Abstract
Structural mass spectrometry (MS) is a field with growing applicability for addressing complex biophysical questions regarding proteins and protein complexes. One of the major structural MS approaches involves the use of chemical cross-linking coupled with MS analysis (CX-MS) to identify proximal sites within macromolecules. Identified cross-linked sites can be used to probe novel protein–protein interactions or the derived distance constraints can be used to verify and refine molecular models. This review focuses on recent advances of “zero-length” cross-linking. Zero-length cross-linking reagents do not add any atoms to the cross-linked species due to the lack of a spacer arm. This provides a major advantage in the form of providing more precise distance constraints as the cross-linkable groups must be within salt bridge distances in order to react. However, identification of cross-linked peptides using these reagents presents unique challenges. We discuss recent efforts by our group to minimize these challenges by using multiple cycles of LC–MS/MS analysis and software specifically developed and optimized for identification of zero-length cross-linked peptides. Representative data utilizing our current protocol are presented and discussed.
Keywords: Mass spectrometry, Structure, Chemical cross-linking, Zero-length cross-linking, Cross-link identification, Molecular modeling
1. Applications of structural MS
Structural mass spectrometry (MS) is a field that uses high-resolution mass spectrometry data to interrogate structural parameters of complex biological molecules. It is typically used as a complement to more established structural methods, such as X-ray crystallography and NMR. Recent studies have demonstrated its usefulness for proteins and protein complexes that were diffi-cult to examine by crystallography or NMR [1–4]. Herein, we will focus on chemical cross-linking combined with mass spectrometry (CX-MS) and advances that have been made in this field, with an emphasis on “zero-length” CX-MS.
1.1. Overview of structural MS techniques
Structural MS techniques can be classified into two broad categories: top-down and bottom-up methods. Top-down methods derive structural insights from MS analyses of intact proteins and protein complexes, or large fragments thereof. Bottom-up methods involve MS analyses of peptide mixtures after proteolytic digestion to identify specific amino acid residues that have been chemically modified or cross-linked. These data are then translated into structural information. For example, ion-mobility MS (IM–MS) [5–7] is a top-down technique that takes advantage of the relationships between the shapes and sizes of protein complexes and their flight time induced by an electric field in the presence of a buffer gas. IM–MS provides collisional cross-sections of protein complexes, which can then be used to distinguish between alternative 3D structure models. Advantages of IM–MS include its abilities to quantify the relative abundance of simultaneously existing conformations and to analyze proteins under native conditions [5]. One limitation of the method is that it requires stable transfer of an intact protein or protein complex into the gas-phase for analysis by MS.
Bottom-up methods are generally more flexible than top-down methods, but suffer from high sample complexity, challenging data analyses of MS/MS data, and difficulty in detecting low-abundance modified peptides. Hydroxyl radical footprinting [8–13], also known as oxidative footprinting, is an example of a bottom-up technique that modifies solvent-accessible amino acid side chains with hydroxyl radicals from hydrogen peroxide or irradiated water molecules. Changes in the pattern of modified residues reflect changes in protein surface topology. These changes can be used to probe protein–protein and protein–ligand binding interfaces or to infer the folding process [14]. However, the fact that almost every amino acid residue can be modified in this fashion [10,15–17] greatly expands the search space for downstream peptide identification. Hydrogen–deuterium exchange (HDX) [18–20] utilizes a similar premise as do hydroxyl radical footprinting, but instead of chemical reactivity, it measures solvent accessibility of backbone hydrogen atoms via exchanges of deuterium atoms from deuterium-containing water (D2O). HDX can be used as either a bottom-up approach, or a top-down approach to visualize complex-wide exchange patterns between conformers [21]. Back-exchange (deuterated site reverting back to hydrogen) is a major concern for this technique, and special sample preparation steps coupled with rapid MS analysis are required to minimize its effects [20].
CX-MS [22–26] is a bottom-up technique that provides structural information in the form of spatial constraints between reactive amino acid side chains. Chemical cross-linking has been used to probe proteins since the 1970s [27], but the difficulty of identifying cross-linked peptides and specific cross-linked residues has limited its use until recently. Interest in chemical cross-linking has increased steadily over the past decade primarily due to improvements in mass spectrometry resolution and mass accuracy, the development of cross-linkers designed specifically for mass spectrometer analysis, and development of specific data analysis tools and strategies. Currently, even structures of large multi-subunit protein complexes and expansive protein–protein interaction networks can be probed effectively with this technique [4,28–31]. Moreover, applications of CX-MS are not limited to the use of tryptic peptides, as it has also been used to analyze intact proteins [26].
Modern non-zero-length cross-linker designs follow a basic template and contain features that belong to four broad aspects: cross-linking reactivity, an optional enrichment tag, an optional isotopic label, and an optional MS-cleavable site [24]. The optional features are generally located in the spacer arm, which is the set of atoms from the reagent that are chemically inserted between the two cross-linked amino acid side chains. Reactivity is defined by the amino acid side chains that are amendable to cross-linking for a given chemistry, with some of the commonly utilized reactivities being amine-to-amine, carboxyl-to-amine, cysteine-to-cysteine, and cysteine-to-non-specific (photo-activated). Some of these cross-linkers also involve enrichment tags such as biotin [32]. Isotopically-labeled cross-linkers are 1:1 mixtures of a single cross-linking reagent that have “heavy” and “light” versions of the spacer arm. By performing a cross-linking reaction with these reagents, all cross-linked peptides should have similar yields of peptide (precursor) ions that differ in mass by an amount equal to the mass difference of the heavy and light linker regions. In an LC–MS/MS experiment, signals corresponding to cross-linked peptides can be differentiated from non-cross-link background by this unique heavy-light pattern of co-eluting peptides. MS-cleavable sites are labile bonds that dissociate upon low-energy fragmentation within the mass spectrometer. These sites are either attached to a reporter ion as a marker of cross-link-specific MS/MS spectra [33], or located within the backbone, or spacer arm, of the cross-linker to allow the two peptides forming the cross-link to be separated for individual MS3 analysis [34]. However, the addition of isotopic tags usually requires a spacer arm of >7 Å and incorporation of enrichment tags, and MS-cleavable tags further increase spacer arm lengths to >20 Å. This allows residues that are located relatively far apart in the structure of the protein or protein complex to be cross-linked and therefore greatly reduces the stringency of the derived distance constraints.
2. Zero-length chemical cross-linking
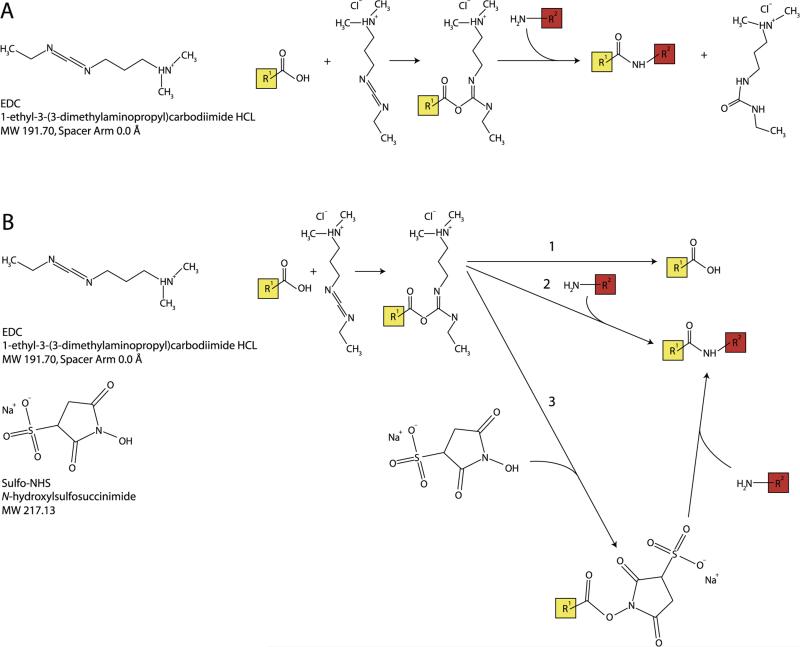
As a systematic in silico analysis showed [23], zero-length cross-links provide the most specific and most powerful distance constraints for refinement and verification of homology models. This is because zero-length cross-linkers do not incorporate atoms from a spacer arm between the reactive sites, which implies that only residues whose side chains are within salt bridge distance can interact. The most common example of a zero-length cross-linker is 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide hydrochloride (EDC). This cross-linker creates an unstable reactive acylisourea ester upon interaction with a carboxyl amino acid residue such as Asp, Glu, or the C-terminal carboxyl group of the protein. The intermediate ester can then interact with a primary amine (Lys or protein N-terminal amine) to form a peptidyl bond with the elimination of a water molecule. Ordinarily, this intermediate species has a lifetime of several seconds; but addition of a second reagent, N-hydroxysuccinimide (NHS) or its water-soluble sulfo-derivative (sulfo-NHS), greatly extends the lifetime of this reactive intermediate, and thus significantly enhances the extent of cross-linking [35,36]. For further details on the chemistry of this reaction, please see Fig. 1.
Fig. 1.
Zero-length cross-linking reaction chemistry. (A) Chemical formula for EDC (left) and forward cross-linking reaction involving an acylisourea intermediate (right) (B) Chemical formulas for EDC and sulfo-NHS (left) and reaction chemistry for zero-length cross-linking reaction, including: (1) the reverse reaction, (2) cross-link formation, and (3) stabilization of the acylisourea intermediate via substitution for an amine-reactive sulfo-NHS ester, followed by cross-link formation.
