Abstract
The transcription factor, Nrf2 (nuclear factor, erythroid derived 2, like 2) belongs to the CNC-bZip protein family, forming a transcriptosome with its direct heterodimer partner, sMaf, and co-factors such as CBP/p300. Nrf2 binds to one or more ARE (Antioxidant Response Element) that are located in the gene regulatory regions of the hundreds of Nrf2 target genes. The ARE is a key enhancer that is activated in response to endogenous or exogenous stresses in order to maintain cellular and tissue homeostasis. Data emanating from gene expression microarray analyses comparing Nrf2-disrupted and wild-type mouse embryonic fibroblasts (MEF) showed that expression of Notch1 and Notch-signaling related genes were decreased in Nrf2-disrupted cells. This observation triggered our research on Nrf2-Notch crosstalk. A functional ARE has been identified upstream of the Notch1 major transcription start site. Furthermore, an Rbpjκ binding site is conserved on the promoters of Nrf2 among animal species. Notch1 is one of the transmembrane Notch family receptors, which drives Notch-signaling together with the Rbpjκ transcription factor. After canonically accepting ligands such as Jags and Deltas, the receptor undergoes cleavage to yield the Notch intracellular domain which translocates to the nucleus. Recent studies using conditional knockout mice indicate that Notch1 as well as Notch2 play important roles postnatally in liver development and in maintenance of hepatic function. In this review, we summarize current understanding of the role of reciprocal transcriptional regulation between Nrf2 and Notch in adult liver from studies using Nrf2, Keap1, and Notch1 genetically engineered mice.
Keywords: Nrf2, Antioxidant Response Element (ARE), Notch, Rbpjκ, liver regeneration
Introduction
Molecular pathways that contribute to basic biological functions are often conserved across species. Both canonical Notch and Nrf2 signaling pathways are evolutionally conserved from C. elegans to higher vertebrate animals[1, 2]. Interestingly, both are transcription factors. Nrf2 and Notch signaling pathways were discovered and described independently. However, the use of conditional knock-out and other constructs of genetically engineered mice have begun to unveil both the presence and functional significance of Nrf2-Notch reciprocal signaling interactions, principally in adult tissues.
Notch
In the earliest phase of Notch research, genetic models employed lower species such as Drosophila and C. elegans. Indeed, a gene locus was discovered from the phenotype of a mutant fly with an indentation in the wings. The gene in the locus responsible for this phenotype, which was later called “Notch”, was considered to play a role in cell fate decisions during Drosophila embryogenesis. Moreover, a deletion mutation of this locus resulted in excessive differentiation to neuronal tissue. Subsequent molecular biological analyses revealed that the Notch gene encoded a single-pass transmembrane protein that functioned as a receptor for ligands on the cell surfaces of neighboring cells. This ligand-receptor interaction was verified consequently to enable the fate decisions of the signal-receiving cells to become non-neuronal cells by restraining neuronal differentiation; this process leads to “lateral specification” which is essential for normal embryonic development [3].
After the concept of Notch signaling had been established in lower animal models, a gene located at the break point on chromosome 9 in the t(7;9)(q34;q34) translocation in a subset of acute T lymphoblastic leukemias in humans was identified as a Notch homolog. It was named translocation-associated Notch homolog 1 (TAN-1) [4]. This gene is currently called NOTCH1, and this discovery revealed that the Notch genes are highly conserved from nematodes up to humans. Leukemia cells harboring the t(7; 9) translocation express a truncated NOTCH1, which does not include a large part of the extracellular domain. This truncated NOTCH1, consequently, is detected in the intracellular space and acts as a constitutively activated transcription factor. Enhanced Notch signaling, such as transduced by TAN1, has led NOTCH1 to be considered an oncogenic factor.
Nowadays, it is clear that the Notch signaling pathway influences cell fate decisions in animals, such as cell differentiation, survival/apoptosis, and cell cycle in both physiologic and pathologic contexts, while also playing a role in stem cell biology. Many Notch signaling-modifier proteins including Rbpjκ have been identified, and the presence of a non-canonical, Rbpjκ-independent pathway has also been described [5, 6]. This review will focus on Rbpjκ-dependent canonical Notch signaling[7].
Canonical Notch signaling
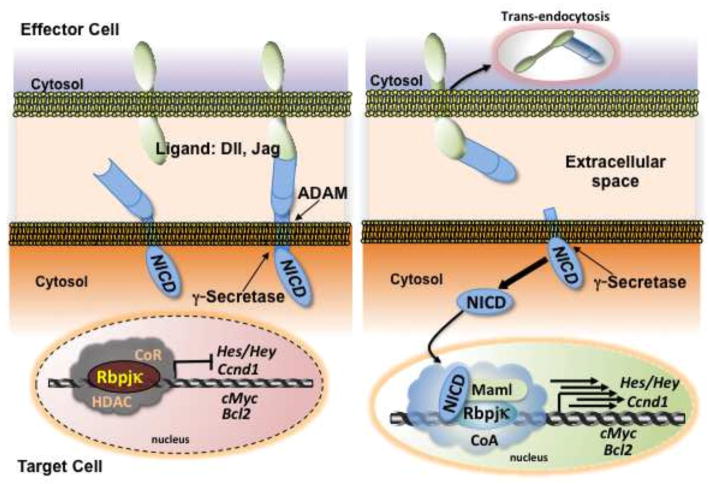
Notch encodes single-pass transmembrane receptors: there are 4 Notch genes (Notch1-Notch4) and 5 specific ligand genes (Jag1, Jag2, Delta-like <Dll> 1, Dll3, Dll4). The ligand genes also encode single-pass transmembrane proteins in mammals. As summarized in figure 1, de novo synthesized Notch protein products firstly undergo intramolecular cleavage to form heterodimers, composed of extracellular and transmembrane subunits localized to the plasma membrane. Once receptor-ligand interactions occur, the Notch molecules in the target cells are processed by two successive proteolytic cleavages. The first cleavage begins extracellularly, close to the transmembrane domain, and is mediated by metalloproteases of the ADAM family [8]. The cleaved extracellular domain of Notch is trans-endocytosed by the ligand presenting neighboring cell. The second cleavage proceeds within the transmembrane domain and is mediated by γ-secretase, which is a multiple protein complex consisting of Presenilin, Nicastrin, Aph1a (Anterior pharynx defective 1 homolog), and Psenen (presenilin enhancer 2 homolog) proteins. The cleavage by γ-secretase permits translocation of the Notch intracellular domain (NICD) fragment into the nucleus, where it binds to Rbpjκ, a direct cis-element binding protein, through the RAM (Rbp-associated molecule) domain and ankyrin repeats. Further, NICD associates with Mastermind-like proteins (MAMLs) through ankyrin repeats and recruits transcriptional activators such as p300, finally converting the Rbpjκ complex from a transcriptional repressor into an activator. Then, target genes begin to be expressed. Hes (hairy and enhancer of split) [9, 10] encodes a basic helix loop helix inhibitory transcription factor that leads to self-renewal of target cells by inhibiting differentiation. Hes is the one of the best characterized of the Notch target genes. The cell cycle promoter CyclinD1, the proliferation-related gene c-Myc, the anti-apoptotic gene Bcl2, the gene for Notch-regulated ankyrin repeat protein (Nrarp), Deltex1, and the pre–T-cell receptor gene (Ptcra) have also been identified as Notch target genes [11]. Thus, mechanistically, Notch transduces the signal received on the plasma membrane of the target cell into the nucleus to contribute to gene expression through collaboration with the specific transcription factor Rbpjκ. Responses are dependent upon cellular context.
