these immunosuppressors in POLE-mutant ECs, we also found substantial increases in cytotoxic differentiation markers and effectors suggesting that the degree of adaptive immune resistance in these cancers may be insufficient to fully suppress CD8+ T cell cytotoxicity (49). Collectively, our data suggest a complex interaction between the antigenic landscape of POLE-mutant ECs and the immune response. In this regard, molecular analysis of recurrences from the few patients with POLE-mutant ECs who do experience relapse may provide insights into mechanisms of immune escape. Lastly, these patients may be good candidates for immune checkpoint inhibitor therapy, as might those patients with POLE proofreading-mutant tumors of other histologies, for which outcomes are more uncertain.
Interestingly, while as anticipated we observed moderately increased T cell infiltration in MSI tumors (25), this was not associated with the marked increase in cytotoxic effector markers seen in POLE mutants. While some studies have reported improved prognosis of MSI ECs, this is inconsistent (50), in contrast to the clear association of MSI with favorable outcome in early colorectal cancer (26). Comparison of the immune response between MSI tumors of both types may provide insights into this discordance.
Our study has limitations. Due to differing sample preservation (FFPE vs fresh frozen) we were unable to validate either the IHC or RNAseq analysis between series, although the results from both analyses were highly concordant. Furthermore, the retrospective nature of our study meant that we were unable to investigate the repertoire and antigen response of T cells in patients with POLE-mutant cancers. This and other functional analyses will require prospective investigation.
In summary, we have demonstrated that ultramutated POLE proofreading-mutant ECs are characterized by a robust intratumoral T cell response, which correlates with, and may be caused by an enrichment of antigenic neo-peptides. Our study provides a plausible mechanism for the excellent prognosis of these cancers, and further evidence of the link between somatic mutation and immunoediting in cancers.
Supplementary Material
TRANSLATIONAL RELEVANCE.
The recently described subgroup of tumors with mutations in the proofreading domain of the DNA polymerase POLE are notable for both their striking mutation burden and, in the case of endometrial cancer (EC), their excellent prognosis. The demonstration that POLE proofreading-mutant ECs display an enhanced anti-tumor T cell response may explain the favorable outcome of these tumors, and suggests that similar investigation in POLE proofreading-mutant cancers of other types may be informative. Furthermore, as both tumor mutation burden and T cell infiltration have been shown to predict response to immune checkpoint inhibitors, our results also raise the possibility that these drugs may benefit patients with recurrent POLE proofreading-mutant ECs, and patients with POLE-mutant tumors of other histologies, for whom clinical outcomes are less certain.
ACKNOWLEDGMENTS
The authors are grateful to Enno Dreef, Natalja ter Haar and Reinhardt van Dijk (LUMC) for excellent technical assistance.
Financial support:
This work was supported by the Dutch Cancer Society Grant (UL2012-5719) (IvG, ES, RN, CdK, EO, CLC, VTHBMS, TB); Cancer Research UK (C6199/A10417) (IMPT), the European Union Seventh Framework Programme (FP7/207-2013) grant 258236 collaborative project SYSCOL (IPMT), the Oxford NIHR Comprehensive Biomedical Research Centre and NIHR senior fellowship (PK), the Oxford Martin School and core funding to the Wellcome Trust Centre for Human Genetics from the Wellcome Trust (090532/Z/09/Z) (LFM, CP, IPMT, DNC) and fellowship funding to PK (WT091663MA). The UMCG Imaging and Microscopy Center (UMIC), is sponsored by NWO-grants 40-00506-98-9021(TissueFaxs) and 175-010-2009-023 (Zeiss 2p). DNC is funded by a Health Foundation/Academy of Medical Sciences Clinician Scientist Fellowship, and has received funding from the Oxford Cancer Research Centre. The results published here include data generated by The Cancer Genome Atlas project funded by the NCI and NIH (cancergenome.nih.gov); we would like to thank the many patients and researchers who contributed to this study.