2.1. Advantages of zero-length CX-MS
Zero-length cross-links offer several key advantages over their longer counterparts. As mentioned above, the primary advantage is more precision on the derived distance constraints, which provide more valuable information for structural model building. This was effectively demonstrated by an in silico analysis that showed that substantially fewer zero-length cross-links were required to accurately reproduce a known protein structure via a modeling experiment, and to discard erroneous structures generated by modeling [23]. Moreover, because the reaction can only occur between residues that are roughly within salt bridge distance of each other, residue interactions captured by this method are more likely to describe direct contact sites rather than simply sites that are in close proximity [37]. In addition, zero-length cross-linking reagents are less likely to generate “dead-end” products, which occur when only one of the reactive groups is able to react with a site on a protein and the second site reacts either with water or a quenching reagent, compared to other cross-linkers. For EDC, this is because the intermediate ester is relatively unstable and reverts back to the unmodified carboxyl if it does not react with an amine. As a result, EDC dead-end adducts have only been observed at high concentrations of cross-linker [38]. In contrast, dead-end products are common and can often have higher stoichiometries than actual cross-links for other cross-linking chemistries. Dead-end products occur when only one of the reactive groups is able to react with a site on a protein and the second site either reacts with water or a quenching reagent. This substantially complicates analysis of non-zero-length cross-link experiment data. Zero-length cross-link experiments are also less likely to generate “self-linked” products that occur when interacting reactive groups are on the same peptide [37], as both reactive groups would have to be on the same peptide and directly interact in a salt bridge. Dead-end and self-linked products of non-zero-length cross-linkers are problematic because they must be accounted for in database searches.
2.2. Disadvantages of zero-length CX-MS
The major challenge with zero-length cross-linkers for CX-MS experiments is the difficulty of identifying sub-stoichiometric cross-linked peptides in the presence of the far more numerous and more abundant linear peptides. This is because adding MS-cleavable bonds to generate MS3 fragmentation spectra [34] or direct incorporation of differential isotopic labels as part of the cross-linking reaction are not feasible as discussed above. An alternative isotopic labeling method that can be used with zero-length cross-linkers involves tagging the N- and C-termini of peptides following the cross-linking reaction, which helps distinguish cross-linked peptides as they contain two N- and two C-termini instead of one each [39]. However, with the exception of strategies that introduce an isotopic tag during proteolysis, such as 18O [40], interpretation of N- or C-terminal tags can be complicated by the presence of internal reactive groups in linear peptides. Another challenge, common to all CX-MS experiments, is that the number of possible cross-links increases exponentially with the amount of unique sequences in the target protein or protein complex, potentially making the search space unmanageably large [23]. In this regard, the limiting factor is the amount of unique primary sequence rather than the molecular weight of the total protein complex. The sequence complexity problem is particularly challenging for zero-length cross-linking experiments due to the absence of an MS-based method to differentiate modified and unmodified peptides.
As with most other types of cross-linkers, the specificity of zero-length cross-linkers such as EDC is a double-edged sword. Because its cross-linking reaction only involves lysine, aspartic acid, glutamic acid residues, and unmodified protein termini, EDC cannot be used to interrogate regions that lack any of these reactive residues. Furthermore, the reactive groups need to be accessible to the aqueous phase. Hence, regions lacking any reactive groups, buried within a protein or macromolecular complex, or buried within the lipid bilayer will be refractory to cross-linking. In addition, very large and very hydrophobic cross-linked peptides are generally very difficult to detect in a mass spectrometer tuned for peptide analysis. Therefore, regions of protein that contain reactive sites but no conveniently located proteolytic sites will be difficult to interrogate by CX-MS, although use of an alternative protease may minimize this problem.
2.3. Structural studies using zero-length CX-MS
Due in large part to the disadvantages mentioned above, zero-length CX-MS has typically been de-emphasized. In fact, a survey of CX-MS reviews published since 2012 reveals that zero-length CX-MS is either not mentioned [31,41–45] or only superficially discussed [24,46]. However, despite all the caveats, several studies have productively utilized zero-length CX-MS in various capacities.
One of these studies took a qualitative approach to zero-length CX-MS by utilizing it as a low-impact stabilizing reagent for protein macrocomplexes, which were then analyzed by matrix-assisted laser desorption ionization (MALDI) time of flight (TOF)-MS [47]. This study did not identify specific cross-linked peptides. It used cross-linking to stabilize quaternary protein complexes of up to 388 kDa that contained around 126.6 kDa in unique protein sequence. Another study combined MALDI-TOF MS with zero-length cross-linking in an effort to elucidate insights on the interactions composing the photosystem II (PSII) in green algae [48]. This study found that the proteins known as PsbO, PsbP, and PsbQ form close interactions with each other as well as with several other proteins in PSII. In addition, one specific interaction between PsbP's N-terminus and PsbQ's C-terminus was identified via manual analysis of uncross-linked and cross-linked MS/MS spectra.
Several studies focused on mapping protein–protein interactions and used zero-length cross-links to determine the precise interaction sites between two purified proteins known to associate with each other [49–51]. One study focused on the interaction between the cytochrome P450 2B6 enzyme and NADPH-cytochrome P450 reductase [49]. This study used 18O labeling and de novo MS/MS sequencing to identify and analyze cross-links between a synthetic peptide mimicking the C-helix of cytochrome P450 2B6 and the connecting domain of the 76.7 kDa P450 reductase. Another study used Fourier transform ion cyclotron resonance (FT-ICR) MS analysis to probe the interface between calmodulin and adenylyl cyclase 8 [51]. A similar technique with a variety of cross-linkers, including EDC, was later used to map the interaction between calmodulin and the skeletal muscle myosin light chain kinase M13 [52]. The resulting cross-links were manually identified with the aid of the General Protein Mass Analysis for Windows (GPMAW) [53] and the Automatic Spectrum Assignment Program (ASAP) [54] software packages, along with manual verification. These cross-links revealed an interaction between a 26-residue peptide corresponding to the N-terminus of M13 and the EF-hand 2 domain in calmodulin, which is in agreement with an NMR structure of the complex.
Zero-length CX-MS was also utilized in combination with data from other structural techniques to assist homology modeling of the interaction interface between proteins. One such study explored the interface between cytochrome P450 2E1 and cyto-chrome b5 using FT-ICR MS, and was able to generate a model of the interaction surface of the roughly 70 kDa 1:1 complex [55]. In another study, the binding interface of the transiently PSII-associated protein known as Psb27 was probed via zero-length CX-MS [56], and the resulting cross-links were identified using MassMatrix [57]. This experiment showed that Psb27 associates with the chlorophyll binding protein CP43 at two distinct sites, and a model of for the 61 kDa complex (with 61 kDa of unique sequence) was constructed. An interesting study combined HDX and zero-length CX-MS to re-examine the dimerization interface of the 14-3-3ζ protein [58]. These analyses uncovered novel contacts in the 56 kDa dimer interface and resolved ambiguous salt bridge interactions that were previously noted.
Several groups have attempted to improve zero-length CX-MS data analysis and to reduce the reliance on manual verification. One study proposed an algorithm, named Popitam, which was based on the premise that a fragmentation pattern of a zero-length cross-linked peptide can be approximated as that of a mixture of two peptides, each having a modification of unknown size. This allows MS/MS spectra of cross-linked peptides to be semi-automatically annotated via conventional peptide identification schemes [59]. The algorithm was tested on a model system of the cytochrome P450 2E1-cytochrome b5 complex. A later study by the same group employed this protocol as part of a de novo protein modeling experiment in conjunction with previously published cryo-EM data in order to determine the structure of the roughly 20 kDa gpE viral capsid protein for bacteriophage lambda [60]. Another zero-length CX-MS analysis strategy modified the database search algorithm of a popular mass spectra data analysis software package, known as SEQUEST [61]. The modified algorithm considered all possible products from a cross-linking reaction and subsequent tryptic digest (cross-linked peptides, adducts, and linear peptides), generated theoretical spectra, then matched them to the observed spectra [62].
As illustrated above, applications of zero-length CX-MS were mostly limited to fairly small proteins or protein complexes. Many of these studies also either relied on prior knowledge of the interaction interface or reported protein–protein interactions without identifying specifically cross-linked residues. Those that did identify specific cross-linked residues typically involved extensive manual analysis of MS/MS spectra.
Our group also performed several earlier zero-length CX-MS studies of red cell spectrin using recombinant protein constructs that preserved functionally important interfaces. We initially used a combination of the SEQUEST [61], GPMAW [53], and ASAP [54] software packages, along with extensive manual curation of MS/MS spectra, to develop a structure for the heterodimer initiation site. The recombinant proteins used here were approximately 100 kDa in size, with 100 kDa of unique sequence [63]. A subsequent study utilized Rosetta Elucidator (Rosetta Biosoftware, Seattle, WA), GPMAW [53], and the homology modeling program MODELLER [64] to determine a model of the spectrin tetramerization interface using a “mini-spectrin” recombinant construct. This construct formed a 180 kDa “tetramer” complex with 90 kDa of unique sequence [65]. Useful structural insights were obtained in these studies, but similar limitations to the above studies by other groups were encountered. That is, the methods used were tedious and time-consuming, and the number of high-confidence cross-links identified were relatively modest. These limitations led to development of a new data analysis pipeline specifically tailored for zero-length CX-MS analysis, which is described in the next section.