Figure 1.
Canonical Notch signaling. Rbpjκ forms a negative regulatory complex on the promoter of Notch target genes and represses their expression. The Notch receptor on the target cell is activated following binding to a ligand presented by a neighboring effector cell. Endocytosis and membrane trafficking regulate ligand and receptor availability at the cell surface. Notch is then cleaved by ADAM and γ-secretase and the cleaved Notch intracellular domain (NICD) migrates to the nucleus. When NICD binds with Rbpjκ, it forms an active transcriptional complex, allowing target genes to be expressed.
Pleiotropic biological effects by Notch signaling
The Notch pathway in mammals can exert pleiotropic effects in each tissue that expresses Notch. As such, Notch-signaling networks regulate a wide range of events in embryonic and post-natal development, including proliferation, apoptosis, border formation and cell fate decisions. There are four major events regulated through Notch. (i) Prevention of differentiation: Notch maintains stem cells and/or transit amplifying cells in an undifferentiated state in the intestinal crypt by inhibiting expression of differentiation promoting genes [12, 13]. (ii) Binary cell fate decisions: in the lymphoid system Notch specifies the T cell lineage at the expense of the B cell lineage from bi-potent early thymocyte progenitors [14]. Equipotent precursor cells provide two alternative cell fates depending on whether an uncommitted progenitor cell receives a strong Notch signal or not [15]. Hepato-cholangiogenesis in the liver may be included in this context. (iii) Induction of differentiation: this phenomenon follows in a different or an opposite context from (i). For example, Notch induces terminal differentiation of transit amplifying cells in the skin. During thymocyte differentiation Notch1 promotes differentiation of pro-T cells into pre-T cells [14]. (iv) Tumorigenesis: constitutive overexpression of Notch within hematopoietic bone marrow cells or in T cell progenitors results in T cell leukemias. Hence, Notch functions as an oncogene in this case [16]. However, Notch seems to function as a tumor suppressor in the skin since the loss of Notch signaling causes the development of basal cell carcinoma-like tumors [17].
Thus, although both the canonical Notch pathway and the Notch-Rbpjκ transcriptional machinery are conserved, the resulting biological responses are rich in variety; sometimes bringing even opposite outcomes in different tissues of mammals. Such diversity in responses is a peculiarity of Notch signaling. Elucidation of Notch signaling crosstalk with other signaling pathways might shed light on these Notch-mediated effects and provide better understanding of their underlying mechanisms.
Keap1-Nrf2-ARE gene expression pathway
Nrf2 is a member of the CNC family transcription factors that includes Nrf1, 2, 3, Bach1, 2 and NF-E2p45 [2]. Expression of Nrf2 target genes is highly and rapidly inducible following exposure of cells to endogenous and exogenous oxidants and electrophilic stresses [18, 19]. Control of this expression is mediated through one or more cis-elements located on the 5′ flanking gene regulatory regions as enhancers, or sometimes, suppressors for gene expression. Initially, this cis-element was called the “electrophile-responsive element” (EpRE) because the first compounds demonstrated to activate this element were electrophiles or compounds easily oxidized to electrophiles [20]. Follow-up studies by others using phenolic antioxidants provided a nomenclature of “antioxidant responsive element” (ARE) [21], which has largely superceded the use of EpRE, despite a poor reflection of the chemistry of pathway activation. The CNC family transcription factors possess a basic leucine zipper domain for direct DNA binding, principally to the ARE. CNC transcription factors, including Nrf2, are conserved among species, as happens with the Notch family. Nrf2 heterodimerizes with small Maf protein (sMaf) to elicit the most potent gene expression mediated through the ARE transcriptosome. Global gene expression analyses in Nrf2 null mouse embryonic fibroblasts or in Nrf2 null mice have revealed that ARE containing genes regulated by the Nrf2 include a wider array of genes that have important biological functions for cell and organism survival. Furthermore, these studies have confirmed that Nrf2 can contribute to the basal and/or induced or repressed expression of its target genes, depending on the particular gene and its cellular context [18].
Keap1 was strategically discovered using the yeast two-hybrid system based on the predicted existence of a negative regulator acting through the Neh2 domain of Nrf2 [22]. Keap1 possesses bric à brac, tram track and broad complex (BTB) domains and β-propeller structures carrying 6-blades contained in the Kelch domain. Keap1, principally localized to the cytoplasm, serves as a scaffold for the degrasome complex for Nrf2 by coupling with the Cul3-Ubiquitin-system [23]. Keap1 null mice died postnatally due to malnutrition caused by constriction of the esophagus and cardia of the stomach due to hyper-keratinization. However, the Keap1 null mice were rescued completely by deleting the Nrf2 gene [24]. Furthermore, the relative expression of most all Nrf2 target genes, such as Nqo1, Gclc and Gclm, which were initially defined by comparisons between Nrf2 null and wild type mice, has a generally inverse relation with the relative expression of the same genes when comparing Keap1 null and wild type mice, due to the constitutive expression of Nrf2 in the Keap1 null mice. Thus, the specificity of Keap1-Nrf2 molecular interaction was elucidated in vivo. ChIP-seq and other approaches have greatly subsequently expanded the list of Nrf2 target genes [25].
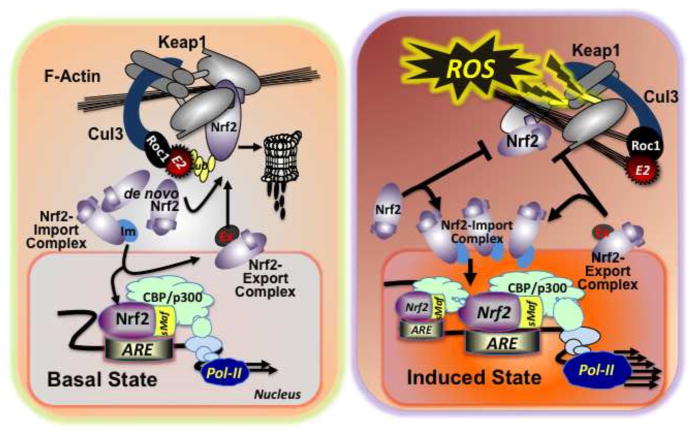
The essence of the Nrf2-ARE signaling system lies in the sequestration of Nrf2 away from the nucleus in the basal state and the translocation and accumulation in the nucleus as a rapid response to stressors including ROS in the induced state [18]. Mechanistically, in the basal state, the majority of de novo synthesized Nrf2 is undergoing proteasomal degradation. When phosphorylation occurs on the Neh6 domain of Nrf2, the Cul1-β-Trcp system can modify it, either in the cytoplasm or nucleus [26]; however, the Keap1-Cul3 system marks de novo synthesized Nrf2 through polyubiquitination leading to rapid proteasomal degradation. In the nucleus, the exportin complex transports Nrf2 to the cytoplasm. In this situation, basal expression of target genes is maintained. Whereas, in the induced state, modification of the Keap1-Cul3 complex through interactions of reactive cysteines by electrophiles or ROS leads to conformational changes disrupting the degrasome function of the complex. This outcome allows newly synthesized Nrf2 access to the nucleus and results in altered expression of target genes [27] (Figure 2).