Footnotes
Conflicts of interest
None
REFERENCES
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.TCGA Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncol. 2014;15:e268–e278. doi: 10.1016/S1470-2045(13)70591-6. [DOI] [PubMed] [Google Scholar]
- 4.Church DN, Briggs SE, Palles C, Domingo E, Kearsey SJ, Grimes JM, et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22:2820–8. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church DN, Stelloo E, Nout RA, Valtcheva N, Depreeuw J, Ter Haar N, et al. Prognostic Significance of POLE Proofreading Mutations in Endometrial Cancer. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/dju402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stelloo E, Bosse T, Nout RA, Mackay HJ, Church DN, Nijman HW, et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod Pathol. 2015 doi: 10.1038/modpathol.2015.43. [DOI] [PubMed] [Google Scholar]
- 7.Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, et al. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;106:17101–4. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.TCGA Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, Schultz N, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–50. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng B, Hoang LN, McIntyre JB, Duggan MA, Nelson GS, Lee CH, et al. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol Oncol. 2014;134:15–9. doi: 10.1016/j.ygyno.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Burnet M. Cancer; a biological approach. I. The processes of control. Br Med J. 1957;1:779–86. doi: 10.1136/bmj.1.5022.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–9. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The Immunoreceptor TIGIT Regulates Antitumor and Antiviral CD8(+) T Cell Effector Function. Cancer Cell. 2014;26:923–37. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–13. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 21.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–66. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 22.Kondratiev S, Sabo E, Yakirevich E, Lavie O, Resnick MB. Intratumoral CD8+ T lymphocytes as a prognostic factor of survival in endometrial carcinoma. Clin Cancer Res. 2004;10:4450–6. doi: 10.1158/1078-0432.CCR-0732-3. [DOI] [PubMed] [Google Scholar]
- 23.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–50. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heemskerk B, Kvistborg P, Schumacher TN. The cancer antigenome. EMBO J. 2013;32:194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shia J, Black D, Hummer AJ, Boyd J, Soslow RA. Routinely assessed morphological features correlate with microsatellite instability status in endometrial cancer. Hum Pathol. 2008;39:116–25. doi: 10.1016/j.humpath.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153–62. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussein YR, Weigelt B, Levine DA, Schoolmeester JK, Dao LN, Balzer BL, et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.143. [DOI] [PubMed] [Google Scholar]
- 28.Nout RA, Smit VT, Putter H, Jurgenliemk-Schulz IM, Jobsen JJ, Lutgens LC, et al. Vaginal brachytherapy versus pelvic external beam radiotherapy for patients with endometrial cancer of high-intermediate risk (PORTEC-2): an open-label, non-inferiority, randomised trial. Lancet. 2010;375:816–23. doi: 10.1016/S0140-6736(09)62163-2. [DOI] [PubMed] [Google Scholar]
- 29.Creutzberg CL, van Putten WL, Koper PC, Lybeert ML, Jobsen JJ, Warlam-Rodenhuis CC, et al. Surgery and postoperative radiotherapy versus surgery alone for patients with stage-1 endometrial carcinoma: multicentre randomised trial. PORTEC Study Group. Post Operative Radiation Therapy in Endometrial Carcinoma. Lancet. 2000;355:1404–11. doi: 10.1016/s0140-6736(00)02139-5. [DOI] [PubMed] [Google Scholar]
- 30.Nout RA, Bosse T, Creutzberg CL, Jurgenliemk-Schulz IM, Jobsen JJ, Lutgens LC, et al. Improved risk assessment of endometrial cancer by combined analysis of MSI, PI3K-AKT, Wnt/beta-catenin and P53 pathway activation. Gynecol Oncol. 2012;126:466–73. doi: 10.1016/j.ygyno.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Reimers MS, Engels CC, Putter H, Morreau H, Liefers GJ, van de Velde CJ, et al. Prognostic value of HLA class I, HLA-E, HLA-G and Tregs in rectal cancer: a retrospective cohort study. BMC Cancer. 2014;14:486. doi: 10.1186/1471-2407-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khalili JS, Hanson RW, Szallasi Z. In silico prediction of tumor antigens derived from functional missense mutations of the cancer gene census. Oncoimmunology. 2012;1:1281–9. doi: 10.4161/onci.21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan F, Duitama J, Al SS, Ayres CM, Corcelli SA, Pawashe AP, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211:2231–48. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Trinh CH, Smith DP, Kalverda AP, Phillips SE, Radford SE. Crystal structure of monomeric human beta-2-microglobulin reveals clues to its amyloidogenic properties. Proc Natl Acad Sci U S A. 2002;99:9771–6. doi: 10.1073/pnas.152337399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–6. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 48.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–59. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speiser DE, Utzschneider DT, Oberle SG, Munz C, Romero P, Zehn D. T cell differentiation in chronic infection and cancer: functional adaptation or exhaustion? Nat Rev Immunol. 2014;14:768–74. doi: 10.1038/nri3740. [DOI] [PubMed] [Google Scholar]
- 50.Diaz-Padilla I, Romero N, Amir E, Matias-Guiu X, Vilar E, Muggia F, et al. Mismatch repair status and clinical outcome in endometrial cancer: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2013;88:154–67. doi: 10.1016/j.critrevonc.2013.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.