3. Strategies and software for in-depth analysis of larger proteins and protein complexes using zero-length cross-linking
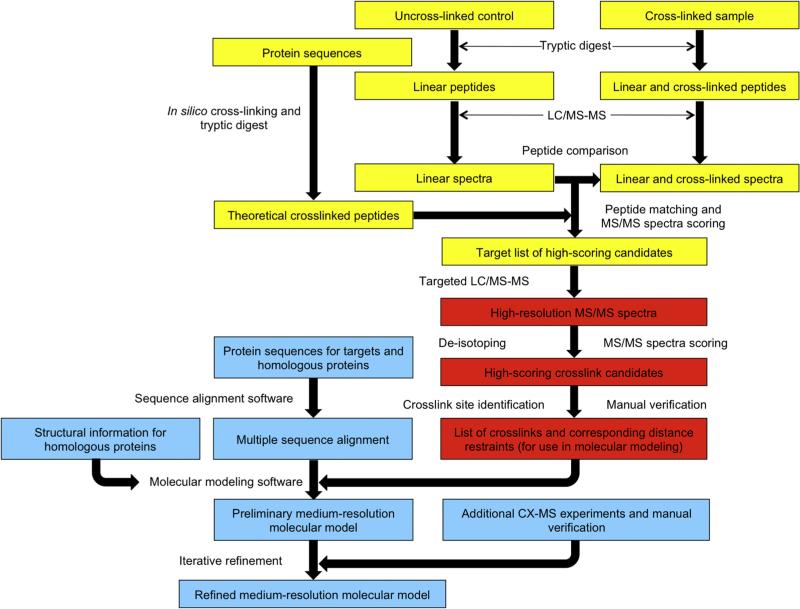
Because zero-length cross-linkers do not possess any of the special properties discussed above that can aid in cross-linked peptide identification, we recently developed an LC–MS/MS data analysis method coupled with a software tool named Zero-Length Cross-link Miner (ZXMiner), which is open-source and publicly available at https://shiek-db.wistar.upenn.edu/proteomics/ZXMiner.zip. Both the data acquisition method and the software were specifically optimized for identification of zero-length cross-links. There are two key components to this strategy (Fig. 2). First, parallel LC–MS/MS analyses of a cross-linked sample and an otherwise identical uncross-linked control are performed, and a label-free comparison of the resulting LC–MS data is performed to identify putative cross-linked precursors. We initially used an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Waltham, MA), which can conduct MS scans in the high-resolution orbitrap and MS/MS scans in the low-resolution ion trap in parallel to achieve a high duty cycle. However, the duty cycle was too low when MS/MS scans were also conducted in the orbitrap in unbiased discovery runs, which resulted in very few identified cross-linked peptides. We therefore used a two-tiered MS analysis approach to address the problem [2,65–67]. Specifically, as shown in Fig. 2, the initial comparison of the control and cross-linked sample is a “discovery run” with high-speed, low-resolution acquisition of MS/MS data in the ion trap. This is followed by a “targeted run” with low-speed, high-resolution acquisition of MS/MS data in the orbitrap. This two-step procedure required a few additional LC–MS/MS runs for most experiments but it allowed acquisition of high-resolution MS/MS data for all putative cross-linked peptides without compromising the method's depth of analysis. Below, we discuss key details of the method, representative results from recently published studies, and further improvements to the pipeline that are on-going.
Fig. 2.
Diagram for the zero-length CX-MS cross-linking protocol, as optimized for a LTQ Orbitrap MS instrument. [66] The protocol is separated into 3 major categories: sample preparation, label-free comparison, and comparison to all possible theoretical cross-linked peptides (in yellow), database search of candidate spectra and cross-link identification (in red), and incorporation of distance restraints into homology modeling experiments (in light blue).
3.1. Label-free comparison to control samples
In ideal cross-linking experiments, each molecule of protein or protein complex should contain only one or at most a few cross-links in order to minimize the risk of over-cross-linking and altering the native structures. As a result, cross-linked peptides typically constitute much less than 1% of the peptides in a tryptic digest of a cross-linked sample. Because uncross-linked control samples should contain all detectable linear peptides, label-free quantitative comparison of LC–MS patterns of a control and cross-linked sample can be used to remove these peptides from further consideration. In an earlier study [66], we used a 10-fold intensity enrichment (cross-link to control) cutoff to compensate for the limited ability of the Elucidator software that was used at that time to distinguish isotopic envelope patterns of low-intensity cross-linked precursors from overlapping non-peptide noise.
To overcome this flaw, we recently developed a label-free comparison software module in ZXMiner that determined the correct precursor masses and charge states of all ions and aligned two LC–MS patterns (manuscript in preparation). This module operates under similar concepts to those utilized by MaxQuant [68], namely using the correlation of intensity profiles across LC–MS scans to improve the detection and de-convolving of isotopic envelopes into monoisotopic species prior to performing label-free comparisons. This is a critical improvement compared to Elucidator, which matches discrete MS signals regardless of whether they are part of a peptide isotopic envelope. The current label-free comparison module greatly diminishes random matches between cross-linked peptide signals and spurious signals in the control sample, as illustrated in Table 1. This increases the specificity of label-free comparison, and greatly decreases the number of candidate cross-linked peptides for subsequent targeted high-resolution MS/MS analyses. As an alternative to our implementation of label-free analysis, any existing label-free analysis software [69,70] that is not coupled to a peptide identification platform can be adopted for cross-linking analysis and should provide similar performance. However, it is expected that label-free algorithms that match de-convoluted monoisotopic masses will out-perform software that match all discrete MS signals.
Table 1.
Distinguishing interference in control samples using label-free comparison software.
| ID | Charge state | m/z | Cross-linked peptide sequences | Intensity in control (Elucidator) | Intensity in control (LFA) |
|---|---|---|---|---|---|
| 1 | 3 | 819.0875 | YEEHLY[E]R-{MSPILGYW[K]IK | 1.9e + 05 | Absent |
| 1 | 4 | 614.5674 | YEEHLY[E]R-{MSPILGYW[K]IK | 3.7e + 05 | Absent |
| 1# | 5 | 495.0544 | YEEHLY[E]R-{M#SPILGYW[K]IK | 4.7e + 03 | Absent |
| 2 | 4 | 877.9311 | FELGLEFPNLPYYIDG[D]VK-HNMLGGCP[K]ER | 3.6e + 05 | Absent |
| 2# | 3 | 1175.5707 | FELGLEFPNLPYYIDG[D]VK-HNM#LGGCP[K]ER | 9.3e + 04 | Absent |
| 3 | 4 | 970.4894 | HNMLGGCP[K]ER-NKKFELGLEFPNLPYYIDG[D]VK | 5.1e + 04 | Absent |
| 3# | 4 | 974.4881 | HNM#LGGCP[K]ER-NKKFELGLEFPNLPYYIDG[D]VK | 2.9e + 04 | Absent |
| 4 | 3 | 1212.9374 | KFELGLEFPNLPYYIDG[D]VK-HNMLGGCP[K]ER | 6.2e + 04 | Absent |
| 4 | 4 | 909.9549 | KFELGLEFPNLPYYIDG[D]VK-HNMLGGCP[K]ER | 4.3e + 05 | Absent |
| 4# | 3 | 1218.2691 | KFELGLEFPNLPYYIDG[D]VK-HNM#LGGCP[K]ER | 5.7e + 04 | Absent |
| 4# | 4 | 913.9536 | KFELGLEFPNLPYYIDG[D]VK-HNM#LGGCP[K]ER | 1.7e + 04 | Absent |
| 5 | 3 | 854.4874 | LLL[E]YLEEK-IEAIPQID[K]YLK | 1.4e + 04 | Absent |
| 5 | 4 | 641.1174 | LLL[E]YLEEK-IEAIPQID[K]YLK | Absent | 6.05e + 06a |
| 6 | 4 | 680.1426 | LLL[E]YLEEK-RIEAIPQID[K]YLK | 7.0e + 03 | Absent |
| 7 | 3 | 912.1363 | LLLEYLE[E]K-YIAD[K]HNMLGGCPK | Absent | Absent |
| 7# | 3 | 917.4679 | LLLEYLE[E]K-YIAD[K]HNM#LGGCPK | 1.0e + 04 | Absent |
| 7# | 4 | 688.3527 | LLLEYLE[E]K-YIAD[K]HNM#LGGCPK | 4.3e + 04 | Absent |
| 8 | 3 | 683.3577 | [D]F[E]TLK-IAYS[K]DFETLK | 1.1e + 05 | Absent |
| 8 | 4 | 512.7701 | [D]F[E]TLK-IAYS[K]DFETLK | 1.2e + 05 | Absent |
| 9 | 5 | 664.5565 | YIAWPLQGWQATFGGG[D]HPPK-I[K]GLVQPTR | 1.9e + 04 | Absent |
| 10 | 4 | 464.7658 | LLL[E]YLEEK-YL[K]SSK | Absent | Absent |
| 11 | 3 | 1023.5399 | LP[E]MLK-[K]FELGLEFPNLPYYIDGDVK | 1.1e + 05 | Absent |
| 11# | 3 | 1028.8716 | LP[E]M#LK-[K]FELGLEFPNLPYYIDGDVK | 2.3e + 05 | Absent |
| 12 | 3 | 1248.3078 | YIAWPLQGWQATFGGGDHPP[K]}-V[D]FLSKLPEMLK | 1.2e + 05 | Absent |
| 13 | 3 | 1012.5001 | MFE[D]R-[K]FELGLEFPNLPYYIDGDVK | 1.7e + 05 | Absent |
Further inspection confirms that a precursor with similar m/z and charge state is present in the control sample. The intensity level of this precursor was 16 times lower than that of the cross-link sample and did not trigger an MS/MS scan. Elucidator could not detect this precursor.