Figure 2.
Keap1-Nrf2-ARE signaling. In the basal state (left), the Keap1-Cul3 system captures de novo synthesized Nrf2 and marks it by polyubiquitination for rapid proteasomal degradation. In the nucleus, exportin forms a complex with Nrf2, and transports Nrf2 to the cytoplasm. In this way, basal levels of target gene expression can be controlled. In the induced state (right), the Keap1-Cul3 degrasome complex is disabled, due to conformational changes in Keap1 evoked by oxidative or electrophilic stressors. This alteration allows de novo synthesized Nrf2 to translocate to the nucleus readily, resulting in accumulation and heterodimerization at AREs and increased or suppressed expression of target genes.
Reciprocal Nrf2-Notch transcriptional regulation in the hepatobiliary system
Nrf2 to Notch1 signaling
Nrf2 and Notch1 expression in MEF
Differential gene expression analyses using microarrays, and more recently ChIP-Seq and RNA-Seq technologies, have been the most effective ways to provide insights into the downstream targets of signaling pathways. In this regard, many microarray analyses have been performed in various tissues of wild-type and Nrf2 null mice to define cytoprotective pathways and to characterize the role of the Keap1-Nrf2-ARE pathway in the pharmacodynamic action of several classes of anticarcinogens [28]. One limitation of using microarray gene expression data from tissues is that the observed differential gene expression patterns are likely to be affected by the presence of heterogeneous cell populations. For this reason our initial gene expression analysis utilized immortalized mouse embryonic fibroblasts (MEF) from Keap1−/−, Nrf2−/−, Keap1−/−::Nrf2−/−, and wild-type mice so as to compare gene expression patterns among homogeneous cell populations [29]. This improved approach led to the observation of differential Notch1 expression that was dependent on the content of Nrf2 in the MEF. Follow-up analyses revealed that the regulatory region of the mouse Notch1 gene possessed a functional ARE on the proximal region of the promoter. Interestingly, Notch-signaling was debilitated in the Nrf2 null MEF. Expression of the direct downstream target genes of Notch signaling, such as Hes/Herp and Nrarp, was dampened in Nrf2 null MEF compared with wild type MEF when co-cultured with human cells constitutively expressing either a delta or jagged Notch-ligand. Hence, the Nrf2-ARE transcriptional machinery on the Notch1 promoter directly influenced the magnitude of Notch-signaling [28].
ARE function in the Notch1 promoter during embryogenesis
Nrf2 null mice are viable, fertile and have not displayed evidence of developmental defects independent of the genetic backgrounds of specific mouse strains [30–32]. By contrast, Notch1 disrupted mice show complete embryonic lethality during organogenesis; most mutants are dead at E10.5 and all die by E11.5 [33]. This phenotypic difference in lethality between Nrf2 null and Notch1 null mice could either mean that the ARE-driven Notch1 gene expression may not contribute to embryonic survival or some other CNC transcription factors, such as Nrf1 and Nrf3, may associate with the ARE to induce Notch1 gene expression. If the ARE on the Notch1 gene regulatory region is functional during embryogenesis and taking into account the lethal phenotype of Nrf1 null mice at mid-late gestation [34, 35], Nrf1 might be implicated in the Notch1-ARE machinery. In addition, Nrf1 and Nrf2 double null mice occur at about 30% of theoretical Mendelian rate at E11.5 but are not observed at E13.5 [36]. This outcome suggests that Nrf2 may compensate in the embryonic form of the Notch1-ARE machinery over a short term in Nrf1 null mice.
Nrf2 and Notch1 coexist in adult hepatocytes
Due to remarkable advances in mouse developmental engineering, such as conditional knockout and conditional, cell-specific gene expression, it has became feasible to generate mouse models with both loss and gain of functions in the Notch or Nrf2 signaling pathways and to study the outcomes in adult tissues. Although both Notch1 and Notch2 gene deletions among the 4 Notch receptor genes are embryonic lethal, the use of conditional gene knockout or overexpression led to the conclusion that Notch1 is dominantly expressed in hepatocytes as evidenced by Notch1-EGFP reporter mice analyses[37]. Furthermore, Notch2 is expressed in cholangiocytes and in the common progenitor cells of both hepatocytes and cholangiocytes as supported by Notch2-LacZ knock-in mouse analyses using an X-gal staining assay [37]. The Nrf2-ARE signaling system is also of vital importance in these cells, regulating an adaptive response against stressors to maintain liver function [18]. Given the huge hepatocyte population in parenchymal cells, Notch1 is apparently the dominant Notch in the liver. Hence, a crosstalk between Notch1 and Nrf2 pathways in hepatocytes can be envisioned to be of essential biological importance.
Nrf2 to Notch1 during liver regeneration
After partial hepatectomy (PH), the existing mature hepatocytes leave their quiescent status and replicate, followed by replication of the non-parenchymal cells of the liver. In this setting, the regenerative response does not require activation of the hepatic progenitor cell compartment, which is proposed to be located in the canals of Hering and is represented by oval cells in rodent liver [38]. When considering the biological importance of Notch and Nrf2 signaling crosstalk, the hypothesis for a role in cell proliferation and tissue repair to maintain liver homeostasis was raised, based on the following observations:
When Notch1 expression was knocked down in the liver of rats by injection of a Notch1-siRNA expression vector 2 days before a two-thirds PH, the proliferation of hepatocytes at days 2 to 4 during the regenerative response was significantly suppressed [39].
A phenotype of a delayed early regrowth phase is observed in the Nrf2 null mice following PH [40].
Nrf2 null alveolar epithelial cells enter cell-cycle arrest [41].
In general, liver of wild-type mice recovers to 75% of the weight of sham control mice 3-days after two-thirds PH. However, in the case of Nrf2 null mice, liver weight returns to only 50% of sham control levels at 3 days post PH. In order to investigate whether a reduction in Notch1 signaling caused this retardation in regrowth of livers of Nrf2 null mice, liver specific NICD expressing mice in the Nrf2 null background were established. This Nrf2 null, NICD overexpressing mouse exhibited a complete rescue from the growth retardation phenotype at 3 days post PH. Thus, Nrf2 to Notch crosstalk is functional in the early phase of liver regeneration when hepatocyte self-renewal occurs [42]. Subsequently, this conjoined molecular signaling has been observed in airway basal stem cells, in turn affecting their proliferation and self-renewal states [43]. It could be speculated that protection from endogenous or exogenous stressors and maintenance of vital homeostasis in adult tissue stem cells and their niches could be mediated, at least partially, by directed Nrf2 to Notch1 signaling.