3.2. Obtaining high-resolution MS/MS spectra
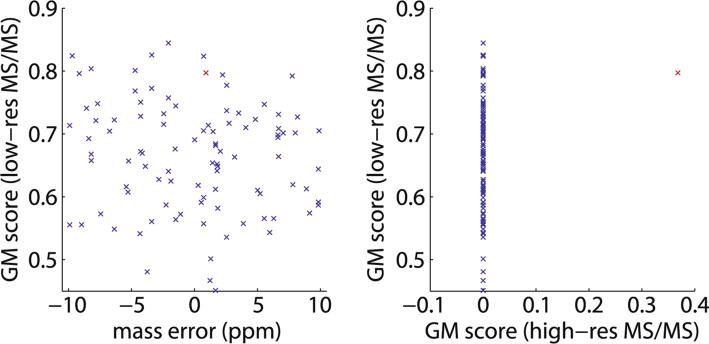
The capability of modern mass spectrometers to produce high-resolution, high-mass-accuracy MS data has been instrumental in advancing the field of proteomics. Until recently, acquisition of high-resolution MS/MS spectra was less common [71,72]. To identify linear peptides, low-resolution MS/MS spectra with mass accuracy of about ±0.5 Da, such as those obtained in a linear ion trap, were reasonably adequate for common search engines such as SEQUEST [61], MASCOT [73], etc. However, because cross-linked peptides are much larger and typically feature higher charge states than their linear counterparts, their MS/MS spectra are much more complex. Multiple fragmented ions, often with different charge states, will occur within a ±0.5 Da mass tolerance window with substantial frequency. Furthermore, the combinatorial expansion in search space when considering cross-linked samples from n peptides to the order of n2 cross-linked peptides greatly increases the frequency where different theoretical cross-linked peptides have precursor masses within 5–10 ppm of one another. Because of these factors, low-resolution MS/MS spectra can be relatively ineffective at distinguishing between alternative cross-linked peptide assignments, as illustrated in Fig. 3.
Fig. 3.
High-resolution MS/MS spectra are required for high-confidence identification of cross-linked peptides. In a zero-length cross-linking study of a large 526 kDa spectrin heterodimer using an Orbitrap XL mass spectrometer, a typical MS/MS scan can be matched to as many as 97 distinct theoretical cross-linked peptides within a 10-ppm mass tolerance range. Red highlights the correct assignment. The geometric mean (GM) score [66] represents the quality of a match between a cross-link sequence and an MS/MS spectrum. (A) Analysis of low-resolution MS/MS data cannot distinguish between the 97 alternative theoretical cross-linked peptides. (B) With high-resolution MS/MS data, only a single correct assignment stands out with a non-zero score.
Our data acquisition strategy overcomes the aforementioned drop in duty cycle that occurs on hybrid ion trap instruments when MS/MS scans are acquired at high resolution by acquiring LC– MS/MS data in two stages. LC–MS data from the discovery analyses was subjected to the label-free comparisons described above and the low-resolution MS/MS associated with putative cross-linked spectra were evaluated using ZXMiner [66] with low stringency scoring parameters. These two analyses significantly narrow down the list of candidate cross-linked peptide precursors. As a result, only a few parallel targeted LC–MS/MS runs were needed to obtain high-resolution, high-accuracy MS/MS spectra for all candidate precursors. In these targeted analyses, both MS and MS/MS scans were conducted in the Orbitrap and for complex samples, lists of candidates were split among multiple LC-MS/MS runs to ensure several attempted MS/MS analyses of all targeted peptides across each chromatographic peak. The number of runs required can be estimated by plotting the expected number of target precursors during each chromatographic time interval (precursor retention times are extracted from the discovery runs). The minimum signal threshold for triggering an MS/MS scan was set at 30,000 ion counts (as compare to the typical setting of 1000 ion counts to trigger MS/MS scans in the ion trap) to compensate for the reduced sensitivity of the orbitrap mass analyzer. Additionally, we highly recommend turning on the monoisotopic precursor selection and screening out precursors with charge states of +1 and +2, as they would rarely correspond to cross-linked tryptic peptides.
3.3. ZXMiner software specifics
Another critical component in the development of an optimized zero-length cross-linking analysis pipeline was the creation of the ZXMiner software. This software tool was developed to address three major goals: increase the throughput of zero-length cross-linking experiments by automating data analysis, increase the number of assigned cross-links, and improve the confidence of cross-link assignments. To the best of our knowledge, only one previously developed cross-link analysis software package, pLink [74], could utilize high-resolution MS/MS data effectively. Additionally, none of the software packages that had been utilized in zero-length CX-MS studies were capable of interpreting the aforementioned data in a high-mass accuracy, high-throughput fashion, which is what prompted us to develop ZXMiner. Indeed, a direct comparison shows that ZXMiner significantly outperforms pLink [74], StavroX [75], Crux [76], and MassMatrix [57] when used for zero-length cross-link detection, as shown in Table 2. An interesting observation is that the actual false discovery rate (FDR) is typically much higher than an estimated FDR for all methods when tested on standard proteins. It is also noteworthy that at the unique sequence level, ZXMiner could identify the largest number of true cross-links with no false positives.
Table 2.
Comparison of GST crosslink identifications using alternative software. Reproduced with permission from [66]. For each peptide sequence, charge states of the cross-linked peptide identified by each software package are indicated. Estimated false discovery rates (FDRs) were derived from the decoy data that each software package provided (not available from MassMatrix). Number of identifications, false positives, and the actual FDRs were calculated directly from this table.
| Unique sequence ID | Peptide sequence | Cα-Cα distance | ZXMiner | pLink | StavroX | Cruxa | Mass matrixb | |
|---|---|---|---|---|---|---|---|---|
| True positives | ||||||||
| 1 | YEEHLYER-{MSPILGYWKIK | 5.5 | 3,4 | 3,4,5 | 3 | 3,4,5 | 3,4 | 5 |
| 1# | YEEHLYER-{M#SPILGYWKIK | 5.5 | 5 | 4,5 | - | 3,4 | N/A | 5 |
| 2 | FELGLEFPNLPYYIDGDVK-HNMLGGCPKER | 8.7 | 4 | 4 | - | - | 3,4 | 4 |
| 2# | FELGLEFPNLPYYIDGDVK-HNM#LGGCPKER | 8.7 | 3 | 3 | - | - | N/A | 3 |
| 3 | HNMLGGCPKER-NKKFELGLEFPNLPYYIDGDVK | 8.7 | 4 | 4,5 | - | - | 4,5 | 4 |
| 3# | HNM#LGGCPKER-NKKFELGLEFPNLPYYIDGDVK | 8.7 | 4 | 4 | - | - | N/A | 5 |
| 4 | KFELGLEFPNLPYYIDGDVK-HNMLGGCPKER | 8.7 | 3,4 | 3,4 | - | - | 3,4 | 3 |
| 4# | KFELGLEFPNLPYYIDGDVK-HNM#LGGCPKER | 8.7 | 3,4 | 3,4 | - | - | N/A | 5 |
| 5 | LLLEYLEEK-IEAIPQIDKYLK | 9.2 | 3,4 | 3,4 | 3 | 3,4 | 3,4 | - |
| 6 | LLLEYLEEK-RIEAIPQIDKYLK | 9.2 | 4 | 4 | - | 4 | - | - |
| 7 | YEEHLYER-{M#SPILGYWK | 10.0 | - | - | - | 3,4 | N/A | N/A |
| 8 | LLLEYLEEK-YIADKHNMLGGCPK | 11.4 | 3 | 3 | 3 | - | 3 | 3 |
| 8# | LLLEYLEEK-YIADKHNM#LGGCPK | 11.4 | 3,4 | 3,4 | - | - | N/A | - |
| 9 | DFETLK-IAYSKDFETLK | 11.6 | 3,4 | 3,4 | 3 | 3 | 3,4 | - |
| 10 | YIAWPLQGWQATFGGGDHPPK}-IKGLVQPTR | 11.8 | 5 | 5 | - | 5 | 5 | 5 |
| 11 | LPEMLK-KFELGLEFPNLPYYIDGDVK | 12.4 | 3 | 3,4 | - | 3,4 | 3,4 | 3 |
| 11# | LPEM#LK-KFELGLEFPNLPYYIDGDVK | 12.4 | 3 | 3 | - | 4 | N/A | 4 |
| 12 | LLLEYLEEK-YLKSSK | 12.4 | 4 | 4 | - | 3,4 | 4 | - |
| 13 | AEISMLEGAVLDIRYGVSR-YIADKHNM#LGGCPK | 13.8c | - | - | - | - | N/A | 4 |
| 14 | YIAWPLQGWQATFGGGDHPPK}-VDFLSKLPEMLK | 14.6d | - | 5 | 5 | - | 4,5 | 5 |
| 14# | YIAWPLQGWQATFGGGDHPPK}-VDFLSKLPEM#LK | 14.6d | 3 | 3 | - | 3,4 | N/A | 5 |
| 15 | MFEDR-KFELGLEFPNLPYYIDGDVK | 15.8c,d | 3 | 3 | 3 | - | - | - |
| 15# | M#FEDR-KFELGLEFPNLPYYIDGDVK | 15.8c,d | - | - | - | 3 | N/A | 3 |
| False positives | ||||||||
| 1 | LLLEYLEEK-LVCFKK | 16.3 | - | - | - | - | 3 | - |
| 2 | IKGLVQPTR-DEGDK | 16.4 | - | - | 3 | 4 | 3 | - |
| 3 | IKGLVQPTR-DEGDKWR | 16.4 | - | - | - | 3 | - | - |
| 4 | DEGDKWR-LPEMLK | 18.5 | - | - | - | 3,4 | 3 | - |
| 5 | DEGDKWRNK-LPEMLKMFEDR | 18.5 | - | - | - | - | 3 | - |
| 6 | VDFLSKLPEMLKMFEDR-ER | 19.0 | - | - | - | - | - | 3 |
| 6# | VDFLSKLPEMLKM#FEDR-ER | 19.0 | - | - | - | 3 | N/A | - |
| 7 | IKGLVQPTR-LLLEYLEEK | 19.1 | - | - | - | - | - | 3 |
| 8 | {MSPILGYWKIK-ERAEISMLEGAVLDIR | 20.9 | - | - | - | 4 | - | - |
| 9 | ERAEISMLEGAVLDIRYGVSR-WRNKK | 21.5 | - | - | - | - | 3 | - |
| 10 | IKGLVQPTR-DFETLK | 22.7 | - | 3 | 3 | - | 3 | 3 |
| 11 | HNMLGGCPKER-LVCFKK | 23.9 | - | - | - | - | 3 | - |
| 12 | LLLEYLEEK-DEGDKWR | 25.2 | - | 3 | - | 3 | - | - |
| 13 | YEEHLYERDEGDKWR-AEISM#LEGAVLDIR | 27.2 | - | - | - | - | N/A | 4 |
| 14 | VDFLSKLPEMLKMFEDR-YIADKHNMLGGCPK | 28.5 | - | - | - | - | 5 | - |
| 15 | IEAIPQIDKYLK-MFEDR | 29.6 | - | - | 3 | - | - | - |
| 16 | LLLEYLEEK-LPEMLKMFEDRLCHK | 33.4 | - | - | - | - | 3 | - |
| 17 | DFETLK-LVCFKK | 34.2 | - | - | - | 3 | - | - |
| Unique precursor level | Estimated FDR: | <1% | <5% | <5% | <5% | <5% | N/A | |
| True positives: | 25 | 30 | 6 | 20 | 19 | 15 | ||
| False positives: | 0 | 2 | 3 | 8 | 9 | 4 | ||
| Actual FDR: | 0% | 6.3% | 33% | 29% | 32% | 21% | ||
| Unique sequence level | True positives: | 13 | 15 | 6 | 10 | 11 | 10 | |
| False positives: | 0 | 2 | 3 | 7 | 9 | 4 | ||
| Actual FDR: | 0% | 12% | 33% | 41% | 45% | 29% |
Crux cannot identify peptides with variable modifications.
MassMatrix cannot identify cross-links involving the protein terminus as one of the cross-linked sites.
Cross-links between subunits.
Cross-links involving flexible regions (as reflected by the elevated B-factors and/or loops).
The inputs for ZXMiner are MS data in mzXML format, a list of candidate MS/MS scans generated by a label-free comparison tool, and databases of pertinent protein sequences in FASTA format. The software first performs an in silico digestion of the proteins with the specified enzyme (usually trypsin), which is then followed by production of a list of all possible theoretical cross-linked peptides. The masses of these theoretical cross-linked peptides are calculated as the sum of the theoretical peptide masses after accounting for the mass change caused by a cross-linking reaction, which is defined by the user. These masses are then compared to those of the candidate cross-linked peptide precursors, resulting in matches between theoretical cross-link sequences and candidate MS/MS spectra. As the LC-MS data was obtained at high mass accuracy, a stringent mass tolerance, typically 5 or 10 ppm, is applied here. For each match, ZXMiner processes the MS/MS spectrum, performs an in silico fragmentation of the putative cross-link sequence to generate a list of theoretical MS/MS ions, and calculates “coverage” scores that represent how well the two sets of data overlap. Individual scores, as well as their geometric mean (GM), which is an overall indicator of cross-link match quality, are reported. When analyzing high-resolution MS/MS spectra, ZXMiner de-convolutes all isotopic envelopes and collapses them into monoisotopic peaks. This use of de-convoluted high-resolution MS/MS data is a critical factor that helps ZXMiner outperform other software packages and achieve a low false discovery rate (FDR) without sacrificing cross-link coverage, as previously shown [66] and illustrated in Table 2.
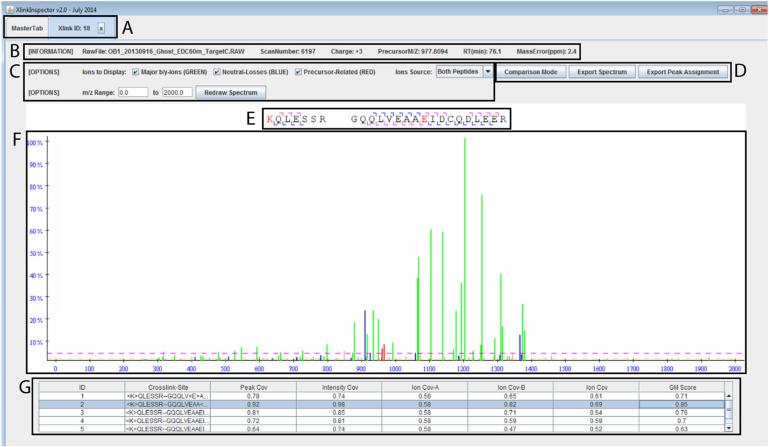
Another important feature of ZXMiner is that the output of raw search results contain the scores of all evaluated MS/MS spectra, cross-linked peptides, and even alternative cross-linked sites on each peptide. Hence, users can re-specify search parameters such as minimum peptide length, minimum scores, or precursor mass tolerance and use ZXMiner to rapidly recalculate the FDR for each identified cross-link without having to re-process LC–MS/MS and sequence data. XlinkInspector, which is part of the ZXMiner package, can be used to inspect the quality of each identified cross-link and confirm the exact cross-linked site (see Fig. 4). In recognition that frequently only a few weak MS/MS ions distinguish between alternative cross-linked sites within a peptide, the final decision regarding alternative cross-link site assignments is left to the user, instead of simply having ZXMiner output the highest-scoring candidates.
Fig. 4.
XlinkInspector is a graphical interface within ZXMiner that aids verification of cross-linked peptide assignment and determination of cross-linked sites. (A) The tabbed interface allows quick navigation between individual cross-link assignments for visual inspection of data quality. (B) The header displays basic information about the selected MS/MS spectrum. (C) This more detailed interface provides flexibility for spectrum annotation and plotting. (D) The list of identified fragmented ions can be exported for further inspection. The annotated spectrum can also be exported in publication-ready Scalable Vector Graphics format (SVG). (E) Visual representation of the identified b- and y-ions on the two peptide sequences. The assigned cross-linked residues are highlighted in red. (F) Color-coded plot of MS/MS data. Identified major b- and y-ions are shown in green. Neutral losses are shown in blue. Precursor-related ions are shown in red. Yellow and pink cutoff lines indicate minimum intensity values required to be designated as a peak or scored in the GM scoring algorithm, respectively. (G) Alternative cross-linked sites are listed along with their respective coverage scores. Annotation of the MS/MS spectrum dynamically changes in panel F when different cross-link sites are selected in panel G.
4. Applications using label-free analysis of cross-links and ZXMiner
The most common use of cross-linked peptide identifications are to uncover or confirm protein–protein interactions and to contribute to structural determination of proteins and protein complexes. For this latter application, high confidence cross-link assignments from ZXMiner are then applied as distance constraints to molecular modeling programs, such as MODELLER [64]. The distance constraint utilized for such experiments is typically tied to the α-carbon atoms of the cross-linked residues in question, as this provides more reliable information for the purposes of model-building. This distance restraint is typically set at 12 Å, but we have seen observed distances of up to 16 Å for high-confidence cross-links in areas likely to exhibit conformational flexibility when evaluating ZXMiner cross-link assignments on solved crystal structures [66]. This upper boundary is further illustrated in Fig. 5, which also highlights the importance of having high-mass accuracy MS/MS spectra. A number of recent studies illustrating the utility of ZXMiner are briefly discussed below.
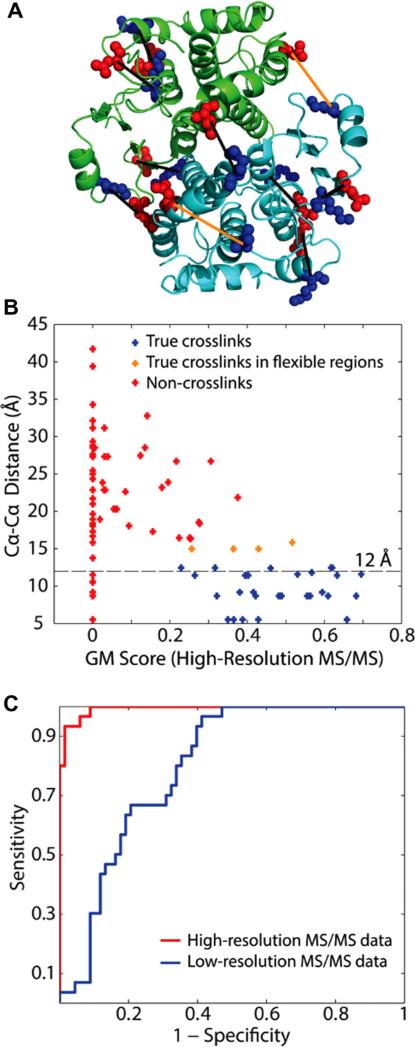
Fig 5.