Notch to Nrf2 signaling
Conserved Rbpjκ recognition sequence in Nrf2 gene regulatory regions
As described in the Introduction, signaling crosstalk fundamental for animal survival is often conserved across species. The cis-elements in the regulatory region of the genes playing pivotal roles in the signaling are also usually conserved among animal species. From this point of view, in silico analyses revealed that Rbpjκ recognition sequences are highly conserved in the Nrf2 promoter regions of many species of animals. Reporter gene assays with mutant Rbpjκ sequences, EMSA and ChIP assay using specific antibodies against NICD or Rbpjκ revealed that at least one Rbpjκ sequence in murine Nrf2 and human Nrf2 gene promoters function as a cis-element at the level of cell culture analyses. When Nrf2 signaling in wild-type MEF was assessed following induction of Notch signaling by a co-culture system with human Notch-ligand presenting cells, the chronological expression of transcripts of Nrf2 and its target genes was increased during the early phase of co-culture as well as that of a representative Notch signaling target gene, Hes1. These studies showed that signaling in the direction of Notch to Nrf2 could be observed at the cellular level.
The immediate next question was to determine whether this signaling flow was biologically relevant in vivo by using mouse models. To this end, Nrf2 expression levels in liver specific NICD overexpressing mice (Rosa NICD/− ::AlbCre) were analyzed. It became evident that Notch to Nrf2 signaling was also functional in vivo [44]. All members of the Notch family can produce NICDs that act as transcriptional co-factors. However, the function of each NICD is not always common as there are diversities in NICD target gene expression among Notch1-3 [45, 46]. The mechanistic details for this diversity in response are not understood. A follow-up question is to determine what kind, if any, diversity exists in the responses of the Nrf2 promoter within the hepatobiliary system, in which the Notch1 intracellular domain is expressed in hepatocytes and the Notch2 intracellular domain in cholangiocytes and their common progenitors.
Morphological and functional similarities in liver specific Notch or Nrf2 overexpressing mice
Since the liver specific NICD overexpression mice have higher levels of hepatic Nrf2 signaling, studies comparing them with liver specific Keap1 null mice were conducted. As seen in liver-specific Keap1 null mice, liver specific NICD overexpressing mice showed striking hepatomegaly. Interestingly, in both mouse constructs, liver size was reduced when the Nrf2 gene was deleted. Another common phenotype was observed in the structure of the intrahepatic bile ducts (IHBD). The density of the IHBD had been reported to increase markedly in liver specific NICD overexpressing mice [47]. Resin casting analysis of the liver specific Keap1 null mice also showed a higher density of branches arborizing from major branches compared to the livers of control mice. When the Nrf2 gene was deleted from liver specific NICD overexpressing mice, the excess micro-branched phenotype and the total cast size of bile ducts returned to controls level. Thus, Notch to Nrf2 signaling probably contributes to total liver size and bile duct formation [44]. Sparks et al reported on ALT activity in a series of Notch-related genetic mutant mice (liver specific Notch2, both Notch1 and Notch2, Rbpjκ knock out mice). They showed that deletion of Notch signaling from parenchymal cells and its progenitor cells at postnatal days led to higher levels of serum ALT [47], which is indicative of damage to hepatocytes in the basal state.
Therefore, to better assess liver functionality, the NICD overexpressing mice were challenged with the hepatotoxin acetaminophen. These mice displayed resistance against acetaminophen, as adjudged by protection against elevated serum ALT levels. However, when the Nrf2 gene was disrupted from these mice, the protective effect against acetaminophen hepatotoxicity evoked by enhanced Notch signaling was eliminated [44]. This result indicated that the cytoprotective effect of Notch signaling might be mediated through Nrf2 signaling. Thus, Notch to Nrf2 directional signaling is likely relevant to cytoprotection in the liver.
Molecular signaling networks in hepatic regeneration
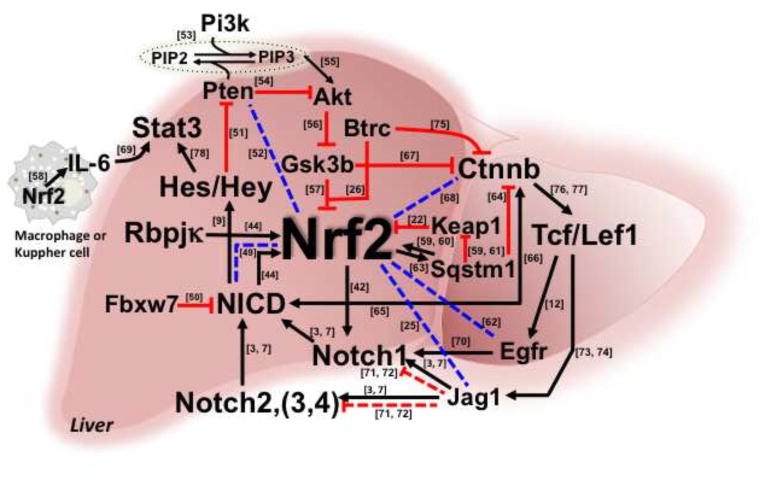
Delays in liver regeneration have been demonstrated by blocking of signals mediated by Nrf2, Notch, TNF, components of complement and IL6, bile acids, serotonin and norepinephrine, among others [48]. There is clearly a multiplicity of pathways affecting the kinetics of cellular proliferation that are involved in the initiation and termination of liver regeneration. The interplay between different transcription factors and signaling molecules are not clearly established. The molecular roles of Nrf2 and/or Notch in the reconstitution of the hepatic environment, replacement and repopulation of parenchylmal and non-parenchymal cells following damage or during regeneration are unknown. It is unlikely that there is a single driver for liver regeneration or indeed other areas of regenerative biology [48]. Indeed, cross-talk between multiple pathways certainly underlie the regenerative response. A network of interactions and responses with Nrf2 artificially set at the nexus is presented in Figure 3. Connections are based on literature reports, but the depth and function of this interactome of signaling factors has not been established. The molecules listed in this figure are confirmed to be expressed in the liver.
Figure 3.
Integrated network of Nrf2 signaling that may affect liver regeneration. Published (solid lines) and hypothetical (broken lines) elements of Nrf2 crosstalk with other signaling networks. Arrows indicate induction and “T” indicates repression. Numbers beside lines link to relevant references.