Analysis of GST cross-links using ZXMiner. Reproduced with permission from [66]. (A) Locations of identified cross-links on the crystal structure of the GST homodimer (PDB ID: 1GTA). Lys residues are highlighted in blue and Glu and Asp are in red. The black lines connect the two α-carbons of each cross-link. Cross-links between residues whose Cα–Cα distances are significantly larger than 12 Å were highlighted in orange. (B) Scatter plot showing the relationship between GM scores derived from high-resolution MS/MS data and Cα–Cα distances for all cross-linked peptide candidates in the GST data set. A few cross-links located in regions likely to exhibit increased flexibility, such as loops or inter-subunit interfaces, slightly exceeded the expected 12 Å maximum Cα–Cα distance. (C) ROC curves showing the superior performance of high-resolution MS/MS data (area under the curve = 0.99) compared with low-resolution data (area under the curve = 0.80).
4.1. Mini-spectrin structure
We employed ZXMiner and zero-length CX-MS to elucidate structural insights for human red cell spectrin tetramers, as well as large conformational differences between “closed” dimers and “open” dimers (2). A 90 kDa fused heterodimer construct with the intact tetramerization site was used as a simplified surrogate for the full-sized 526 kDa heterodomer for these zero-length CX-MS experiments [77]. CX-MS analysis of mini-spectrin tetramers using ZXMiner revealed important additional cross-links not previously identified using older data acquisition and data analysis methods [63,65], which enabled determination of a more refined structure for the spectrin tetramer region (Fig. 6). Analysis of mini-spectrin dimers revealed several distance constraints that were mutually exclusive and were produced by the open and closed dimer forms of the protein that were in equilibrium and could not be independently isolated. Distance constraints derived from these cross-links allowed us to generate the first structures for the open and closed forms of mini-spectrin dimers (Fig. 7) [2]. Moreover, subsequent studies utilizing an L207P mutant of the mini-spectrin construct, which is a mutation previously shown to destabilize tetramer formation and which causes hereditary elliptocytosis in patients with this mutation [78], revealed cross-links outside the expected ranges of molecular flexibility of the wild-type mini-spectrin dimer. Distance constraints derived from these cross-links were used to elucidate a structure of the mutant dimer (Fig. 8), which in turn illustrated the allosteric effects of this mutation. Similar studies were also conducted on an L260P mutant that has similar destabilizing effects on spectrin tetramer formation [79], revealing a similar structural perturbation to that shown in Fig. 8 [67].
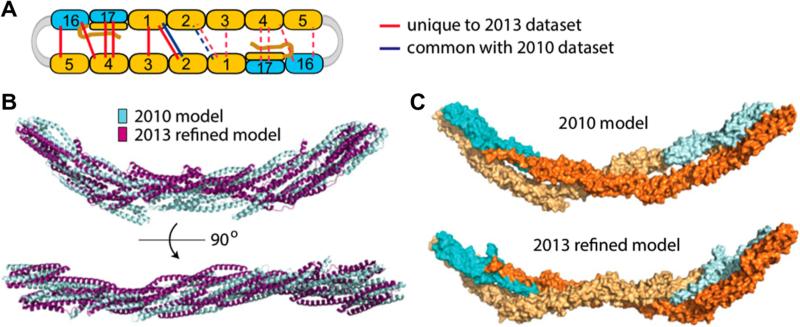
Fig. 6.
Solution structures for mini-spectrin tetramer based on zero-length CX-MS data analysis using ZXMiner. Adapted from [2] with permission. (A) Locations of interdomain cross-links used to model mini-spectrin tetramer. Blue lines indicate cross-links identified previously (65); red lines indicate new cross-links identified using the ZXMiner workflow; dashed lines indicate the same cross-links repeated in the second half of the tetramer. (B) Superimposition of present and previous tetramer structures. (C) Space-filling representations of tetramer models. β-spectrin domains are colored in bright or pale cyan, and α-spectrin domains are colored in bright or pale orange to distinguish the two strands.
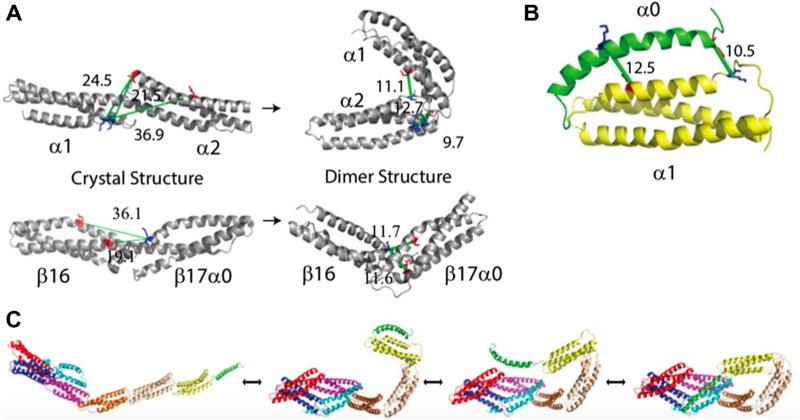
Fig. 7.
Zero-Length CX-MS enables identification and modeling of large changes in conformation for mini-spectrin dimers. Adapted from [2] with permission. (A) Open dimerspecific cross-links indicative of nonhelical connectors before (left) and after (right) structural refinement. Lys residues in blue; Glu/Asp residues in red; green lines are cross-links with labeled Cα–Cα distances. (B) Open dimer model supported by two cross-links between α0 and α1 domains. (C) Structures showing the interconversion between fully extended open dimer to closed dimer.
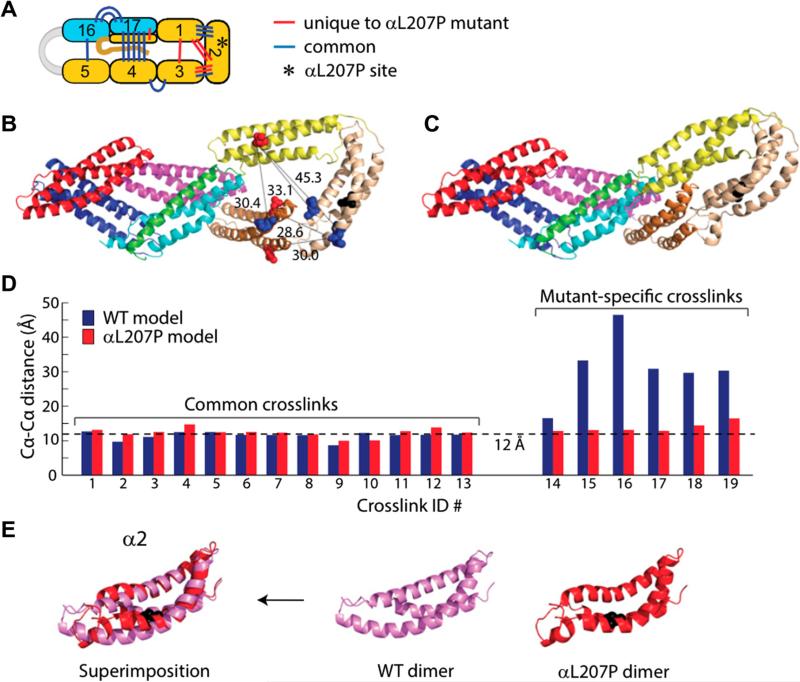
Fig. 8.
Cross-links and structure for the L207P mutant mini-spectrin dimer. Reproduced from [2] with permission. (A) Locations of αL207P inter-domain cross-links; the asterisk indicates the location of the aL207P mutation. (B) Locations of five αL207P mutant-specific cross-links indicative of conformational rearrangements in the α1–α2–α3 region plotted on the WT structure for comparison; blue, Lys; red, Glu/Asp; black, Pro mutation; black lines, cross-links with Cα–Cα distances labeled. (C) Model of the aL207P mutant closed dimer. (D) Cα–Cα distances for inter-domain cross-links identified in the αL207P mutant dimer on the WT and αL207P closed dimer structures. (E) Superimposition of a2 domains from the WT and αL207P closed dimer. The mutated residue is shown in black.
4.2. Solution structure of peroxiredoxin-6 (PRDX6)
Another protein that was analyzed using our zero-length CX-MS strategy was peroxiredoxin-6 (PRDX6). This antioxidant protein is commonly found in alveolar lung tissue, where it plays important roles in mitigating reactive oxygen species (ROS) damage via its antioxidant activity [80] and in lipid homeostasis due to its phospholipase A2 activity [81]. Although a high-resolution crystal structure of the catalytic intermediate of PRDX6 had been reported [82], the protein is conformationally flexible in solution, and this flexibility affects the protein's functional properties, particularly its phospholipase activity [83]. Interestingly, phosphorylation of a Thr residue (Thr-177) modulates phospholipase activity, but this residue is not solvent-exposed in the crystal structure [84]. This strongly suggests that a conformational difference between the reduced and peroxidase intermediate (crystal structure) forms of this enzyme exists. To explore these discrepancies, we performed zero-length CX-MS experiments on purified PRDX6 [85]. Analysis of these cross-links revealed significant differences between the reduced form of the protein and the crystal structure, indicating structural differences associated with catalysis. This study displays the resolving power of zero-length CX-MS when applied to the detection of relatively subtle changes in solution conformations of flexible proteins.
5. Further enhancing zero-length CX-MS
In addition to applying the existing analysis strategy to multiple biological problems, we are further refining the data acquisition methods and ZXMiner software to extend the capacities of this technique to protein complexes that exceed 1 MDa of unique sequence, as summarized below.
5.1. Zero-length CX-MS on more complex samples
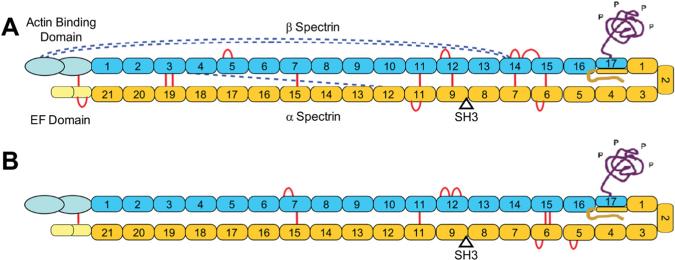
Numerous studies over the past decade have utilized longer-range cross-links to probe large protein complexes. However, as described above, application of zero-length CX-MS to large complexes is more difficult. Although our current method can identify some cross-links in larger protein complexes, the number of identified cross-links per unit mass is modest. For example, we recently probed full-length spectrin using zero-length CX-MS in order to begin to elucidate the structure of the 1052 kDa tetramer [2]. Most of the molecule is comprised of many tandem homologous 12 kDa domains (Fig. 9) and several crystal structures of up to three tandem domains are available for homology modeling. Our prior success in developing structures for mini-spectrin closed dimers, open dimers and tetramers [2] and preliminary analysis of intact spectrin (Fig. 9) suggest that an experimentally verified structure of the entire tetramer could be developed if sufficient distance constraints could be determined throughout the molecule.
Fig. 9.
Schematics of spectrin heterodimers with identified cross-links. Adapted from [66] with permission. Cross-links that fit the known domain structure and lateral alignment of the subunits are indicated by red lines, while those indicative of the protein folding back upon itself are shown by dashed blue lines. (A) Spectrin heterodimer with cross-links identified using purified heterodimers in solution. (B) Cross-links identified using intact membranes and isolated membrane cytoskeletons.
In addition, our goals for zero-length CX-MS include extending its utility to in-depth analysis of cellular structures in order to obtain structural information of protein complexes in the most biologically relevant context possible. A good model system for this purpose is human erythrocyte membranes, which can be readily purified to high homogeneity and which contains a relatively modest number of major proteins that includes spectrin [86]. One of the reasons why we are interested in this system is that, despite extensive biochemical analysis of individual proteins and simple complexes, current interaction and structural models of the membrane are crude with many key unresolved structural and functional questions. In initial pilot studies of intact red cell membranes, one of our primary targets is the protein known as anion exchanger 1 (AE1) or band 3, which is the most abundant red cell membrane protein with about 1.2 million copies per cell [87]. A large portion of the protein is embedded in the lipid bilayer, but about half of the mass is in the cytoplasm [88]. We have identified cross-links between several AE1 cytoplasmic domains, and structural models for the protein are currently being developed. Additionally, the cross-linking data have allowed us to resolve one key controversy concerning this protein, which is whether the transmembrane antiporter is comprised of 12 or 14 transmembrane spans [89], as the cross-linking data is only consistent with the 14-span model.
5.2. Zero-length CX-MS on new instruments with high-speed acquisition of high-resolution MS/MS spectra
One component of our strategy for enabling zero-length cross-linking analyses on very complex biological systems is to adapt the ZXMiner analysis strategy pipeline for use with some of the most recently developed MS/MS instruments, such as the Q Exactive Plus mass spectrometer (Thermo Scientific, Waltham, MA). These instruments are capable of greatly increased acquisition of high-resolution MS/MS spectra and better sensitivity when compared to the LTQ-Orbitrap XL instruments used during development of the current method. This faster data acquisition speed should reduce or eliminate the need for separate discovery and targeted LC–MS/MS runs, thereby reducing both instrument time and the total amount of cross-linked sample required for in-depth analysis. In addition, the increased sensitivity should improve detection of low abundance cross-linked peptides in very complex peptide mixtures.
Our preliminary evaluations of a Q Exactive Plus mass spectrometer revealed that its overall performance exceeded that of the Orbitrap XL, although differences in scores between two positive cross-links and decoys were smaller. This appeared to be primarily due to the difference in fragmentation methods used (HCD for Q Exactive Plus vs. CID for Orbitrap XL). The MS/MS spectra from the Q Exactive Plus also contained variable amounts of ions that did not match any predicted b- or y-ions or neutral losses thereof. In order to address these issues, data acquisition parameters are being systematically tested in order to determine the optimal conditions for CX-MS analysis on this instrument. Representative comparisons between the two instruments, which use the mini-spectrin construct described above, are displayed in Table 3. These results demonstrate significant progress toward reducing instrument time and increasing depth of analysis as measured by the number of unique cross-links assigned with a 0% FDR.
Table 3.
Comparison of LTQ Orbitrap and Q Exactive plus for zero-length CX-MS experiments using mini-spectrin.
| Banda | Instrument | Number of MS runsb | Total MS time (h) | Cross-linked peptides |
|
|---|---|---|---|---|---|
| Totalc | Uniquec | ||||
| Dimer | LTQ Orbitrap | 12 | 24 | 48 | 22 |
| Dimer | Q Exactive + | 3 | 6 | 40 | 23 |
| Tetramer | LTQ Orbitrap | 12 | 24 | 71 | 38 |
| Tetramer | Q Exactive + | 3 | 6 | 107 | 59 |
The mini-spectrin band used for the in-gel digest.
All MS runs consisted of 2-h LC gradients and injection sizes of about 0.5 μg of protein.
Peptides have FDR = 0.0.
6. Summary
In this review, we discuss the current state of the zero-length CX-MS field and compare it to those that utilize other cross-linkers. A data acquisition and data analysis strategy developed specifically for zero-length CX-MS is described. A key component of the pipeline is ZXMiner, a software package that automates all data analysis steps and includes a graphical interface to finalize assignment of cross-link sites when multiple potential reactive sites are present. This approach has been applied to multiple biological systems, including a mini-spectrin with 90 kDa of unique sequence with excellent depth of analysis and intact spectrin with 526 kDa of unique sequence with moderate depth of analysis. Ongoing optimization of data acquisition and data analysis on Q Exactive Plus and Q Exactive HF mass spectrometers are enabling effective analysis of intact red cell membranes and other complex biological problems whose major proteins represent sequence massive search spaces in the 3–5 MDa range.
Acknowledgments
The authors gratefully acknowledge the assistance of the Wistar Institute Proteomics Core and the expert assistance of Peter Hembach. This work was supported by the US National Institute of Health Grants R01HL038794 and R01DK084188 (to D.W.S.), P30CA010815 (NCI core Grant to the Wistar Institute), and T32GM008275 as well as a Philadelphia Health Care Trust Fellowship (to R.R.S.).
References
- 1.Olson AL, Tucker AT, Bobay BG, Soderblom EJ, Moseley MA, Thompson RJ, Cavanagh J. Structure. 2014 doi: 10.1016/j.str.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sriswasdi S, Harper SL, Tang HY, Gallagher PG, Speicher DW. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1801–1806. doi: 10.1073/pnas.1317620111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. Nature. 2014;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 4.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konijnenberg A, Butterer A, Sobott F. Biochim. Biophys. Acta. 2013;1834:1239–1256. doi: 10.1016/j.bbapap.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 7.Lanucara F, Holman SW, Gray CJ, Eyers CE. Nat. Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Chance MR. Anal. Chem. 2011;83:7234–7241. doi: 10.1021/ac200567u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiselar JG, Chance MR. J. Mass Spectrom. 2010;45:1373–1382. doi: 10.1002/jms.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maleknia SD, Downard KM. Chem. Soc. Rev. 2014;43:3244–3258. doi: 10.1039/c3cs60432b. [DOI] [PubMed] [Google Scholar]
- 11.Monroe EB, Heien ML. Anal. Chem. 2013;85:6185–6189. doi: 10.1021/ac400107c. [DOI] [PubMed] [Google Scholar]
- 12.Yan Y, Chen G, Wei H, Huang RY, Mo J, Rempel DL, Tymiak AA, Gross ML. J. Am. Soc. Mass Spectrom. 2014;25:2084–2092. doi: 10.1007/s13361-014-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vahidi S, Stocks BB, Liaghati-Mobarhan Y, Konermann L. Anal. Chem. 2013;85:8618–8625. doi: 10.1021/ac401148z. [DOI] [PubMed] [Google Scholar]
- 14.Poor TA, Jones LM, Sood A, Leser GP, Plasencia MD, Rempel DL, Jardetzky TS, Woods RJ, Gross ML, Lamb RA. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2596–E2605. doi: 10.1073/pnas.1408983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu G, Chance MR. Anal. Chem. 2004;76:1213–1221. doi: 10.1021/ac035422g. [DOI] [PubMed] [Google Scholar]
- 16.Xu G, Takamoto K, Chance MR. Anal. Chem. 2003;75:6995–7007. doi: 10.1021/ac035104h. [DOI] [PubMed] [Google Scholar]
- 17.Xu G, Chance MR. Anal. Chem. 2005;77:2437–2449. doi: 10.1021/ac0484629. [DOI] [PubMed] [Google Scholar]