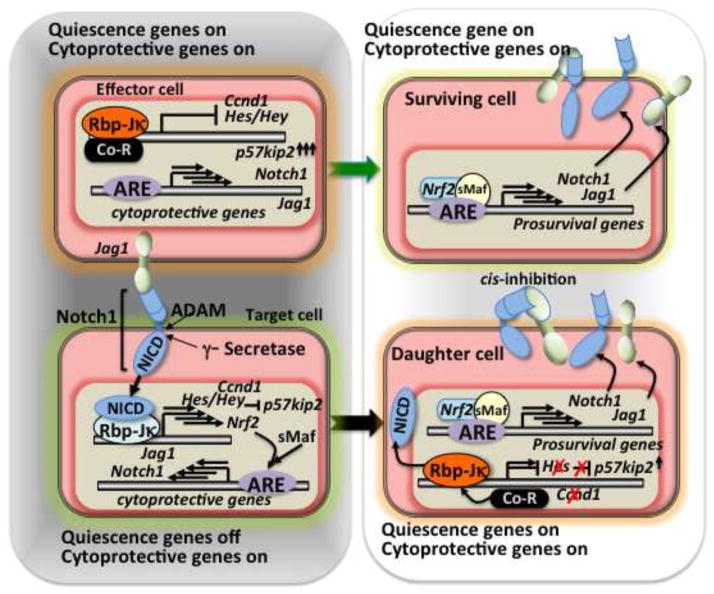
When hepatocytes encounter a regenerative stress condition, both the effector and target cells of Notch signaling likely need to enhance Nrf2 signaling (Figure 4, left). In the case of the effector cells, they need to express quiescence mediating genes such as p57kip2 [79, 80] together with cytoprotective gene expression through Nrf2 signaling (Figure 4, left and right upper). Consequently, the effector cells succeed in providing the ligand to Notch signaling target cells, contributing both to their protection and that of the target cells. The major role of the target hepatocytes is to renew cell number through one round of mitosis [81]. In order to quickly allow for mitosis under a setting of stress, Hes/Hey family genes, which are transcriptional repressors and direct target genes of Notch, might turn off the expression of quiescence gene. Cell cycle promoter genes such as Ccnd1, which is also Notch target gene [82], simultaneously begin to be expressed along with cytoprotective genes through coupled Notch-Nrf2 signaling. Therefore, higher levels of Nrf2 expression might be expected at the transcriptional level (Figure 4, lower left). How signaling for hepatic regeneration is terminated back into the mode of quiescence is unclear. However, the putative Nrf2 target gene [25] Jag1, one of the Notch-ligands expressed in postnatal liver, could play a role in cis-inhibition of Notch signaling [71, 72] (Figure 4, right). Perhaps the biological importance of the Notch-Nrf2 axis in the liver regeneration lies in the early phase of cell division to compensate for the triggering loss of the cells.
Figure 4.
Role of Nrf2-Notch1 crosstalk in the hepatic regeneration.
Left: Early phase of liver regeneration, Right: terminal phase of liver regeneration.
Lessons from genetically engineered mice
Biological significance of Nrf2-Notch crosstalk
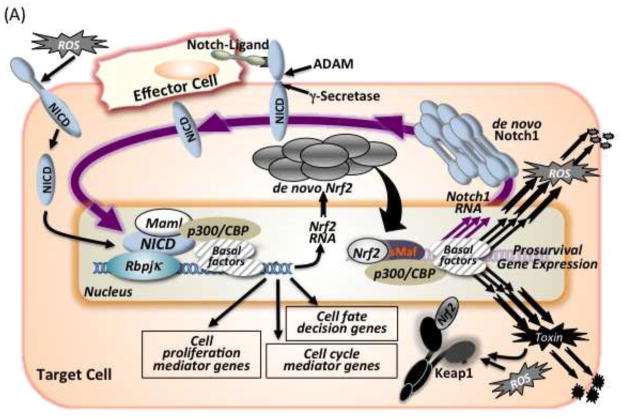
In general, Nrf2 is widely expressed in tissues and various cell types. However, careful analysis of the data by the Kan group [30] indicates that Nrf2 mRNA expression patterns are not held at a constant level. Their in situ hybridization analyses clearly showed differences in expression levels among tissues and cells types. Hence the conclusion “The various cells could express Nrf2 gene opportunely when it is required”. This variable expression pattern makes Nrf2-Notch crosstalk much more suitable for promulgating survival of cells and organisms. As Nrf2 disruption has not shown any apparent phenotype during embryogenesis, Nrf2-Notch crosstalk might be less important or perhaps inactive during gestation. However, by postnatal days Nrf2-Notch crosstalk could be harnessed in tissues that possess the functions of regeneration, repair or compensation of damaged or injured cells, especially in the liver and digestive tract. Furthermore, in those tissues, adult stem cells have been discovered [83]. This crosstalk can probably influence the adaptive defenses of the stem cells and be important for the maintenance of homeostasis of the niche. In addition to hepato-cholagionenesis in adult liver [47, 84, 85], the airways basal stem cell system for proliferating or self-renewal in the lung [43], enterogenesis in the intestinal crypt [86], adult neurogenesis occurring in the subventricular zone and/or the dentate gyrus of the hippocampus in the central nervous system [15, 87, 88], dermatogenesis in the skin [17, 89, 90], osteogenesis in bone [91, 92], cell fate determination in hematopoiesis [93, 94] and angiogenesis in various tissues [95] might all be valuable settings for further evaluation (Figure 5A).
Figure 5.
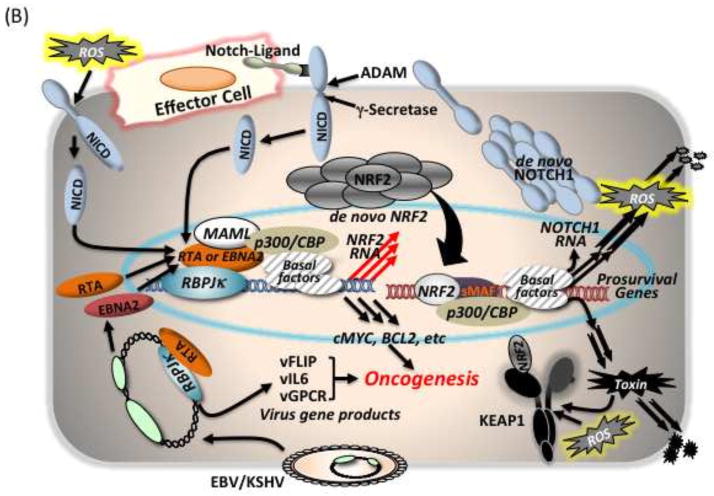
Notch-Nrf2 axis. (A) Possible links for biological events occurring in postnatal tissues through this axis. (B) Scheme for hijacking of the NOTCH-NRF2 axis in humans by viral infections such as EBV or KSHV, thereby facilitating oncogenesis. Infected cells express EBNA2 or RTA, which are viral genome products exhibiting NICD mimicry structures for association with Rbpjκ. This mimicry enhances expression of NOTCH target genes such as cMYC, BCL2 and possibly NRF2. In the case of KSHV, RTA can recruit cellular RBPJκ onto its gene promoters for expression of viral genes such as vFLIP, vIL6 and vGPCR. These viral and abused cellular gene products together might contribute to oncogenesis. NRF2 signaling consequently supports the survival of the virus infected cell.