- 18.Konermann L, Pan J, Liu YH. Chem. Soc. Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 19.Wales TE, Engen JR. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 20.Wei H, Mo J, Tao L, Russell RJ, Tymiak AA, Chen G, Iacob RE, Engen JR. Drug Discov. Today. 2014;19:95–102. doi: 10.1016/j.drudis.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Abzalimov RR, Bobst CE, Kaltashov IA. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20087–20092. doi: 10.1073/pnas.1315029110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Back JW, de Jong L, Muijsers AO, de Koster CG. J. Mol. Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 23.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Mol. Cell. Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paramelle D, Miralles G, Subra G, Martinez J. Proteomics. 2013;13:438–456. doi: 10.1002/pmic.201200305. [DOI] [PubMed] [Google Scholar]
- 25.Rappsilber J. J. Struct. Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinz A. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 27.Fasold H, Klappenberger J, Meyer C, Remold H. Angewandte Chemie. 1971;10:795–801. doi: 10.1002/anie.197107951. (International Ed., in English) [DOI] [PubMed] [Google Scholar]
- 28.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitner A, Joachimiak LA, Bracher A, Monkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, Ludtke S, Chiu W, Hartl FU, Aebersold R, Frydman J. Structure. 2012;20:814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, Malmstrom L, Aebersold R. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 31.Walzthoeni T, Leitner A, Stengel F, Aebersold R. Curr. Opin. Struct. Biol. 2013;23:252–260. doi: 10.1016/j.sbi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Petrotchenko EV, Serpa JJ, Borchers CH. Mol. Cell. Proteomics. 2011;10(M110):001420. doi: 10.1074/mcp.M110.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Back JW, Hartog AF, Dekker HL, Muijsers AO, de Koning LJ, de Jong L. J. Am. Soc. Mass Spectrom. 2001;12:222–227. doi: 10.1016/S1044-0305(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 34.Kao A, Chiu CL, Vellucci D, Yang Y, Patel VR, Guan S, Randall A, Baldi P, Rychnovsky SD, Huang L. Mol. Cell. Proteomics. 2011;10(M110):002212. doi: 10.1074/mcp.M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grabarek Z, Gergely J. Anal. Biochem. 1990;185:131–135. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 36.Staros JV, Wright RW, Swingle DM. Anal. Biochem. 1986;156:220–222. doi: 10.1016/0003-2697(86)90176-4. [DOI] [PubMed] [Google Scholar]
- 37.Zybailov BL, Glazko GV, Jaiswal M, Raney KD. J. Proteomics Bioinform. 2013;6:001. doi: 10.4172/jpb.S2-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruce JE. Proteomics. 2012;12:1565–1575. doi: 10.1002/pmic.201100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Shafey A, Tolic N, Young MM, Sale K, Smith RD, Kery V. Protein Sci. 2006;15:429–440. doi: 10.1110/ps.051685706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Anal. Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 41.Budayeva HG, Cristea IM. Adv. Exp. Med. Biol. 2014;806:263–282. doi: 10.1007/978-3-319-06068-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merkley ED, Cort JR, Adkins JN. J. Struct. Funct. Genomics. 2013;14:77–90. doi: 10.1007/s10969-013-9160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serpa JJ, Parker CE, Petrotchenko EV, Han J, Pan J, Borchers CH. Eur. J. Mass Spectrom. 2012;18:251–267. doi: 10.1255/ejms.1178. (Chichester, Eng) [DOI] [PubMed] [Google Scholar]
- 44.Hyung SJ, Ruotolo BT. Proteomics. 2012;12:1547–1564. doi: 10.1002/pmic.201100520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stengel F, Aebersold R, Robinson CV. Mol. Cell. Proteomics. 2012;11(R111):014027. doi: 10.1074/mcp.R111.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pacholarz KJ, Garlish RA, Taylor RJ, Barran PE. Chem. Soc. Rev. 2012;41:4335–4355. doi: 10.1039/c2cs35035a. [DOI] [PubMed] [Google Scholar]
- 47.Lepvrier E, Doigneaux C, Moullintraffort L, Nazabal A, Garnier C. Anal. Chem. 2014;86:10524–10530. doi: 10.1021/ac502561e. [DOI] [PubMed] [Google Scholar]
- 48.Nagao R, Suzuki T, Okumura A, Niikura A, Iwai M, Dohmae N, Tomo T, Shen JR, Ikeuchi M, Enami I. Plant Cell Physiol. 2010;51:718–727. doi: 10.1093/pcp/pcq042. [DOI] [PubMed] [Google Scholar]
- 49.Bumpus NN, Hollenberg PF. J. Inorg. Biochem. 2010;104:485–488. doi: 10.1016/j.jinorgbio.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marekov LN. Curr. Protoc. Protein Sci. 2007;19(19):16. doi: 10.1002/0471140864.ps1916s47. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt A, Kalkhof S, Ihling C, Cooper DM, Sinz A. Eur. J. Mass Spectrom. 2005;11:525–534. doi: 10.1255/ejms.748. (Chichester, Eng) [DOI] [PubMed] [Google Scholar]
- 52.Kalkhof S, Ihling C, Mechtler K, Sinz A. Anal. Chem. 2005;77:495–503. doi: 10.1021/ac0487294. [DOI] [PubMed] [Google Scholar]
- 53.Hojrup P. In: Ion Formation from Organic Solvents. Hedin A, et al., editors. John Wiley & Sons; 1990. pp. 61–66. [Google Scholar]
- 54.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Q, Doneanu CE, Shaffer SA, Adman ET, Goodlett DR, Nelson SD. J. Biol. Chem. 2006;281:20404–20417. doi: 10.1074/jbc.M601785200. [DOI] [PubMed] [Google Scholar]
- 56.Liu H, Huang RY, Chen J, Gross ML, Pakrasi HB. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18536–18541. doi: 10.1073/pnas.1111597108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H, Freitas MA. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haladova K, Mrazek H, Jecmen T, Halada P, Man P, Novak P, Chmelik J, Obsil T, Sulc M. J. Struct. Biol. 2012;179:10–17. doi: 10.1016/j.jsb.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzner RA, Larson Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Anal. Chem. 2008;80:8799–8806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh P, Nakatani E, Goodlett DR, Catalano CE. J. Mol. Biol. 2013;425:3378–3388. doi: 10.1016/j.jmb.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 61.Eng JK, McCormack AL, Yates JR. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 62.McIlwain S, Draghicescu P, Singh P, Goodlett DR, Noble WS. J. Proteome Res. 2010;9:2488–2495. doi: 10.1021/pr901163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li D, Tang HY, Speicher DW. J. Biol. Chem. 2008;283:1553–1562. doi: 10.1074/jbc.M706981200. [DOI] [PubMed] [Google Scholar]
- 64.Sali A, Blundell TL. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 65.Li D, Harper SL, Tang HY, Maksimova Y, Gallagher PG, Speicher DW. J. Biol. Chem. 2010;285:29535–29545. doi: 10.1074/jbc.M110.145573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sriswasdi S, Harper SL, Tang HY, Speicher DW. J. Proteome Res. 2014;13:898–914. doi: 10.1021/pr400953w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harper SL, Sriswasdi S, Tang HY, Gaetani M, Gallagher PG, Speicher DW. Blood. 2013;122:3045–3053. doi: 10.1182/blood-2013-02-487702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cox J, Mann M. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 69.Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Mol. Cell. Proteomics. 2013;12:549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu W, Smith JW, Huang CM. J. Biomed. Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frank AM, Savitski MM, Nielsen ML, Zubarev RA, Pevzner PA. J. Proteome Res. 2007;6:114–123. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan C, Park BH, McDonald WH, Carey PA, Banfield JF, VerBerkmoes NC, Hettich RL, Samatova NF. BMC Bioinformatics. 2010;11:118. doi: 10.1186/1471-2105-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 74.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Nat. Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 75.Gotze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, Krauth F, Fritzsche R, Kuhn U, Sinz A. J. Am. Soc. Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 76.Park CY, Klammer AA, Kall L, MacCoss MJ, Noble WS. J. Proteome Res. 2008;7:3022–3027. doi: 10.1021/pr800127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harper SL, Li D, Maksimova Y, Gallagher PG, Speicher DW. J. Biol. Chem. 2010;285:11003–11012. doi: 10.1074/jbc.M109.083048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gallagher PG, Tse WT, Coetzer T, Lecomte MC, Garbarz M, Zarkowsky HS, Baruchel A, Ballas SK, Dhermy D, Palek J, et al. J. Clin. Invest. 1992;89:892–898. doi: 10.1172/JCI115669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Glele-Kakai C, Garbarz M, Lecomte MC, Leborgne S, Galand C, Bournier O, Devaux I, Gautero H, Zohoun I, Gallagher PG, Forget BG, Dhermy D. Br. J. Haematol. 1996;95:57–66. doi: 10.1046/j.1365-2141.1996.d01-1869.x. [DOI] [PubMed] [Google Scholar]
- 80.Manevich Y, Fisher AB. Free Radic. Biol. Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 81.Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. J. Lipid Res. 2007;48:2306–2318. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 82.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Nat. Struct. Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 83.Rahaman H, Zhou S, Dodia C, Feinstein SI, Huang S, Speicher D, Fisher AB. Biochemistry. 2012;51:5521–5530. doi: 10.1021/bi300380h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Y, Feinstein SI, Manevich Y, Chowdhury I, Pak JH, Kazi A, Dodia C, Speicher DW, Fisher AB. Biochem. J. 2009;419:669–679. doi: 10.1042/BJ20082061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rivera-Santiago R, Harper S, Zhou S, Sriswasdi S, Feinstein SI, Fisher AB, Speicher DW. Biochem. J. 2015 doi: 10.1042/BJ20141463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baines AJ. Biochem. Soc. Trans. 2009;37:796–803. doi: 10.1042/BST0370796. [DOI] [PubMed] [Google Scholar]
- 87.van den Akker E, Satchwell TJ, Williamson RC, Toye AM. Molecules Dis. 2010;45:1–8. doi: 10.1016/j.bcmd.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 88.Choi I. Curr. Top. Membr. 2012;70:77–103. doi: 10.1016/B978-0-12-394316-3.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirai T, Hamasaki N, Yamaguchi T, Ikeda Y. Biochem. Cell Biol. 2011;89:148–156. doi: 10.1139/o10-160. [DOI] [PubMed] [Google Scholar]