Possible endogenous trigger of the Notch-Nrf2 axis: ROS
Reactive oxygen species (ROS) are formed as byproducts of the normal metabolism of oxygen and have important roles in cell signaling and homeostasis. They are also inducers of Nrf2-ARE signaling. Recently, it was reported that ROS could activate the Notch pathway in fibroblasts derived from a systemic sclerosis model mouse. In these mice, skin fibrosis is induced by intradermal generation of HOCl that oxidizes skin proteins. This stress leads to the stimulation and proliferation of fibroblasts and to the overproduction of collagen and then to fibrosis [96]. The Keap1-Nrf2-ARE pathway likely contributes to this process. Simultaneously, intradermal ROS can induce the synthesis of the metalloprotease ADAM17, which is a trigger protease in the first step of Notch activation and cleaves the ectodomain of the Notch receptor. ROS-induced ADAM17 could be another major factor for activation of the Notch pathway in systemic sclerosis [97, 98]. In this way, ROS might function as a dual trigger of Nrf2-Notch crosstalk.
Dark side of activation of both signaling pathways (oncogenesis)
It is estimated that 15% of all human tumors worldwide are caused by viruses [99]. Viruses are usually not complete carcinogens, and the known human cancer viruses display different roles in transformation. However, infections are considered to increase the risk of cancer. In general, viral gene expression and replication are closely linked to the differentiation state of the infected cell. The Notch pathways have been established as attractive targets for virus interaction and manipulation as in the cases of the γ-herpes viruses (Epstein-Barr virus <EBV> and Kaposi’s sarcoma-associated herpes virus <KSHV> [100]) as well as HTLV-1 (Human T-cell leukemia virus type 1) [101]. The EBV product, EBNA2, which is one of the first viral genes expressed after infection, can function as a transcription co-factor with Rbpjκ instead of endogenous NICD, due to its structural mimicry [102–104]. Then, EBV can manipulate cellular NICD target gene expression and also contribute to the expression of EBV genes by recruiting cellular Rbpjκ onto its promoter regions. In parallel, the KSHV product, replication and transcription activator protein (RTA) [105], is also known to exploit Notch signaling in a similar manner as EBV in host cells. The establishment of a lifelong persistent infection in the host is a critical strategy for the survival of the virus.
Independently, many studies on human cancers have reported an augmentation of NRF2 signaling. As the prototypic NRF2 target genes fall into the categories of detoxification and antioxidant enzymes, increased NRF2 signaling may be a spontaneous “cell decision” in cancer cells and provide them an apparent advantage encompassing enhanced cell proliferation and metabolic switching as well as in survival against cytotoxic cancer drugs and irradiation. Thus, the hijacking of the NOTCH-NRF2 axis might be one of the key pathways to promote cancer. The NICD mimicry EBV-product, EBNA2, might induce NRF2 gene expression in human cells, as could RTA expressed from KSHV genome following infection [106]. This means that both EBV and KSHV might be able to express the cell survival factor NRF2 in infected cells at least at the RNA level. In the case of KSHV, it was demonstrated and seemed to be important that the latent KSHV protein, vFLIP (Fas-associated death domain-like IL-1-converting enzyme-inhibitory protein) increased SQSTM1 expression and led to stabilization of NRF2 [107]. Thus, KSHV could dually activate the NRF2 pathway during oncogenesis by hijacking canonical NOTCH signaling transcriptionally and manipulating the autophagic pathway of KEAP1 degradation through induction of SQSTM1 post-translationally (Figure 5B).
An extensive study of somatic point mutations in 4742 human cancers and their matched normal-tissue samples (‘tumor–normal pairs’) across 21 tumor types has been reported recently[108]. NRF2, KEAP1 and NOTCH1 appear in the list as frequently mutated genes in selected tumor types and were deemed to be significant in both the combined sets of tumors and in individual tumor types. This observation leads to the speculative notion that the outcome from aberrant NRF2-NOTCH crosstalk by mutation in these genes might enhance tumorigenesis and progression to cancer. Examination of the exclusiveness of mutations in both pathways within a tumor might further buttress of an important role for aberrant crosstalk. Exclusive mutations in NOTCH1 and NRF2 may prevail in head and neck squamous cell carcinomas [109]. In addition, NOTCH signaling already has been shown to participate in another transcriptional network that includes WNT signaling [77, 110], which also participates in oncogenesis.
Possible down-regulation mechanisms of Nrf2-Notch crosstalk
Through Notch-Nrf2 crosstalk studies, it has been clearly shown that NICD can activate the Nrf2 pathway and Nrf2 can activate the Notch1 pathway in the liver. This interaction leads to a positive feedback loop system; a similar genetically-driven linkage may play a role in oncogenesis. Presumably, a system for down-regulation of this crosstalk should exist for the maintenance of homeostasis in healthy cells. Perhaps target gene products of each signaling program contribute to a negative feedback mechanism for regulation of Notch and/or Nrf2 signaling. These factors may act as direct transcriptional repressors for Notch genes and/or Nrf2 gene expression. Furthermore, epigenetic factors such as histone acetyltransferases, histone deacetylases and DNA methyltransferases which modify chromatin structure, may gradually alter gene expression leading to cellular homeostatic recovery. Post-translational modifications that affect Nrf2 and/or Notch (NICD) degradation machinery may allow for a quicker negative feedback response than modulation at the transcriptional level. In any case, these processes may be linked to anti-oncogenic pathways, leading to diminished proliferation and prompt cell elimination.
Conclusions
Nrf2-ARE signaling and Notch signaling can be regulated by reciprocal transcriptional machinery, at least postnatally. Namely, Notch1 is an Nrf2 target gene and Nrf2 is a Notch-Rbpjκ target gene. Nrf2-Notch crosstalk influences the expression of defense systems against endogenous and exogeneous stressors leading to cytoprotection and enhances maintenance of cellular homeostasis and tissue organization through actions on cell proliferation kinetics and cell fate determinations of stem cell renewal and cell specification of differentiation. These actions may vary amongst tissues and particularly within specific regions, such as the niche where adult tissue stem cells or progenitor cells reside. At this point, it is clear that Nrf2-Notch crosstalk can produce a positive feedback gene expression response. Detailed mechanisms for the down regulation of this crosstalk remain to be characterized. Given the importance of this crosstalk on cell fate, pharmacological interventions targeting aspects of this transcription factor collaboration through agents that interfere with Notch signaling or inducers or inhibitors that influence Nrf2 signaling are likely to have roles in the prevention of tumorigenesis and suppression of tumor growth. A fuller appreciation of the role of cellular and tissue context will be required however, to assure the assiduous use of such strategies.
Highlights.
Reciprocal Nrf2-Notch transcriptional regulation in the hepatobiliary system
Molecular signaling networks in hepatic regeneration
Biological significance of Nrf2-Notch crosstalk
Acknowledgments
This work is supported by National Institutes of Health grants, CA0949076 and CA197222 to TWK. DVC is supported by a Marie Curie IOF (PIOF-GA-2012-329442) within the 7th European Community Framework Programme.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, Conlon RA. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 2.Alam J. The Mammalian Cap and Collar Family of Transcription Factors. Antioxid Redox Signal. 2006;8:39–42. doi: 10.1089/ars.2006.8.39. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 4.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 5.Andersen P, Uosaki H, Shenje LT, Kwon C. Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. 2012;22:257–265. doi: 10.1016/j.tcb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayaz F, Osborne BA. Non-canonical notch signaling in cancer and immunity. Front Oncol. 2014;4:345. doi: 10.3389/fonc.2014.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol. 2009;29:5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 10.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan X, Apte U, Micsenyi A, Kotsagrelos E, Luo JH, Ranganathan S, Monga DK, Bell A, Michalopoulos GK, Monga SP. Epidermal growth factor receptor: a novel target of the Wnt/beta-catenin pathway in liver. Gastroenterology. 2005;129:285–302. doi: 10.1053/j.gastro.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 14.Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Lee PP, Wilson CB, Held W, MacDonald HR, Radtke F. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 15.Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 16.Koch U, Radtke F. Notch in T-ALL: new players in a complex disease. Trends Immunol. 2011;32:434–442. doi: 10.1016/j.it.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 18.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 19.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc Natl Acad Sci U S A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 22.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 25.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh K, Mimura J, Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 28.Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2002;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 30.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 32.Skoko JJ, Wakabayashi N, Noda K, Kimura S, Tobita K, Shigemura N, Tsujita T, Yamamoto M, Kensler TW. Loss of Nrf2 in mice evokes a congenital intrahepatic shunt that alters hepatic oxygen and protein expression gradients and toxicity. Toxicol Sci. 2014;141:112–119. doi: 10.1093/toxsci/kfu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huppert SS, Le A, Schroeter EH, Mumm JS, Saxena MT, Milner LA, Kopan R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 34.Farmer SC, Sun CW, Winnier GE, Hogan BL, Townes TM. The bZIP transcription factor LCR-F1 is essential for mesoderm formation in mouse development. Genes Dev. 1997;11:786–798. doi: 10.1101/gad.11.6.786. [DOI] [PubMed] [Google Scholar]
- 35.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. Embo J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 37.Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, Radtke F, Schmid RM, Siveke JT. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607–616. doi: 10.1002/hep.22381. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 39.Kohler C, Bell AW, Bowen WC, Monga SP, Fleig W, Michalopoulos GK. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 2004;39:1056–1065. doi: 10.1002/hep.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. Embo J. 2008;27:212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy NM, Kleeberger SR, Bream JH, Fallon PG, Kensler TW, Yamamoto M, Reddy SP. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene. 2008;27:5821–5832. doi: 10.1038/onc.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, Kensler TW. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul MK, Bisht B, Darmawan DO, Chiou R, Ha VL, Wallace WD, Chon AT, Hegab AE, Grogan T, Elashoff DA, Alva-Ornelas JA, Gomperts BN. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent Notch signaling. Cell Stem Cell. 2014;15:199–214. doi: 10.1016/j.stem.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakabayashi N, Skoko JJ, Chartoumpekis DV, Kimura S, Slocum SL, Noda K, Palliyaguru DL, Fujimuro M, Boley PA, Tanaka Y, Shigemura N, Biswal S, Yamamoto M, Kensler TW. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol. 2014;34:653–663. doi: 10.1128/MCB.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu K, Chiba S, Saito T, Kumano K, Hamada Y, Hirai H. Functional diversity among Notch1, Notch2, and Notch3 receptors. Biochem Biophys Res Commun. 2002;291:775–779. doi: 10.1006/bbrc.2002.6528. [DOI] [PubMed] [Google Scholar]
- 46.Bellavia D, Checquolo S, Campese AF, Felli MP, Gulino A, Screpanti I. Notch3: from subtle structural differences to functional diversity. Oncogene. 2008;27:5092–5098. doi: 10.1038/onc.2008.230. [DOI] [PubMed] [Google Scholar]
- 47.Sparks EE, Huppert KA, Brown MA, Washington MK, Huppert SS. Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology. 2010;51:1391–1400. doi: 10.1002/hep.23431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kraman M, McCright B. Functional conservation of Notch1 and Notch2 intracellular domains. Faseb J. 2005;19:1311–1313. doi: 10.1096/fj.04-3407fje. [DOI] [PubMed] [Google Scholar]
- 50.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 51.Kim SJ, Lee HW, Baek JH, Cho YH, Kang HG, Jeong JS, Song J, Park HS, Chun KH. Activation of nuclear PTEN by inhibition of Notch signaling induces G2/M cell cycle arrest in gastric cancer. Oncogene. 2015 doi: 10.1038/onc.2015.80. [DOI] [PubMed] [Google Scholar]
- 52.Rojo AI, Rada P, Mendiola M, Ortega-Molina A, Wojdyla K, Rogowska-Wrzesinska A, Hardisson D, Serrano M, Cuadrado A. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxid Redox Signal. 2014;21:2498–2514. doi: 10.1089/ars.2014.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toker A, Cantley LC. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 55.Delcommenne M, Tan C, Gray V, Rue L, Woodgett J, Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci U S A. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pap M, Cooper GM. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J Biol Chem. 1998;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 57.Rojo AI, Sagarra MR, Cuadrado A. GSK-3beta down-regulates the transcription factor Nrf2 after oxidant damage: relevance to exposure of neuronal cells to oxidative stress. J Neurochem. 2008;105:192–202. doi: 10.1111/j.1471-4159.2007.05124.x. [DOI] [PubMed] [Google Scholar]
- 58.Wruck CJ, Streetz K, Pavic G, Gotz ME, Tohidnezhad M, Brandenburg LO, Varoga D, Eickelberg O, Herdegen T, Trautwein C, Cha K, Kan YW, Pufe T. Nrf2 induces interleukin-6 (IL-6) expression via an antioxidant response element within the IL-6 promoter. J Biol Chem. 2011;286:4493–4499. doi: 10.1074/jbc.M110.162008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 60.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu M, Zou Y, Nambiar SM, Lee J, Yang Y, Dai G. Keap1 modulates the redox cycle and hepatocyte cell cycle in regenerating liver. Cell Cycle. 2014;13:2349–2358. doi: 10.4161/cc.29298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petherick KJ, Williams AC, Lane JD, Ordonez-Moran P, Huelsken J, Collard TJ, Smartt HJ, Batson J, Malik K, Paraskeva C, Greenhough A. Autolysosomal beta-catenin degradation regulates Wnt-autophagy-p62 crosstalk. Embo J. 2013;32:1903–1916. doi: 10.1038/emboj.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, Taga T. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol. 2008;28:7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ross DA, Kadesch T. The notch intracellular domain can function as a coactivator for LEF-1. Mol Cell Biol. 2001;21:7537–7544. doi: 10.1128/MCB.21.22.7537-7544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yost C, Farr GH, 3rd, Pierce SB, Ferkey DM, Chen MM, Kimelman D. GBP, an inhibitor of GSK-3, is implicated in Xenopus development and oncogenesis. Cell. 1998;93:1031–1041. doi: 10.1016/s0092-8674(00)81208-8. [DOI] [PubMed] [Google Scholar]
- 68.Brigelius-Flohe R, Kipp AP. Selenium in the redox regulation of the Nrf2 and the Wnt pathway. Methods Enzymol. 2010;527:65–86. doi: 10.1016/B978-0-12-405882-8.00004-0. [DOI] [PubMed] [Google Scholar]
- 69.Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 70.Kitade M, Factor VM, Andersen JB, Tomokuni A, Kaji K, Akita H, Holczbauer A, Seo D, Marquardt JU, Conner EA, Lee SB, Lee YH, Thorgeirsson SS. Specific fate decisions in adult hepatic progenitor cells driven by MET and EGFR signaling. Genes Dev. 2013;27:1706–1717. doi: 10.1101/gad.214601.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.del Alamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 2011;21:R40–47. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 72.Wang R, Liu K, Chen L, Aihara K. Neural fate decisions mediated by trans-activation and cis-inhibition in Notch signaling. Bioinformatics. 2011;27:3158–3165. doi: 10.1093/bioinformatics/btr551. [DOI] [PubMed] [Google Scholar]
- 73.Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133:4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- 74.Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, Lopez-Bigas N, Bellora N, Alba MM, Torres F, Dunach M, Sanjuan X, Gonzalez S, Gridley T, Capella G, Bigas A, Espinosa L. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marikawa Y, Elinson RP. beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech Dev. 1998;77:75–80. doi: 10.1016/s0925-4773(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 76.Willert K, Shibamoto S, Nusse R. Wnt-induced dephosphorylation of axin releases beta-catenin from the axin complex. Genes Dev. 1999;13:1768–1773. doi: 10.1101/gad.13.14.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson MD, Monga SP. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 78.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 79.Jia J, Lin M, Zhang L, York JP, Zhang P. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol Cell Biol. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyaoka Y, Miyajima A. To divide or not to divide: revisiting liver regeneration. Cell Div. 2013;8:8. doi: 10.1186/1747-1028-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 84.Ishimura N, Bronk SF, Gores GJ. Inducible nitric oxide synthase up-regulates Notch-1 in mouse cholangiocytes: implications for carcinogenesis. Gastroenterology. 2005;128:1354–1368. doi: 10.1053/j.gastro.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 85.Fiorotto R, Raizner A, Morell CM, Torsello B, Scirpo R, Fabris L, Spirli C, Strazzabosco M. Notch signaling regulates tubular morphogenesis during repair from biliary damage in mice. J Hepatol. 2013;59:124–130. doi: 10.1016/j.jhep.2013.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 87.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 88.Taupin P, Gage FH. Adult neurogenesis and neural stem cells of the central nervous system in mammals. J Neurosci Res. 2002;69:745–749. doi: 10.1002/jnr.10378. [DOI] [PubMed] [Google Scholar]
- 89.Vauclair S, Nicolas M, Barrandon Y, Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev Biol. 2005;284:184–193. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 90.Fuchs E. Skin stem cells: rising to the surface. J Cell Biol. 2008;180:273–284. doi: 10.1083/jcb.200708185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hinoi E, Fujimori S, Wang L, Hojo H, Uno K, Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J Biol Chem. 2006;281:18015–18024. doi: 10.1074/jbc.M600603200. [DOI] [PubMed] [Google Scholar]
- 92.Hinoi E, Takarada T, Fujimori S, Wang L, Iemata M, Uno K, Yoneda Y. Nuclear factor E2 p45-related factor 2 negatively regulates chondrogenesis. Bone. 2007;40:337–344. doi: 10.1016/j.bone.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 93.Kim JH, Thimmulappa RK, Kumar V, Cui W, Kumar S, Kombairaju P, Zhang H, Margolick J, Matsui W, Macvittie T, Malhotra SV, Biswal S. NRF2-mediated Notch pathway activation enhances hematopoietic reconstitution following myelosuppressive radiation. J Clin Invest. 2014;124:730–741. doi: 10.1172/JCI70812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murakami S, Shimizu R, Romeo PH, Yamamoto M, Motohashi H. Keap1-Nrf2 system regulates cell fate determination of hematopoietic stem cells. Genes Cells. 2014;19:239–253. doi: 10.1111/gtc.12126. [DOI] [PubMed] [Google Scholar]
- 95.Gridley T. Notch signaling in the vasculature. Curr Top Dev Biol. 2010;92:277–309. doi: 10.1016/S0070-2153(10)92009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Servettaz A, Goulvestre C, Kavian N, Nicco C, Guilpain P, Chereau C, Vuiblet V, Guillevin L, Mouthon L, Weill B, Batteux F. Selective oxidation of DNA topoisomerase 1 induces systemic sclerosis in the mouse. J Immunol. 2009;182:5855–5864. doi: 10.4049/jimmunol.0803705. [DOI] [PubMed] [Google Scholar]
- 97.Kavian N, Servettaz A, Mongaret C, Wang A, Nicco C, Chereau C, Grange P, Vuiblet V, Birembaut P, Diebold MD, Weill B, Dupin N, Batteux F. Targeting ADAM-17/notch signaling abrogates the development of systemic sclerosis in a murine model. Arthritis Rheum. 2010;62:3477–3487. doi: 10.1002/art.27626. [DOI] [PubMed] [Google Scholar]
- 98.Kavian N, Servettaz A, Weill B, Batteux F. New insights into the mechanism of notch signalling in fibrosis. Open Rheumatol J. 2012;6:96–102. doi: 10.2174/1874312901206010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Butel JS. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 100.Hayward SD, Liu J, Fujimuro M. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci STKE. 2006;2006:re4. doi: 10.1126/stke.3352006re4. [DOI] [PubMed] [Google Scholar]
- 101.Pancewicz J, Taylor JM, Datta A, Baydoun HH, Waldmann TA, Hermine O, Nicot C. Notch signaling contributes to proliferation and tumor formation of human T-cell leukemia virus type 1-associated adult T-cell leukemia. Proc Natl Acad Sci U S A. 2010;107:16619–16624. doi: 10.1073/pnas.1010722107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zimber-Strobl U, Strobl LJ, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm GW. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. Embo J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sakai T, Taniguchi Y, Tamura K, Minoguchi S, Fukuhara T, Strobl LJ, Zimber-Strobl U, Bornkamm GW, Honjo T. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J Virol. 1998;72:6034–6039. doi: 10.1128/jvi.72.7.6034-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liang Y, Ganem D. Lytic but not latent infection by Kaposi’s sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc Natl Acad Sci U S A. 2003;100:8490–8495. doi: 10.1073/pnas.1432843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gjyshi O, Bottero V, Veettil MV, Dutta S, Singh VV, Chikoti L, Chandran B. Kaposi’s sarcoma-associated herpesvirus induces Nrf2 during de novo infection of endothelial cells to create a microenvironment conducive to infection. PLoS Pathog. 2014;10:e1004460. doi: 10.1371/journal.ppat.1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gjyshi O, Flaherty S, Veettil MV, Johnson KE, Chandran B, Bottero V. Kaposi’s Sarcoma-Associated Herpesvirus Induces Nrf2 Activation in Latently Infected Endothelial Cells through SQSTM1 Phosphorylation and Interaction with Polyubiquitinated Keap1. J Virol. 2015;89:2268–2286. doi: 10.1128/JVI.02742-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smalley MJ, Dale TC. Wnt signalling in mammalian development and cancer. Cancer Metastasis Rev. 1999;18:215–230. doi: 10.1023/a:1006369223282. [DOI] [PubMed] [Google Scholar]